Abstract
Antibody-dependent enhancement (ADE) is a complex phenomenon mediated by antibodies, frequently pre-existing non-neutralizing or sub-neutralizing antibodies. In the course of infectious diseases, ADE may be responsible for worsening the clinical course of the disease by increasing the virulence of pathogens (ADE of infection) or enhancing disease severity (ADE of disease). Here we reviewed the mechanisms thought to be behind the ADE phenomenon and its potential relationship with COVID-19 severity. Since the early COVID-19 epidemics, ADE has been mentioned as a possible mechanism involved in severe COVID-19 disease and, later, as a potential risk in the case of infection after vaccination. However, current data do not support its role in disease severity, both after infection and reinfection.
Keywords: Antibody-dependent enhancement, Neutralizing antibodies, SARS-CoV-2, Therapies, Vaccines
1. Introduction
The interaction between humans and viruses (and its outcome) is determined by several factors, which mainly comprise innate and adaptive immunity [1]. Both types of immunity can contribute to the control of viral infections and virus clearance, but also to determine virus pathogenicity and tissue damage.
Virus surface is an antigenic structure that usually triggers cellular and humoral immune responses aimed at eradicating the virus in the host. Among humoral defenses, antibodies, produced after a viral infection, contribute to several levels of antiviral defense, neutralizing the virus and reducing its infectivity by the inhibition of its entry into the host cells. However, in some circumstances, antibodies may potentiate the viral infection of host cells, leading to increased infectivity and promoting inflammation and tissue injury.
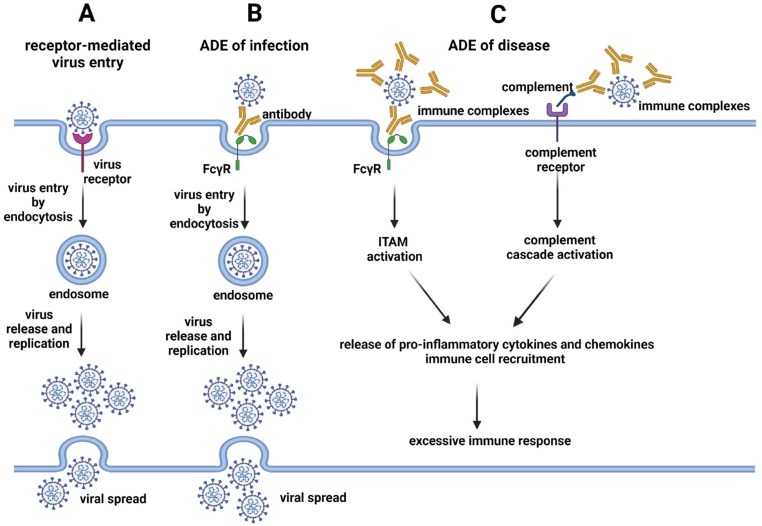
Antibody-dependent enhancement (ADE) is a complex phenomenon mediated by antibodies, frequently pre-existing non-neutralizing or sub-neutralizing antibodies (nNAbs), through their fragment crystallizable (Fc) region in their carboxyl-terminal domain. In the course of infectious diseases, ADE may be responsible for worsening the clinical course of the disease by increasing the virulence of the pathogens (ADE of infection) or enhancing disease severity (ADE of disease) (Fig. 1 ).
Fig. 1.
(A) Viruses initiate infection through the interaction of viral proteins with virus-specific receptors on host cells. Viruses enter the host cells mainly through endocytosis, virions are uncoated, viral genome is replicated, viral proteins are produced and new virions are assembled for their final spread. (B) In ADE of infection, pre-existing non-neutralizing or sub-neutralizing antibodies bind to the virus and interact with the fragment crystallizable gamma receptors (Fcγ) mainly present on the surface of myeloid cells, facilitating the internalization of the virus into host cells by endocytosis. If the virus can productively infect these cells, it replicates and spread like in (A), increasing the production of viral particles per cell or/and increasing the number of infected cells. (C) In ADE of disease, pre-existing non-neutralizing or sub-neutralizing antibodies can form immune complexes with viral particles. These immune complexes may interact with the Fcγ receptors of host myeloid cells, activating the immunoreceptor tyrosine-based activation motifs (ITAMs) of these receptors. Alternatively, immune complexes, through the activated complement components, interact with the complement receptors of host cells, activating the complement cascade. In both cases, the activation results in the release of pro-inflammatory cytokines and chemokines and recruitment of immune cells, exacerbating the immune response.
ADE of infection has been reported in vitro and in vivo for several viruses, but ADE of infection observed in vitro does not predict ADE of disease in humans [2]. In ADE of infection, pre-existing specific antibodies against a virus serotype do not completely neutralize a different serotype of the same virus, rather facilitating its entrance and replication in phagocytic cells harboring fragment crystallizable gamma (FcγR) and/or complement receptors, like monocytes, macrophages or granulocyte cells, in this way acting as a “Trojan horse” further allowing the virus to get into permissive cells [3]. Entry of viruses in the host cells occurs through clathrin-coated vesicles, mainly using FcγRII receptor [4] (Fig. 1 B). By binding to the antibody Fc-portion, FcγRs provide a link between the specificity of the adaptive immune system and the effector functions triggered by innate immune effector cells.
ADE of disease is the result of an excessive immune response to pathogens when the antibody-pathogen complex, through the effector functions of the Fc, is able to potentiate the disease by initiating a strong immune cascade that results in a severe pathology. Antibodies still act as a “Trojan horse”, allowing the virus to get into cells and exacerbate the immune response. The outcome of ADE of disease depends on the balance of the activation of the inhibitory FcγR signaling through the immunoreceptor tyrosine-based activation (ITAMs) and the immunoreceptor tyrosine-based inhibitory (ITIM) motifs. One factor that modulates FcγR signaling is the glycosylation state of the IgG Fc domains within immune complexes [5] (Fig. 1 C).
Also based on the main pathogenic mechanism involved, two different types of ADE can develop and are named extrinsic and intrinsic ADE [6]. Extrinsic ADE refers to the increase in the number of cells that are infected due to the binding of the antibody-virus complex to the FcγRs, facilitating the virus internalization into host cells. Extrinsic ADE seems to be mediated by IgG type antibodies. The balance of FcγRs that are engaged by immune complexes determines the degree of the inflammatory effector cell response [5]. Intrinsic ADE, mainly mediated by IgM type antibodies, refers to the increased production of viral particles per cell as a consequence of the internalized nNab-virus complexes that modulate the innate antiviral response in order to increase virus production. The ADE phenomenon can prompt the massive release of inflammatory and vasoactive mediators that ultimately contribute to disease severity [6]. The two enhancement pathways, IgG- and IgM-related, are independent but not mutually exclusive and can act synergistically.
nNabs can be naturally acquired by previous infections, passive maternal immunity or acquired with vaccination. ADE was first observed in vitro sixty years ago by Hawkes et al., who described an increased infectivity of several viruses, belonging to the Flaviviridae family, which was mediated by specific antisera produced in fowls [7]. Later, Halstead et al. described this process in patients during Dengue fever reinfection. Dengue fever is a mosquito-borne illness caused by the Dengue virus, an RNA virus belonging to the Flaviviridae family with 4 serotypes [8]. The first Dengue fever event can clinically manifest as a mild febrile disease, whilst, during a secondary infection, whether caused by different Dengue virus serotypes, the symptoms can be more severe and characterized by severe thrombocytopenia and increased vascular permeability causing disseminated hemorrhage. The serotype-specific antibodies produced during the first infection cross-react with other virus serotypes but without completely neutralizing them, in turn allowing easier infection of monocytes through antibody-receptor-mediated uptake of the virus.
Apart from some Flavivirus like Dengue virus, ADE has been described for few other human viruses, like Human Immunodeficiency Virus-1 (HIV1) [9], West Nile virus [10], Zika virus [11] or Ebola virus [12]. ADE has also been described for some respiratory infections like Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), Middle East Respiratory Syndrome Coronavirus (MERS-CoV) or Human Influenza virus [13], [14], [15], [16], [17], [18], [19] and it has been reported both in vivo and in vitro [20]. In these respiratory infections, nNabs enhance viral entry into lung phagocytic cells, disseminating the infection and initiating a powerful immune cascade that results in lung pathology.
Recently, ADE has also been described for multiple bacterial infections in both animal models and humans [21].
2. SARS-CoV-2 and ADE: mechanisms and evidence
Since the emergence of the SARS-CoV-2 pandemic in the Chinese city of Wuhan in December 2019, concern of ADE for SARS-CoV-2 were raised based on preliminary data on previous outbreaks of coronaviruses like SARS-CoV in 2002 and MERS-CoV in 2012 and 2015 [18], [20], [22], [23], [24]. ADE was related to both viruses, as an alternative mechanism of infection of host cells and the cause of abnormal cytokine production. Among the four major structural SARS-CoV2 proteins, the Spike envelope glycoprotein (S) has been identified as the most important antigen inducing neutralizing and protective antibodies. Entry of SARS-CoV-2 into the cells of infected individuals has been shown early on to depend on the receptor-binding domain (RBD) of the S protein. Accordingly, much of the research on SARS-CoV-2 has focused on the epitopes of the RBD recognized by antibodies to learn more about viral pathogenesis, to find an effective treatment and build a vaccine.
Neutralizing antibodies bind to viral protein epitopes that interact with host cell receptors which are crucial for viral infection. Therefore, antibodies against SARS-CoV-2 RBD are considered as neutralizing [25]. RBD interacts with the human receptor angiotensin-converting enzyme 2 (ACE2) to enter susceptible cells. ACE2 is mainly expressed in the cells of the respiratory tract and its expression is regulated by interferons [26]. ACE2 and FcγR concomitant expression in respiratory cells is scarce, so the ADE mechanism caused by SARS-Cov-2 via the dual-receptor mechanism in lung could be mediate by cells of the immune system expressing FcγR and ACE2 [2], [27].
The mechanism of the damaging inflammatory response characteristic of severe coronavirus disease 19 (COVID-19) is still unclear. Discordant data have been published regarding ADE-mediated interleukin-6 (IL-6) aberrant production as the cause of severe COVID-19 [28], [29], [30]. Furthermore, Multisystem Inflammatory Syndrome in children (MIS-C) has been hypothesized to be mediated by ADE although this speculation remains to be demonstrated [31]. Recent studies have suggested that non-neutralizing afucosilated IgG-FcγR interactions could trigger an abnormal cytokine response in myeloid cells in the lung of patients with severe COVID-19 [32]. In contrast, BNT162b2 SARS-CoV-2 messenger RNA vaccine-elicited IgG are highly fucosylated and enriched in Fc sialylation. Interestingly, these Fc glycoforms seem to have reduced inflammatory potential [32]. Previously, the presence of afucosylated Fc glycans (≥10%) on maternal anti-Dengue virus IgG has been shown to be a risk factor for susceptibility of infants to disease during primary Dengue virus infections [33]. IgG glycosylation is a mechanism not completely understood but it seems regulated mainly by heritable factors and related to better clinical prognosis of infections [34].
The virus-(neutralizing or non-neutralizing)antibody complex binds to the FcγR on the surface of immune cells such as monocytes and macrophages, possibly allowing SARS-CoV-2 entry also without the usage of ACE2 (Fig. 1 B). The family of FcγRs is broadly expressed in immune cells, however, only FcIIγR and FcIIIγR have been described to be involved in ADE [30]. The concern about a potential role of ADE for severe COVID-19 pathogenesis was early raised during pandemic, but currently there is no data confirming this hypothesis. Positive correlation between the titer of antibodies against SARS-CoV-2 [35], [36] and COVID-19 severity initially supported this hypothesis, posing a problem for vaccine development and therapeutic approaches in patients infected with different SARS-CoV-2 variants [20]. By the use of convalescent–phase plasma from COVID-19 patients, Maemura et al. have recently shown that SARS-CoV-2 induces antibodies that can elicit ADE of infection in vitro as for 6 months after infection, but these antibodies do not facilitate or exacerbate abnormal cytokine production by macrophages [30]. Convalescent plasma from patients recovered from COVID-19 was initially used as therapy in several trials with suboptimal results, but ADE was not considered as a cause of progression to severe disease or all-cause mortality in treated patients [37], [38].
Cross-reactivity of pre-existing antibodies with SARS-CoV-2 proteins may have important consequences both on virus neutralization and on ADE. Cross-reactivity may be determined by the presence of glycans on viral proteins. Glycosylation of viral proteins is observed for envelope protein (Env) of HIV-1, hemagglutinin protein (HA) of influenza virus, coronavirus Spike protein (S) and envelope proteins of many other viruses [39]. Since glycosylation of viral proteins mainly occurs through the host-glycosylation pathway [39], it is possible that different viruses present similar glycosylation pattern and antibodies elicited by one virus may cross-react with another virus. An antibody targeting the glycosylated Env protein of HIV-1 (2G12) has recently been shown to also neutralize influenza virus, due to the presence of similar oligomannose-type clusters on HA [40]. The same 2G12 antibody, together two further antibodies directed against glycans of the HIV-1 Env glycoprotein (PTG126 and PTG128) have been shown to display high cross-reactivity with the S protein of SARS-CoV-2, but they are incapable of neutralizing pseudoviruses expressing SARS-CoV-2 S protein and most probably do not mediate ADE [41].
Epidemiological studies have shown the presence of pre-existing antibodies cross-reactive with SARS-CoV-2 in the serum of sub-Saharan African patients. This presence has been related to a lower COVID-19 morbidity/mortality in this populations [42], [43]. Plasmodium and human immunodeficiency virus (HIV), both microorganisms being endemic in Africa, have been shown to induce in patients nNAbs that can cross-react with SARS-CoV-2, but these antibodies do not correlate with more severe forms of COVID-19 disease [44], [45], [46]. Polyclonal plasma from HIV-infected children cross-reacts with SARS-CoV-2 but, to our knowledge, there are no data in the literature on their possible involvement in ADE [47]. Moreover, cross-reactivity of SARS-CoV-2 antigen/antibodies can lead to HIV-antibody false positive results in chemiluminescent HIV immunoassays [48].
More recently, pre-existing IgG targeting several seasonal coronavirus-conserved SARS-CoV-2 spike subdomains have been correlated with differential COVID-19 disease outcomes and authors suggested that the severity of disease depend on epitope targeting by humoral recall response. Pre-existing IgG targeting the S2′FP region of SARS-CoV-2 are predictors of more severe COVID-19 than IgG targeting HR2, 5′fHR” or RDB region [49]. More studies are necessary to understand if ADE is involved in these effects.
Influenza vaccination is associated with reduced susceptibility to or decreased COVID-19 severity in a recent meta-analysis [50] and sera from convalescent COVID-19 patients and from SARS-CoV-2 spike immunized mice showed cross-reaction with the Influenza virus HA protein in vitro [51].
Overall, these studies suggest that cross-reactive non-neutralizing responses across several infections and SARS-CoV-2 infection do not drive ADE phenomena in COVID-19.
3. Omicron and ADE: foreseen risks
The RBD and the N-terminal domain (NTD) of SARS-CoV-2 S protein have accumulated mutations overtime as evidenced by several variants of concern (VOC) and the sequential waves of infection around the world. Currently, the epidemic curve of SARS-CoV-2 is silently raising again with the surge of a new SARS-CoV-2 variant (Omicron BA.2-lineage).
Omicron, the last identified VOC, contains 15 mutations in the RBD and 11 mutations in the NTD, respectively, and its RBD binds to ACE2 with a ∼2.4 fold enhanced affinity compared to the original Wuhan-Hu-1 strain, possibly explaining only partially the increased transmissibility of the variant [52], [53].
Currently, no variant of SARS-CoV-2 has accumulated as many mutations as the Omicron variant. This variability is likely related to immune evasion, as evidenced by the increase in reinfections during the last wave and the detection of new infections in vaccinated patients, mainly attributed to Omicron variant. The antigenic shift led to severe damping of the neutralizing activity of antibodies which are present in patients with a previous SARS-CoV-2 infection (possibly with a different variant) or in vaccinated individuals. Several studies in infected or vaccinated individuals based on the humoral antibody SARS-CoV-2 responses have reported a reduction in the neutralizing antibody titers to the Omicron variant in comparison to the wild-type SARS-CoV-2 [54], [55]. Moreover, SARS-CoV-2 reinfections seem to be less severe than the first one both in children [56] and adults [57] and even in solid organ transplant (SOT) patients [58].
Among all therapeutic monoclonal antibodies (mAbs) approved against SARS-CoV-2, the only ones with neutralizing activity against the Omicron variant are sotrovimab and the cilgavimab/tixagevimab cocktail. Even these mAbs experienced respectively a 2–3-fold and 12–200-fold reduced potency against the Omicron variant, using both pseudovirus and authentic virus assays [59]. LY-CoV1404, a new mAb recently approved by the U.S. Food and Drug Administration (FDA) has demonstrated to potently neutralize almost all circulating SARS-CoV-2 variants, including B.1.1.7, B.1.351, B.1.617.2, B.1.427/B.1.429, P.1, B.1.526, B.1.1.529 and the BA.2 subvariant [60]. Available data fail to demonstrate that mAbs or convalescent plasma from COVID-19 patients can facilitate SARS-CoV-2 D614 infection in human blood monocyte-derived macrophages inducing ADE in vitro [61].
Today, with the neutralizing antibody response induced by vaccination or previous SARS-CoV-2 infection weakening over time, whether non-neutralizing or sub-neutralizing antibodies would lead to enhanced viral invasion or proinflammatory damage upon reinfection is becoming a serious concern. The risk of SARS-CoV-2 reinfection and COVID-19 hospitalization in individuals after a previous SARS-Cov-2 infection or after SARS-CoV-2 vaccination remained low and it seems less severe [62].
The initial alarm raised about the possible emergence of ADE with the use of convalescent sera for the treatment of COVID-19 [63], and then after vaccination, is today weakening. Recent research has described a reduced cross-neutralization against VOCs (especially for Omicron BA.2) in plasma of patients infected with Omicron BA.1 and without history of vaccination, with opposite results reported in vaccinated patients [64]. While authors comprehensibly highlight the potential implications of the study for the introduction of omicron-based vaccines, theoretically the reduced cross-reactive neutralization could rise the risk of ADE, which is not currently confirmed in the many data available about both SARS-CoV-2 reinfections and infections after vaccine [65].
Probably, the mechanism of ADE is not completely known and there are no markers to certainly predict or identify the possible benefit or risk of the presence of non– or sub-neutralizing antibodies coming from natural infection, vaccines or specific mAbs. Clinical data has not yet stablished a role for ADE in human COVID-19 pathology. It has been suggested that assays verifying early antibody quality/quantity, the expression of cognate FcγR on phagocytic cells or the presence of Fc glycoforms within antibodies could be used to anticipate COVID-19 outcomes.
In the meantime, vaccines and mAbs are considered optimal therapies, regardless the potential role of ADE in COVID-19 severity, which so far seems not to be confirmed in the most recent VOC waves.
Funding
None.
CRediT authorship contribution statement
Isabella Zanella: Conceptualization, Writing – original draft, Writing – review & editing. Melania Degli Antoni: Writing – review & editing. Valentina Marchese: Writing – review & editing. Francesco Castelli: Writing – review & editing. Eugenia Quiros-Roldan: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Rouse B.T., Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat. Rev. Immunol. 2010;10(7):514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584(7821):353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 3.Okuya K., Hattori T., Saito T., Takadate Y., Sasaki M., Furuyama W., Marzi A., Ohiro Y., Konno S., Hattori T., Takada A., Medina R.A. Multiple routes of antibody-dependent enhancement of SARS-CoV-2 infection. Microbiol. Spectr. 2022;10(2) doi: 10.1128/spectrum.01553-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molfetta R., Quatrini L., Gasparrini F., Zitti B., Santoni A., Paolini R. Regulation of fc receptor endocytic trafficking by ubiquitination. Front Immunol. 2014;18(5):449. doi: 10.3389/fimmu.2014.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bournazos S., Wang T.T., Dahan R., Maamary J., Ravetch J.V. Signaling by antibodies: recent progress. Annu. Rev. Immunol. 2017;35(1):285–311. doi: 10.1146/annurev-immunol-051116-052433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narayan R., Tripathi S. Intrinsic ADE: the dark side of antibody dependent enhancement during dengue infection. Front Cell Infect Microbiol. 2020;2(10) doi: 10.3389/fcimb.2020.580096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkes R.A. Enhancement of the infectivity of arboviruses by specific antisera produced in domestic fowls. Aust. J. Exp. Bio. Med. Sci. 1964;42(4):465–482. doi: 10.1038/icb.1964.44. [DOI] [PubMed] [Google Scholar]
- 8.Halstead S.B., Crowe J.E., Jr., Boraschi D., Rappuoli R. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol. Spectr. 2014;2(6) doi: 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- 9.Jr Robinson W.E., Montefiori D.C., Gillespie D.H., Mitchell W.M. Complement-mediated, antibody-dependent enhancement of HIV-1 infection in vitro is characterized by increased protein and RNA syntheses and infectious virus release. J. Acquir. Immune Defic. Syndr. 1989;2:33–42. [PubMed] [Google Scholar]
- 10.Gollins S.W., Porterfield J.S. Flavivirus infection enhancement in macrophages: an electron microscopic study of viral cellular entry. J. Gen. Virol. 1985;66(9):1969–1982. doi: 10.1099/0022-1317-66-9-1969. [DOI] [PubMed] [Google Scholar]
- 11.Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V.M., Malasit P., Rey F.A., Mongkolsapaya J., Screaton G.R. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016;17(9):1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takada A., Feldmann H., Ksiazek T.G., Kawaoka Y. Antibody-dependent enhancement of Ebola virus infection. J. Virol. 2003;77(13):7539–7544. doi: 10.1128/jvi.77.13.7539-7544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura M., Webster R.G., Ennis F.A. Antibodies to HA and NA augment uptake of influenza A viruses into cells via Fc receptor entry. Virology. 1991;182(1):211–219. doi: 10.1016/0042-6822(91)90664-w. [DOI] [PubMed] [Google Scholar]
- 14.Winarski K.L., Tang J., Klenow L., Lee J., Coyle E.M., Manischewitz J., Turner H.L., Takeda K., Ward A.B., Golding H., Khurana S. Antibody-dependent enhancement of influenza disease promoted by increase in hemagglutinin stem flexibility and virus fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 2019;116(30):15194–15199. doi: 10.1073/pnas.1821317116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., Tang H., Nishiura K., Peng J., Tan Z., Wu T., Cheung K.W., Chan K.H., Alvarez X., Qin C., Lackner A., Perlman S., Yuen K.Y., Chen Z. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., Kase R., Sekiguchi S., Morita K., Hishima T., Suzuki H., Karamatsu K., Yasutomi Y., Shida H., Kidokoro M., Mizuno K., Matsushima K., Kohara M. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 2008;181(9):6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- 17.Tseng C.-T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., Peters C.J., Couch R.B., Poehlmann S. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS ONE. 2012;7(4):e35421. doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal A.S., Tao X., Algaissi A., Garron T., Narayanan K., Peng B.-H., Couch R.B., Tseng C.-T. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum. Vaccin. Immunother. 2016;12(9):2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricke DO D.O. Two different antibody-dependent enhancement (ADE) risks for SARS-CoV-2 antibodies. Front. Immunol. 2021:12:640093. doi: 10.3389/fimmu.2021.640093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5(10):1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 21.Torres V.V.L., Coggon C.F., Wells T.J. Antibody-dependent enhancement of bacterial disease: prevalence. Mech. Treatment Infect. Immun. 2021;89(4):e00054–e121. doi: 10.1128/IAI.00054-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor A., Foo S.-S., Bruzzone R., Vu Dinh L., King N.J.C., Mahalingam S. Fc receptors in antibody-dependent enhancement of viral infections. Immunol. Rev. 2015;268(1):340–364. doi: 10.1111/imr.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh C.S., Yang J.Y., Liu W.T., Huang J.C., Chen Y.M.A., Wang S.F. SARS coronavirus has antibody-dependent enhancement (ADE) effect through the autologous antibodies against envelope spikes on Fcγ receptor expressing cells. J. Virus. Erad. 2016;2:48. doi: 10.1016/S2055-6640(20)31216-4. [DOI] [Google Scholar]
- 24.Lee N., Chan P.K.S., Ip M., Wong E., Ho J., Ho C., Cockram C.S., Hui D.S. Anti-SARS-CoV IgG response in relation to disease severity of severe acute respiratory syndrome. J. Clin. Virol. 2006;35(2):179–184. doi: 10.1016/j.jcv.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales-Núñez J.J., Muñoz-Valle J.F., Torres-Hernández P.C., Hernández-Bello J. Overview of neutralizing antibodies and their potential in COVID-19. Vaccines. 2021;9(12):1376. doi: 10.3390/vaccines9121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth M.H., Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.-E., Barbry P., Leslie A., Kiem H.-P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J., Banovich N., Barbry P., Brazma A., Desai T., Duong T.E., Eickelberg O., Falk C., Farzan M., Glass I., Haniffa M., Horvath P., Hung D., Kaminski N., Krasnow M., Kropski J.A., Kuhnemund M., Lafyatis R., Lee H., Leroy S., Linnarson S., Lundeberg J., Meyer K., Misharin A., Nawijn M., Nikolic M.Z., Ordovas-Montanes J., Pe’er D., Powell J., Quake S., Rajagopal J., Tata P.R., Rawlins E.L., Regev A., Reyfman P.A., Rojas M., Rosen O., Saeb-Parsy K., Samakovlis C., Schiller H., Schultze J.L., Seibold M.A., Shalek A.K., Shepherd D., Spence J., Spira A., Sun X., Teichmann S., Theis F., Tsankov A., van den Berge M., von Papen M., Whitsett J., Xavier R., Xu Y., Zaragosi L.-E., Zhang K. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Deng T., Zhang Y., Niu W., Nie Q., Yang S., Liu P., Pei P., Chen L., Li H., Cao B. ACE2 can act as the secondary receptor in the FcγR-dependent ADE of SARS-CoV-2 infection. iScience. 2022;25(1):103720. doi: 10.1016/j.isci.2021.103720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu J., Sasaki T., Yamanaka A., Ichihara Y., Koketsu R., Samune Y., Cruz P., Sato K., Tanga N., Yoshimura Y., Murakami A., Yamada M., Itoi K., Nakayama E.E., Miyazaki K., Shioda T. The potential of COVID-19 patients' sera to cause antibody-dependent enhancement of infection and IL-6 production. Sci. Rep. 2021;11(1):23713. doi: 10.1038/s41598-021-03273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen X.-R., Li Q., Li H.-L., Wang X., Wang Q., Zheng X.-S., Geng R., Zhang Y.-L., Li B., Jiang R.-D., Liu M.-Q., Zhu Y., Zhang W., Yang X.-L., Peng K., Zhou P. Antibody-dependent enhancement of SARS-CoV-2 infection of human immune cells. In vitro assessment provides insight in COVID-19 pathogenesis. Viruses. 2021;13(12):2483. doi: 10.3390/v13122483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.T. Maemura, M. Kuroda, T. Armbrust, S. Yamayoshi, P.J. Halfmann, Y. Kawaoka, Antibody-dependent enhancement of SARS-CoV-2 infection is mediated by the IgG receptors FcγRIIA and FcγRIIIA but does not contribute to aberrant cytokine production by macrophages, mBio 12 (5) (2021) e0198721. doi: 10.1128/mBio.01987-21. Epub 2021 Sep 28. PMID: 34579572; PMCID: PMC8546849. [DOI] [PMC free article] [PubMed]
- 31.Panaro S., Cattalini M. The spectrum of manifestations of severe acute respiratory syndrome-coronavirus 2 (SARS-CoV2) infection in children: what we can learn from multisystem inflammatory syndrome in children (MIS-C) Front. Med. (Lausanne) 2021;8 doi: 10.3389/fmed.2021.747190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty S., Gonzalez J.C., Sievers B.L., Mallajosyula V., Chakraborty S., Dubey M., Ashraf U., Cheng B.-L., Kathale N., Tran K.Q.T., Scallan C., Sinnott A., Cassidy A., Chen S.T., Gelbart T., Gao F., Golan Y., Ji X., Kim-Schulze S., Prahl M., Gaw S.L., Gnjatic S., Marron T.U., Merad M., Arunachalam P.S., Boyd S.D., Davis M.M., Holubar M., Khosla C., Maecker H.T., Maldonado Y., Mellins E.D., Nadeau K.C., Pulendran B., Singh U., Subramanian A., Utz P.J., Sherwood R., Zhang S., Jagannathan P., Tan G.S., Wang T.T. Early non-neutralizing, afucosylated antibody responses are associated with COVID-19 severity. Sci. Transl. Med. 2022;14(635) doi: 10.1126/scitranslmed.abm7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thulin N.K., Brewer R.C., Sherwood R., Bournazos S., Edwards K.G., Ramadoss N.S., Taubenberger J.K., Memoli M., Gentles A.J., Jagannathan P., Zhang S., Libraty D.H., Wang T.T. Maternal anti-dengue IgG fucosylation predicts susceptibility to dengue disease in infants. Cell Rep. 2020;31(6):107642. doi: 10.1016/j.celrep.2020.107642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klarić L., Tsepilov Y.A., Stanton C.M., Mangino M., Sikka T.T., Esko T., Pakhomov E., Salo P., Deelen J., McGurnaghan S.J., Keser T., Vučković F., Ugrina I., Krištić J., Gudelj I., Štambuk J., Plomp R., Pučić-Baković M., Pavić T., Vilaj M., Trbojević-Akmačić I., Drake C., Dobrinić P., Mlinarec J., Jelušić B., Richmond A., Timofeeva M., Grishchenko A.K., Dmitrieva J., Bermingham M.L., Sharapov S.Z., Farrington S.M., Theodoratou E., Uh H.-W., Beekman M., Slagboom E.P., Louis E., Georges M., Wuhrer M., Colhoun H.M., Dunlop M.G., Perola M., Fischer K., Polasek O., Campbell H., Rudan I., Wilson J.F., Zoldoš V., Vitart V., Spector T., Aulchenko Y.S., Lauc G., Hayward C. Glycosylation of immunoglobulin G is regulated by a large network of genes pleiotropic with inflammatory diseases. Sci. Adv. 2020;6(8) doi: 10.1126/sciadv.aax0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen W., Zhang J., Qin X., Wang W., Xu M., Wang L.-F., Xu C., Tang S., Liu P., Zhang L., Liu X., Zhang Y., Yi C., Hu Z., Yi Y. SARS-CoV-2 neutralizing antibody levels are correlated with severity of COVID-19 pneumonia. Biomed. Pharmacother. 2020;130:110629. doi: 10.1016/j.biopha.2020.110629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurano M., Ohmiya H., Kishi Y., Okada J., Nakano Y., Yokoyama R., Qian C., Xia F., He F., Zheng L., Yu Y., Jubishi D., Okamoto K., Moriya K., Kodama T., Yatomi Y. Measurement of SARS-CoV-2 antibody titers improves the prediction accuracy of COVID-19 maximum severity by machine learning in non-vaccinated patients. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.811952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening covid-19: a randomized clinical trial. JAMA. 2020;324:460–470. doi: 10.1001/jama.2020.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agarwal A., Mukherjee A., Kumar G., Chatterjee P., Bhatnagar T., Malhotra P. Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) BMJ. 2020;371 doi: 10.1136/bmj.m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe Y., Bowden T.A., Wilson I.A., Crispi M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta Gen. Subj. 2019;1863(10):1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C.-C., Watanabe Y., Wu N.C., Han J., Kumar S., Pholcharee T., Seabright G.E., Allen J.D., Lin C.-W., Yang J.-R., Liu M.-T., Wu C.-Y., Ward A.B., Crispin M., Wilson I.A., Zhou T. A cross-neutralizing antibody between HIV-1 and influenza virus. PLoS Pathog. 2021;17(3):e1009407. doi: 10.1371/journal.ppat.1009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mannar D., Leopold K., Subramaniam S. Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry. Sci. Rep. 2021;11(1):12448. doi: 10.1038/s41598-021-91746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tso F.Y., Lidenge S.J., Peña P.B., Clegg A.A., Ngowi J.R., Mwaiselage J., Ngalamika O., Julius P., West J.T., Wood C. High prevalence of pre-existing serological cross-reactivity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in sub-Saharan Africa. Int. J. Infect. Dis. 2021;102:577–583. doi: 10.1016/j.ijid.2020.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overton E.T., Weir I.R., Zanni M.V., Fischinger S., MacArthur R.D., Aberg J.A., Fitch K.V., Frank M., Albrecht H., Goodenough E., Rhame F.S., Fichtenbaum C.J., Bloomfield G.S., Malvestutto C., Supparatpinyo K., McCallum S., Douglas P.S., Alter G., Ribaudo H., Grinspoon S.K. Asymptomatic SARS-CoV-2 infection is common among ART-treated people with HIV. J. Acquir. Immune Defic. Syndr. 2022 doi: 10.1097/QAI.0000000000003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osei S.A., Biney R.P., Anning A.S., Nortey L.N., Ghartey-Kwansah G. Low incidence of COVID-19 case severity and mortality in Africa; Could malaria co-infection provide the missing link? BMC Infect. Dis. 2022;22(1):78. doi: 10.1186/s12879-022-07064-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Achan J., Serwanga A., Wanzira H., Kyagulanyi T., Nuwa A., Magumba G., Kusasira S., Sewanyana I., Tetteh K., Drakeley C., Nakwagala F., Aanyu H., Opigo J., Hamade P., Marasciulo M., Baterana B., Tibenderana J.K. Current malaria infection, previous malaria exposure, and clinical profiles and outcomes of COVID-19 in a setting of high malaria transmission: an exploratory cohort study in Uganda. Lancet Microbe. 2022;3(1):e62–e71. doi: 10.1016/S2666-5247(21)00240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perween R., PraveenKumar M., Shrivastava T., Parray H.A., Singh V., Singh S., Chiranjivi A., Jakhar K., Sonar S., Tiwari M., Reema, Panchal A.K., Sharma C., Rathore D.K., Ahamed S., Samal S., Mani S., Bhattacharyya S., Das S., Luthra K., Kumar R. The SARS CoV-2 spike directed non-neutralizing polyclonal antibodies cross-react with Human immunodeficiency virus (HIV-1) gp41. Int. Immunopharmacol. 2021;101:108187. doi: 10.1016/j.intimp.2021.108187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra N., Kumar S., Singh S., Bansal T., Jain N., Saluja S., Kumar R., Bhattacharyya S., Palanichamy J.K., Mir R.A., Sinha S., Luthra K., Subbarao K. Cross-neutralization of SARS-CoV-2 by HIV-1 specific broadly neutralizing antibodies and polyclonal plasma. PLoS Pathog. 2021;17(9):e1009958. doi: 10.1371/journal.ppat.1009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S.S. Tan, K.L. Chew, S. Saw, R. Jureen, S. Sethi, Cross-reactivity of SARS-CoV-2 with HIV chemiluminescent assay leading to false-positive results, J. Clin. Pathol. 74 (9) (2021) 614, doi: 10.1136/jclinpath-2020-206942. [DOI] [PMC free article] [PubMed]
- 49.Garrido J.L., Medina M.A., Bravo F., McGee S., Fuentes-Villalobos F., Calvo M., Pinos Y., Bowman J.W., Bahl C.D., Barria M.I., Brachman R.A., Alvarez R.A. IgG targeting distinct seasonal coronavirus- conserved SARS-CoV-2 spike subdomains correlates with differential COVID-19 disease outcomes. Cell Rep. 2022;39(9):110904. doi: 10.1016/j.celrep.2022.110904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su W., Wang H., Sun C., Li N., Guo X., Song Q., Liang Q., Liang M., Ding X., Sun Y. The association between previous influenza vaccination and COVID-19 infection risk and severity: a systematic review and meta-analysis. Am. J. Prev. Med. 2022 doi: 10.1016/j.amepre.2022.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murugavelu P., Perween R., Shrivastava T., Singh V., Ahmad Parray H., Singh S., Chiranjivi A.K., Thiruvengadam R., Singh S., Yadav N., Jakhar K., Sonar S., Mani S., Bhattacharyya S., Sharma C., Vishwakarma P., Khatri R., Kumar Panchal A., Das S., Ahmed S., Samal S., Kshetrapal P., Bhatnagar S., Luthra K., Kumar R. Non-neutralizing SARS CoV-2 directed polyclonal antibodies demonstrate cross-reactivity with the HA glycans of influenza virus. Int. Immunopharmacol. 2021;99:108020. doi: 10.1016/j.intimp.2021.108020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K.R., Dillen J.R., Powell A.E., Chen A., Maher C., Yin L.i., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N.M., Logue J., Iqbal N.T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippà P.E., Franzetti-Pellanda A., Garzoni C., Halfmann P.J., Kawaoka Y., Hebner C., Purcell L.A., Piccoli L., Pizzuto M.S., Walls A.C., Diamond M.S., Telenti A., Virgin H.W., Lanzavecchia A., Snell G., Veesler D., Corti D. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602(7898):664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiecco G., Storti S., Degli Antoni M., Focà E., Castelli F., Quiros-Roldan E. Omicron genetic and clinical peculiarities that may overturn SARS-CoV-2 pandemic: a literature review. Int. J. Mol. Sci. 2022:23(4):1987. doi: 10.3390/ijms23041987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., Wang Z., Gaebler C., Caskey M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2022;386(6):599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iketani S., Liu L., Guo Y., Liu L., Chan J.-W., Huang Y., Wang M., Luo Y., Yu J., Chu H., Chik K.-H., Yuen T.-T., Yin M.T., Sobieszczyk M.E., Huang Y., Yuen K.-Y., Wang H.H., Sheng Z., Ho D.D. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604(7906):553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mensah A.A., Campbell H., Stowe J., Seghezzo G., Simmons R., Lacy J., Bukasa A., O'Boyle S., Ramsay M.E., Brown K., Ladhani S.N. Risk of SARS-CoV-2 reinfections in children: a prospective national surveillance study between January, 2020, and July, 2021, in England. Lancet Child Adolesc. Health. 2022;6(6):384–392. doi: 10.1016/S2352-4642(22)00059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall V.J., Foulkes S., Charlett A., Atti A., Monk E.J.M., Simmons R., Wellington E., Cole M.J., Saei A., Oguti B., Munro K., Wallace S., Kirwan P.D., Shrotri M., Vusirikala A., Rokadiya S., Kall M., Zambon M., Ramsay M., Brooks T., Brown C.S., Chand M.A., Hopkins S., Andrews N., Atti A., Aziz H., Brooks T., Brown C.S., Camero D., Carr C., Chand M.A., Charlett A., Crawford H., Cole M., Conneely J., D'Arcangelo S., Ellis J., Evans S., Foulkes S., Gillson N., Gopal R., Hall L., Hall V.J., Harrington P., Hopkins S., Hewson J., Hoschler K., Ironmonger D., Islam J., Kall M., Karagiannis I., Kay O., Khawam J., King E., Kirwan P., Kyffin R., Lackenby A., Lattimore M., Linley E., Lopez-Bernal J., Mabey L., McGregor R., Miah S., Monk EJM, Munro K., Naheed Z., Nissr A., O'Connell A.M., Oguti B., Okafor H., Organ S., Osbourne J., Otter A., Patel M., Platt S., Pople D., Potts K., Ramsay M., Robotham J., Rokadiya S., Rowe C., Saei A., Sebbage G., Semper A., Shrotri M., Simmons R., Soriano A., Staves P., Taylor S., Taylor A., Tengbe A., Tonge S., Vusirikala A., Wallace S., Wellington E., Zambon M., Corrigan D., Sartaj M., Cromey L., Campbell S., Braithwaite K., Price L., Haahr L., Stewart S., Lacey E.D., Partridge L., Stevens G., Ellis Y., Hodgson H., Norman C., Larru B., Mcwilliam S., Winchester S., Cieciwa P., Pai A., Loughrey C., Watt A., Adair F., Hawkins A., Grant A., Temple-Purcell R., Howard J., Slawson N., Subudhi C., Davies S., Bexley A., Penn R., Wong N., Boyd G., Rajgopal A., Arenas-Pinto A., Matthews R., Whileman A., Laugharne R., Ledger J., Barnes T., Jones C., Botes D., Chitalia N., Akhtar S., Harrison G., Horne S., Walker N., Agwuh K., Maxwell V., Graves J., Williams S., O'Kelly A., Ridley P., Cowley A., Johnstone H., Swift P., Democratis J., Meda M., Callens C., Beazer S., Hams S., Irvine V., Chandrasekaran B., Forsyth C., Radmore J., Thomas C., Brown K., Roberts S., Burns P., Gajee K., Byrne T.M., Sanderson F., Knight S., Macnaughton E., Burton BJL, Smith H., Chaudhuri R., Hollinshead K., Shorten R.J., Swan A., Shorten R.J., Favager C., Murira J., Baillon S., Hamer S., Gantert K., Russell J., Brennan D., Dave A., Chawla A., Westell F., Adeboyeku D., Papineni P., Pegg C., Williams M., Ahmad S., Ingram S., Gabriel C., Pagget K., Cieciwa P., Maloney G., Ashcroft J., Del Rosario I., Crosby-Nwaobi R., Reeks C., Fowler S., Prentice L., Spears M., McKerron G., McLelland-Brooks K., Anderson J., Donaldson S., Templeton K., Coke L., Elumogo N., Elliott J., Padgett D., Mirfenderesky M., Cross A., Price J., Joyce S., Sinanovic I., Howard M., Lewis T., Cowling P., Potoczna D., Brand S., Sheridan L., Wadams B., Lloyd A., Mouland J., Giles J., Pottinger G., Coles H., Joseph M., Lee M., Orr S., Chenoweth H., Auckland C., Lear R., Mahungu T., Rodger A., Penny-Thomas K., Pai S., Zamikula J., Smith E., Stone S., Boldock E., Howcroft D., Thompson C., Aga M., Domingos P., Gormley S., Kerrison C., Marsh L., Tazzyman S., Allsop L., Ambalkar S., Beekes M., Jose S., Tomlinson J., Jones A., Price C., Pepperell J., Schultz M., Day J., Boulos A., Defever E., McCracken D., Brown K., Gray K., Houston A., Planche T., Pritchard Jones R., Wycherley D., Bennett S., Marrs J., Nimako K., Stewart B., Kalakonda N., Khanduri S., Ashby A., Holden M., Mahabir N., Harwood J., Payne B., Court K., Staines N., Longfellow R., Green M.E., Hughes L.E., Halkes M., Mercer P., Roebuck A., Wilson-Davies E., Gallego L., Lazarus R., Aldridge N., Berry L., Game F., Reynolds T., Holmes C., Wiselka M., Higham A., Booth M., Duff C., Alderton J., Jory H., Virgilio E., Chin T., Qazzafi M.Z., Moody A.M., Tilley R., Donaghy T., Shipman K., Sierra R., Jones N., Mills G., Harvey D., Huang YWJ, Birch J., Robinson L., Board S., Broadley A., Laven C., Todd N., Eyre D.W., Jeffery K., Dunachie S., Duncan C., Klenerman P., Turtle L., De Silva T., Baxendale H., Heeney J.L. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN) Lancet. 2021;397(10283):1459–1469. doi: 10.1016/S0140-6736(21)00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. Morris, S. Anjan, S. Pallikkuth, P. Frattaroli, S. Courel, A. Fernandez, et al., Reinfection with SARS-CoV-2 in solid-organ transplant recipients: Incidence density and convalescent immunity prior to reinfection, Transpl. Infect. Dis. (2022) e13827. doi: 10.1111/tid.13827. [DOI] [PMC free article] [PubMed]
- 59.F. Touret, C. Baronti, B. Pastorino, In vitro activity of therapeutic antibodies against SARS-CoV-2 Omicron BA.1 and BA.2 (2022) available online at Research Square, doi: 10.21203/rs.3.rs-1415749/v1 (preprint). [DOI] [PMC free article] [PubMed]
- 60.Westendorf K., Žentelis S., Wang L., Foster D., Vaillancourt P., Wiggin M., Lovett E., van der Lee R., Hendle J., Pustilnik A., Sauder J.M., Kraft L., Hwang Y., Siegel R.W., Chen J., Heinz B.A., Higgs R.E., Kallewaard N.L., Jepson K., Goya R., Smith M.A., Collins D.W., Pellacani D., Xiang P., de Puyraimond V., Ricicova M., Devorkin L., Pritchard C., O’Neill A., Dalal K., Panwar P., Dhupar H., Garces F.A., Cohen C.A., Dye J.M., Huie K.E., Badger C.V., Kobasa D., Audet J., Freitas J.J., Hassanali S., Hughes I., Munoz L., Palma H.C., Ramamurthy B., Cross R.W., Geisbert T.W., Menachery V., Lokugamage K., Borisevich V., Lanz I., Anderson L., Sipahimalani P., Corbett K.S., Yang E.S., Zhang Y.i., Shi W., Zhou T., Choe M., Misasi J., Kwong P.D., Sullivan N.J., Graham B.S., Fernandez T.L., Hansen C.L., Falconer E., Mascola J.R., Jones B.E., Barnhart B.C. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39(7):110812. doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y.-T., Allen R.D., Kim K., Shafee N., Gonzalez A.J., Nguyen M.N., Valentine K.M., Cao X., Lu L., Pai C.-I., Johnson S., Kerwin L., Zhou H., Zhang Y., Shresta S. SARS-CoV-2 monoclonal antibodies with therapeutic potential: Broad neutralizing activity and No evidence of antibody-dependent enhancement. Antiviral. Res. 2021;195:105185. doi: 10.1016/j.antiviral.2021.105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nordström P., Ballin M., Nordström A. Risk of SARS-CoV-2 reinfection and COVID-19 hospitalisation in individuals with natural and hybrid immunity: a retrospective, total population cohort study in Sweden. Lancet Infect. Dis. 2022;S1473–3099(22):00143–148. doi: 10.1016/S1473-3099(22)00143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130(4):1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson S.I., Madzorera V.S., Spencer H., Manamela N.P., van der Mescht M.A., Lambson B.E., Oosthuysen B., Ayres F., Makhado Z., Moyo-Gwete T., Mzindle N., Motlou T., Strydom A., Mendes A., Tegally H., de Beer Z., Roma de Villiers T., Bodenstein A., van den Berg G., Venter M., de Oliviera T., Ueckermann V., Rossouw T.M., Boswell M.T., Moore P.L. SARS-CoV-2 Omicron triggers cross-reactive neutralization and Fc effector functions in previously vaccinated, but not unvaccinated, individuals. Cell Host Microbe. 2022;30(6):880–886.e4. doi: 10.1016/j.chom.2022.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall V., Foulkes S., Insalata F., Kirwan P., Saei A., Atti A., Wellington E., Khawam J., Munro K., Cole M., Tranquillini C., Taylor-Kerr A., Hettiarachchi N., Calbraith D., Sajedi N., Milligan I., Themistocleous Y., Corrigan D., Cromey L., Price L., Stewart S., de Lacy E., Norman C., Linley E., Otter A.D., Semper A., Hewson J., D’Arcangelo S., Chand M., Brown C.S., Brooks T., Islam J., Charlett A., Hopkins S. Protection against SARS-CoV-2 after Covid-19 Vaccination and Previous Infection. N. Engl. J. Med. 2022;386(13):1207–1220. doi: 10.1056/NEJMoa2118691. [DOI] [PMC free article] [PubMed] [Google Scholar]