Abstract
While there have been extensive analyses characterizing cellular and humoral responses across the severity spectrum in COVID-19, outcome predictors within severe COVID-19 remain less comprehensively elucidated. Furthermore, properties of antibodies (Abs) directed against viral antigens beyond spike and their associations with disease outcomes remain poorly defined. We perform deep molecular profiling of Abs directed against a wide range of antigenic specificities in severe COVID-19 patients. The profiles included canonical (spike [S], receptor-binding domain [RBD], and nucleocapsid [N]) and non-canonical (orf3a, orf8, nsp3, nsp13, and membrane [M]) antigenic specificities. Notably, multivariate Ab profiles directed against canonical or non-canonical antigens are equally discriminative of survival in severe COVID-19. Intriguingly, pre-pandemic healthy controls have cross-reactive Abs directed against nsp13, a protein conserved across coronaviruses. Consistent with these findings, a model built on Ab profiles for endemic coronavirus antigens also predicts COVID-19 outcome. Our results suggest the importance of studying Abs targeting non-canonical severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and endemic coronavirus antigens in COVID-19.
Keywords: systems immunology, antibody profiling, COVID-19, non-canonical antigens, endemic coronaviruses, cross-reactivity, nsp13
Graphical abstract
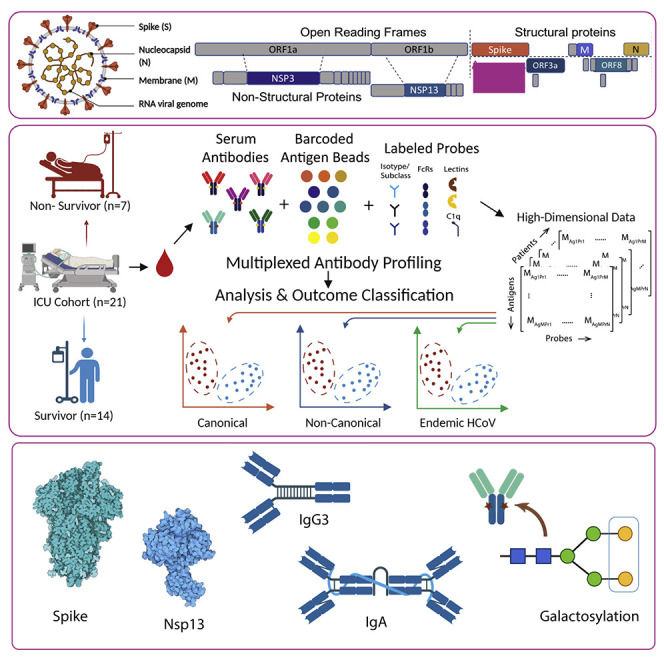
Peddireddy et al. perform deep molecular profiling of antibodies in severe COVID-19 patients and identify multivariate humoral signatures of outcome bifurcation involving canonical and non-canonical SARS-CoV-2, as well as endemic CoV antigenic specificities. Our results suggest the importance of Abs targeting non-canonical SARS-CoV-2 antigens, as well as those directed against endemic coronaviruses in favorable outcomes of severe COVID-19.
Introduction
The continued spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) remains a significant threat globally, in spite of deployment of effective vaccines, due to newly emerging variants. A critical challenge is posed by the high symptomatic heterogeneity and unpredictable course of disease progression in COVID-19 (Rodebaugh et al., 2021). Progression from asymptomatic infection or mild symptoms to severe disease has been broadly linked to advanced age and certain comorbidities (Ng et al., 2021). However, for those with severe COVID-19 disease, there is still a lack of personalized predictors of the course of disease and its outcomes. Although significant effort has been dedicated to establishing the immunological underpinnings of COVID-19 (Carvalho et al., 2021), the immunological drivers of mortality and survival outcomes within severe COVID-19 patients remain unclear. Recently, we identified dysregulated monocyte states as key predictors of outcomes within severe COVID-19 (Cillo et al., 2021).
The humoral response directed against selected SARS-CoV-2 antigens, e.g., spike (S) and nucleocapsid (N), or their sub-domains, e.g., receptor-binding domain (RBD) of S, which taken together we term as canonical antigens here, have been extensively studied (Atyeo et al., 2020; Bartsch et al., 2021; Zohar et al., 2020). Relative to those with asymptomatic infection or with mild symptoms, antibody titers against canonical antigens are higher in patients with severe disease, leading to early concerns about antibodies contributing to disease pathology, potentially via mechanisms like antibody-dependent enhancement (ADE) (Iwasaki and Yang, 2020; Lee et al., 2020) or via the antibody-mediated activation of inflammatory pathways, especially since proinflammatory antibody Fc structures have been found to correlate with disease severity (Bye et al., 2021; Chakraborty et al., 2020; Hoepel et al., 2021; Larsen et al., 2021). Vaccine studies, meanwhile, have shown that titers of vaccine-elicited neutralizing antibodies directed against the S antigen are a key correlate of protection (Khoury et al., 2021; Sadarangani et al., 2021). Recently, longitudinal profiling of antibodies against canonical antigens, after natural infection, has revealed distinct temporal trajectories of immunoglobulin (Ig) subclasses and non-neutralizing functions of these antibodies that track with disease severity and outcome (Zohar et al., 2020). However, it remains to be determined which of these features of antibodies (Abs) directed against canonical antigens are predictive of recovery from severe COVID-19 disease.
Beyond the canonical antigens, the SARS-CoV-2 genome is predicted to encode up to 25 additional proteins (Gordon et al., 2020), which we term here as non-canonical antigens. It has been observed that cellular and humoral immune responses directed against these non-canonical targets also arise upon SARS-CoV-2 infection (Grifoni et al., 2020; Shrock et al., 2020). Ab responses against some non-canonical antigens have been shown to be serological markers of COVID-19 at early and late time points of illness (Hachim et al., 2020). However, it remains to be determined whether Abs directed against non-canonical antigens versus those directed against canonical antigens can independently or combinatorially predict the outcomes of severe COVID-19 disease. Given the prolonged exposure to a high viral burden in patients with severe COVID-19, Abs against non-canonical antigens may play a role in protection or exacerbation of disease. In the context of other viral infections, generation of Abs directed against non-neutralizing targets has been linked to either protective, neutral, or detrimental effects (Lamere et al., 2011; To et al., 2012). Indeed, in the context of COVID-19 as well, there is evidence pointing to the lack of selective targeting of S versus N antigens being linked to disease severity in COVID-19 (Zohar et al., 2020). In addition, it is notable that a number of these non-canonical SARS-CoV-2 protein antigens are known to share high sequence similarity with the corresponding proteins in endemic human coronaviruses (eHCoVs) (Hicks et al., 2021). Whether prior eHCoV exposure and the associated immune memory affect outcome after SARS-CoV-2 infection remains an unsettled debate, and both protective and pathological effects have been reported in recent literature (Aydillo et al., 2021; Guo et al., 2021). Thus, tracking the Ab responses against these antigens could provide important insights in formulating improved SARS-CoV-2 as well as pan-coronavirus vaccines beyond the S-based formulations in current use.
To address these questions, we developed a highly multiplexed, sample-sparing SARS-CoV-2 humoral profiling platform that measures biophysical properties of antigen-specific Abs directed against a broad set of canonical and non-canonical antigens as well as eHCoV antigens, including their isotypes, subclasses, Fc receptor binding, and glycosylation from corresponding serum samples. Importantly, we found that Abs directed against canonical and non-canonical antigens were independently equally predictive of disease outcomes. Notably, pre-pandemic healthy controls were found to have Abs against specific non-canonical antigens with high similarity to those in eHCoVs. Finally, eHCoV-specific Abs were themselves also predictive of outcome in severe COVID-19. Thus, our results suggest the importance of Abs targeting non-canonical SARS-CoV-2 and endemic CoV antigens in favorable outcomes of severe COVID-19.
Results
Multivariate antibody responses against canonical antigens are predictive of severe COVID-19 outcomes
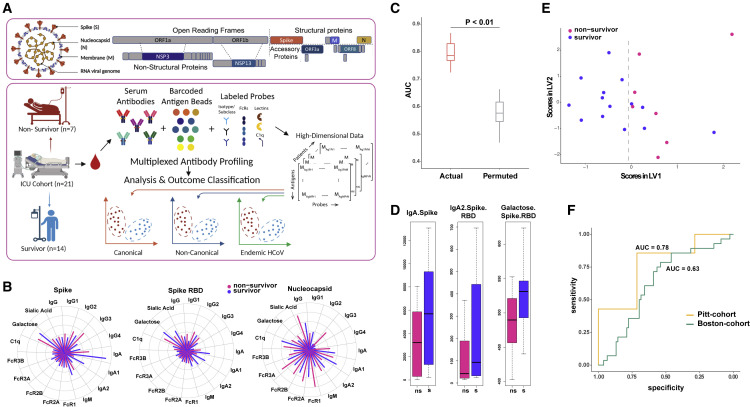
Using our highly multiplexed, SARS-CoV-2 Ab profiling platform, we characterized and quantified serum Abs directed against canonical antigens for 21 severe (14 survivors and 7 non-survivors) COVID-19 patients from blood drawn soon after their intensive care unit (ICU) admission (Figure 1A, Pittsburgh UPMC cohort). Demographic details for these patients have been previously described (Bain et al., 2021; Cillo et al., 2021). Briefly, COVID-19 was diagnosed in these subjects based on reference-standard nasopharyngeal swab SARS-CoV-2 qPCR. Patients were admitted to the ICU a median of 6 days after symptom onset, and serum was collected from these patients within 24 h post-enrollment in the study. Of a wide range of measured clinical covariates, higher age and higher BMI showed trends of being associated with higher mortality and the administration of glucocorticoids trended to being associated with survival; however, none of these factors were of univariate significance given the sample size of the cohort (Bain et al., 2021; Cillo et al., 2021). Further, a multivariate model built using clinical features was not significantly predictive (in a k-fold cross-validation framework with permutation testing) either, demonstrating that these clinical features have limited utility in predicting outcome bifurcation. This underscored the need for high dimensional profiling immune system features, such as deep Ab profiling.
Figure 1.
Multivariate antibody responses against canonical antigens are predictive of severe COVID-19 outcomes
(A) Conceptual overview of the SARS-CoV-2 antibody profiling platform used to characterize and quantify serum Abs directed against canonical and non-canonical antigens.
(B) Polar plots illustrating measured Ab features against canonical Ag specificities—spike, spike RBD, and nucleocapsid.
(C) Performance of LASSO model to discriminate between survivors and non-survivors built using deep humoral profiles against canonical Ag specificities. Model performance is measured in a k-fold cross-validation framework with permutation testing. Actual denotes the performance of the model, built on real data. Permuted denotes performance of the model on shuffled data in a matched cross-validation framework (negative control).
(D) LASSO-selected features from model built using deep humoral profiles against canonical Ag specificities. ns, non-survivor; s, survivor.
(E) PLS-DA using only the LASSO-selected features from the model in (D) to discriminate between survivors and non-survivors.
(F) Performance of the model (built using the Pitt cohort) on an orthogonal (Boston) cohort.
Unlike well-characterized differences in S Ab titers of COVID-19 patients across the severity spectrum, we wished to determine whether Ab profiles at the point of ICU admission could predict bifurcation of subsequent outcomes—survival versus death (Figure 1A). Furthermore, unlike previous studies that have focused on temporal differences (Zohar et al., 2020), we focused on Ab profiles measured at the point of ICU admission, as it represents a clinically relevant and actionable time point. Further, as noted above, there were not significant differences in other characteristics, including age, treatment, and viral loads, between the survivors and non-survivors.
We observed that both survivors and non-survivors had significantly higher Abs across canonical specificities than pre-pandemic healthy controls, confirming the quality and specificity of our assay (Figures S1A–S1O). However, the univariate differences between survivors and non-survivors were not striking (Figures 1B and S1A–S1O). Examination of pairwise relationships also did not reveal significant differences in the underlying correlation structure (Figure S1P). No single feature was discriminative by clinical outcomes. Therefore, we pursued a multivariate machine-learning approach that incorporates different quantitative and qualitative aspects of the Ab response to determine whether it could discriminate patients by clinical outcome. We used a two-step machine-learning approach, as previously described (Ackerman et al., 2018; Das et al., 2020; Lu et al., 2020; Sadanand et al., 2018; Suscovich et al., 2020), to identify a minimal set of predictive Ab features that could discriminate between survivors and non-survivors. Our approach comprised feature selection using the least absolute shrinkage and selection operator (LASSO): the use of L1 regularization on high-dimensional data (i.e., data where the number of Ab features far exceeds the number of subjects) that helps prevent overfitting (Ackerman et al., 2018; Das et al., 2020; Lu et al., 2020; Sadanand et al., 2018; Suscovich et al., 2020). This was followed by classification using the down-selected features. We found that a model generated using this approach was significantly predictive of outcome as measured in a k-fold cross-validation framework with permutation testing (Figure 1C; STAR Methods). We also evaluated the model using three additional metrics—precision, recall, and F1 score. Based on all three metrics, the model performed significantly better than a negative-control model (based on permutation testing), across folds and replicates of k-fold cross-validation (Figures S1Q–S1S). Interestingly, a multivariate model that includes clinical features in addition to the deep Ab profiles does not perform better than the above model built on deep Ab profiles. This suggests that the clinical features do not add anything additional in terms of discrimination beyond the deep Ab profiles.
The model was based on three features—anti-S IgA, anti-S RBD IgA2, and RBD-directed Ab galactosylation (Figure 1D). These features were stable across folds and replicates of k-fold cross-validation (Figure S1T). These sensitivity analyses demonstrate that these are indeed the most stable features that serve as a minimal set of robust predictive biomarkers of outcome bifurcation. The same features also did not show clear trends in stratifying acute respiratory distress syndrome (ARDS) survivors and non-survivors (Figure S1U). While given the small size of this cohort, we are underpowered to make definite conclusions; these results suggest there are both similarities and differences in Ab features that can stratify COVID-19 survivors and non-survivors versus non-COVID ARDS survivors and non-survivors.
As an orthogonal way to visualize the stratification achieved by these Ab features, we performed partial least squares discriminant analyses (PLS-DA) using just these three down-selected features. The PLS-DA demonstrated that these three markers were able to stratify the survivors and the non-survivors (Figure 1E). Notably, all three Ab features were higher in survivors compared with non-survivors (Figure 1D), suggesting that higher levels of IgA Abs directed against the S protein or its RBD and the increased levels of galactosylation of RBD-specific Abs are associated with favorable outcomes.
To validate the robustness of the uncovered Ab features, we tested the performance of our model on an independent cohort of severe ICU patients (Boston Massachusetts General Hospital [MGH] cohort; Zohar et al., 2020). The Boston cohort had patients in three categories—moderate (n = 82), severe (n = 76), and deceased (n = 35). However, they measured fewer features (probes and antigens) for each sample. Critically, we had no role in the study design or recruitment strategy for this cohort. Our model generated using the Pittsburgh cohort remained significantly predictive for the Boston cohort Figure 1F), albeit with a slightly decreased performance. We attributed this reduction in performance to the availability of fewer features in the Boston cohort dataset, specifically the lack of antigen-specific Ab glycosylation measurements, one of the three discriminating features for our model. The other two features (anti-S IgA and anti-RBD IgA2) exhibited identical univariate trends across the two cohorts and datasets (Figure S1V). Thus, the multivariate cross-prediction provides a lower-bound estimate of the performance of our model on an orthogonal cohort. Overall, our results demonstrate that a model built using Ab profiles corresponding to canonical specificities is robust, both to cross-validation and cross-prediction with a distinct cohort. More importantly, they demonstrate that, within severe COVID-19 patients, outcome bifurcation can be accurately predicted at the point of ICU admission based on IgA rather than IgG Abs directed against S and its RBD as well as RBD-specific Ab galactosylation.
Multivariate antibody responses against non-canonical antigens independently predict severe COVID-19 outcomes
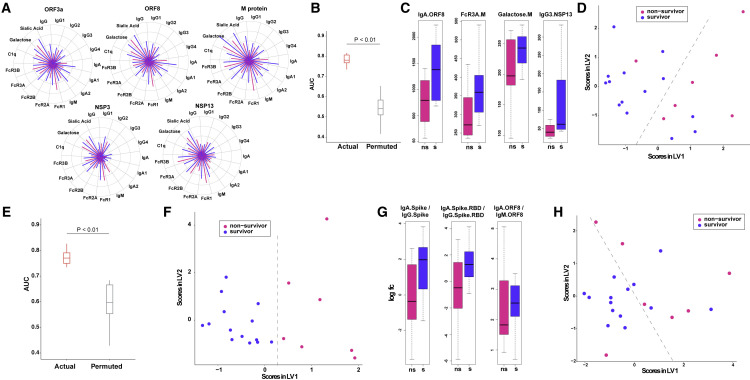
Next, we sought to examine the levels of Ab responses directed against non-canonical antigens (nsp3, nsp13, orf3a, and orf8) in patients with severe COVID-19 and whether they were independently predictive of outcome. We detected Ab responses to these non-canonical antigens both in the survivors and non-survivors, with no significant univariate differences between them (Figures 2A and S2A–S2O), similar to what was observed for canonical Ab specificities. Examination of pairwise relationships also did not reveal significant differences in the underlying correlation structure (Figure S2P). So we constructed a multivariate model, as described above, using only the Ab responses corresponding to non-canonical specificities. Strikingly, this model was also significantly predictive of outcomes (Figure 2B), and the performance of this model was as good as that of our previous model built using canonical specificities (Figures 1C and 2B). Model performance remained significantly better than a negative-control model, across folds and replicates of k-fold cross-validation with various evaluation metrics (Figures S2Q–S2S).
Figure 2.
Multivariate antibody responses against non-canonical antigens independently predict severe COVID-19 outcomes
(A) Polar plots illustrating measured antibody responses against non-canonical antigenic specificities ORF3a, ORF8, NSP3, NSP13, and M.
(B) Performance of LASSO model to discriminate between survivors and non-survivors built using deep humoral profiles against non-canonical Ag specificities. Model performance is measured in a k-fold cross-validation framework with permutation testing (negative control).
(C) LASSO-selected features from model built using deep humoral profiles against non-canonical Ag specificities.
(D) PLS-DA using only the LASSO-selected features from the model in (C) to discriminate between survivors and non-survivors.
(E) Performance of LASSO model to discriminate between survivors and non-survivors built using deep humoral profiles against canonical and non-canonical Ag specificities. Model performance is measured in a k-fold cross-validation framework with permutation testing (negative control).
(F) LASSO-selected features from combined canonical and non-canonical antigenic specificities.
(G) Post hoc feature selection based on the ratios of IgA to IgG and IgA to IgM.
(H) PLS-DA using only the ratios of IgA to IgG and IgA to IgM to discriminate between survivors and non-survivors.
The model selected four features: anti-orf8 IgA, anti-nsp13 IgG3, anti-membrane (M) Ab FcR3A binding, and anti-M Ab galactosylation (Figure 2C). As in the earlier analyses, these features were stable across folds and replicates of k-fold cross-validation (Figure S2T), demonstrating their robustness. The same features also did not show clear trends in stratifying ARDS survivors and non-survivors (Figure S2U). Analogous to our previous analyses, a PLS-DA visualization also demonstrated that these four features were able to stratify the survivors and the non-survivors (Figure 2D). Importantly, these four Ab features, unlike many others (Figures S2A–S2O), were higher in survivors. Our findings thus address an important question regarding higher Ab titers, especially against non-neutralizing, non-canonical target antigens being potentially associated with worse outcomes in severe COVID-19 (Lee et al., 2020). The results suggest that higher Ab titers with particular isotypes directed against specific canonical as well as non-canonical antigens are associated with favorable outcomes in severe disease.
As noted above, IgA Abs for both canonical (S and S-RBD) and non-canonical (orf8) antigens were higher in survivors compared with non-survivors (Figure 1, Figure 2D and 2C). Further, increased galactosylation of both RBD- and M-specific Abs was associated with favorable outcomes (Figure 1, Figure 2D and 2C). Thus, similar Ab profiles for both canonical and non-canonical specificities were associated with survival. This was next corroborated by constructing a predictive model by combining canonical and non-canonical specificities. As anticipated, the predictive performance of the model remained unchanged (Figure 2E), and it highlighted a subset of the Ab features revealed by those based on canonical and non-canonical Ag specificities alone (Figure S2V). Next, we examined whether the ratios of IgA/IgG or IgA/IgM Abs directed against S, RBD, and orf8 (antigens identified in the earlier analyses) were predictive of outcomes. This was a post hoc analysis (STAR Methods) where we focused on ratios of IgA/IgG Abs for features identified in the canonical and non-canonical models (Figure 2G). A multivariate PLS-DA visualization using just these three ratios discriminated between survivors and non-survivors (Figure 2H). These results reinforce the importance of IgA Abs directed against both canonical and non-canonical specificities as important predictors of outcome in severe COVID-19. Overall, our results demonstrate that Abs with particular non-canonical antigen specificities are independently as informative as those Abs directed against the canonical S protein in predicting severe COVID-19 disease mortality outcomes.
Antibodies directed against endemic CoV antigens as predictors of severe COVID-19 outcome
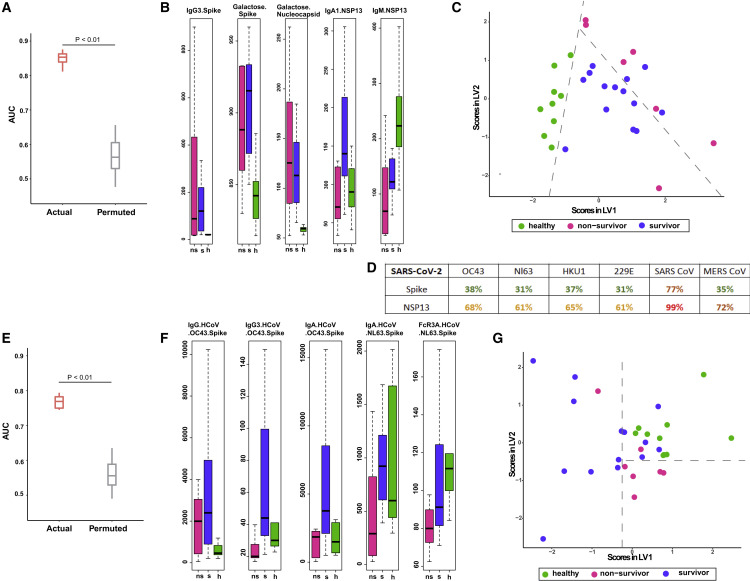
In the process of analyzing corresponding Ab profiles in pre-pandemic healthy controls, we noticed significant levels of reactivities to specific non-canonical SARS-CoV-2 antigens, particularly nsp13 and nsp3 (Figures S2A–S2O and S3A–S3P). This was unlike Abs directed against canonical antigens, including S, which were, on the other hand, very close to or at baseline in these controls (Figures S2A–S2O and S3A–S3O). Given this surprising finding of Ab reactivity to nsp13 and nsp3 in sera of pre-pandemic individuals, we hypothesized that these Abs may have been generated by prior infections of such individuals with eHCoVs and their cross-reactivity to SARS-Cov-2 nsp13 and nsp3 was a consequence of the greater conservation of these proteins across coronaviruses. To test this hypothesis, we analyzed the sequence similarity of nsp13 and S to corresponding antigens in SARS- and Middle East respiratory syndrome (MERS)-CoV and eHCoVs (Figure 3D). While S shares low sequence similarity to SARS-CoV and eHCoV S antigens, nsp13 has high sequence similarity to corresponding SARS- and MERS-CoV and eHCoV antigens (Figure 3D). Thus, Abs directed against endemic eHCoV nsp13 may cross-react with their SARS-CoV-2 homologs.
Figure 3.
Antibodies directed against endemic CoV antigens as predictors of severe COVID-19 outcome
(A) Performance of LASSO model to discriminate between survivors, non-survivors, and healthy controls built using deep humoral profiles against canonical and non-canonical Ag specificities. Model performance (three way) is measured in a k-fold cross-validation framework with permutation testing. Actual denotes the performance of the model, built on real data. Permuted denotes performance of the model on shuffled data in a matched cross-validation framework (negative control).
(B) LASSO-selected features from model built using deep humoral profiles against canonical and non-canonical Ag specificities. h, healthy.
(C) PLS-DA using only the LASSO-selected features from the model in (B) to discriminate between healthy controls, survivors, and non-survivors.
(D) Sequence similarities of SARS-CoV-2 spike and NSP13 with corresponding homologs in OC43, NL63, HKU1, 229E, SARS CoV, and MERS CoV.
(E) Performance of LASSO model to discriminate between survivors, non-survivors, and healthy controls built using deep humoral profiles against non-SARS-CoV-2 Ag specificities. Model performance is measured in a k-fold cross-validation framework with permutation testing (negative control) for outcome prediction between survivors, non-survivors, and healthy controls (three way).
(F) LASSO-selected features from model built using deep humoral profiles against non-SARS-CoV-2 Ag specificities.
(G) PLS-DA using only the LASSO-selected features from the model in (F) to discriminate between healthy controls, survivors, and non-survivors.
Given the possibility of pre-existing, cross-reactive Abs to SARS-Cov-2 antigens, we constructed a multivariate three-way machine-learning model that could discriminate not only between survivors and non-survivors but also pre-pandemic healthy controls. The three-way model performed even better (using all four metrics—area under the ROC curve [AUC], precision, recall, and F1 score) than the previous two-way models that discriminated between survivors and non-survivors (Figures 3A and S3Q–S3S). The LASSO model selected five features that reflected the intriguing trends described above. These features included anti-S IgG3, S-specific and N-specific Ab galactosylation, and anti-nsp13 IgA1 and IgM (Figure 3B). Analogous to our earlier analyses, a PLS-DA visualization also demonstrated that these five features were able to stratify survivors, non-survivors, and healthy controls (Figure 3C). Strikingly, while anti-S IgG3 was close to baseline in pre-pandemic healthy controls, the anti-nsp13 IgM was highest in pre-pandemic healthy controls and anti-nsp13 IgA1 was highest in survivors followed by healthy controls (Figure 3B). This raises the possibility that boosting of a pre-existing cross-reactive memory B cell response to nsp13 may account for the increased levels of these IgA Abs in severe COVID-19 patients and in turn their association with favorable outcomes.
To further test the possibility that boosting of pre-existing Abs to eHCoVs that may cross-react with SARS-CoV-2 antigens can associate with severe COVID-19 disease outcomes, we analyzed Abs directed against eHCoV antigens (OC43 S and NL63 S) and used that dataset to build a multivariate machine learning (ML) model. Remarkably, a multivariate model built only on Abs directed against OC43 S and NL63 S was significantly predictive of outcomes and was also independently able to accurately discriminate between survivors, non-survivors, and pre-pandemic healthy controls (Figure 3E). The model performance remained significantly better than a negative-control model (based on permutation testing) across folds and replicates of k-fold cross-validation when precision, recall, and F1 score were used as the evaluation metric (Figures S3T–S3V). Interestingly, while both survivors and non-survivors had significantly higher IgG Abs to OC43 S than healthy controls, IgA, IgG3 Abs, and Ab binding to Fc receptor 3A were higher in survivors and healthy controls compared with non-survivors (Figure 3F). IgA and IgG3 Abs against OC43 S were also found to be higher in survivors relative to healthy controls. A PLS-DA visualization also demonstrated that these five markers were able to stratify survivors, non-survivors, and healthy controls (Figure 3G). These findings suggest that pre-existing Abs to canonical and non-canonical antigens of eHCoVs and their boosting, especially of particular isotypes and subclasses, by SARS-CoV-2 infection may have a beneficial role in favorable outcomes in severe COVID-19.
Discussion
Our study represents the deepest humoral profile of Abs against canonical specificities and the first comprehensive profile of Abs directed against non-canonical antigens as well as those directed against eHCoV antigens, in the context of severe COVID-19 disease. Prior studies focusing on longitudinal profiling provide insights into how trajectories are predictive (Zohar et al., 2020), while profiling at a single early time point in this study provides insights specific to an actionable time point. Critically, we found that multivariate models incorporating Ab responses against both canonical and non-canonical antigens were both discriminative of outcome. Interestingly, similar molecular features of Abs for both canonical and non-canonical specificities drove outcome bifurcation with survivors having more IgA and IgG3 isotypes, as well as higher Ab galactosylation. Further, the SARS-CoV-2 canonical-antigen-specific features that stratified non-survivors and survivors were specific only to SARS-CoV-2 antigens, i.e., the same Flu hemagglutinin (HA)- or Ebola-GP-specific features were not able to discriminate survivors and non-survivors, suggesting context-specific antigen specificities for the uncovered features. Interestingly, pre-pandemic healthy controls were found to have humoral responses against SARS-CoV-2 proteins with high sequence similarity with endemic coronaviruses, particularly nsp13. Notably, higher levels of IgA Abs against nsp13 were found to be correlated with protection in severe COVID-19. Humoral profiles of eHCoV Abs were also predictive of outcome bifurcation, with higher levels of IgA and IgG3 Abs against OC43 and NL63 S being associated with survival.
While previous studies have focused solely on a small subset of SARS-CoV2 antigens, our study provides the first concrete evidence that Ab responses against different subsets of antigens (canonical, non-canonical, and endemic) are independently and equally discriminative of outcome. It has been speculated that higher Ab titers, especially against non-neutralizing targets, could be reflective of more severe disease and could also potentially be tied to ADE (Lee et al., 2020). Instead, our results suggest IgA and IgG3 Abs directed against canonical as well as non-canonical SARS-CoV-2 antigens could have a beneficial role in disease outcomes. IgA Abs function at mucosal surfaces and have previously been tied to vaccine-induced protection (Ackerman et al., 2018). In addition, IgG3 is known to be a particularly potent inducer of Ab effector functions via higher affinity association with Fc receptors (FcRs). Further, complementary to earlier findings of aberrant glycosylation, specifically afucosylation of S-specific Abs being correlated to disease severity (Larsen et al., 2021), we find here that differential glycosylation, specifically higher galactosylation, of Abs specific to both canonical and non-canonical antigens is associated with survival in severe COVID-19.
While there has been speculation regarding the roles of cross-reactive B and T cells as well as Abs in driving outcome bifurcation for COVID-19 (Anderson et al., 2021; Le Bert et al., 2020; Shrock et al., 2020; Loyal et al., 2021), our study identifies for the first time a range of multivariate humoral profiles of Abs that can be robustly associated with outcome bifurcation in severe COVID-19. A recent study reported that not everyone exposed to SARS-CoV-2 necessarily develops seropositivity, suggesting that some individuals clear sub-clinical infections (Swadling et al., 2021). In these individuals, pre-existing, nsp13-specific T cells are expanded (Swadling et al., 2021). These findings strongly complement those described herein and collectively suggest that pre-existing nsp13-specific B cell responses are also likely boosted on exposure to SARS-CoV-2 and in turn associate with favorable outcomes.
Although our study was performed during the first wave before the emergence of viral variants, including Delta and Omicron, we note that these and other variants of concern (VOCs) vary mostly in their surface proteins, primarily the S protein (and the RBD epitope within the S protein). These changes have a major impact on the binding affinity of S- and RBD-specific Abs, e.g., monoclonals that worked as neutralizing Abs for the Delta variant had significantly lowered neutralization efficacy for Omicron. By focusing on non-canonical and non-envelope proteins that are not impacted by these viral variants, we demonstrate that a model built using only features corresponding to non-canonical specificities is significantly predictive of outcome bifurcation. Furthermore, we analyze both Fab specificities and Fc properties; the latter features are not expected to be modulated by the viral variants. Finally, we show that we can predict outcome bifurcation even with profiles of Abs corresponding to endemic CoV specificities. Thus, our overall findings regarding Ab features driving severe COVID-19 outcomes are predicted to be stable across the spectrum of viral variants.
Our approach can now be extended to identify humoral correlates of breakthrough infections in vaccinated individuals and disease severity in unvaccinated individuals. Previous studies focusing on differences in the nature, quality, and durability of natural and vaccine-induced humoral responses have focused on canonical antigens. Our study demonstrates the importance of looking at non-canonical specificities. These considerations are of direct relevance in the current context, and the identified humoral correlates are likely to be more robust across VOCs for the reasons described above. Our characterization of the protective nature of Ab responses against a broad panel of antigens has implications for the formulation of improved second-generation SARS-CoV-2 vaccine as well as pan-coronavirus vaccine design.
Limitations of the study
Our study is focused on the identification of humoral correlates of outcome bifurcation in severe COVID-19 in unvaccinated subjects, with a key emphasis on Abs directed against non-canonical SARS-CoV-2 and endemic CoV antigens. The same correlates may not directly predict outcomes in mild and moderate COVID-19 patients, especially in vaccinated and boosted individuals. However, the same framework can be directly extrapolated to identify corresponding humoral correlates.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IgG Fc-PE | Southernbiotech | Cat#9040-09 |
| IgG1 Fc-PE | Southernbiotech | Cat#9052-09 |
| IgG2 Fc-PE | Southernbiotech | Cat#9070-09 |
| IgG3 Hinge-PE | Southernbiotech | Cat#9210-09 |
| IgG4 Fc-PE | Southernbiotech | Cat#9200-09 |
| IgM | Southernbiotech | Cat#9020-09 |
| IgA | Southernbiotech | Cat#2050-09 |
| IgA1 | Southernbiotech | Cat#9130-09 |
| IgA2 | Southernbiotech | Cat#9140-09 |
| Biological samples | ||
| Serum samples from SARS-CoV2 patients and healthy controls | University of Pittsburgh and University of Pittsburgh Medical Center | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| HA (Michigan) | ImmuneTech | Cat #1920 |
| HA phuket | Immunetech | Cat #1171 |
| HA Singapore | Immunetech | Cat #2123 |
| Ebola Zaire | ImmuneTech | Cat#1805 |
| BSA | Thermofisher scientific | Cat #29130 |
| PBS | Corning | Cat #MT21040CMX |
| NHS | Pierce | Cat#PI24510 |
| EDC | Pierce | Cat#PG82079 |
| SARS- CoV-2 Spike | ImmuneTech | Cat #IT-002-032p |
| SARS- CoV-2 Spike RBD | ImmuneTech | Cat #IT-002-036p |
| SARS- CoV-2 Nucleocapsid | SinoBiological | Cat #IT-002-036p |
| SARS- CoV-2 ORF3a | Bioworld | Cat #NCP0026P |
| SARS- CoV-2 ORF8 | Bioworld | Cat #NCP0025P |
| SARS- CoV-2 NSP3 | mybiosource | Cat #MBS156024 |
| SARS- CoV-2 NSP13 | mybiosource | Cat #MBS2563852 |
| SARS- CoV-2 M protein | mybiosource | Cat #MBS156019 |
| HCoV-OC43 | SinoBiological | Cat #40607-V08B |
| HCoV-NL63 | SinoBiological | Cat #40604-V08B |
| CD64 | Acrobiosystems | Cat #FCA-H82E8-25ug |
| CD32a | Acrobiosystems | Cat #CDA-H82E6-25ug |
| CD32b | Acrobiosystems | Cat # CDB-H82E0-25ug |
| CD16a | Acrobiosystems | Cat # CDA-H82E9-25ug |
| CD16b | Acrobiosystems | Cat # CDB-H82Ea-25ug |
| Lectin kit I | Vector labs | Cat# RLK-2200 |
| Native human C1q protein | Abcam | Cat #ab96363 |
| Deposited data | ||
| Antibody-omic data | This study | https://github.com/jishnu-lab/Covid19AbOmics |
| Software and algorithms | ||
| R Statistical Computing Environment | R Statistical Computing Environment | https://www.r-project.org/ |
Resource availability
Lead contact
Requests for data and code used for the study should be directed to and will be fulfilled by the Lead Contact Jishnu Das (jishnu@pitt.edu).
Materials availability
-
•
This study did not generate new unique reagents.
Experimental model and subject details
Cohort details
After obtaining written informed consent from patients or legally authorized representatives acting on their behalf, subjects with acute hypoxemic respiratory failure and symptoms of COVID-19 were recruited in a prospective, observational cohort study (University of Pittsburgh Institutional Review Board study number 20040036). The patients were hospitalized in ICUs at two hospitals (Presbyterian and Shadyside) within the University of Pittsburgh Medical Center system. All patients underwent at least one nasopharyngeal swab testing for SARS-CoV-2 qPCR, which may have been repeated at the discretion of the treating physicians when the first test was negative and significant clinical suspicion for COVID-19 remained. In this study, we profiled subjects from this cohort (14 survivors and 7 non-survivors). No a-priori power calculations were used to determine cohort size and the 2:1 survival:death ratio reflects an inherent feature of the cohort. Survivors had a mean age of 65.4 years (with a standard deviation of 9.4 years). Non-survivors had a mean age of 70.9 years (with a standard deviation of 12.2 years). There were 10 males and 4 females within survivors, and 2 males and 5 females within non-survivors.
Method details
Sample preparation
Serum was obtained as described earlier and stored at −80C until used. Upon thawing before use, all samples were transferred to a 96 well U-bottom plate at an appropriate dilution for the intended probe (1:500 for IgG2, IgG3, IgG4, IgA, IgA1, IgA2, IgM, RCA I, SNA, C1Q & 1:2000 for IgG, IgG1, FcRs).
Multiplexed antigen-specific antibody profiling
A multiplexed antigen-specific Ab profiling workflow was developed based on a protocol reported earlier (Brown et al., 2017). Briefly, pooled barcoded antigen-coupled beads were incubated with samples and then with fluorescently labeled probes. Following set of probes were used: 1) Antigen-specific subclass/isotype titers were measured using PE-labeled mouse-anti-human IgG1, IgG2, IgG3, IgG4, total IgG, IgA1, IgA2, IgM. 2) Antigen FcR/complement binding profiles were measured using biotinylated FcRs (FcR1, FcR2A, FcR2B, FcR3A, FcR3B) tetramerized with streptavidin-PE or PE-labeled complement (C1q). 3) Antigen-specific glycosylation profiles were measured using lectin-binding using PE-labeled lectins (SNA for sialic acid, RCA1 for galactosylation).
All antigens including SARS-CoV-2 Antigens (Spike [ImmuneTech IT-002-032p], Spike RBD [ImmuneTech IT-002-036p], Nucleocapsid [SinoBiological IT-002-036p], Orf3a [Bioworld NCP0026P], ORF8 [Bioworld NCP0025P], NSP3 [mybiosource MBS156024], NSP13 [mybiosource MBS2563852], M protein [mybiosource MBS156019]) and eHCoV antigens (HCoV-OC43 [SinoBiological 40607-V08B], HCoV-NL63 [SinoBiological 40604-V08B]) were coupled to Luminex MagPlex magnetic microsphere beads of different regions at a ratio of about 8ug of antigen per million beads using an EDC-NHS chemistry and then blocked with and stored until use in storage buffer (1XPBS, 0.1% BSA, 0.1% Tween) at 4C. PE coupled anti-Igs (Southern Biotech) were diluted from the stock vials to a concentration of 1ug/ml in 1XPBS. Biotinylated FcRs (Acro Biosystems) were reacted with Streptavidin-PE (Thermo Fisher) at a 4:1 molar ratio for 20 min and then diluted to a concentration of 1ug/ml in 1XPBS. Rhodamine/Cy-3 coupled lectins (Vector Labs) were diluted in lectin buffer to a concentration of 20ug/ml.
Conjugated beads were diluted in assay buffer (1XPBS, 0.1% BSA) to make a working bead solution and added at a 1:9 sample:bead volume ratio in wells of a 96 well flat bottom plate, and incubated for 1 h. Sample-bound beads were then washed twice in a wash buffer (PBS, 0.1% Tween) using a magnetic plate separator and resuspended in the appropriate probe buffer. Diluted probe solution was then added to the wells at a1:9 bead:probe volume ratio and incubated for 30 min. All incubation steps were performed at room temperature on a plate shaker. Probe-bound beads were then washed twice with wash buffer and resuspended in Luminex MagPix drive fluid before reading on a Luminex MagPix instrument. All assays were performed in duplicate and a correlation coefficient of R2 > 0.8 was verified for technical replicability. An arithmetic mean of the two measured MFI values from the replicates is then used as the readout.
Quantification and statistical analyses
Machine learning models to discriminate by outcome using humoral responses against different specificities (canonical, non-canonical and endemic CoV antigens)
Data was pre-processed to remove features with low values (mean MFI <50). This was done in an unsupervised setting to avoid any biases. All features were centered and scaled (i.e., z-scored) to have a mean 0 and standard deviation 1.
We used a two-step machine learning model to identify a minimal set of predictive biomarkers of outcome. This comprised feature selection on the high-dimensional data (features >> number of subjects) using the least absolute shrinkage and selection operator (LASSO) (Tibshirani, 1996), followed by classification using the down-selected features using support vector machines(SVM) (Breiman, 2001). The use of LASSO (L1 regularization) helps prevent over-fitting on high dimensional data. This two-step procedure is similar to what has been successfully used earlier for high-dimensional humoral immune measurements (Ackerman et al., 2018; Lu et al., 2020; Sadanand et al., 2018).
The performance of the models were evaluated in a rigorous 10-fold cross validation framework, and the significance of the models was quantified using permutation testing (Ojala and Garriga, 2010). The overall framework is analogous to what has been previously described (Ackerman et al., 2018; Lee et al., 2020a; Sadanand et al., 2018). Briefly, the dataset was split into 10 subsets – 9 subsets are used for training while the 10th one is used for testing. Each subset served as the test set once, therefore each individual was in the test fold exactly once for each cross-validation run. For each test fold, LASSO-based feature selection was performed using the nine training folds. The coefficient for the LASSO penalty term (i.e., lambda for regularization) was determined via a second internal cross-validation using only the fold-specific training dataset. A fold-specific linear support vector machine (SVM) model was built using the LASSO-selected features and training data for that fold. This fold-specific classifier was subsequently used to predict the labels for the individuals in the test set for that fold. This process was repeated for each of the ten folds to generate a set of predicted outcomes for each individual. This was then compared to the true set of outcome labels to calculate a classification accuracy for that cross-validation replicate. We performed 100 independent ten-fold cross-validation replicates, to account for different ways in which the training and test folds can be split. This is a stringent and appropriate way of performing cross-validation, as both steps involved in the model (feature selection and subsequent classification using the selected features) are performed in a cross-validation setting with data held out. The significance of model performance was evaluated using permutation testing (Ojala and Garriga, 2010), by randomly shuffling the data with respect to the arm labels, within the cross-validation framework described above (i.e., a cross-validation framework matched to the actual model). The model is deemed significantly predictive if it meets an exact p value (based on a permutation test) threshold of <0.01 and an effect size threshold of 0.15 in AUC i.e., it has an AUC that is at least 0.15 better than the negative control model (we calculate the difference in medians across replicates of k-fold cross-validation).
To visualize the modules selected by the LASSO model on the whole dataset, we applied a partial least squares discriminant analysis (PLS-DA). PLS-DA is a supervised dimension reduction method, which transforms a new set of features that are a linear combination of the original features and then fits a linear model via least squares using these new features. We carried out separate PLS-DA analyses using down-selected features from applying LASSO on the whole dataset. PLS-DA was applied for both 2 groups (between survivors and non-survivors) and 3 groups (among survivors, non-survivors, and healthy controls).
We analyzed the canonical, non-canonical and non-SARS-CoV2 specificities separately. We start with humoral responses for canonical (S/RBD/N) antigenic specificities to generate a multivariate machine learning model for outcome (survivors vs non-survivors) bifurcation. We applied a similar approach on the non-canonical (orf3a/orf8/nsp3/nsp13/M) antigenic specificities for outcome bifurcation. We then combined both the canonical and non-canonical antigens to generate a multivariate machine learning model for outcome bifurcation between survivors and non-survivors. Then we also included the healthy controls along with the survivors and non-survivors to generate a multivariate machine learning model for outcome prediction (3-way). We also built a multivariate machine learning model using endemic coronavirus antigenic specificities.
Validation-cohort
We sought to validate the robustness of biomarkers, corresponding to humoral responses against canonical Ag specificities, identified in the Pittsburgh cohort. Specifically, we examined their predictive power in an orthogonal Boston cohort (Zohar et al., 2020). Model training (both down-selection of features and model fitting) was performed using only our cohort (Pittsburgh), and the model generation process was completely blinded to this second orthogonal validation cohort (Boston), ensuring that this is a true cross-prediction. Then we tested the performance of the model learned using the Pittsburgh cohort on the Boston cohort.
PLS-DA based visualization using IgA/IgG and IgA/IgM ratios
We performed a post-hoc greedy feature selection based on the ratios of IgA to IgG and IgA to IgM. We took the Spike, RBD, and Orf8 specificities for the ratios. We then performed a PLS-DA on the selected ratios and to exhibit the discriminating power of these feature-ratios between survivors and non-survivors.
Implementation of LASSO and PLS
LASSO was implemented using glmnet in R. If no feature was selected by LASSO in a specific fold for a given replicate, we randomly selected 5 features (only for that fold in that replicate) and used an ordinary least squares estimator. PLS-DA was implemented using the plsr function in R.
Acknowledgments
This study was supported by a COVID-19 pilot grant from the UPMC ITTC to J.D. and H.S. It was also partially supported by NIH grants DP2AI164325 to J.D. and U01AI141990 to H.S. The authors also acknowledge support from the University of Pittsburgh Center for Research Computing through the high-performance computing resources provided.
Author contributions
J.D., A. Sarkar, and H.S. conceptualized, designed, and supervised all aspects of the study. S.P.P. performed the experiments with help from G.M.V. S.A.R. did all computational analyses with inputs from A.R.C. A. Somasundaram, C.J.W., W.B., B.J.M., B.M., J.S.L., P.R., A.R., T.C.B., D.A.A.V., G.D.K., and A.M. contributed to cohort design, recruitment, and sample collection. J.D., A. Sarkar, and H.S. wrote the manuscript with input from all authors.
Declaration of interests
J.D. is a consultant for SeromYx Systems.
Published: June 8, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111020.
Supplemental information
Data and code availability
-
•
Detailed code, associated datasets and documentation are available at https://github.com/jishnu-lab/Covid19AbOmics. A corresponding stable release can be accessed at https://doi.org/10.5281/zenodo.6585876.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- Ackerman M.E., Das J., Pittala S., Broge T., Linde C., Suscovich T.J., Brown E.P., Bradley T., Natarajan H., Lin S., et al. Route of immunization defines multiple mechanisms of vaccine-mediated protection against SIV. Nat. Med. 2018;24:1590–1598. doi: 10.1038/s41591-018-0161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J., Garcia-Sastre A. Immunological imprinting of the antibody response in COVID-19 patients. Nat. Commun. 2021;12:3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain W., Yang H., Shah F.A., Suber T., Drohan C., Al-Yousif N., DeSensi R.S., Bensen N., Schaefer C., Rosborough B.R., et al. COVID-19 versus non-COVID-19 acute respiratory distress syndrome: comparison of demographics, physiologic parameters, inflammatory biomarkers, and clinical outcomes. Ann Am Thorac. Soc. 2021;18:1202–1210. doi: 10.1513/AnnalsATS.202008-1026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch Y.C., Fischinger S., Siddiqui S.M., Chen Z., Yu J., Gebre M., Atyeo C., Gorman M.J., Zhu A.L., Kang J., et al. Discrete SARS-CoV-2 antibody titers track with functional humoral stability. Nat. Commun. 2021;12:1018. doi: 10.1038/s41467-021-21336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- Brown E.P., Dowell K.G., Boesch A.W., Normandin E., Mahan A.E., Chu T., Barouch D.H., Bailey-Kellogg C., Alter G., Ackerman M.E. Multiplexed Fc array for evaluation of antigen-specific antibody effector profiles. J. Immunol. Methods. 2017;443:33–44. doi: 10.1016/j.jim.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bye A.P., Hoepel W., Mitchell J.L., Jegouic S., Loureiro S., Sage T., Vidarsson G., Nouta J., Wuhrer M., de Taeye S., et al. Aberrant glycosylation of anti-SARS-CoV-2 spike IgG is a prothrombotic stimulus for platelets. Blood. 2021;138:1481–1489. doi: 10.1182/blood.2021011871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat. Rev. Immunol. 2021;21:245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Gonzalez J., Edwards K., Mallajosyula V., Buzzanco A.S., Sherwood R., Buffone C., Kathale N., Providenza S., Xie M.M., et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Preprint at medRxiv. 2020 doi: 10.1101/2020.05.15.20103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo A.R., Somasundaram A., Shan F., Cardello C., Workman C.J., Kitsios G.D., Ruffin A.T., Kunning S., Lampenfeld C., Onkar S., et al. People critically ill with COVID-19 exhibit peripheral immune profiles predictive of mortality and reflective of SARS-CoV-2 lung viral burden. Cell Rep. Med. 2021;2:100476. doi: 10.1016/j.xcrm.2021.100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J., Devadhasan A., Linde C., Broge T., Sassic J., Mangano M., O'Keefe S., Suscovich T., Streeck H., Irrinki A., et al. Mining for humoral correlates of HIV control and latent reservoir size. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e5. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wang Y., Kang L., Hu Y., Wang L., Zhong J., Chen H., Ren L., Gu X., Wang G., et al. Cross-reactive antibody against human coronavirus OC43 spike protein correlates with disease severity in COVID-19 patients: a retrospective study. Emerg. Microbes. Infect. 2021;10:664–676. doi: 10.1080/22221751.2021.1905488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R., Poon L.L.M., et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- Hicks J., Klumpp-Thomas C., Kalish H., Shunmugavel A., Mehalko J., Denson J.P., Snead K.R., Drew M., Corbett K.S., Graham B.S., et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal betacoronaviruses. J. Clin. Immunol. 2021;41:906–913. doi: 10.1007/s10875-021-00997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepel W., Chen H.J., Geyer C.E., Allahverdiyeva S., Manz X.D., de Taeye S.W., Aman J., Mes L., Steenhuis M., Griffith G.R., et al. High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abf8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Lamere M.W., Moquin A., Lee F.E., Misra R.S., Blair P.J., Haynes L., Randall T.D., Lund F.E., Kaminski D.A. Regulation of antinucleoprotein IgG by systemic vaccination and its effect on influenza virus clearance. J. Virol. 2011;85:5027–5035. doi: 10.1128/JVI.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M.D., de Graaf E.L., Sonneveld M.E., Plomp H.R., Nouta J., Hoepel W., Chen H.J., Linty F., Visser R., Brinkhaus M., et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2021;371 doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374 doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.L., Das J., Grace P.S., Fortune S.M., Restrepo B.I., Alter G. Antibody Fc glycosylation discriminates between latent and active tuberculosis. J. Infect. Dis. 2020 doi: 10.1093/infdis/jiz643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W.H., Tipih T., Makoah N.A., Vermeulen J.G., Goedhals D., Sempa J.B., Burt F.J., Taylor A., Mahalingam S. Comorbidities in SARS-CoV-2 patients: a systematic review and meta-analysis. mBio. 2021;12 doi: 10.1128/mBio.03647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala M., Garriga G.C. Permutation tests for studying classifier performance. J. Mach. Learn. Res. 2010;11:1833–1863. [Google Scholar]
- Rodebaugh T.L., Frumkin M.R., Reiersen A.M., Lenze E.J., Avidan M.S., Miller J.P., Piccirillo J.F., Zorumski C.F., Mattar C. Acute symptoms of mild to moderate COVID-19 are highly heterogeneous across individuals and over time. Open Forum Infect. Dis. 2021;8:ofab090. doi: 10.1093/ofid/ofab090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanand S., Das J., Chung A.W., Schoen M.K., Lane S., Suscovich T.J., Streeck H., Smith D.M., Little S.J., Lauffenburger D.A., et al. Temporal variation in HIV-specific IgG subclass antibodies during acute infection differentiates spontaneous controllers from chronic progressors. AIDS. 2018;32:443–450. doi: 10.1097/QAD.0000000000001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadarangani M., Marchant A., Kollmann T.R. Immunological mechanisms of vaccine-induced protection against COVID-19 in humans. Nat. Rev. Immunol. 2021;21:475–484. doi: 10.1038/s41577-021-00578-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370 doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suscovich T.J., Fallon J.K., Das J., Demas A.R., Crain J., Linde C.H., Michell A., Natarajan H., Arevalo C., Broge T., et al. Mapping functional humoral correlates of protection against malaria challenge following RTS,S/AS01 vaccination. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb4757. [DOI] [PubMed] [Google Scholar]
- Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J. Roy. Stat. Soc. B. 1996;58:267–288. [Google Scholar]
- To K.K., Zhang A.J., Hung I.F., Xu T., Ip W.C., Wong R.T., Ng J.C., Chan J.F., Chan K.H., Yuen K.Y. High titer and avidity of nonneutralizing antibodies against influenza vaccine antigen are associated with severe influenza. Clin. Vaccine Immunol. 2012;19:1012–1018. doi: 10.1128/CVI.00081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e2. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Detailed code, associated datasets and documentation are available at https://github.com/jishnu-lab/Covid19AbOmics. A corresponding stable release can be accessed at https://doi.org/10.5281/zenodo.6585876.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.