Abstract
The radiation-resistant bacterium Deinococcus radiodurans is known as the world’s toughest bacterium. The S-layer of D. radiodurans, consisting of several proteins on the surface of the cellular envelope and intimately associated with the outer membrane, has therefore been useful as a model for structural and functional studies. Its main proteinaceous unit, the S-layer deinoxanthin-binding complex (SDBC), is a hetero-oligomeric assembly known to contribute to the resistance against environmental stress and have porin functional features; however, its precise structure is unknown. Here, we resolved the structure of the SDBC at ∼2.5 Å resolution by cryo-EM and assigned the sequence of its main subunit, the protein DR_2577. This structure is characterized by a pore region, a massive β-barrel organization, a stalk region consisting of a trimeric coiled coil, and a collar region at the base of the stalk. We show that each monomer binds three Cu ions and one Fe ion and retains one deinoxanthin molecule and two phosphoglycolipids, all exclusive to D. radiodurans. Finally, electrophysiological characterization of the SDBC shows that it exhibits transport properties with several amino acids. Taken together, these results highlight the SDBC as a robust structure displaying both protection and sieving functions that facilitates exchanges with the environment.
Keywords: cell envelope deinoxanthin, SDBC, S-layer, porins, SlpA, DR_2577, phosphoglycolipids, transport properties, amino acid import
Abbreviations: SDBC, S-layer deinoxanthin-binding complex; S-layer, surface layer
Bacterial surface layers (S-layers) are proteinaceous repetitive assemblies characterized by crystalline regularity. These structures coat the entire cell body and represent the outermost barrier in the cell envelopes of many prokaryotes (1, 2). The structural organization of these assemblies is putatively considered to contribute to primary protection, frequently supporting the resistance features in extremophiles (3, 4, 5). In spite of their interesting structures, enigmatic functions, and exploitable properties, only a few S-layers were extensively studied (6, 7) and for none of them, the proteinaceous features of their specific units are deeply described. The radiation-resistant bacterium Deinococcus radiodurans has been one of the most studied model organism for understanding the S-layers' architecture at low-middle resolution by several techniques (8, 9, 10, 11). In the last decade, a detailed characterization of the protein components and their individual assemblies was facilitated by the development of gentle isolation procedures. This new approach allowed us to observe in greater detail the functional and structural features of the S-layer from this bacterium (11, 12). The D. radiodurans S-layer was found to result from the repetitive assembly of three complexes (11). The main component of this S-layer is the protein DR_2577 (SlpA), which binds the xanthophyll carotenoid deinoxanthin and was found to be part of a hetero-oligomeric complex resulting in the main assembly named S-layer deinoxanthin-binding complex (SDBC). The SDBC was extensively characterized both functionally and biochemically providing important information with respect to its stability, oligomerization, and spectral properties (13, 14, 15, 16, 17). In addition to DR_2577, this complex was found to be composed of other five different subunits, namely DR_2310, DR_0505, DR_A0283, DR_A0282, and DR_A0281 (18). Here, we report the high-resolution structure of the SDBC solved at ∼2.5 Å resolution by cryo-EM single-particle analysis. The structure shows in quasiatomic details the features of the largest known porin so far, which appears to assemble as a trimer forming three pores of ∼64 Å diameter each. From the structural analysis, it emerges evidently the seamlessness between the S-layer and the outer membrane of this bacterium. Differently from other porins, this complex shows an extended (∼120 Å) trimeric coiled-coil structure, the stalk region, departing from the center of symmetry between the three pores and toward the periplasm, possibly interacting with the inner membrane. Finally, at the base of the stalk is observed a large dynamic region, the collar. This last region appears to be extremely dynamic so that in the present analysis it remained unassigned and putatively ascribable to one or more of the other known SDBC subunits. The typical β-barrel organization is characterized by four metal-binding sites for each pore (DR_2577 monomer), and interestingly, the external interface between monomers is characterized by the presence of two unique lipids of D. radiodurans with a deinoxanthin molecule (14) located nearby and almost facing the periplasmic side. Functional characterization of the SDBC confirms its transport properties and, apart from the previously shown nonselective permeation of ions (11, 18), here we show its activity with different types of amino acids. The present findings provide direct evidence for the active functionality of this protein complex and its S-layer, showing it as a sieved barrier able to protect while allowing exchange and communication with the environment.
Results
SDBC organization
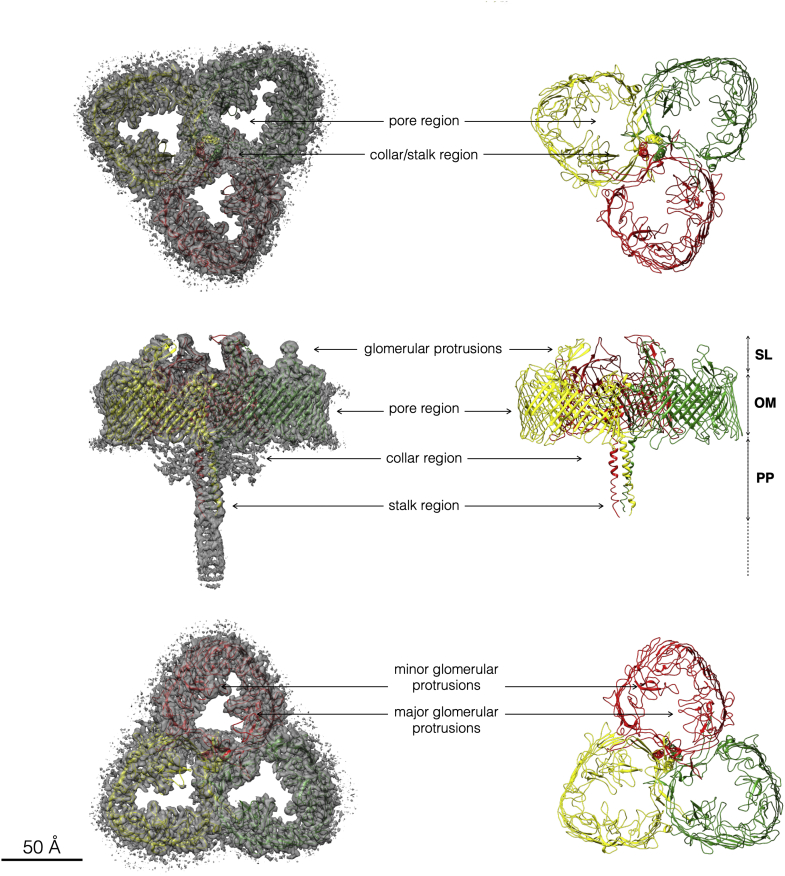
The structure of the SDBC was solved at ∼2.5 Å resolution and allowed the fitting of three copies of the protein DR_2577, for which were assigned 949 residues of the sequence, from serine 219 to phenylalanine 1167 (Fig. 1). The overall SDBC structure is organized in distinct parts, which differ structurally and dimensionally allowing to define three regions. The main part of the pore region is characterized by a dominant β-barrel structure. This region consists of 30 β-sheets that twist and coil identifying a toroidal structure with a width of ∼76 Å and a pore diameter of ∼64 Å (Fig. 1). From the side view, the external face of each pore appears to retain native lipids providing clear evidence of the extended immersion of this part of the complex in the outer membrane. At the S-layer side, each DR_2577 monomer has two glomerular protrusions, a major and a minor, which partially occlude the top side of the pore (Fig. 1). A characteristic property of these glomerular regions is the tendency to be partially disordered with respect to the rest of the complex. At the threefold symmetry axis, a trimeric coiled coil defines the stalk region (Fig. 1). This region departs from the central area of the trimeric assembly extending for ∼120 Å into the periplasmic space and possibly reaching the inner membrane (Fig. 1). Each of the three DR_2577 monomers contributes with one alpha-helix at the N terminus side (aa 218–254) to the trimeric coiled-coil. This region is also very dynamic (18) and its distal part, even if present in the map, remained unassigned for the first 218 amino acids of the DR_2577 sequence (Fig. 1). Finally, a collar region of ∼69 Å length, also carrying a threefold symmetry, is localized at the periplasmic side and sits at the base of the stalk centered in the axis of symmetry (Fig. 1). This region of the SDBC is also dynamic, and although the density map clearly indicates a typical β-sandwich organization, the assignment of some of the specific SDBC subunits and their fitting into the final model was so far not possible (Fig. S1). As previously reported (18), both the distal part of the stalk and the collar are dynamic regions clearly identified in the 2D classification, but it was not possible to resolve them, therefore are not present in the model here shown.
Figure 1.
SDBC map at 2.5 Å resolution and resolved model. The SDBC map and the fitted model reveal a trimeric organization of the protein DR_2577 (chain A in yellow, chain B in green, and chain C in red). The images show the different regions of this complex named pore, collar, and stalk. At the upper part of each pore region two glomerular protrusions emerge, a major and a minor one. A scale bar represents 50 Å. OM, outer membrane; PP, periplasm; SDBC, S-layer deinoxanthin-binding complex; SL, S-layer.
Cofactors binding
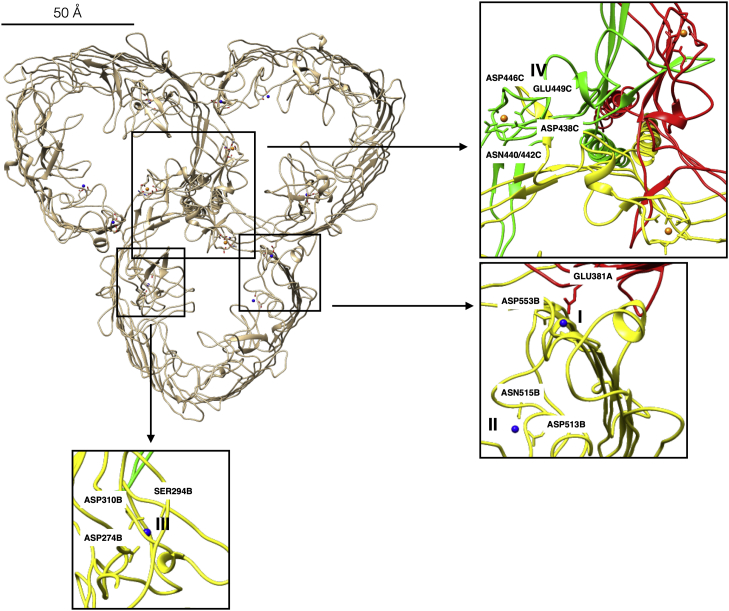
Each monomeric component of the SDBC has different binding sites for cofactors. In particular, four metal-binding sites (I, II, III, and IV) for each monomer are localized in different regions of the protein DR_2577 (Fig. 2). The site I binds Cu and is at the interface between two monomers involving the residues Asp553 of one monomer and Glu381 of the other one (Fig. 2, inset). Of the remaining metal-binding sites, II (Asp515 and Asp513) and III (Asp274, Asp310, and Ser294) bind Cu while the site IV (Asp338, Asn440, Asn442, Asp446, and Glu449) binds Fe. These three sites are exclusive for each monomer, hence are not shared at their interfaces (Fig. 2, insets). It is worth mentioning that site III sits into the major glomerular region of each monomer (Fig. 2, inset). Coordination and distances for each of the four binding sites supported the assignment of Cu in sites I, II, III, and Fe in site IV (Fig. S2 and Table S1). The presence of these sites might be important for stabilizing the complex as previously reported (16).
Figure 2.
Metal-binding sites of the SDBC. The four metal-binding sites for each DR_2577 monomer are shown; in blue are depicted the three Cu atoms (I, II, III) and in orange the single Fe atom (IV). Related interacting residues are indicated. In the upper inset is shown the interface between the three monomers at the symmetry axis where the three binding sites for Fe (IV) are present. The inset in the middle shows the shared Cu-binding site (I) at the interface between two monomers and another Cu-binding site (II) in the same area but not shared. The inset in the bottom shows the last Cu-binding site (III) characterized to be part of the major glomerular region. A scale bar represents 50 Å. SDBC, S-layer deinoxanthin-binding complex.
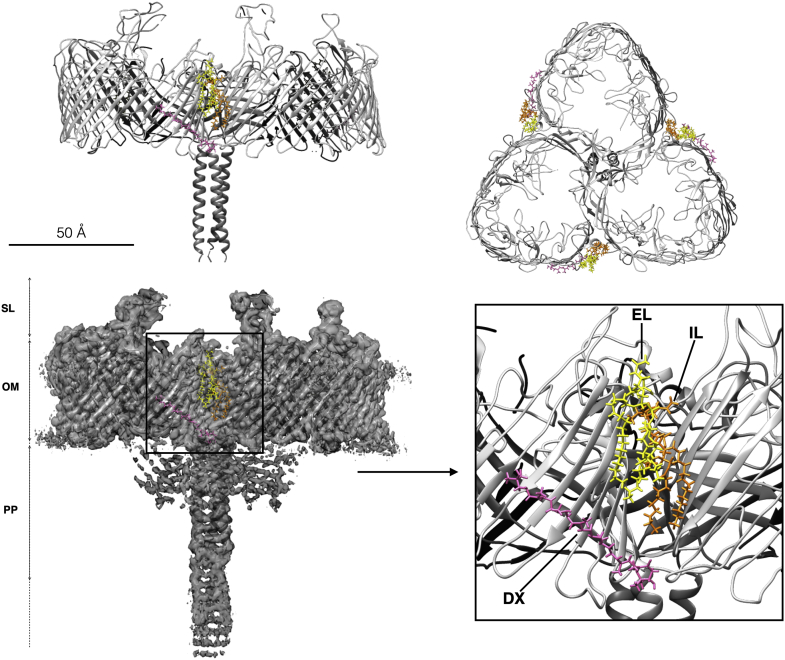
While the metal-binding sites are mainly in the internal part of DR_2577, the other cofactors, characteristic of this complex are localized at the external surface of the protein and at the interface between the monomers and the outer membrane. In this region are also present two molecules, one internal and one external, of the α-galactosyl-phosphatidylglyceroylalkyl-amine that differ from each other for the length of their alkyl groups and are typical of D. radiodurans. At a distance of 7.5 Å from the external lipid it is localized a deinoxanthin molecule sitting into a transversal groove of the β-barrel organization almost facing the periplasm (Fig. 3).
Figure 3.
Deinoxanthin and lipid-binding sites of the SDBC. The images show side and top views of the model, and a side view of the map with details of the deinoxanthin (DX) and the lipid-binding sites. DX is shown in pink and the two different lipids are shown in yellow, external lipid (EL), and orange, the internal one (IL). The inset shows a detail of these sites. A scale bar represents 50 Å. OM, outer membrane; PP, periplasm; SDBC, S-layer deinoxanthin-binding complex; SL, S-layer.
The SDBC function
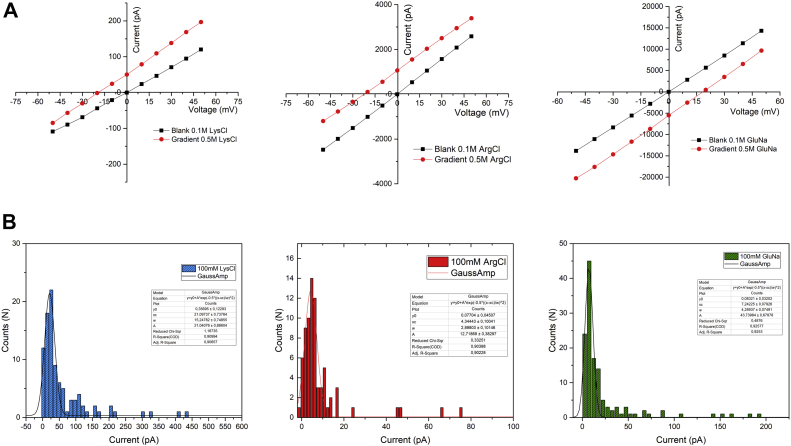
The SDBC was reconstituted in lipid bilayer membranes and its transport properties were assessed. These functional tests not only confirmed previous analyses that showed nonselective transport properties using monovalent and divalent cations (11, 18) but also highlighted the SDBC transport properties for different types of charged amino acids. After assessing the functional integrity of the complex in 100 mM KCl as previously reported (11, 18), we performed the same measurements but in presence of three different amino acids, lysine, arginine, and glutamic acid. For these three amino acids, the SDBC was found to have a single conductance unit G of 0.42 nS, 0.086 nS, 0.14 nS, respectively; a reversal potential Vrev (Δsalt = 400 mM) of −19.9 mV, −21.1 mV, +15.9 mV, respectively, and corresponding permeability ratios PCat+/PAn− of 1:3.1, 1:3.6, and PAn−/PCat+ of 1:2.6, respectively (Fig. 4). In all cases, results showed a clear transport activity indicating important properties of amino acids' translocation for this S-layer complex.
Figure 4.
Electrophysiological assay. In the image are shown the transport properties of the SDBC inserted into a lipid bilayer for charged amino acids. A, from left to right, are shown the reversal potential curves for LysCl, ArgCl, and GluNa for a Δ(Salt) = 400 mM. B, from left to right, are shown the channel conductances associated to a sequence of stepwise insertions and the related membrane currents for the three different amino acids LysCl, ArgCl, and GluNa at a concentration equal to 100 mM. The insets for each plot shown in (B) provide all the processing and statistical parameters used for calculating the single-channel conductance (y0 = offset, xc = center, w = width, A = amplitude). SDBC, S-layer deinoxanthin-binding complex.
Discussion
The structural characterization of the SDBC at high resolution allowed to indicate specific functional features of the SDBC. The main protein subunit of this complex, the DR_2577, was known to bind the xanthophyll carotenoid deinoxanthin, a feature that provides the protein and the SDBC with the ability to protect the bacterium against UV radiations (14) and desiccation (15) by quenching mechanisms. It was also reported that when the DR_2577 is knocked out, the stability of the entire cell envelope is compromised (19), and the resulting mutants lose the capabilities of resistance to desiccation and UV exposure (14). Recently, in situ studies have shown how the SDBC complex is embedded in the organization of the cell envelope (11). This observation not only explains the dismantling of the cell envelope in DR_2577 KO mutants (19), which appears coarse and disaggregated, but also agrees with the structural observation reported here, which indicates the SDBC as an integral building block between outer membrane and S-layer (Fig. 1).
The SDBC is known to retain metal ions (16), and in the present report four metal-binding sites were identified and assigned to Cu and Fe (Fig. 2). In this respect, it must be mentioned the strong limit on metal assignment for cryo-EM unlikely X-ray crystallography (20).
The oligomerization properties of DR_2577 and the resulting SDBC assembly were shown to be influenced by the presence of iron (Fe3+) and copper (Cu2+) (16). Accordingly, here we find one of the Cu-binding sites (I) localized and kept in place by residues at the interface between monomers, suggesting a direct stabilizing effect on the assembly (Fig. 2, inset). The intramonomeric metal-binding sites, two assigned to Cu (II and III) and one to Fe (IV), might also be involved in the structural stabilization of the SDBC but any other function (e.g., contribution to protection mechanisms against oxidative stress) cannot be excluded. It is worth mentioning that in the structure, the appearance of other weak densities could putatively indicate the presence of other ligands; however, these densities are unassigned due to a limit in the current resolution.
Each DR_2577 monomer binds deinoxanthin, a xanthophyll typical of D. radiodurans. Deinoxanthin is positioned into a transversal groove of the β-barrel organization, in proximity but aside from the monomer interface and almost at the periplasmic side (Fig. 3). While in the present structure the resolution of the maps allow to clearly assign a single binding site for this cofactor, similar grooves surrounding the SDBC at low values of sigma show elongated densities, which could be deinoxanthin molecules, but they were difficult to assign due to poor definition of the map (Fig. S3). This observation may also indicate a partial loss of the deinoxanthin molecules during solubilization, suggesting that this xanthophyll may be organized as a belt surrounding the whole complex. Deinoxanthin was reported to be important for contributing to the extreme resistance of this bacterium. In particular, it was shown that it acts not only as an antioxidant scavenger for reactive oxygen species and radicals by a chemoprotective mechanism but also by a photoprotective one where the high frequency photons in the UVC are quenched as heat and fluorescence (14). These mechanisms explained how through the SDBC the bacterium has the ability to grow under UV light. The protein DR_2577 has an exceptionally high content in phenylalanine and tyrosine residues that, in agreement with previous observation, act as antennas able to absorb UVC photons possibly involved in the mechanism of quenching (14). Here, we observe the majority of these residues localized at the external surface of the complex near to the deinoxanthin and also to the empty grooves where it was not possible to clearly assign densities to the deinoxanthin (Figs S3 and S4).
It is also very interesting the stable presence in the structure of specific phosphoglycolipids described a few decades ago as unique for this bacterium (21, 22, 23, 24) and to which was also assigned a putative role in the resistance features of this bacterium. In the present structure, they appear tightly associated along the interface between monomers and in close proximity with the deinoxanthin molecule. The high occupancy of these molecules and their position strongly suggest a possible role in sustaining the xanthophyll function, but currently, this relationship remains to be demonstrated.
Finally, the transport properties of the SDBC for charged molecules, such as amino acids rather than just atomic ions (Fig. 4), are confirmed by the dimensions of the pores, which compared to common porins, such as OmpF (25), are interestingly larger (∼64 Å for DR_2577 versus ∼20 Å for OmpF). This fact points toward the physiological importance of the SDBC in assisting and allowing the exchange of substances, including nutrients, with the surrounding environment. In this context, the presence of metal ions might also influence the transport of charged molecules by modulating the electrostatics of the pore surface (26, 27). Further experiments are currently ongoing to shed a light on these dynamics.
The results shown here not only elucidate the structural and functional details of a peculiar S-layer protein assembly but also opens up to applications in biomedicine for exploiting similar structures in pathogenic bacteria as new antimicrobial targets and in nanotechnologies as new nanoporous molecular machines exploitable for next generation sequencing.
Experimental procedures
Cell culturing and SDBC purification
D. radiodurans (strain R1; ATCC 13939) was cultivated in tryptone glucose yeast extract broth at 30 °C for 24 h as describe in (12). The cell envelope isolation and SDBC purification were performed according to (11, 18) with no modifications. In these studies, all chromatography columns were subjected to the ReGenFix procedure (https://www.regenfix.eu/) for regeneration and calibration prior use.
Cryo-EM
The whole sample preparation and data acquisition were done at CEITEC MU. The 3.5 μl of SDBC sample (5 mg/ml) was applied to freshly glow-discharged transmission electron microscopy grids (Protochips, Cu, 300 mesh, R1.2/1.3) and vitrified into liquid ethane using ThermoScientific Vitrobot Mark IV (4 °C, 100% rel. humidity, 30 s waiting time, 3 s blotting time). The grids were subsequently mounted into the Autogrid cartridges and loaded to a FEI Talos Arctica (ThermoScientific) transmission electron microscope for screening prior to data acquisition. The SDBC data were collected using a FEI Titan Krios operated at 300 kV using the SerialEM software (https://bio3d.colorado.edu/SerialEM/; 28). The microscope was aligned for fringe-free imaging. The data were collected on the K2 direct electron detection camera (Gatan) positioned behind the Gatan Imaging Filter at the calibrated pixel size of 0.818 Å/px (nominal magnification of 1,650,00×). The energy selecting slit was set to 10 eV. The data from a 5.0 s exposure were saved into 40 frames containing overall dose of 55 e/Å2. The dataset comprised 9594 movies in total. The image defocus was set to to vary between −0.5 μm and −1 μm.
Data analysis
The movies were first corrected for the drift during data acquisition using program MotionCor2 (29) and the contrast transfer function (CTF) parameters were estimated using GCTF (30). The images with astigmatism larger than 2500 Å and estimated resolution based on the contrast transfer function fit analysis worse than 4.0 Å were excluded from subsequent data analysis. A set of 20 micrographs was randomly selected from dataset for manual particle picking using e2boxer.py program (31). The manually picked particles were used for generation of the model for automated particle picking using program CrYOLO (32). The overall number of 670,560 particles were extracted from the dose-weighted micrographs using Relion3.1 (33) and imported in CryoSPARC (https://cryosparc.com/; 34) for the reference-free 2D classification. The damaged particles were further removed using the ab initio reconstruction task in CryoSPARC. Finally, 252,122 particles were selected for the model refinement using the C3 symmetry, which was followed by defocus refinement and another round of 3D refinement, which resulted in 2.54 Å cryo-EM map (according to the FSC(0.143) criteria). Finally, another round of the 3D refinement was performed without imposing the symmetry, which provided 2.88 Å cryo-EM map. A de novo model was built using Coot software (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/; 35), and the resulting model was refined with the Phenix software (http://www.phenix-online.org/; 36). Metal assignment was performed using the metal-binding site validation server (CheckMyMetal) (20). Model visualization and fittings were done using the Chimera software (https://www.cgl.ucsf.edu/chimera/; 37).
Electrophysiology assay
Electrophysiology measurements were performed according to (11). Bilayer lipid membranes were formed in a 100 μm diameter aperture of a 25 μm thick Teflon partition separating a two-compartment Teflon chamber (2.5 ml each). The Teflon membrane was pretreated with a hexadecane/hexane solution (1% v/v), and both chambers were filled with a different buffer solution depending on the condition tested (100 mM ArgCl, 10 mM Hepes, pH 7.0; 100 mM GluNa, 10 mM Hepes, pH 7.0; 100 mM LysCl, 10 mM Hepes, pH 6.0). Lipids (10 μl of DPhPC dissolved in n-pentane at a final concentration of 5 mg/ml) were added in proximity of the aperture allowing the bilayers formation. After bilayer formation, the SDBC sample was added into the cis-side of the chamber, which was considered the virtual ground. For reversal potential (Vrev) measurements calomel electrodes were used. First, a current–voltage (I-V) recording of this multichannel state was captured by varying the Vm from +50 to −50 in decrements of ΔVm = 10 mV and its profile was treated as the blank (no salt gradient). Next, identical I-V measurements were performed but changing the salt concentration on the trans-side of the membrane with Δ(Salt) = 400 mM. The reversal potential was determined by comparing this I-V profile with that of the blank, and the values were then fitted to Goldman–Hodgkin–Katz (Equation 1), where the selectivity of the channel is expressed as the ratio of the permeabilities of the anion and the cation (PC/PA or PA/PC).
| (1) |
For the measurements, a MultiClamp 700A computer controlled current and voltage clamp was used. Output signals were filtered by a low-pass Bessel filter at 10 kHz and saved at a sampling frequency of 50 kHz using an Axon Digidata 1440A digitizer (Molecular Devices, LLC). Data acquisition and analyses were performed using, a homemade LabVIEW (National Instruments), Clampfit 10.0.3 (Molecular Devices, LLC), and Origin Lab 2018 (Northampton).
Resource availability
Primary data are available upon reasonable request to the lead contact.
Data availability
The SDBC maps, the coordinates files, FSC data, and other relevant information about data acquisition and processing have been deposited in the Electron Microscopy Data Bank: for the SDBC with C3 symmetry, 2.54 Å resolution, EMD-14715 and PDB ID-7ZGY; for the SDBC with C1 symmetry, 2.88 Å resolution, EMD-14714 and PDB ID-7ZGX. All data will be made publicly available upon acceptance of the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge the Cryo-electron microscopy and tomography core facility CEITEC MU of CIISB (Brno, Czech Republic), Instruct-CZ Centre supported by MEYS CR (LM2018127) and iNEXT-Discovery, project number 871037, funded by the Horizon 2020 program of the European Commission. We are grateful to Dr J. Nováček for the support at the cryo-EM core facility.
Author contributions
D. P. and D. F. conceptualization; D. P. and D. F. methodology; D. P., D. F., D. dS; and M. W. validation; D. P., D. F., D. dS; S. K., and L. I. formal analysis; D. P., D. F., and L. I. investigation; D. P. and D. F. resources; D. P. and D. F. data curation; D. P., D. F., D. dS., L. I., S. K., P. H., and M. W. writing–original draft; D. P. and D. F. writing–review & editing; D. P. and D. F. visualization; D. P. supervision; D. P. and D. F. project administration; D. P. and S. K. funding acquisition.
Funding and additional information
This work was supported by the National Science Center (Poland) with the Sonata BIS 7 Program (2017) Grant PRO-2017/26/E/NZ1/00344 and the Harmonia 10 Program (2018) Grant PRO-2018/30/M/NZ1/00284 (both to D. P., D. F., and P. H.), the Czech Science Foundation Grant 1825144Y (to S. K.).
Edited by Mike Shipston
Contributor Information
Domenica Farci, Email: domenica.farci@umu.se.
Dario Piano, Email: dario.piano@unica.it.
Supporting information
References
- 1.Bahl H., Scholz H., Bayan N., Chami M., Leblon G., Gulik-Krzywicki T., et al. Molecular biology of S-layers. FEMS Microbiol. Rev. 1997;20:47–98. doi: 10.1111/j.1574-6976.1997.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 2.Messner P., Allmaier G., Schäffer C., Wugeditsch T., Lortal S., König H., et al. Biochemistry of S-layers. FEMS Microbiol. Rev. 1997;20:25–46. doi: 10.1111/j.1574-6976.1997.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 3.Gentner N.E., Mitchel R.E. Ionizing radiation-induced release of a cell surface nuclease from Micrococcus radiodurans. Radiat. Res. 1975;61:204–215. [PubMed] [Google Scholar]
- 4.Sleytr U.B., Sàra M. Bacterial and archaeal S-layer proteins: Structure–function relationship and their biotechnological applications. Trends Biotechnol. 1997;15:20–26. doi: 10.1016/S0167-7799(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 5.Kumar J., Ghosh P., Kumar A. Ultraviolet-B radiation stress-induced toxicity and alterations in proteome of Deinococcus radiodurans. Microb. Physiol. 2021;31:1–15. doi: 10.1159/000512018. [DOI] [PubMed] [Google Scholar]
- 6.Baranova E., Fronzes R., Garcia-Pino A., Van Gerven N., Papapostolou D., Péhau-Arnaudet G., et al. SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature. 2012;487:119–122. doi: 10.1038/nature11155. [DOI] [PubMed] [Google Scholar]
- 7.Bharat T.A.M., Kureisaite-Ciziene D., Hardy G.G., Yu E.W., Devant J.M., Hagen W.J.H., et al. Structure of the hexagonal surface layer on Caulobacter crescentus cells. Nat. Microbiol. 2017;2:17059. doi: 10.1038/nmicrobiol.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumeister W., Barth M., Hegerl R., Guckenberger R., Hahn M., Saxton W.O. Three-dimensional structure of the regular surface layer (HPI layer) of Deinococcus radiodurans. J. Mol. Biol. 1986;187:241–250. doi: 10.1016/0022-2836(86)90231-7. [DOI] [PubMed] [Google Scholar]
- 9.Müller D.J., Schoenenberger C.A., Schabert F., Engel A. Structural changes in native membrane proteins monitored at subnanometer resolution with the atomic force microscope: a review. J. Struct. Biol. 1997;119:149–157. doi: 10.1006/jsbi.1997.3878. [DOI] [PubMed] [Google Scholar]
- 10.Lister T.E., Pinhero P.J. In vivo atomic force microscopy of surface proteins on Deinococcus radiodurans. Langmuir. 2001;17:2624–2628. [Google Scholar]
- 11.Farci D., Kereïche S., Pangeni S., Haniewicz P., Bodrenko I.V., Ceccarelli M., et al. Structural analysis of the architecture and in situ localization of the main S-layer complex in Deinococcus radiodurans. Structure. 2021;29:1279–1285.e3. doi: 10.1016/j.str.2021.06.014. [DOI] [PubMed] [Google Scholar]
- 12.Farci D., Bowler M.W., Kirkpatrick J., McSweeney S., Tramontano E., Piano D. New features of the cell wall of the radio-resistant bacterium Deinococcus radiodurans. Biochim. Biophys. Acta. 2014;1838:1978–1984. doi: 10.1016/j.bbamem.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Farci D., Bowler M.W., Esposito F., McSweeney S., Tramontano E., Piano D. Purification and characterization of DR_2577 (SlpA) a major S-layer protein from Deinococcus radiodurans. Front. Microbiol. 2015;6:414. doi: 10.3389/fmicb.2015.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farci D., Slavov C., Tramontano E., Piano D. The S-layer protein DR_2577 binds the carotenoid deinoxanthin and under desiccation conditions protect against UV-radiation in Deinococcus radiodurans. Front. Microbiol. 2016;7:155. doi: 10.3389/fmicb.2016.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farci D., Slavov C., Piano D. Coexisting properties of thermostability and Ultraviolet radiation resistance in the main S-layer complex of Deinococcus radiodurans. Photochem. Photobiol. Sci. 2018;17:81–88. doi: 10.1039/c7pp00240h. [DOI] [PubMed] [Google Scholar]
- 16.Farci D., Guadalupi G., Bierła K., Lobinski R., Piano D. The role of iron and copper on the oligomerization dynamics of DR_2577, the main S-layer protein of Deinococcus radiodurans. Front. Microbiol. 2019;10:1450. doi: 10.3389/fmicb.2019.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adamec F., Farci D., Bína D., Litvín R., Khan T., Fuciman M., et al. Photophysics of deinoxanthin, the keto-carotenoid bound to the main S-layer unit of. Deinococcus Radiodurans. Photochem. Photobiol. Sci. 2020;19:495–503. doi: 10.1039/d0pp00031k. [DOI] [PubMed] [Google Scholar]
- 18.Farci D., Aksoyoglu M.A., Farci S.F., Bafna J.A., Bodrenko I., Ceccarelli M., et al. Structural insights into the main S-layer unit of Deinococcus radiodurans reveal a massive protein complex with porin-like features. J. Biol. Chem. 2020;295:4224–4236. doi: 10.1074/jbc.RA119.012174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothfuss H., Lara J.C., Schmid A.K., Lidstrom M.E. Involvement of the S-layer proteins Hpi and SlpA in the maintenance of cell envelope integrity in Deinococcus radiodurans R1. Microbiology. 2006;152:2779–2787. doi: 10.1099/mic.0.28971-0. [DOI] [PubMed] [Google Scholar]
- 20.Handing K.B., Niedzialkowska E., Shabalin I.G., Kuhn M.L., Zheng H., Minor W. Characterizing metal-binding sites in proteins with X-ray crystallography. Nat. Protoc. 2018;13:1062–1090. doi: 10.1038/nprot.2018.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson R., Hansen K. Structure of a novel phosphoglycolipid from Deinococcus radiodurans. J. Biol. Chem. 1985;260:12219–12223. [PubMed] [Google Scholar]
- 22.Huang Y., Anderson R. Phosphatidylglyceroylalkylamine, a novel phosphoglycolipid precursor in Deinococcus radiodurans. J. Bacteriol. 1991;173:457–462. doi: 10.1128/jb.173.2.457-462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y., Anderson R. Structure of a novel glucosamine-containing phosphoglycolipid from Deinococcus radiodurans. J. Biol. Chem. 1989;264:18667–18672. [PubMed] [Google Scholar]
- 24.Huang Y., Anderson R. Glucosyl diglyceride lipid structures in Deinococcus radiodurans. J. Bacteriol. 1995;177:2567–2571. doi: 10.1128/jb.177.9.2567-2571.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saint N., Lou K.L., Widmer C., Luckey M., Schirmer T., Rosenbusch J.P. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J. Biol. Chem. 1996;271:20676–20680. [PubMed] [Google Scholar]
- 26.Singh P.R., Ceccarelli M., Lovelle M., Winterhalter M., Mahendran K.R. Antibiotic permeation across the OmpF channel: modulation of the affinity site in the presence of magnesium. J. Phys. Chem. B. 2012;116:4433–4438. doi: 10.1021/jp2123136. [DOI] [PubMed] [Google Scholar]
- 27.Ferrara L.G.M., Wallat G.D., Moynié L., Dhanasekar N.N., Aliouane S., Acosta-Gutiérrez S., et al. MOMP from Campylobacter jejuni is a trimer of 18-stranded β-barrel monomers with a Ca2+ ion bound at the constriction zone. J. Mol. Biol. 2016;428:4528–4543. doi: 10.1016/j.jmb.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Met. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludtke S.J., Baldwin P.R., Chiu W. Eman: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 32.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheres S.H. Relion: implementation of a bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Met. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SDBC maps, the coordinates files, FSC data, and other relevant information about data acquisition and processing have been deposited in the Electron Microscopy Data Bank: for the SDBC with C3 symmetry, 2.54 Å resolution, EMD-14715 and PDB ID-7ZGY; for the SDBC with C1 symmetry, 2.88 Å resolution, EMD-14714 and PDB ID-7ZGX. All data will be made publicly available upon acceptance of the manuscript.