Abstract
COVID survivors frequently experience lingering neurological symptoms that resemble cancer-therapy-related cognitive impairment, a syndrome for which white matter microglial reactivity and consequent neural dysregulation is central. Here, we explored the neurobiological effects of respiratory SARS-CoV-2 infection and found white-matter-selective microglial reactivity in mice and humans. Following mild respiratory COVID in mice, persistently impaired hippocampal neurogenesis, decreased oligodendrocytes, and myelin loss were evident together with elevated CSF cytokines/chemokines including CCL11. Systemic CCL11 administration specifically caused hippocampal microglial reactivity and impaired neurogenesis. Concordantly, humans with lasting cognitive symptoms post-COVID exhibit elevated CCL11 levels. Compared with SARS-CoV-2, mild respiratory influenza in mice caused similar patterns of white-matter-selective microglial reactivity, oligodendrocyte loss, impaired neurogenesis, and elevated CCL11 at early time points, but after influenza, only elevated CCL11 and hippocampal pathology persisted. These findings illustrate similar neuropathophysiology after cancer therapy and respiratory SARS-CoV-2 infection which may contribute to cognitive impairment following even mild COVID.
Keywords: COVID-19, long COVID, cognitive impairment, neuroinflammation, microglia, hippocampal neurogenesis, oligodendrocytes, myelin, H1N1 influenza
Graphical abstract
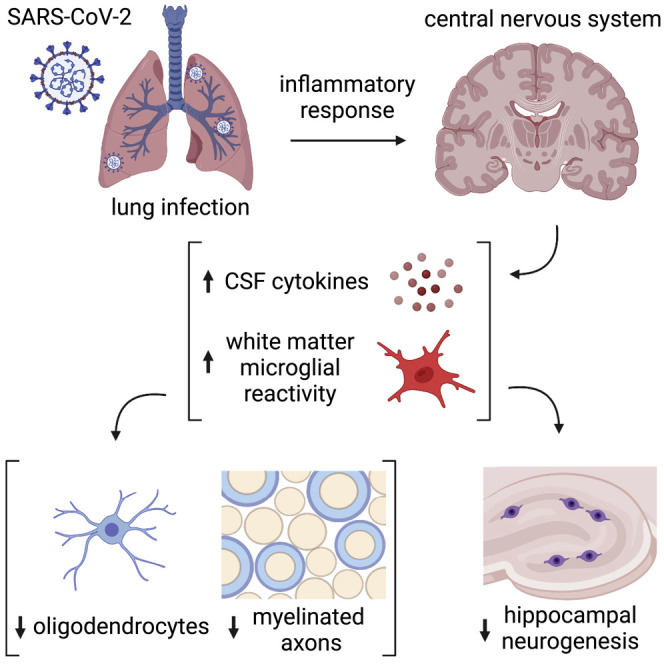
Mild respiratory COVID causes neuroinflammation and multi-lineage cellular dysregulation in the central nervous system, a phenomenon mirroring cancer-therapy-related cognitive impairment.
Introduction
The COVID-19 (coronavirus disease 2019) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has to date resulted in over 500 million documented COVID cases worldwide. Neurological symptoms are emerging as relatively common sequelae, with persistent cognitive impairment affecting approximately one in four COVID survivors (Nasserie et al., 2021). While more common in individuals who had experienced severe COVID requiring hospitalization, even those with mild symptoms in the acute phase may experience lasting cognitive dysfunction (Becker et al., 2021; Nasserie et al., 2021). Colloquially known as “COVID fog,” this syndrome of COVID-associated cognitive impairment is characterized by impaired attention, concentration, speed of information processing, memory, and executive function (Becker et al., 2021; Nasserie et al., 2021). Together with increased rates of anxiety, depression, disordered sleep, and fatigue, this syndrome of cognitive impairment contributes substantially to the morbidity of “long COVID” and in many cases prevents people from returning to their previous level of occupational function (Davis et al., 2021). Given the scale of SARS-CoV-2 infection, this syndrome of persistent cognitive impairment represents a major public health crisis (Nath, 2020).
The syndrome of cognitive symptoms that COVID survivors frequently experience closely resembles the syndrome of cancer-therapy-related cognitive impairment (CRCI; Wefel et al., 2015), commonly known as “chemo fog.” Neuroinflammation is central to the pathophysiology of CRCI (for review, see Gibson and Monje [2021]), raising the possibility of shared pathophysiological mechanisms. Microglia, brain-resident macrophages, become persistently reactive following exposure to certain chemotherapy drugs, after cranial radiation, or after systemic inflammatory challenge with low-dose lipopolysaccharide (Geraghty et al., 2019; Gibson et al., 2019; Monje et al., 2002, 2003, 2007). A distinct subpopulation of microglia that reside in white matter (Hammond et al., 2019) are selectively activated by systemic insults such as exposure to the chemotherapy drug methotrexate (Gibson et al., 2019). Reactive microglia and brain-infiltrating macrophages impair mechanisms of cellular homeostasis and plasticity such as the ongoing generation of myelin-forming oligodendrocytes (Gibson et al., 2019), myelin plasticity (Geraghty et al., 2019), and new neuron generation in the hippocampus (Monje et al., 2002, 2003, 2007). Local microglial cytokine secretion contributes to at least part of this dysregulation (Monje et al., 2003). Elevated circulating cytokine/chemokine levels, particularly CCL11, can also limit neurogenesis and contribute to cognitive impairment (Villeda et al., 2011). In addition to these direct effects of inflammatory mediators on cellular plasticity in the brain, microglia also induce neurotoxic astrocyte reactivity through cytokine signaling (Liddelow et al., 2017). Astrocytes can assume a range of reactive states (Hasel et al., 2021; Sadick et al., 2022) that can induce further pathophysiology, with certain states of reactive astrocytes inducing oligodendrocyte and neuronal cell death (Liddelow et al., 2017; Guttenplan et al., 2021). Additionally, this sub-state of reactive astrocytes are poor supporters of synaptic connections—further adding to network dysfunction (Liddelow et al., 2017). This complex cellular dysregulation is thought to contribute importantly to cognitive impairment, and anti-inflammatory strategies correct this multicellular dysregulation and rescue cognition in disease states such as those occurring after exposure to neurotoxic cancer therapies (Geraghty et al., 2019; Gibson et al., 2019; Monje et al., 2003) and in aging (Villeda et al., 2011).
While severe COVID can cause multi-organ disease and numerous potential mechanisms affecting the nervous system (Lee et al., 2021; Nath and Smith, 2021; Remsik et al., 2021; Chen et al., 2021), even mild COVID could result in a detrimental neuroinflammatory response, including reactivity of these exquisitely sensitive white matter microglia. Given this context, we hypothesized that the inflammatory response to even mild COVID may induce elevation in neurotoxic cytokines/chemokines, a pattern of white matter microglial reactivity, and consequent dysregulation of myelin-forming oligodendrocytes and hippocampal neural precursor cells.
Results
Mild respiratory COVID causes prominent neuroinflammation
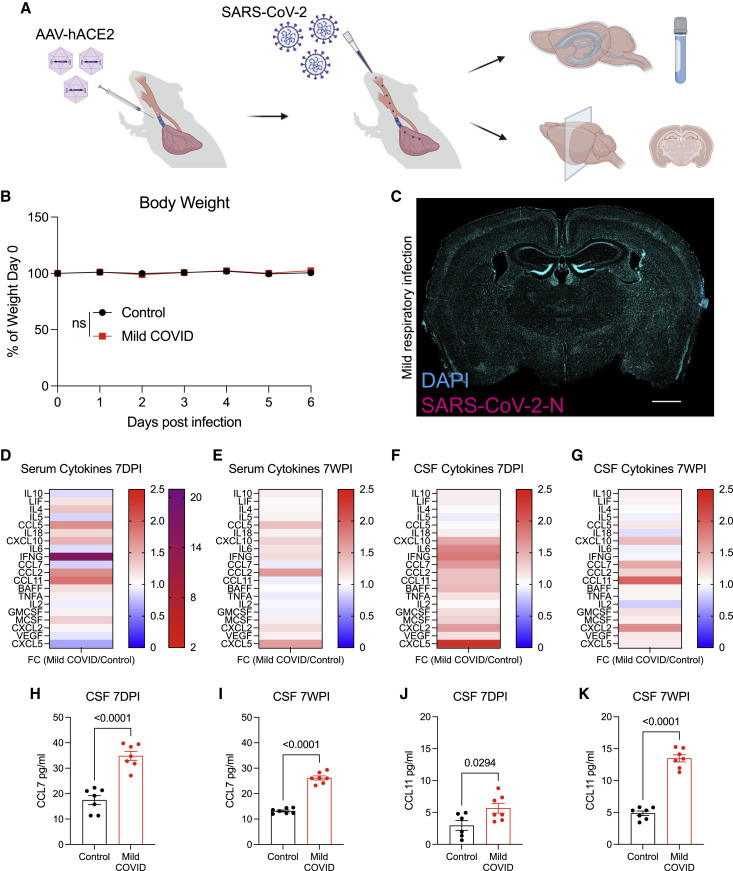
To test the effects of mild COVID, we used a mouse model of mild SARS-CoV-2 infection limited to the respiratory system that clears within 1 week (Israelow et al., 2020; Song et al., 2021). SARS-CoV-2 infection requires expression of its viral entry receptor, human ACE2. In this model, human ACE2 is delivered via AAV vector to the trachea and lungs. Two weeks following intratracheal ACE2-AAV delivery, SARS-CoV-2 is delivered intranasally (Figure 1 A). Control mice received intratracheal AAV expressing human ACE2, but only mock infection intranasally. Mice exhibit no weight loss (Figure 1B) or observable sickness behavior; the challenges of working with a biosafety level-3 mouse model limited the feasibility of further behavioral assessments. No virus was detected in brain (Figure 1C), unlike in the case of a neuroinvasive model (Song et al., 2021) of SARS-CoV-2 infection (Figure S1 A). As expected, SARS-CoV-2 was present in the lungs of infected mice, and lung histopathology demonstrated minimal interstitial infiltrates without evidence of alveolar damage (Figures S1B and S1C). Despite the lack of evident symptoms/signs of illness in this respiratory infection model, we found prominently elevated cytokine profiles in serum and in cerebrospinal fluid (CSF) at 7-day and 7-week time points following respiratory infection (Figures 1D–1G). At 7 days post-infection, elevated CSF cytokines and chemokines include IFN-γ, IL6, TNF-α, CXCL10, CCL7, CCL2, CCL11, GMCSF, and BAFF, among others (Figures 1F and 1H; Table S1). Of these, CXCL10, CCL7, CCL2, CCL11, and GMCSF remain elevated, and IL10 and CCL5 become elevated in CSF by 7 weeks post-infection (Figures 1G and 1I; Table S1). Notably, CSF levels of CCL11, a chemokine associated with cognitive impairment in aging (Villeda et al., 2011), were further increased at 7 weeks compared with 7 days post-infection (Figures 1J and 1K; Table S1). In the same mice, serum CCL11 levels were elevated at 7 days post-infection and—in contrast to persistent elevation in CSF—normalized in serum by 7 weeks post-infection (Figures 1D, 1E, S6C, and S6D; Table S1). These results indicate that isolated respiratory infection with SARS-CoV-2 can result in profound and prolonged changes in cytokines within the CNS.
Figure 1.
Mild respiratory COVID mouse model exhibits CSF cytokine/chemokine elevations
(A) Schematic of experimental paradigm for respiratory system-restricted SARS-CoV-2 infection in mice and experimental workflow. Created with biorender.com.
(B) Body weight (% of day 0 weight) of control and mild COVID mice. Data shown as mean ± SEM; n = 24 mice per group; ns, p > 0.05 by two-way ANOVA with multiple comparisons.
(C) Confocal micrograph of SARS-CoV-2 nucleocapsid protein (SARS-CoV-2-N) 7 days post-infection (7DPI). SARS-CoV-2-N, magenta; DAPI, cyan. Scale bars, 1 mm.
(D and E) Cytokine analyses of serum in control and mild COVID mice 7 days post-infection (D) and 7 weeks post-infection (7WPI) (E). Data shown as fold change (FC) median fluorescence intensity compared with control group; n = 5–7 (CD1 strain) mice per group.
See Table S1 for individual statistics.
(F and G) Cytokine analyses of CSF in control and mild COVID mice 7 days post-infection (F) and 7 weeks post-infection (G). Data shown as fold change (FC) median fluorescence intensity compared with control group; n = 6–7 mice (CD1 strain) per group.
See Table S1 for individual statistics.
(H and I) CSF levels of CCL7 from mice 7 days post-infection (H) and 7 weeks post-infection (I). n = 7 mice per group.
(J and K) CCL11 levels in CSF of mice 7 days post-infection (J) and 7 weeks post-infection (K). n = 6 mice per control group and n = 7 mice per mild COVID group in (J). n = 7 mice per group in (K).
Data shown as mean ± SEM; each dot represents an individual mouse; p values shown in figure panels; ns, p > 0.05; two-tailed unpaired t test.
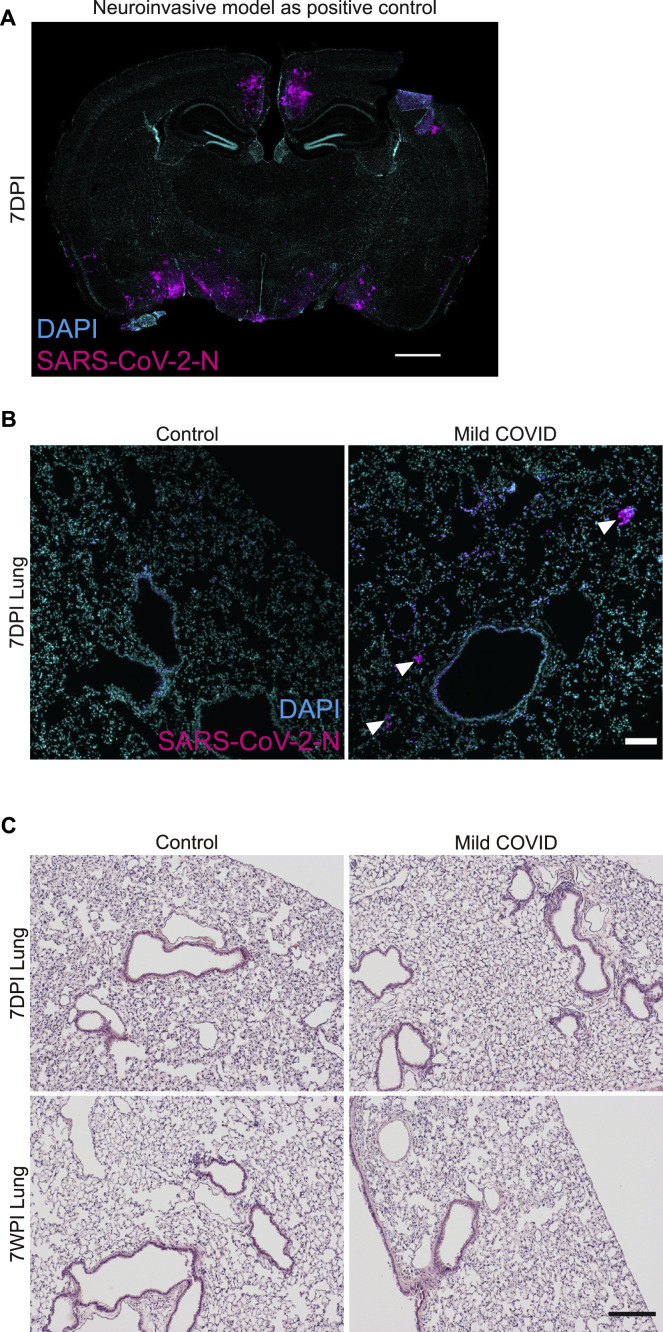
Figure S1.
SARS-CoV-2 mouse models, related to Figure 1
(A) Neuroinvasive SARS-CoV-2 mouse model as a positive control for detection of virus in the brain. Confocal micrograph of coronal section of mouse brain, illustrating SARS-CoV-2 nucleocapsid protein (SARS-CoV-2-N) 7 days post-infection (7DPI) (SARS-CoV-2-N, magenta; DAPI, cyan). Scale bars, 1 mm.
(B) Mild respiratory COVID mouse model, detection of virus in the lung. Representative confocal micrographs of SARS-CoV-2 nucleocapsid protein (SARS-CoV-2-N, magenta; DAPI, cyan) in mouse lung 7 days post-infection, a time point at which the viral infection is largely cleared. Arrowheads highlight SARS-CoV-2-N nucleocapsid protein immunostaining (magenta). Scale bars, 100 μm.
(C) Mild respiratory COVID mouse model, hematoxylin and eosin (H&E) histology in mouse lung tissue at 7 days and 7 weeks post-mild respiratory SARS-CoV-2 infection (7WPI). Minimal interstitial infiltrates with patchy lymphoid aggregates were observed without evidence of alveolar damage. Scale bars, 200 μm.
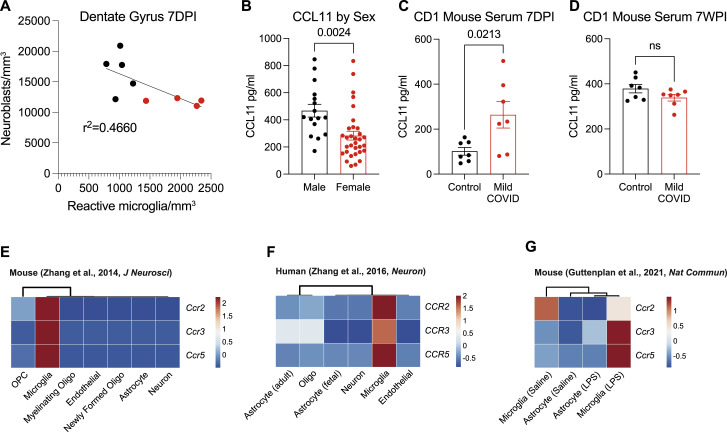
Figure S6.
Microglial reactivity, CCL11 levels, and microglial CCL11 receptor expression, related to Figure 5
(A) Correlation between neuroblasts (DCX+) and activated microglia (IBA1+ CD68+) in the dentate gyrus of CD1 strain mice 7 days post-infection (7DPI). Line fitted with simple linear regression (n = 5 mice per control group; n = 4 mice per mild COVID group), related to Figure 4.
(B) Plasma levels of people experiencing long COVID with “brain fog” broken down by sex (n = 16 subjects per male group; n = 32 subjects per female group), related to Figure 5.
(C and D) Serum levels of CCL11 from CD1 strain mice 7 days post-infection and 7 weeks post-infection (7WPI) (D) n = 7 mice per group, related to Figure 1.
(E–G) CCR2, CCR3, and CCR5 transcriptional expression in mouse neural cell types (E), human neural cells (F), and mouse reactive microglia and astrocytes after lipopolysaccharide (LPS) stimulation (G). (E–G) Data are expressed as row-scaled FPKM. Rows are centered; unit variance scaling is applied to rows. Columns are clustered using correlation distance and average linkage.
Data in (B–D) shown as mean ± SEM; each dot represents an individual mouse or human subject; p values shown in figure panels; ns p > 0.05; two-tailed unpaired t test.
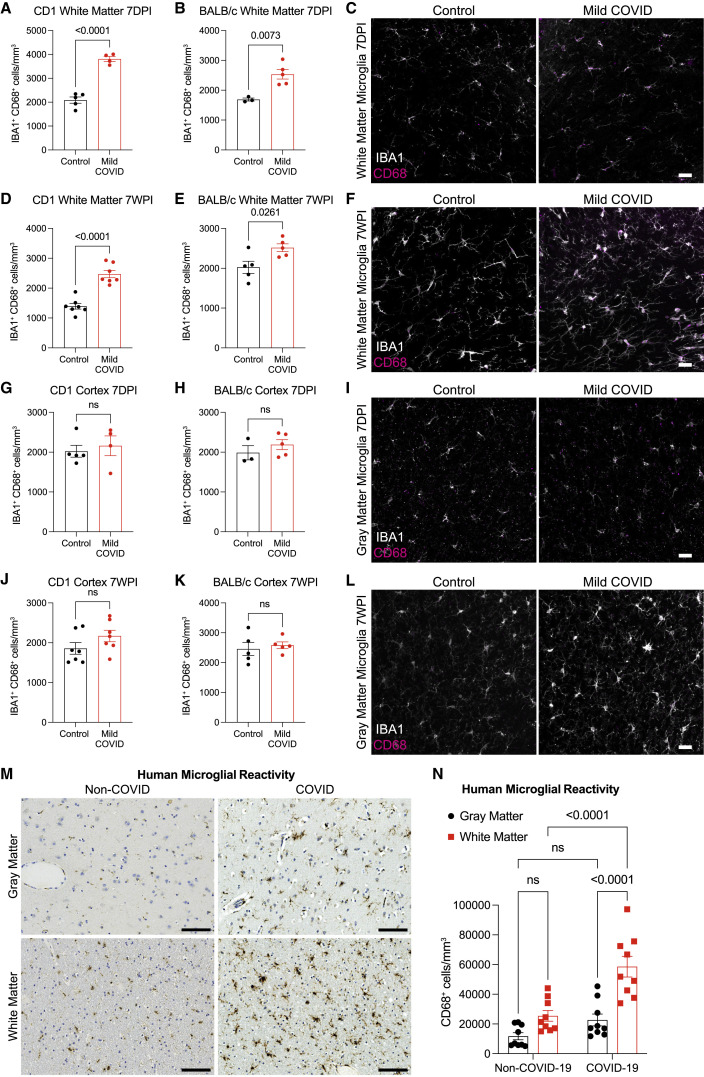
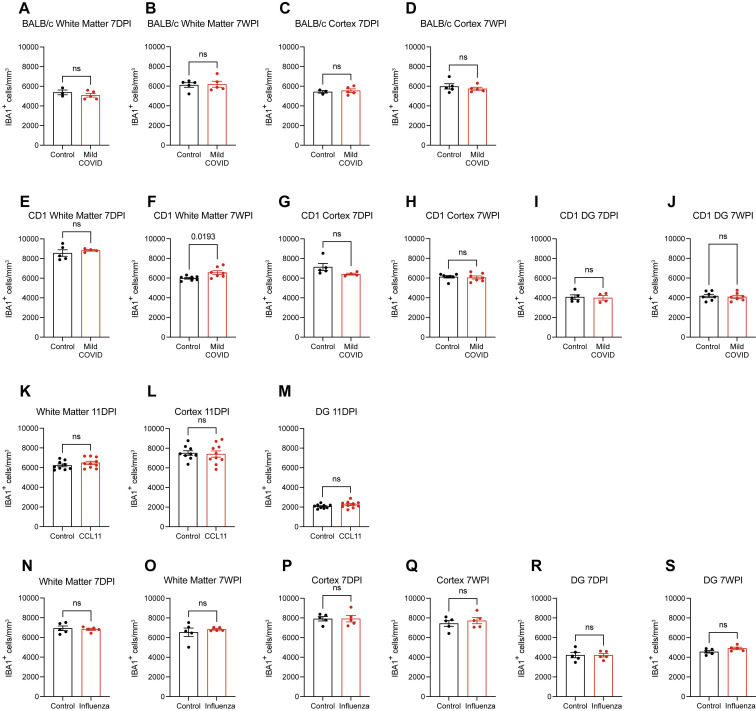
We next examined microglial/macrophage reactivity in the mild COVID mouse model compared with control mice. As hypothesized, we found increased microglial/macrophage reactivity in subcortical white matter, as assessed by IBA1 and CD68 co-positivity (Figure 2 ). This pattern of white-matter-specific microglial/macrophage reactivity was evident in two different mouse strains (CD1 and BALB/c) at 7 days post-infection (Figures 2A–2C) and persisted to 7 weeks post-infection (Figures 2D–2F). In contrast to the subcortical white matter, microglial/macrophage reactivity was not observed in the cortical gray matter (Figures 2G–2L). While microglial/macrophage reactivity was evident in white matter by increased expression of the marker CD68, overall numbers of microglia were largely unchanged (Figure S2 ).
Figure 2.
White matter microglial reactivity after mild respiratory COVID
(A and B) Activated microglia (IBA1+ CD68+) quantification 7 days post-infection in the cingulum of the corpus callosum of (A) CD1 (n = 5 control, 4 mild COVID mice) and (B) BALB/c (n = 3 control, n = 5 mild COVID mice).
(C) Representative confocal micrographs of reactive microglia (IBA1, white; CD68, magenta) in the cingulum of the corpus callosum of BALB/c mice 7 days post-infection.
(D and E) Reactive microglia (IBA1+ CD68+) quantification 7 weeks post-infection in the cingulum of the corpus callosum of (D) CD1 (n = 7 mice/group) and (E) BALB/c (n = 5 mice/group) mice.
(F) Representative confocal micrographs of reactive microglia (IBA1, white; CD68, magenta) in the cingulum of the corpus callosum of BALB/c mice 7-weeks post-infection.
(G and H) Reactive microglia (IBA1+ CD68+) quantification 7 days post-infection in the cortical gray matter of (G) CD1 (n = 5 control, 4 mild COVID mice) and (H) BALB/c (n = 3 control, n = 5 mild COVID mice).
(I) Representative confocal micrographs of reactive microglia (IBA1, white; CD68, magenta) in the cortical gray matter of BALB/c mice 7 days post-infection.
(J and K) Reactive microglia quantification (IBA1+ CD68+) 7 weeks post-infection in the cortical gray matter of (J) CD1 (n = 7 mice/group) and (K) BALB/C (n = 5 mice/group).
(L) Representative confocal micrographs of reactive microglia (IBA1, white; CD68, magenta) in the cortical gray matter of BALB/c mice 7 weeks post-infection.
Data shown as mean ± SEM; each dot represents an individual mouse; p values shown in figure panels; ns p > 0.05 by two-tailed, unpaired t test. (C, F, I, and L) Scale bars, 50 μm.
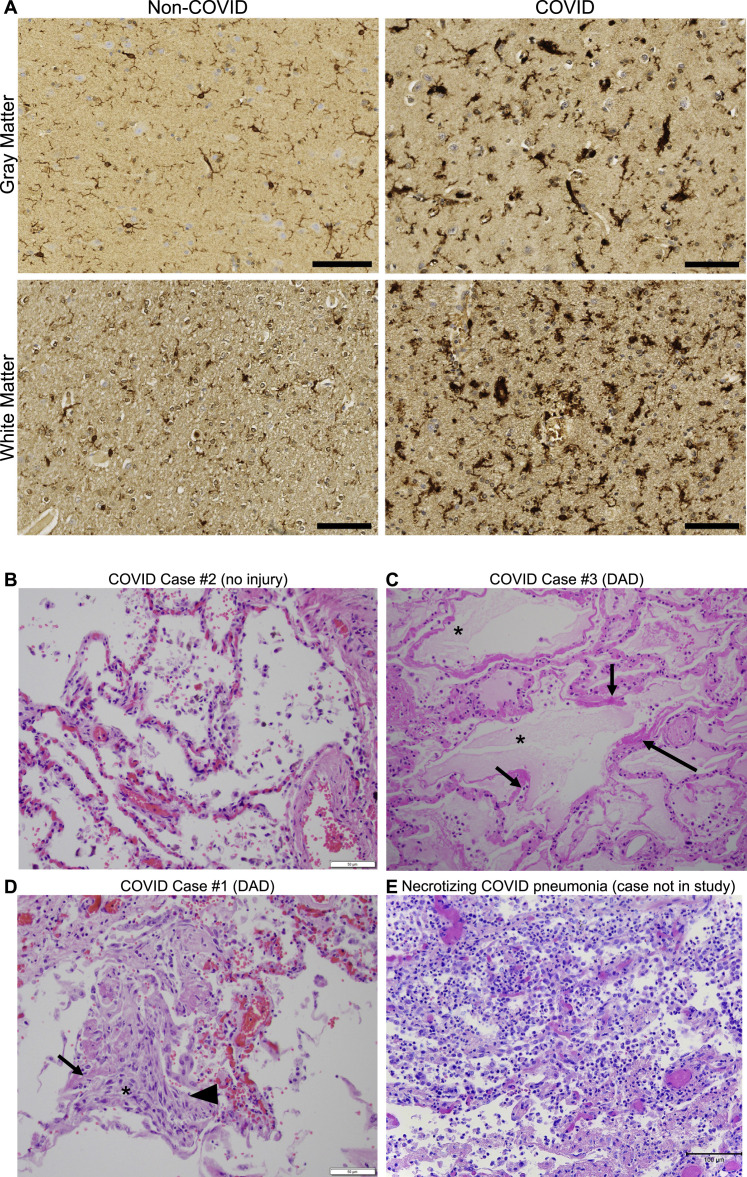
(M) Representative micrographs of CD68 immunostaining (brown) in the gray (cerebral cortex) or subcortical white matter of human subjects with COVID or in non-COVID control subjects.
(N) Reactive microglia (CD68+ cells) quantification (n = 9 human subjects for each group).
Data shown as mean ± SEM; ns, p > 0.05 by two-way ANOVA with multiple comparisons; each dot represents a mouse or human subject. Scale bars, 100 μm.
See also Figures S2 and S3, Table S2, and Data S1.
Figure S2.
Total microglia counts, related to Figures 2, 4, 5, and 7
(A–S) Total microglial counts (IBA1+ cells), assessed at 7-days or 7-weeks following mild respiratory COVID in cortex or subcortical white matter of BALB/c (A–D) and CD1 (E–H) mouse strains and in hippocampal white matter of the dentate gyrus (DG) of CD1 mice (I and J), or assessed in cortex, subcortical white matter and hippocampal white matter at the end of a CCL11 systemic administration paradigm (11 days post-injection [DPI], K–M), or assessed in cortex, subcortical white matter and hippocampal white matter following (N–S).
Data shown as mean ± SEM; each dot represents an individual mouse; p values shown in figure panels; ns p > 0.05; two-tailed unpaired t test.
White matter microglial reactivity in humans after COVID
We had the opportunity to examine human cortex and subcortical white matter samples (Figures 2M, 2N, and S3A) from a cohort of nine individuals (7 male, 2 female, age 24–73) found to be SARS-CoV-2-positive by nasal swab PCR at the time of death during the spring of 2020 (March–July 2020), when the original SARS-CoV-2 strain was dominant. Five of these subjects died suddenly and were found to have SAR-CoV-2 infection, and four subjects died within days to weeks after onset of symptoms; of these 9 individuals, only two (COVID cases #1 and #2; Figure S3; Table S2) required ICU admission (Lee et al., 2021). As each of these individuals died for reasons that were or may have been related to COVID (Lee et al., 2021), these cases cannot be considered “mild.” However, in contrast to most cases available for brain tissue examination, these individuals did not die during hospitalization and did not exhibit fulminant pulmonary disease (Figures S3B–S3E ; Table S2). Lung examination of these cases revealed infection-associated lung injury that ranged from none to moderate (diffuse alveolar damage), without evidence of the necrotizing pneumonia that characterizes severe pulmonary COVID (Figures S3B–S3E; Table S2). The control group represents a cohort of 9 individuals without SARS-CoV-2 infection (9 male, age 43–84). Similar to the mouse model, humans with COVID exhibit robustly elevated CD68+ microglial/macrophage reactivity in subcortical white matter compared with cortical gray matter and compared with individuals without SARS-CoV-2 infection (Figures 2M, 2N, and S3A).
Figure S3.
Human histopathology, related to Figure 2
(A) Representative micrographs of pan-microglial marker IBA1 immunostaining (brown) in the cerebral cortex (gray matter) or subcortical white matter of human subjects with or without COVID. Scale bars, 100 μm.
(B–E) Representative micrographs of lung tissue from human subjects with COVID.
(B) COVID Case #2 with no definite pneumonia or diffuse alveolar damage. Scale bar, 50 μm.
(C) COVID Case #3. Lung showing intra-alveolar fluid (∗), and early hyaline membrane formation (arrows), indicative of acute alveolar damage (200× magnification).
(D) COVID Case #1. Lung showing diffuse alveolar damage including hyaline membranes (arrow), organizing pneumonia (∗) and reactive pneumocytes (arrowhead). Scale bars, 50 μm.
(E) For comparison, an example of necrotizing COVID pneumonia (from a case not represented in this study) with alveolar spaces obliterated by significant infiltrative immune cells, chiefly neutrophils and macrophages. Vascular congestion present. Scale bars, 100 μm.
Figure 3.
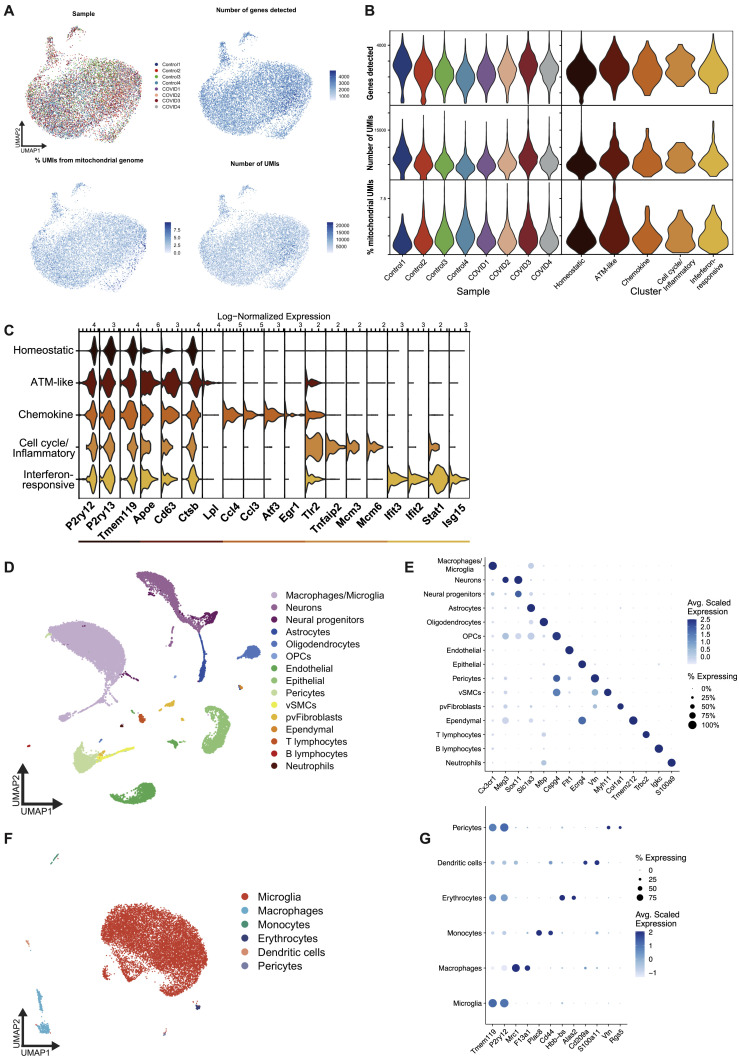
Microglia exhibit heterogeneous transcriptional changes after mild respiratory COVID
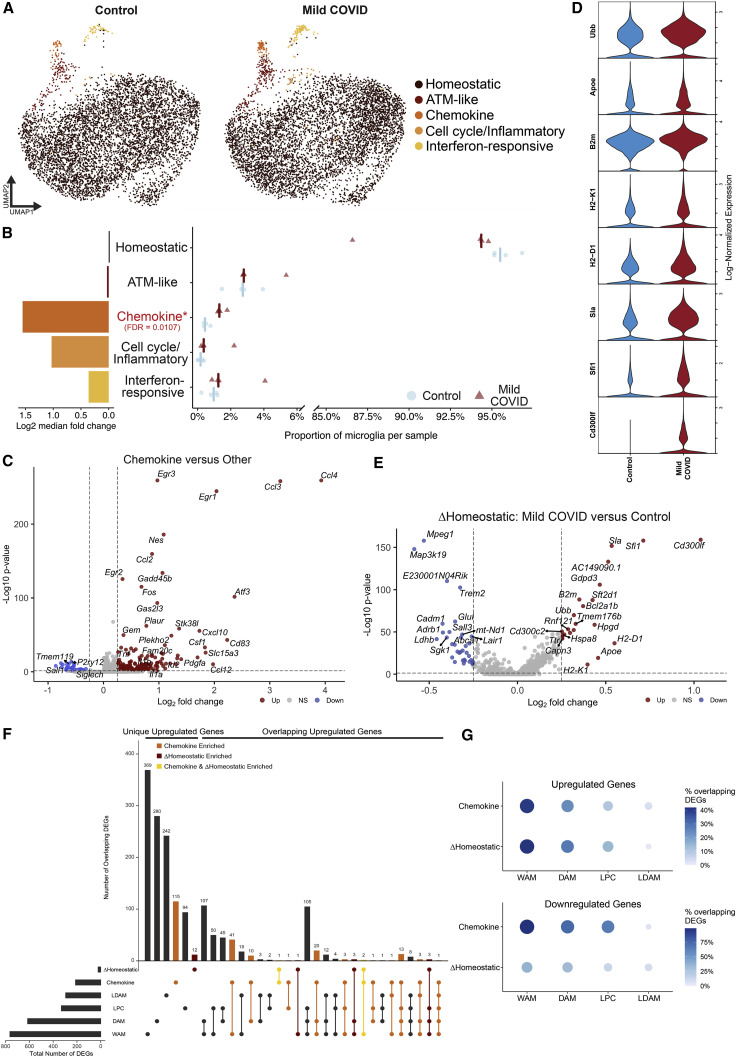
(A) UMAP plot of microglia colored by cluster, divided between mild COVID and control samples (n = 4 mice/group, 5,983 single microglial cells from mild COVID mice and 5,967 single microglial cells from control mice). ATM, axon tract microglia (Hammond et al., 2019).
(B) Right: quantification of the proportion of microglia per sample from each microglia cluster, comparing between mild COVID and control samples; left: bar plot showing the log2 fold change between median proportions in mild COVID samples versus control samples. Positive fold change indicates higher proportion in mild COVID samples. FDR refers to false discovery rate calculated from moderated t test using Benjamini and Hochberg correction for multiple comparisons. See STAR Methods for differential abundance testing statistics.
(C) Volcano plot illustrating results of differential expression testing between chemokine cluster microglia and all other microglia.
(D) Violin plots illustrating expression of eight genes upregulated in homeostatic cluster microglia from mild COVID samples compared with controls.
(E) Volcano plot illustrating results of differential expression testing between homeostatic cluster microglia from mild COVID samples compared with controls. These differentially expressed genes are abbreviated ΔHomeostatic.
For (C and E), plots depict results of two-sided Wilcoxon rank-sum test. p values shown are adjusted for multiple comparisons using Bonferroni correction. Genes were deemed differentially expressed if the magnitude of the average log2 fold change exceeded 0.25 and the adjusted p value was less than 0.05. Upregulated genes are highlighted in red; downregulated genes are highlighted in blue.
(F) Upset plot comparing the intersections between upregulated genes from the chemokine microglia cluster gene signature, and the genes upregulated in the homeostatic microglia cluster in mild COVID compared with control, and the genes upregulated in white matter-associated microglia (WAM) (Safaiyan et al., 2021), disease-associated microglia (DAM) (Safaiyan et al., 2021), lipid-droplet-accumulating microglia (LDAM) (Marschallinger et al., 2020), and microglia following demyelinating injury from lysolecithin (LPC) (Hammond et al., 2019).
(G) Dot plots depicting the percentage of the upregulated or downregulated genes from the chemokine or ΔHomeostatic signatures which are also up- or downregulated, respectively, in the compared signatures.
See also Figures S4 and S5, Tables S2 and S3, and Data S1.
Single-cell transcriptomic profiling of microglia after mild respiratory COVID in mice
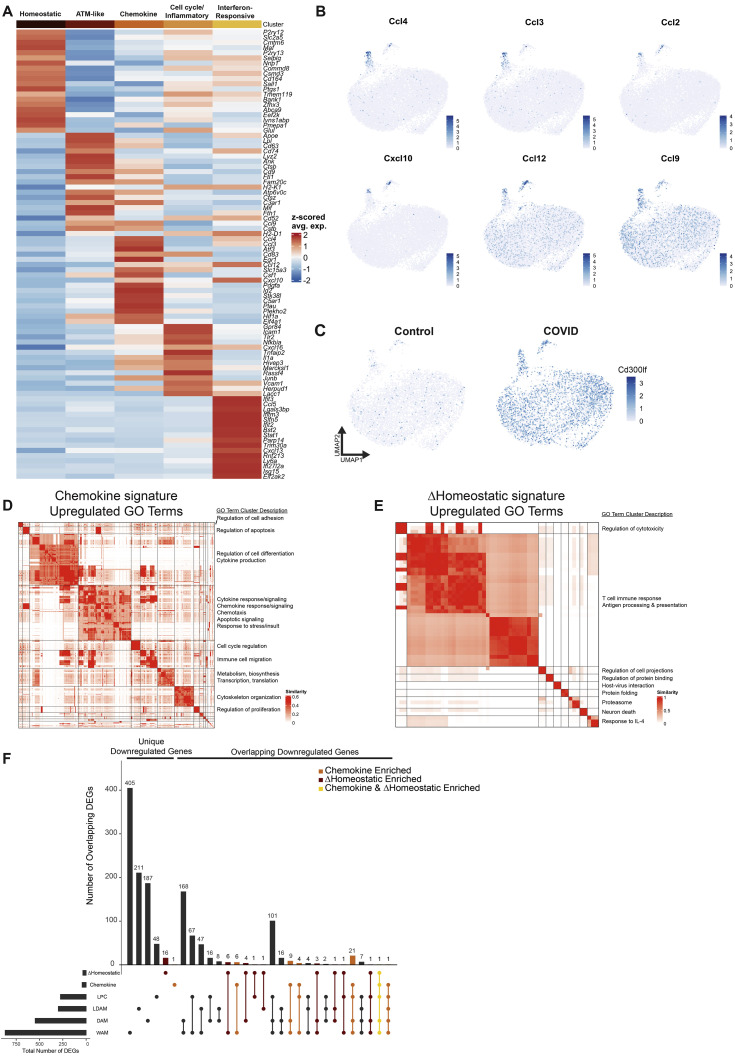
To characterize the state of microglia and other myeloid cells following mild respiratory COVID, we performed single-cell RNA sequencing of cortical and white matter microglia 7 days following mild respiratory SARS-CoV-2 infection (5,983 single microglial cells from mild COVID mice and 5,967 microglial cells from control mice; Figures 3 , S4 , and S5). This revealed five distinct microglial states: a homeostatic cluster defined by higher expression of microglia-specific genes such as P2ry12 and Tmem119; a cluster resembling previously described axon tract microglia (ATM; Hammond et al., 2019) with increased expression of Apoe, Cd63, and Lpl; a chemokine-expressing cluster (e.g., Ccl2, Ccl4, Ccl3, and Cxcl10); a cluster enriched in inflammation-related genes like Tlr2, Il1a, and Tnfaip2 as well as cell-cycle-related genes (Mcm3/4/6); and a cluster expressing interferon-responsive genes (e.g., Ifit3, Ifit2, and Isg15; Figures 3A and S4A). While each of these microglial states was detectable in both control mice and mice following mild respiratory SARS-CoV-2 infection, post-COVID mice exhibited an increased abundance of the chemokine-enriched microglia (Figure 3B), a small but striking subpopulation of cells characterized by expression of several chemokines and cytokines like Tnf, Il1a, and Il1b (Figures 3C and S4B; Table 3), consistent with an inflammatory reactive response.
Figure S4.
Single-cell RNA sequencing of microglia after mild respiratory COVID in mice, related to Figure 3
(A) Heatmap of Z-scored average expression of the top 20 positive marker genes by average log2 fold change for each microglia cluster.
(B) UMAP plots overlaid with log-normalized expression of chemokine genes, illustrating expression in the chemokine microglia cluster.
(C) Split UMAP plot overlaid with log-normalized expression of Cd300lf, which is upregulated in microglia following mild respiratory SARS-CoV-2 infection.
(D) Similarity matrix illustrating clustering of all gene ontology biological process terms, which were significantly enriched in the chemokine gene signature based on semantic similarity.
(E) Similarity matrix of clustering of GO terms enriched in the ΔHomeostatic gene signature.
For (D and E), significance of GO term enrichment was tested using over-representation permutation test. Threshold of significance was Benjamini and Hochberg corrected p value < 0.01. See Table S3 for complete results. GO term clusters were manually annotated with the descriptions at the right-hand side.
(F) Upset plot comparing the intersections between downregulated genes from the chemokine microglia cluster gene signature, and the genes downregulated in the homeostatic microglia cluster in COVID samples versus control, and the genes downregulated in white matter-associated microglia (WAM; Safaiyan et al., 2021), disease-associated microglia (DAM; Safaiyan et al., 2021), lipid-droplet-accumulating microglia (LDAM; Marschallinger et al., 2020), and microglia following demyelinating injury from lysolecithin (LPC; Hammond et al., 2019).
Figure S5.
Microglia single-cell RNA sequencing quality controls, related to Figure 3
(A) UMAP plots overlaid with sample metadata and common quality control metrics.
(B) Violin plots of common quality control metrics across samples and microglia clusters.
(C) Violin plots of log-normalized expression of top marker genes for each microglia cluster.
(D) UMAP plot depicting first round of clustering and cell type annotation, used to identify microglia for downstream analysis. Cell types were annotated based on marker gene expression, some of which is shown in (E).
(E) Dot plot showing the average scaled expression of cell type markers across the original clusters, demonstrating the validity of the cell type labels. The size of each dot corresponds to the proportion of cells in the indicated group which express the gene.
(F) UMAP showing subclustered cells from the macrophages/microglia cluster depicted in (D) and (E), with newly updated cell type labels. This second round of clustering was performed to remove contaminating cells of other types before a third round of clustering to identify microglia states.
(G) Dot plot showing expression of cell type marker genes used in the annotation of the clusters shown in (F). Note that while some non-microglial clusters appear to express the microglia-specific markers Tmem119 and P2ry12, this is driven by a small number of outlier microglia which clustered with these non-microglial cell types. The final microglia cluster does not express canonical markers of these other cell types, indicating that the cluster is highly pure. Analysis of this group of cells is what appears in Figure 3.
We also found transcriptional changes within the homeostatic microglia cluster in post-COVID mice compared with controls, with increased expression of genes commonly upregulated in inflammation and involved in antigen processing and presentation, like B2m, H2-D1, H2-K1, as well as the cell surface glycoprotein Cd300lf (Figures 3D, 3E, and S4C). Members of this family are involved in regulation of immune responses and may regulate phagocytosis of apoptotic cells and maintain immune homeostasis (Alvarez-Errico et al., 2004; Izawa et al., 2014); microglial expression of Cd300lf contributes to cerebral malaria resistance (Keswani et al., 2020). The homeostatic microglia cluster in post-COVID mice exhibit downregulation of characteristic microglial genes such as Trem2 and Sall3, as well as Adrb1, a gene known to be downregulated in lipid-droplet-accumulating microglia (LDAM) in aging white matter (Marschallinger et al., 2020; Figure 3E). Further investigation of these COVID-related transcriptional signatures using gene ontology highlighted upregulation of cytokine production, chemotaxis, and regulation of apoptosis (Figure S4D, chemokine sub-state), and upregulation of cytotoxicity, immune responses, and antigen processing and presentation (Figure S4E, homeostatic signature)—suggesting that even microglia without a “full” reactive response have putative altered function.
We next compared the chemokine and altered homeostatic gene signatures to previously described pathology-specific reactive microglial states, including the aforementioned LDAM, disease-associated microglia (DAM) observed in Alzheimer’s disease (Keren-Shaul et al., 2017), white-matter-associated microglia (WAM) observed in aging (Safaiyan et al., 2021), and microglia following a focal demyelinating injury induced by lysolecithin injection (Hammond et al., 2019). We found a high degree of overlap between the upregulated and downregulated genes from both the chemokine and altered homeostatic gene signatures and the other microglial states, with both signatures in post-COVID microglia showing the greatest similarity to the WAM and DAM states (Figures 3F, 3G, and S4F). The similarity of these gene expression changes to white matter aging and injury is consistent with increased microglial reactivity in the subcortical white matter following SARS-CoV-2 infection. However, the previously described microglial states in other disease contexts, neither individually nor in combination, completely encompass the respiratory COVID-related transcriptional changes, highlighting aspects of a distinct state of microglial/myeloid cell reactivity following mild respiratory COVID.
Mild respiratory COVID impairs hippocampal neurogenesis
Reactive microglia/macrophages and other aspects of inflammatory responses to systemic illness (Monje et al., 2003) or aging (Villeda et al., 2011) can impair the generation of new neurons in the hippocampus, an ongoing mechanism of neural plasticity thought to support healthy memory function (Clelland et al., 2009; Zhang et al., 2008). In particular, cytokines/chemokines can directly inhibit hippocampal neurogenesis (Monje et al., 2003), including interleukin-6 (IL6; Monje et al., 2003) and the chemokine CCL11 (also called eotaxin-1; Villeda et al., 2011). The CSF cytokine profiles following mild respiratory SARS-CoV-2 infection described above highlight elevated IL6 in CSF at 7 days and persistently elevated CCL11 in CSF at 7 weeks (Figures 1F and 1G).
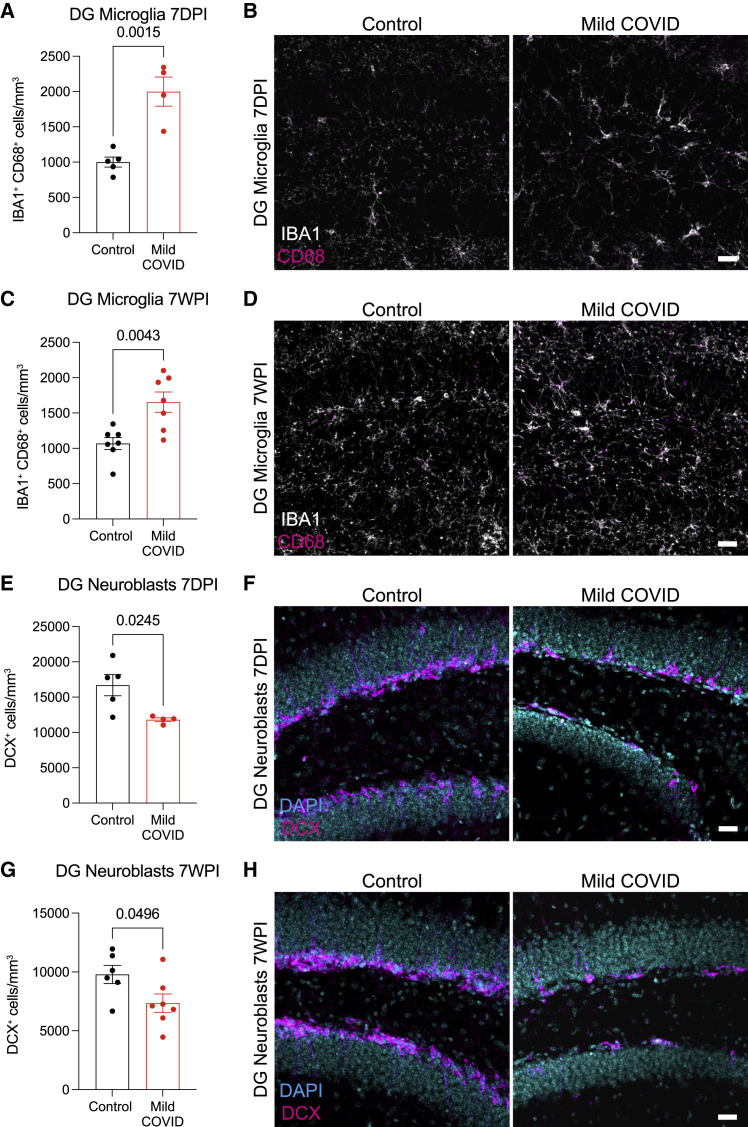
Examining the mouse hippocampus following mild respiratory COVID, we found robustly increased microglial/macrophage reactivity in hippocampal white matter at 7 days post-infection (Figures 4A and 4B) that persists until at least 7 weeks post-infection (Figures 4C and 4D). Consistent with previous observations that reactive microglia/macrophages can inhibit hippocampal neurogenesis (Monje et al., 2003), a stark decrease in new neuron generation was evident, as assessed by Doublecortin-positive cell quantification, at 7 days post-infection (Figures 4E and 4F) and persisted until at least 7 weeks post-infection (Figures 4G and 4H). Considering the number of newly generated hippocampal neurons as a function of the reactive microglial load, we found an inverse correlation between neurogenesis and reactive microglia/macrophages in the hippocampus (Figure S6A).
Figure 4.
Decreased hippocampal neurogenesis after mild respiratory COVID
(A) Reactive microglia (IBA1+ CD68+) quantification 7 days post-infection (7DPI) in the dentate gyrus (hilar white matter) (n = 5 control, n = 4 mild COVID mice).
(B) Representative confocal micrographs of reactive microglia (IBA1, white; CD68, magenta) in the dentate gyrus 7 days post-infection in control and mild COVID mice.
(C) Reactive microglia (IBA1+ CD68+) quantification 7 weeks post-infection (7WPI) in the dentate gyrus of control and mild COVID mice (n=7 mice/group).
(D) Representative confocal micrographs of reactive microglia (IBA1, white; CD68, magenta) in the dentate gyrus of control and mild COVID mice 7 weeks post-infection.
(E) Neuroblast (DCX+) quantification 7 days post-infection in the dentate gyrus of control (n = 5) and mild COVID (n = 4) mice.
(F) Representative confocal micrographs of neuroblasts (DCX, magenta; DAPI, cyan) in the dentate gyrus of control and mild COVID mice 7 days post-infection.
(G) Neuroblast (DCX+) quantification 7 weeks post-infection in the dentate gyrus of control (n = 6) and mild COVID (n = 7) mice.
(H) Representative confocal micrographs of neuroblasts (DCX, magenta; DAPI, cyan) in the dentate gyrus of control and mild COVID mice 7 weeks post-infection.
Data shown as mean ± SEM; each dot represents an individual mouse (A, C, E, and G). (A, C, E, and G) unpaired two-tailed t test. p values shown in figure panels. Scale bars, 50 μm. DG, dentate gyrus.
Elevated CCL11 levels in humans with cognitive symptoms following COVID
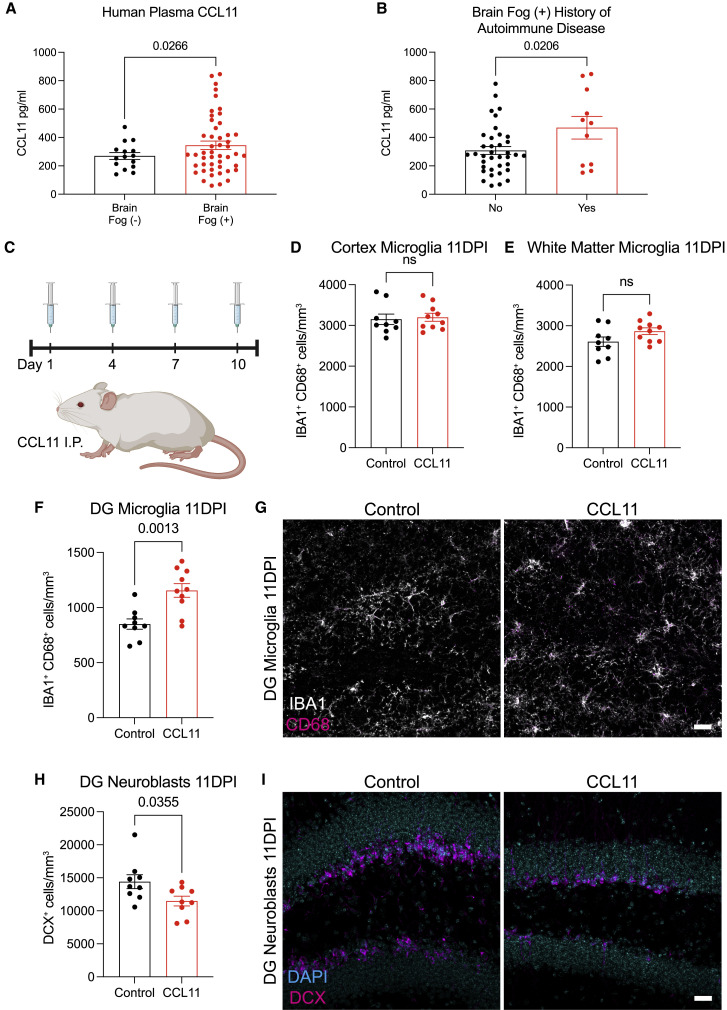
To further explore the relevance of elevated CCL11 levels to cognitive sequelae of COVID infection in humans, we next examined circulating CCL11 cytokine levels in the plasma of people suffering from long COVID with and without cognitive symptoms. These individuals, who were infected in the spring of 2020 when the original strain of the virus was dominant and prior to the availability of vaccines, chiefly experienced a relatively mild course of SARS-CoV-2 infection, without hospitalization in more than 90% of subjects. We found elevated CCL11 levels in the plasma of people with long COVID exhibiting cognitive deficits, or “brain fog” (n = 48 subjects, 16 male/32 female, mean age 46.1 ± 14.6 years) compared with those with long COVID lacking cognitive symptoms (n = 15 subjects, 4 male/11 female, mean age 46.7 ± 14.1 years; Figure 5 A). To help discern the variability in CCL11 levels among COVID patients with brain fog, we performed a multiple linear regression. This determined that two variables, patient sex and history of autoimmune disease, significantly accounted for variability in serum CCL11 levels. Subsequent comparisons revealed that those with a history of autoimmune disease had higher CCL11 levels than those patients who did not (Figure 5B) and that—despite a higher incidence of cognitive symptoms and long COVID reported in female patients overall (Seeßle et al., 2022; Sudre et al., 2021; Tabacof et al., 2022)—male patients had significantly higher levels of CCL11 than female patients in this cohort (Figure S6B). These variables did not interact with one another, suggesting each was an independent predictor of CCL11 levels. Patient’s age, body mass index, and duration of time since symptom onset did not account for variability of CCL11 levels.
Figure 5.
Elevated CCL11 levels associated with cognitive impairment induce hippocampal dysregulation
(A) Plasma levels of CCL11 in people experiencing long COVID with (n = 48 human subjects, brain fog [+]) and without (n = 15 human subjects, brain fog [−]) cognitive symptoms.
(B) History of autoimmune disease in patients reporting “brain fog” (n = 37 human subjects without history of autoimmune disease, n = 11 human subjects with history of autoimmune disease).
(C) Timeline of CCL11 challenge in CD1 strain mice. Brains were collected 24 h after last injection (day 11 post-injection, 11DPI). Created with biorender.com.
(D and E) Reactive microglia (IBA1+ CD68+) quantification 11 days post-CCL11 injection in the cortical gray matter (D) and cingulum of the corpus callosum (E) of mice (n = 9 control, n = 10 CCL11-treated mice).
(F) Reactive microglia (IBA1+ CD68+) quantification 11 days post-CCL11 injection in the dentate gyrus of mice (n = 9 control, n = 10 CCL11-treated mice).
(G) Representative confocal micrographs of activated microglia (IBA1, white; CD68, magenta) in the dentate gyrus of mice 11 days post-CCL11 injection.
(H) Neuroblast (DCX+) quantification 11 days post-injection in the dentate gyrus of mice (n = 9 mice/group).
(I) Representative confocal micrographs of neuroblasts (DCX, magenta; DAPI, cyan) in the dentate gyrus of mice 11 days post-CCL11 injection.
Data shown as mean ± SEM. (A, B, D–F, and H) unpaired two-tailed t test. ns, p > 0.05. p values shown in figure panels. Each dot represents one mouse or human subject. Scale bars, 50 μm. DG, dentate gyrus.
See also Figures S2 and S6 and Data S1.
CCL11 causes specific reactivity of hippocampal microglia and impairs neurogenesis
CCL11, associated with cognitive impairment in aging, has previously been shown to inhibit neurogenesis (Villeda et al., 2011). CCL11 receptors CCR2, CCR3, and CCR5 are chiefly expressed in microglia (Bennett et al., 2016; Zhang et al., 2014, 2016), and microglial CCR2, CCR3, and CCR5 gene expression increases in inflammatory states (Guttenplan et al., 2021; Hasel et al., 2021; Sadick et al., 2022; Figures S6E–S6G). A direct effect of CCL11 on hippocampal neuroblasts has also been reported (Villeda et al., 2011). Given the prominent elevation of CCL11 in mouse models of mild respiratory COVID and the elevated CCL11 levels in people with cognitive symptoms after COVID, we next tested the effects of systemic CCL11 on microglial reactivity (Figures 5C–5I). Using the systemic CCL11 paradigm described by Villeda and colleagues (Villeda et al., 2011), we administered 4 doses of CCL11 intraperitoneally over a 10-day period (Figure 5C). Systemic CCL11 induced microglial/macrophage reactivity specifically in hippocampal white matter (Figures 5F and 5G), and not in the cortex or subcortical white matter (Figures 5D and 5E). Overall numbers of microglia were unchanged (Figures S2K–S2M). As expected (Villeda et al., 2011), we also found decreased hippocampal neurogenesis following systemic CCL11 administration (Figures 5H and 5I).
Mild respiratory COVID impairs myelinating oligodendrocytes
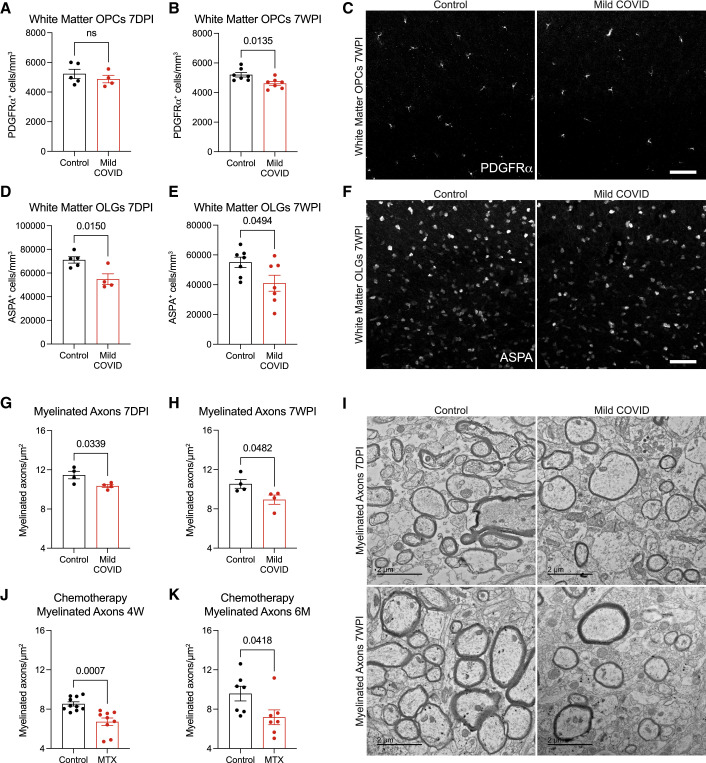
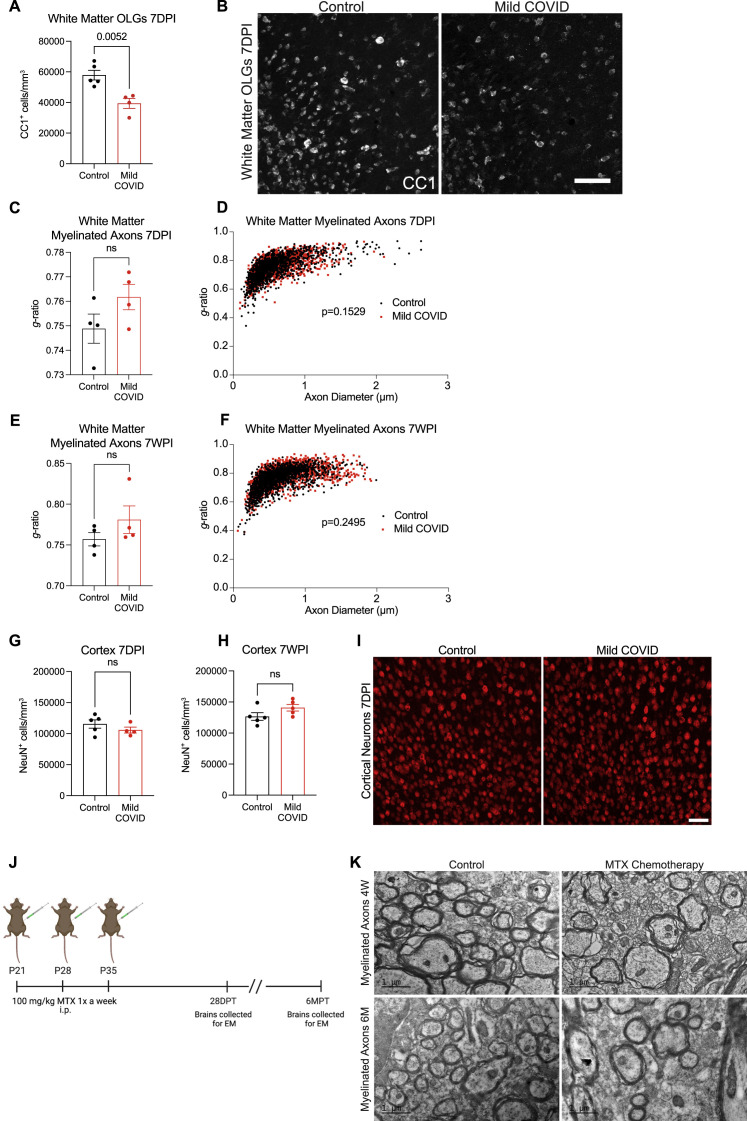
We next explored the effects of mild respiratory COVID on oligodendroglial lineage cells. Reactive microglia/macrophages can alternatively promote (Miron et al., 2013) or impair (Gibson et al., 2019) oligodendrogenesis, depending on the precise microglial cell state. In disease states such as CRCI, reactive microglia cause a dysregulation of oligodendroglial lineage cells (Gibson et al., 2019) and loss of myelin plasticity (Geraghty et al., 2019). Examining oligodendroglial lineage cells in subcortical white matter (cingulum of the corpus callosum), we found maintenance of the oligodendrocyte precursor cell population marked by PDGFRα at 7 days after mild respiratory SARS-CoV-2 infection (Figure 6 A). However, by 7 weeks following infection, a mild decrease (∼10%) in the number of oligodendrocyte precursor cells was evident (Figures 6B and 6C). Mature oligodendrocytes, assessed by ASPA and CC1 immunoreactivity, exhibited a greater magnitude of depletion (Figures 6D–6F, S7A, and S7B), with loss of approximately one third of oligodendrocytes occurring by 7 days post-infection (Figure 6D). This depletion in mature oligodendrocytes persisted until at least 7 weeks post-infection (Figure 6E).
Figure 6.
Oligodendrocyte and myelin loss after mild respiratory COVID
(A and B) Oligodendrocyte precursor cell (PDGFRα+) quantification in the cingulum of the corpus callosum of CD1 strain mice 7 days (n = 5 control, n = 4 mild COVID mice) (A) and 7 weeks (n = 7 mice/group) (B) post-infection (7DPI and 7WPI, respectively).
(C) Representative confocal micrographs of oligodendrocyte precursor cells (PDGFRα, white) in the cingulum of the corpus callosum of mice 7 weeks post-infection. Scale bar, 50 μm.
(D and E) Oligodendrocyte (ASPA+) quantification in the cingulum of the corpus callosum of mice 7 days post-infection (n = 5 control, n = 4 mild COVID mice) (D) and 7 weeks post-infection (n = 7 mice/group) (E).
(F) Representative confocal micrographs of oligodendrocytes (ASPA, white) in the cingulum of the corpus callosum of mice 7 weeks post-infection. Scale bars, 50 μm.
(G and H) Quantification of myelinated axons in the cingulum of the corpus callosum of mice 7 days post-infection (G) and 7 weeks post-infection (H) (n = 4 mice/group).
(I) Representative transmission electron microscopy (EM) images at the level of the cingulum of the corpus callosum in cross-section for (G and H). Myelinated axons evident as electron-dense myelin sheaths encircling axons, viewed in cross-section. Scale bars, 2 μm.
(J and K) Comparison to methotrexate chemotherapy-related cognitive impairment model: quantification of myelinated axons in the cingulum of the corpus callosum at 4 weeks post-methotrexate chemotherapy treatment (n = 10 vehicle control, n = 9 MTX-treated mice; J) and 6 months post-methotrexate chemotherapy treatment (n = 7 mice/group; K).
Data shown as mean ± SEM; each dot represents an individual mouse; unpaired, two-tailed t test. p values shown in figure panels. OPCs, oligodendrocyte precursor cells; OLGs, oligodendrocytes; MTX, methotrexate.
See also Figures S7 and Data S1.
Figure S7.
Myelinated axon deficits after mild respiratory COVID or methotrexate chemotherapy, related to Figure 6
(A) Oligodendrocyte (CC1+) quantification in the cingulum of the corpus callosum of mild COVID and control mice 7 days post-infection (n = 5 mice per control group; n = 4 mice per mild COVID group).
(B) Representative confocal micrographs of oligodendrocytes (CC1, white) in the cingulum of the corpus callosum of mild COVID and control mice 7 days post-infection. Scale bars, 50 μm.
(C) Cumulative g ratios of myelinated axons per animal at 7 days post-infection (n = 4 mice per group).
(D) Scatter plots of g ratio relative to axon diameter 7 days post-infection. Black dots, control axons; red dots, axons from mice with mild respiratory COVID.
(E) Cumulative g ratios of myelinated axons per animal at 7 weeks post-infection (n = 4 mice per group).
(F) Scatter plots of g ratio relative to axon diameter 7 weeks post-infection. Black dots, control axons; red dots, axons from mice with mild respiratory COVID.
(G) Deep layer cortical neuron (NeuN+) density quantified in frontal (M2) cortex of mild COVID and control mice 7 days post-infection (n = 5 mice per control group; n = 4 mice per mild COVID group).
(H) Deep layer cortical neuron (NeuN+) density quantified in frontal (M2) cortex of mild COVID and control mice 7 weeks post-infection (n = 5 mice per control group; n = 5 mice per mild COVID group).
(I) Representative confocal micrographs of deep layer cortical neurons (NeuN, red) in mild COVID and control mice 7 days post-infection. Scale bars, 50 μm.
(J) Schematic of experimental paradigm for methotrexate (MTX) chemotherapy administration in mice. 100 mg/kg MTX was administered via intraperitoneal (i.p.) injection on post-natal day (P)21, P28, and P35 and brains were collected for electron microscopy (EM) 28 days after the final MTX dose and 6 months after the final MTX dose. DPT, days post-treatment; MPT, months post-treatment; EM, electron microscopy. Schematic created with biorender.com.
(K) Representative transmission electron microscopy (EM) images at the level of the cingulum of the corpus callosum in cross-section at 4 weeks and 6 months following MTX exposure, related to Figures 6J and 6K. Myelinated axons evident as electron-dense myelin sheaths encircling axons, viewed in cross-section. Scale bars, 1 μm.
p values shown in (D) and (F) were obtained by comparing mean g ratios per mouse between groups in (C) and (E), respectively. Data in (A), (C), (E), (G), and (H) shown as mean ± SEM; each dot represents an individual mouse; unpaired, two-tailed t test; ns, p > 0.05; p value shown in figure panel.
Consistent with the loss of oligodendrocytes described above, electron microscopy ultrastructural analyses revealed a frank loss of myelin, the insulating ensheathment of axons by oligodendrocyte processes that increases the speed of action potential conductance (Smith, and Koles, 1970, Waxman, 1980) and provides metabolic support to axons (Fünfschilling et al., 2012). We found a decrease in myelinated axon density in subcortical white matter (cingulum of the corpus callosum) evident by 7 days post-infection (Figures 6G–6I). The pattern of decreased myelinated axon density was not focal or plaque-like as occurs in inflammatory demyelinating disorders such as multiple sclerosis. This loss of myelin persisted until at least 7 weeks following mild respiratory SARS-CoV-2 infection and was similar in magnitude to the loss of myelinated axons seen 4 weeks (Figures 6J, S7J, and S7K) and 6 months (Figures 6K, S7J, and S7K) after methotrexate chemotherapy exposure. In contrast to methotrexate chemotherapy exposure (Geraghty et al., 2019; Gibson et al., 2019), changes in myelin sheath thickness relative to axon diameter (g ratio) were not conclusively identified in the remaining myelin sheaths after mild respiratory COVID (Figures S7C–S7F). This observed decrease in myelinated cortical projection axons after mild respiratory COVID was not accounted for by changes in the number of cortical neurons (Figures S7G–S7I). Such persistent loss of myelin in subcortical projections would be predicted to impair neural circuit function and axon health, adding to the numerous deleterious neurobiological consequences of SARS-CoV-2 infection.
Comparison to H1N1 influenza mild respiratory infection
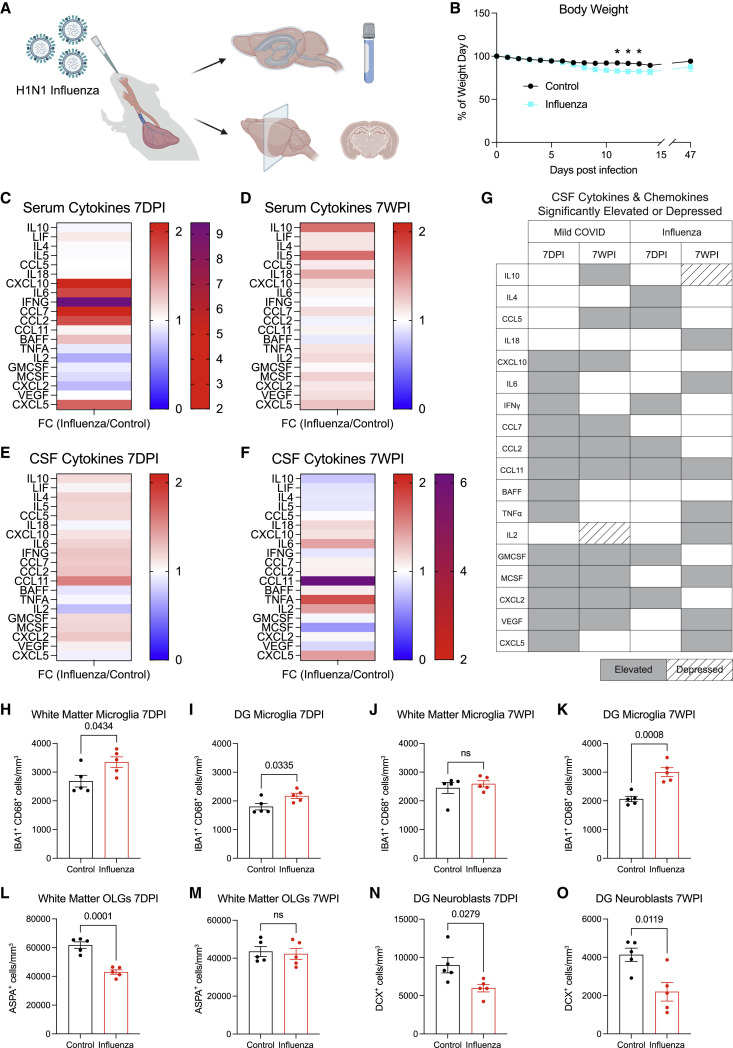
Other respiratory viral infections such as influenza are also associated with cognitive and other neurological sequelae. The H1N1 strain of influenza, which is not neuroinvasive in mouse models, causes acute elevation in CNS cytokines, hippocampal microglial reactivity, and impaired performance in cognitive behavioral tests (Hosseini et al., 2018; Jurgens et al., 2012). We examined a mouse model of respiratory H1N1 influenza, which causes only minimal sickness behavior with mild (<10%) weight loss (Figures 7A and 7B) and elevation in serum cytokines/chemokines (Figures 7C and 7D; Table S4). We compared the neuroinflammatory response to mild respiratory H1N1 influenza with SARS-CoV-2 infection (Figures 7E–7G). Examination of CSF cytokines at 7 days revealed elevation in several cytokines/chemokines similar to mice with mild respiratory COVID, including IFN-γ, CCL11, CCL2, and GMCSF, as well as distinct elevations in IL4 and CCL5 (Figure 7G). By 7 weeks after influenza infection, the only chemokine that remained elevated—and in fact increased from the 7 day time point—is CCL11 (Figures 7E–7G). However, a number of additional cytokines/chemokines were elevated at 7 weeks after influenza that were not elevated at 7 days, including TNF-α, IL6, IL2, CXCL5, and IL18, among others (Figures 7E–7G; Table S4). Together, a comparison of these cytokine profiles after mild respiratory infection with influenza or SARS-CoV-2 illustrates overlapping but distinct neuroinflammatory profiles, with the notable shared feature of persistently elevated CCL11.
Figure 7.
Comparison of neuroinflammatory response and cellular deficits following mild respiratory H1N1 influenza
(A) Schematic of experimental paradigm for respiratory H1N1 influenza infection in CD1 strain mice and experimental workflow. Created with biorender.com.
(B) Body weight (% of day 0 weight) of control and influenza mice. Data shown as mean ± SEM; n = 5 mice/group; ∗ p < 0.05 by two-way ANOVA with multiple comparisons.
(C and D) Cytokine analyses of serum in control and influenza mice 7 days post-infection (7DPI) (C) and 7 weeks post-infection (7WPI) (D). Data shown as fold change (FC) median fluorescence intensity compared with control group; n = 4–5 mice/group.
See Table S4 for individual statistics.
(E and F) Cytokine analysis of CSF in control and influenza mice 7-days post-infection (E) and 7-weeks post-infection (F). Data shown as fold change (FC) median fluorescence intensity compared with control group; n = 5 mice/group.
See Table S4 for individual statistics.
(G) Chart illustrating CSF cytokines and chemokines that were significantly elevated or depressed in mild COVID and influenza cohorts at 7 days post-infection and 7 weeks post-infection.
(H and I) Reactive microglia (IBA1+ CD68+) quantification 7 days post-influenza infection in the cingulum of the corpus callosum (H) and dentate gyrus (I) of mice (n = 5 mice/group).
(J and K) Reactive microglia (IBA1+ CD68+) quantification 7 weeks post-influenza infection in the cingulum of the corpus callosum (J) and dentate gyrus (K) of mice (n = 5 mice/group).
(L and M) Oligodendrocyte (ASPA+) quantification in the cingulum of the corpus callosum of mice 7 days post-influenza infection (L) and 7 weeks post-influenza infection (M) (n = 5 mice/group).
(N and O) Neuroblast (DCX+) quantification 7 days post-influenza infection (N) and 7 weeks post-influenza infection in the dentate gyrus of mice (n = 5 mice/group).
Data shown as mean ± SEM; each dot represents an individual mouse (H–O). Unpaired two-tailed t test in (H–O). ns, p > 0.05. p values shown in figure panels. DG, dentate gyrus.
See also Figures S2, Table S4 and Data S1.
We next examined microglial/macrophage reactivity following respiratory influenza infection. We found the same pattern of white matter-selective microglial/macrophage reactivity in the subcortical and hippocampal white matter (Figures 7H and 7I) at 7 days post-infection. By 7 weeks, microglial reactivity had normalized in the subcortical white matter (Figure 7J). In contrast, microglial/macrophage reactivity persisted at 7 weeks in hippocampal white matter (Figure 7K), a finding that is consistent with the persistently elevated CSF CCL11 levels (Figure 7F), which we demonstrate above specifically increases reactivity of hippocampal microglia/macrophages (Figures 5F and 5G). As above, overall numbers of microglia were unchanged (Figures S2N–S2S). Concordant with this pattern of microglial/macrophage reactivity, oligodendrocytes were decreased in the subcortical white matter at 7 days (Figure 7L) but recovered as microglial reactivity resolved by 7 weeks (Figure 7M). Hippocampal neurogenesis was deficient at both 7-day and 7-week time points, consistent with the persistent hippocampal microglial/macrophage reactivity (Figures 7N and 7O). In comparison to SARS-CoV-2, mild respiratory infection with H1N1 influenza results in similar hippocampal pathology, but without the lasting effects on subcortical white matter integrity seen after respiratory COVID.
Discussion
Taken together, the findings presented here illustrate that even mild respiratory infection with SARS-CoV-2 can result in persistent neuroinflammatory changes and consequent dysregulation of neural cell types important for healthy cognitive function. Such neuroinflammatory changes—particularly white-matter-selective/enriched microglial reactivity—are associated with impaired hippocampal neurogenesis, dysregulation of the oligodendroglial lineage, and myelin loss in other disease contexts (Geraghty et al., 2019; Gibson et al., 2019; Guttenplan et al., 2021; Liddelow et al., 2017; Monje et al., 2003; Villeda et al., 2011). Various immune challenges induce diverse immunological responses that differentially involve particular subsets of immune cells and induce disease-specific cytokine and chemokine profiles systemically and in the CNS. The differences in neuroinflammatory profiles observed here in mouse models of SARS-CoV-2 and H1N1 influenza respiratory infection underscore this principle. SARS-CoV-2 infection induces a broad inflammatory response—well beyond the typical type-1 immune response seen with other respiratory viral infections (Lucas et al., 2020). Concordantly, we find here that even mild respiratory COVID can induce prominent elevation of multiple cytokines and chemokines together with lasting reactivity of white matter microglia in subcortical and hippocampal regions. By comparison, H1N1 influenza elicited a partially overlapping but also distinct CSF cytokine/chemokine profile and more restricted pattern of persistent cellular changes.
Systemic inflammation-induced neuroinflammation, consequent glial dysregulation and impairment of neural function is a principle that extends beyond COVID. Other viral syndromes such as influenza can cause microglial reactivity, elevated CNS cytokine levels, and impair cognitive function, even in the absence of neuroinvasion (Hosseini et al., 2018; Jurgens et al., 2012). However, the oligodendroglial deficits observed in this study were more persistent following COVID than H1N1 influenza and were concordant with the lasting changes that follow methotrexate chemotherapy. Systemic exposure of rats to low levels of bacterial liposaccharide (LPS)—sufficient to cause mild-moderate sickness behavior but not sepsis—similarly causes microglial reactivity and impairment in hippocampal neurogenesis (Monje et al., 2003). People recovering from critical illness in general experience high rates of long-term cognitive impairment, and persistent cognitive impairment is an important component of post-intensive care syndrome (PICS; Pandharipande et al., 2013). The particularly immunogenic nature of SARS-CoV-2 (Lucas et al., 2020) and the persistence of the neuroinflammatory changes observed here, together with the magnitude of the population infected during this pandemic, contribute to the emerging crisis of lasting cognitive impairment associated with COVID.
Elevated levels of CCL11 found in mice after respiratory infection with either SARS-CoV-2 or H1N1 influenza, and in humans with cognitive symptoms after COVID, can cause deficient hippocampal neurogenesis as shown here and previously (Villeda et al., 2011), and induces microglial reactivity specifically in the hippocampus. Whether CCL11 elevation at higher and more consistent levels than used in our intermittent systemic CCL11 administration paradigm can cause microglial reactivity in additional neuroanatomical regions such as subcortical white matter remains to be determined, but this finding implies particular vulnerability of the hippocampus to the neuroinflammatory consequences of respiratory infection, consistent with the recent reports of limbic system structural changes in COVID survivors (Douaud et al., 2022). This region/circuit-specific effect of CCL11 highlights the potential of varying cytokine/chemokine profiles to exert disease-specific effects on the nervous system and for different infections to therefore cause differing prevalence of certain cognitive or neuropsychiatric symptoms.
Consistent with the clinical similarities in the “COVID fog” and “chemo fog” syndromes of cognitive impairment, the findings here illustrate numerous pathophysiological similarities including depletion of myelinating oligodendrocytes and myelin loss. Myelin modulates the speed of neural impulse conduction (Smith and Koles, 1970; Waxman, 1980), provides metabolic support to axons (Fünfschilling et al., 2012), and exhibits adaptive plasticity (Gibson et al., 2014); even small changes in myelination can exert profound effects on neural circuit dynamics and consequently on cognitive function (Steadman et al., 2020; Noori et al., 2020; Pajevic et al., 2014). Subcortical white matter microglial/macrophage reactivity and associated loss of oligodendrocytes and myelinated axons are similar in mouse models of SARS-CoV-2 respiratory infection and methotrexate chemotherapy-related cognitive impairment. While methotrexate directly stimulates reactivity of microglia (Gibson et al., 2019), the mechanisms mediating respiratory infection-induced subcortical white matter microglial reactivity remain to be fully elucidated and may involve cytokine/chemokine signaling mechanisms or cellular neural-immune interactions that require further study.
In addition to the neuropathophysiological changes resulting from even relatively mild acute respiratory infection and consequent neuroinflammation, more severe COVID can result in additional neuropathological consequences (Nath and Smith, 2021). These include microvascular thrombi, neuron loss, cortical inflammation, and even direct brain infection in some cases (Lee et al., 2021; Remsik et al., 2021; Song et al., 2021; Yang et al., 2021). It is thus not surprising that the neurological sequelae of SARS-CoV-2 infection are proving to be both common and debilitating (Nasserie et al., 2021; Mazza et al., 2021; Qureshi et al., 2022; Becker et al., 2021; Tabacof et al., 2022). While the inflammation-induced multicellular dysregulation described here may be potentially reversible with therapeutic interventions as discussed below, these more severe possible complications of COVID such as stroke can cause irreversible damage to the brain.
Limitations of the study
Here, we studied the neurobiological consequences of respiratory system infection in adulthood with the original strain of SARS-CoV-2 in the absence of vaccination. The incidence and severity of cognitive impairment following COVID caused by newer SARS-CoV-2 variants such as the Omicron variant, or as a result of breakthrough infection in vaccinated individuals, remains to be fully determined. Emerging data indicate that the risk of cognitive impairment may be decreased in breakthrough infections of fully vaccinated individuals (Antonelli et al., 2022). If and how respiratory-system-infection-induced neuroinflammation in vaccinated individuals with adaptive immune responses to SARS-CoV-2 differs from the unvaccinated context is an important topic for future work. Furthermore, the neurobiological effects of respiratory COVID in childhood and in old age require dedicated study.
As occurs after cancer therapies and in other disease states, the microglial reactivity present after mild respiratory SARS-CoV-2 infection may also induce neurotoxic astrocyte reactivity (Liddelow et al., 2017; Gibson et al., 2019; Guttenplan et al., 2021), as well as aberrant microglial pruning of synapses (Hinkle et al., 2019; Hosseini et al., 2018; Vasek et al., 2016). In the context of severe COVID, astrocytes in the human cerebral cortex exhibit a transcriptional program consistent with neurotoxic reactivity (Yang et al., 2021). Whether this astrocyte-mediated neurotoxicity occurs after mild respiratory COVID remains to be determined. The oligodendrocyte loss observed here could be caused reactive astrocyte toxic lipids, which is known to cause oligodendrocyte cell death (Liddelow et al., 2017; Guttenplan et al., 2021). Our single-cell transcriptomic studies did not contain sufficient numbers of cells to draw conclusions about the transcriptional state of astrocytes or cell types other than microglia. Incorporating more animals and additional techniques such as single nucleus sequencing and spatial transcriptomics will be required to further explore the state of astrocytes, remaining oligodendrocytes, vascular, and other cell types following respiratory COVID. Furthermore, the microglial gene expression changes evident following respiratory COVID will require spatial validation and functional testing in future work.
Preclinical studies in disease models that share numerous features with the cellular dysregulation observed here after mild respiratory COVID have identified anti-inflammatory and neuro-regenerative strategies that restore neural plasticity and rescue cognition (Ayoub et al., 2020; Geraghty et al., 2019; Gibson et al., 2019; Monje et al., 2003; Villeda et al., 2011). Such insights from diseases such as CRCI may elucidate therapeutic strategies to restore healthy cognitive function following COVID. For example, microglial depletion strategies can restore similar multicellular deficits and rescue impaired cognitive performance in mouse models of CRCI (Acharya et al., 2016; Gibson et al., 2019), but it remains to be determined if similar microglial-targeting interventions will prove beneficial for cognitive impairment associated with COVID. Future work, incorporating cognitive testing in mice following respiratory COVID, will be needed to test possible therapeutic interventions aimed at restoring neural homeostasis and plasticity and mitigating the neurobiological sequelae of COVID.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SARS-CoV-2 nucleocapsid | GeneTex | Cat#GTX135357; RRID AB_2868464 |
| Rabbit anti-IBA1 | Wako Chemicals | Cat#019-19741; RRID: AB_839504 |
| Rat anti-CD68 | Abcam | Cat#ab53444; RRID: AB_869007 |
| Rabbit anti-DCX | Abcam | Cat#ab18723; RRID: AB_732011 |
| Rabbit anti-ASPA | EMD Millipore | Cat#ABN1698; RRID: AB_2827931 |
| Goat anti-PDGFRa | R&D Systems | Cat#AF1062; RRID: AB_2236897 |
| Mouse anti-CC1 | EMD Millipore | Cat#OP80; RRID: AB_2057371 |
| Chicken anti-NeuN | EMD Millipore | Cat#ABN91; RRID: AB_11205760 |
| Alexa 647 donkey anti-rabbit IgG | Jackson Immunoresearch | Cat# 711-605-152; RRID: AB_2492288 |
| Alexa 488 donkey anti rat IgG | Jackson Immunoresearch | Cat# 712-545-153; RRID: AB_2340684 |
| Alexa 594 donkey anti-rabbit IgG | Jackson Immunoresearch | Cat# 711-585-152; RRID: AB_2340621 |
| Alexa 488 donkey anti goat IgG | Jackson Immunoresearch | Cat# 705-545-147; RRID: AB_2336933 |
| Alexa 647 donkey anti-mouse IgG | Jackson Immunoresearch | Cat# 715-605-150; RRID: AB_2340862 |
| DAPI | Thermo Fisher Scientific | Cat# 62247 |
| Prolong Gold Mounting Medium | Life Technologies | Cat# P36930 |
| Normal Donkey Serum | Jackson Immunotherapy | Cat# 017-000-121 |
| Mouse anti-CD68 | Thermo Fisher Scientific | Cat# 14-0688-82; RRID: AB_11151139 |
| PowerVision polyHRP anti-mouse IgG | Leica Biosystems | Cat# PV6114; RRID: AB_1307589 |
| PowerVision polyHRP anti-rabbit IgG | Leica Biosystems | Cat# PV6119; RRID: AB_1307590 |
| Bacterial and virus strains | ||
| AAV-CMV-hACE2 | Janelia Viral tools | N/A |
| Biological samples | ||
| SARS-Related Coronavirus 2, Isolate USA-WA1/2020 | BEI resources | Cat# NR-52281 |
| Chemicals, peptides, and recombinant proteins | ||
| 8% Glutaraldehyde | Electron Microscopy Services | Cat#16000 |
| 16% Paraformaldehyde | Electron Microscopy Services | Cat#15700 |
| 0.1 M sodium cacodylate | Electron Microscopy Services | Cat#12300 |
| 3,3′- diaminobenzidine (DAB) | Vector Laboratories | Cat# SK-4100 |
| Hematoxylin Solution | DAKO | Cat# CS70030-2 |
| EcoMount Mounting Medium | Biocare Medical | Cat# EM897L |
| Critical commercial assays | ||
| Immune Monitoring 48-Plex Mouse ProcartaPlex Panel | Thermo Fisher Scientific | Cat# EPX480-20834-901 |
| Human Cytokine/Chemokine 71-Plex Discovery Assay Array | Eve Technologies | Cat# HD71 |
| Recombinant Mouse CCL11/Eotaxin protein | R&D Systems | Cat# 420-ME-020/CF |
| Deposited data | ||
| Single Cell sequencing dataset | NCBI Sequencing Read Archive (SRA) BioProject | NCBI: PRJNA833593 |
| Experimental models: Organisms/strains | ||
| Mouse: CD1 | Charles River | Strain #022 |
| Mouse: Balb/cJ | Jackson Laboratory | Strain #000651 |
| Software and algorithms | ||
| FIJI | NIH | https://imagej.net/software/fiji/ |
| Zen | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| Imaris image analysis software | Oxford Instruments | https://imaris.oxinst.com/packages |
| Graphpad Prism | Dotmatics | https://www.graphpad.com/ |
| R (v4.1.1) | R Core Team | https://www.r-project.org/ |
| Suerat (v4.1.0) | Hao et al. (2021) | https://github.com/satijalab/seurat |
| Harmony (v0.1.0) | Korsunsky et al. (2019) | https://github.com/immunogenomics/harmony |
| Speckle (v0.0.3) | Phipson et al. (2021) | https://doi.org/10.1101/2021.11.28.470236 |
| clusterProfiler package (v4.0.5) | Wu et al. (2021) | N/A |
| simplifyEnrichment package (v1.2.0) | Gu and Hübschmann (2021) | https://doi.org/10.1101/2020.10.27.312116 |
| UpsetR package (v1.4.0) | Conway et al. (2017) | https://github.com/hms-dbmi/UpSetR/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Michelle Monje (mmonje@stanford.edu)
Materials availability
Plasmids generated in this study have been deposited with Addgene plasmid # 158957; http://n2t.net/addgene:158957; RRID: Addgene_158957.
Data and code availability
-
•
Single cell RNA sequencing data are publicly available on NCBI Sequencing Read Archive (SRA): BioProject ID PRJNA833593 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA833593/). Additional supplemental items, including the source data for all figures in this paper (Data S1) are available on Mendeley Data: https://data.mendeley.com/datasets/jw25y27vrm/3. Microscopy images reported in this paper will be shared by the lead contact upon request.
-
•
All original code is available on GitHub: https://github.com/liddelowlab/Mild_COVID_Microglia-Cell_2022.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental method and subject details
Mouse models
6- to 12-week-old female CD1 and BALB/c mice were purchased from Charles River and Jackson Laboratory, respectively, and housed at Yale University. All experiments were performed in CD1 mice, with exception of assessment of microglial reactivity in both CD1 and BALB/c mice. All procedures used in this study complied with federal guidelines and the institutional policies of the Yale School of Medicine Animal Care and Use Committee. For the CCL11 study, 8-week-old CD1 male and female mice were purchased from Charles River and housed at Stanford University; all procedures complied with Institutional Animal Care and Use Committee protocol at Stanford.
Human subjects
Human brain samples were collected as part of routine autopsy procedures in New York, Iowa, Maryland and California (see Table S2). Human blood samples from subjects with long-COVID were collected with informed consent as part of an IRB-approved protocol at Mount Sinai School of Medicine.
AAV-hACE preparation
AAV-hACE2 was constructed using InfusionTM (Takara) cloning per manufacturers protocol. In short, pAAV-EF1a-mCherry-IRES-Cre (a gift from Karl Deisseroth; Addgene plasmid # 55632; http://n2t.net/addgene:55632; RRID: Addgene_55632) was linearized by restriction digest with EcoRI and BamHI to remove mCherry-IRES-Cre. PCR amplification of hACE2 containing MYC and Flag tag (Origene CAT#: RC208442) with 15BP overhang was performed and used in the infusion reaction. Plasmid sequence was confirmed by sangar sequencing of the insert. AAV-hACE2-cMYC-Flag was deposited with Addgene (Addgene plasmid #158957; http://n2t.net/addgene:158957; RRID: Addgene_158957). AAV viral particle preparations were performed by HHMI virus core with AAV9 capsid.
Mild respiratory COVID model
Mice were infected intratracheally with AAV-hACE2 as previously described (Song et al., 2021). Briefly, animals were anaesthetized using a mixture of ketamine (50 mg/kg) and xylazine (5 mg/kg), injected intraperitoneally. The rostral neck was shaved and disinfected. A 5-mm incision was made, the salivary glands were retracted, and the trachea was visualized. Using a 500-μl insulin syringe, a 50-μl bolus injection of 1011 genome copies (GC) of AAV- hACE2 was injected into the trachea. The incision was closed with VetBond skin glue. Following intramuscular administration of analgesic (meloxicam and buprenorphine, 1 mg/kg), animals were placed in a heated cage until full recovery. For subsequent SARS-CoV-2 infection, mice were anesthetized using 30% vol/vol isoflurane diluted in propylene glycol. Using a pipette, 50μl of SARS-CoV-2 (3 × 107 PFU/ml) was delivered intranasally. Littermate control mice were transduced with AAV-hACE2 but not infected with SARS-CoV-2.
Neuroinvasive COVID model positive control
Mice were anesthetized using ketamine and xylazine. For intracisternal magna injection of AAV-hACE2, the dorsal neck was shaved and sterilized. A 2-cm incision was made at the base of the skull, and the dorsal neck muscles were separated using forceps. After visualization of the cisterna magna, a Hamilton syringe with a 15°, 33-gauge needle was used to puncture the dura. 3μl of AAV-hACE2 (3 × 1012 viral particles/mouse) was administered per mouse at a rate of 1μl/min. Upon completion of the injection, the needle was left in to prevent backflow for an additional 3 min. The skin was stapled and disinfected, and the same postoperative procedures were performed as for intratracheal injections. Intratracheal injection of AAV-hACE2 was performed at the same time as intracisternal magna injection. For subsequent SARS-CoV-2 infection, mice were anesthetized using 30% vol/vol isoflurane diluted in propylene glycol. Using a pipette, 50μl of SARS-CoV-2 (3 × 107 PFU/ml) was delivered intranasally.
Generation of SARS-CoV-2 stocks
To generate SARS-CoV-2 viral stocks, Huh7.5 cells were inoculated with SARS-CoV-2 isolate USA-WA1/2020 (NR-52281; BEI Resources). To generate stocks, VeroE6 cells overexpressing hACE2 and transmembrane serine protease 2 (TMPRSS2) [provided by B. Graham National Institutes of Health-Vaccine Research Center (NIH-VRC)] were infected at a multiplicity of infection of 0.01 for 2 to 3 until sufficient cytopathic effect was noted. After incubation, the supernatant was clarified by centrifugation (500g × 5 min) and filtered through a 0.45-μm filter. To concentrate virus, filtered supernatants were applied to Amicon Ultra-15 centrifugal filter (Ultracel 100k) and spun at 2000 rpm for 15 min. The supernatant was then aliquoted for storage at −80°C. Viral titers were measured by standard plaque assay using Vero E6 cells.
H1N1 influenza respiratory infection model
CD1 mice were anesthetized by 30% v/v isoflurane diluted in propylene glycol and inoculated intranasally with 50 pfu of A/PR8/34 (H1N1) in 20μL PBS as previously described (Ichinohe et al., 2011). Body weights of mice were monitored.
CCL11 administration mouse model
CD1 mice were weighed and injected intraperitoneally with recombinant mouse CCL11 (10μg/kg in PBS, R&D systems 420-ME) or PBS vehicle control four times over a ten-day period at D1, D4, D7, and D10, and then perfused 24 hours after the final injection on D11, as described in Villeda et al. (2011).
Methotrexate chemotherapy model
Electron microscopy images from animals used in the Gibson et al. (2019) study were analyzed for myelinated axon density at 4 week and 24 weeks following the last exposure to methotrexate (100mg/kg) which was administered at postnatal day (P) 21, P28 and P35 intraperitoneally (Gibson et al., 2019).
Method details
Perfusion and immunohistochemistry
Mice were anesthetized with isoflurane and transcardially perfused with 10ml 0.1M PBS followed by 10ml 4% paraformaldehyde (PFA). Brains were fixed in 4% PFA overnight at 4oC, before being transferred to 30% sucrose for cryoprotection. Brains were embedded in Tissue-Tek (Sakura) and sectioned coronally at 40μm using a sliding microtome (Microm HM450; Thermo Scientific). For immunohistochemistry, a 1 in 6 series of 40μm coronal sections was incubated in 3% normal donkey serum in 0.3% Triton X-100 in TBS blocking solution at room temperature for one hour. Rabbit anti-SARS-CoV-2 nucleocapsid (1:250, GeneTex GTX135357), rabbit anti-IBA1 (1:1000, Wako Chemicals 019-19741), rat anti-CD68 (1:200, Abcam ab53444), rabbit anti-DCX (1:500, Abcam ab18723), rabbit anti-ASPA (1:250, EMD Millipore ABN1698), goat anti-PDGFRα (1:500; R&D Systems AF1062), mouse anti-CC1 (1:100; EMD Millipore OP80), chicken anti-NeuN (1:500; Millipore Sigma ABN 91) were diluted in 1% normal donkey serum in 0.3% Triton X-100 in TBS and incubated overnight at 4oC (CC1 was incubated for 7 days at 4oC). Sections were then rinsed three times in 1X TBS and incubated in secondary antibody solution (Alexa 647 donkey anti-rabbit IgG, 1:500 (Jackson Immunoresearch); Alexa 488 donkey anti rat IgG, 1:500 (Jackson Immunoresearch); Alexa 594 donkey anti-rabbit IgG, 1:500 (Jackson Immunoresearch); Alexa 488 donkey anti goat IgG, 1:500 (Jackson Immunoresearch); Alexa 647 donkey anti-mouse IgG, 1:500 (Jackson Immunoresearch) in 1% blocking solution at room temperature protected from light for two hours. Sections were rinsed 3 times in TBS, incubated with DAPI for 5 minutes (1:1000; Thermo Fisher Scientific) and mounted with ProLong Gold mounting medium (Life Technologies).
Confocal imaging and quantification
All cell counting was performed by experimenters blinded to experimental conditions on a Zeiss LSM700 or LSM800 scanning confocal microscope (Zeiss). Images were taken at 20X magnification and analyzed using FIJI software. For microglia imaging, three consecutive sections ranging approximately from bregma +1.2 to +0.8 were selected for cortex and corpus callosum, and four consecutive sections ranging approximately from bregma -1.8 to -2.4 were selected for the hilus of the dentate gyrus. For each section, the superficial cortex, deep cortex, cingulum, genu of the corpus callosum, and hilus of the dentate gyrus were identified, and two 219.5 x 219.5 μm fields per slice were selected in those areas for quantification. All CD68+ and IBA1+ cells were counted in those regions. For analysis of immature neurons, four consecutive sections ranging approximately from bregma -1.8 to -2.4 were selected for the dentate gyrus, and four 219.5 x 219.5 μm fields per slice were selected in those areas for quantification. All DCX+ cells within the inner layer of the dentate gyrus were counted in those regions. For analysis of oligodendrocytes (ASPA+ or CC1+) and oligodendrocyte precursor cells (PDGFRα+) four to five consecutive sections ranging from bregma +1.2 to +0.8 were stained. The cingulum of the corpus callosum in each hemisphere was imaged with one 319.5 x 319.5 μm field and cells were quantified using the “Spots” function in Imaris image analysis software (Oxford Instruments). Each image was manually inspected to ensure accurate oligodendrocyte counts. False positive and false negative counts were manually removed or added, respectively. For NeuN cortical imaging, four consecutive sections ranging approximately from bregma +1.2 to +0.8 were selected for deep cortex directly above the cingulum region and two 419.5 x 419.5 μm fields per slice were selected in that area for quantification. NeuN+ cells were also quantified using the “Spots” function in Imaris image analysis software. False positive and false negative counts were manually removed or added, respectively.
Electron microscopy
7-days or 7-weeks after infection (as described above), or 4 weeks or 6 months following methotrexate administration as in Gibson et al. (2019), mice were sacrificed by transcardial perfusion with Karnovsky’s fixative: 2% glutaraldehyde (Electron Microscopy Sciences (EMS 16000) and 4% paraformaldehyde (EMS 15700) in 0.1 M sodium cacodylate (EMS 12300), pH 7. Samples were prepared as detailed previously in Gibson et al. (2019). Axons in the cingulum of the corpus callosum were analyzed for g-ratios calculated by dividing the shortest axonal diameter by the total diameter (diameter of axon/diameter of axon plus myelin sheath), as well as for myelinated axon density (total number of myelinated axons in each frame) by experimenters who were blinded to experimental conditions and genotypes. A minimum of 300 axons were scored per group.
Mouse CSF and serum cytokine analysis
The Luminex multiplex assay was performed by the Human Immune Monitoring Center at Stanford University. Mouse 48-plex Procarta kits were purchased from Thermo Fisher, Santa Clara, California, USA, and used according to the manufacturer’s recommendations with modifications as described below. Briefly: beads were added to a 96-well plate and washed in a Biotek Elx405 washer. Samples were added to the plate containing the mixed antibody-linked beads and incubated overnight at 4°C with shaking. Cold (4°C) and room temperature incubation steps were performed on an orbital shaker at 500-600 RPM. Following the overnight incubation plates were washed in a Biotek Elx405 washer and then biotinylated detection antibody added for 60 minutes at room temperature with shaking. Plate was washed as described above and streptavidin-PE was added. After incubation for 30 minutes at room temperature, washing was performed as above and reading buffer was added to the wells. CSF samples were measured as singlets, while serum samples were measured in duplicate. Plates were read on a FM3D FlexMap instrument with a lower bound of 50 beads per sample per cytokine. Custom Assay Chex control beads were purchased from Radix Biosolutions, Georgetown, Texas, and were added to all wells.
Isolation of patient plasma and cytokine/chemokine measurements
Patient plasma was collected with informed consent and IRB approval as previously described (Lucas et al., 2020). Patient whole blood was collected in sodium heparin-coated vacutainers. Blood samples were gently agitating at room temperature until delivered to Yale from Mount Sinai Client. All blood samples were processed on the day of collection. Plasma samples were collected following centrifugation of whole blood at 400g without brake at room temperature for 10 minutes. The undiluted plasma was then transferred to 15 ml polypropylene conical tubes, aliquoted, and stored at −80 °C. Plasma samples were shipped to Eve Technologies (Calgary, Alberta, Canada) on dry ice, and levels of cytokines and chemokines were measured using the Human Cytokine Array/Chemokine Array 71-403 Plex Panel (HD71). All samples were measured at the time of the first thaw.
Human immunohistochemistry
Human brain tissue samples were prepared and analyzed as previously described (Lee et al., 2021). Human brains obtained at the time of autopsy in New York and in Iowa during the spring of 2020 were fixed in formalin. Five-micron thickness sections were obtained from formalin-fixed paraffin-embedded (FFPE) blocks. Slides were deparaffinized and then rehydrated using xylene and graded ethanol. Preparation of slides for immunohistochemistry involved antigen retrieval by Heat-Induced Antigen Retrieval (HIAR) methods in 10 mM citrate buffer at pH 6.0 or 10 mM Tris/EDTA buffer at pH 9.0, or Enzyme-Induced Antigen Retrieval (EIAR) methods in proteinase K. Peroxidase blocking was done by incubating tissues in 0.3% to 3.0% hydrogen peroxide for 10 minutes at room temperature. Protein blocking was performed using protein block solution (DAKO) for 30 minutes at room temperature. Sections were exposed to primary antibodies and incubated overnight at room temperature. Antibodies included IBA1 (Wako 019-19741) and CD68 (Thermo Fisher Scientific 14-0688-82). Following 1X tris-buffered saline (TBS) wash containing 0.05% TritonX-100, sections were incubated with PowerVision polymeric horseradish peroxidase (poly-HRP) anti-mouse IgG (Leica Biosystems) or PowerVision polyHRP anti-rabbit IgG for 2 hours at room temperature. Antibody binding was visualized with 3,3′- diaminobenzidine (DAB; Vector Laboratories). Sections were then counterstained with hematoxylin (DAKO). Slides were coverslipped using EcoMount mounting medium (Biocare Medical). A whole slide scanner (Aperio AT2, Leica Biosystems) was used for image acquisition.
Human lung histology was assessed on standard clinical H&E images by board-certified pathologists.
Mouse lung histology
Lung tissues were fixed with 4% PFA overnight followed by soaking in 70% ethanol. Fixed tissue was embedded in paraffin blocks and sectioned by the Yale Pathology Tissue Services. Hematoxylin and eosin (H&E) staining was performed by the Yale Pathology Tissue Services. A blinded pulmonary pathologist reviewed the slides and identified immune cell infiltration and other related pathologies. H&E images were captured using light microscopy (BX51; Olympus).
scRNA-seq (digestion, FACS, 10X, sequencing)
Perfusion buffer, dissection buffer and digestion buffer were freshly prepared as previously described (Marsh et al., 2022). Mice were anesthetized with isoflurane and perfused intracardially with 10ml cold perfusion buffer. Brains were quickly dissected and placed into an adult mouse brain slicer matrix within dissection buffer. A 3mm cerebrum slice was cut and contained cortex and corpus callosum followed by chopping in digestion buffer. Chopped tissue was incubated for 45 minutes at 37°C and triturated by pipetting every 15 minutes. After incubation, the cell suspension was then passed through a 70μm filter. Cells were washed with complete DMEM and spun to pellet cells. Cell pellets were resuspended in 40% Percoll in PBS and followed by spinning for 25 minutes at 650g. The supernatant containing myelin debris was discarded and the pellet was resuspended in FACS buffer (HBSS+1% FBS+1mM EDTA) with inhibitory cocktail (Actinomycin D+Triptolide+Anisomycin).
For FACS sorting, the cells were resuspended in FC blocker solution (2.4G2, BioxCell) and incubated on ice for 10 mins. Cocktails of desired staining antibodies including live-dead Aqua, PE-anti-CD11b (M1/70, BioLegend) and Alexa Fluor 647 anti-CD45 (30-F11, BioLegend) were added directly to cell suspension for 20 minutes incubation at 4°C. Prior to sort, cell were washed and resuspend with FACS buffer. CD45 and CD11b analysis was used as a quality control prior to sorting. Live cells (live-dead Aqua negative) were sorted on a BD FACSAria (BD Biosciences).
Single-cell suspensions were loaded onto the Chromium Controller (10x Genomics) for droplet formation. scRNA-seq libraries were prepared using the Chromium Single Cell 3′ Reagent Kit (10x Genomics) and Single Cell V(D)J Kit, according to manufacturer’s protocol. Samples were sequenced on the NovaSeq 6000 with 28-bp read 1, 8-bp i7 index and 98-bp read 2 for the gene-expression library. Sequencing results were demultiplexed into Fastq files using the Cell Ranger (10x Genomics, 6.1.2) mkfastq function. Samples were aligned to mm10-2.2.0 10x genome. All procedures except sequencing were processed at Yale BSL-3 facility.
Quantification and statistical analysis
For all quantifications, experimenters were blinded to sample identity and condition.
For fluorescent immunohistochemistry and confocal microscopy, images were taken at 20X and cells considered co-labeled when markers co-localized within the same plane. For cortical/white matter microglia staining three sections per mouse, eight frames in standardized locations per section (24 images per mouse) were counted, for hippocampal microglia staining, four sections per mouse, 2 frames in standardized location per section (8 images per mouse) were counted. For hippocampal neuroblast staining, four sections per mouse, 4 frames in standardized location per section (16 images per mouse) were counted. For oligodendrocyte staining four to five sections per mouse, 2 frames in standardized location per section (8-10 images per mouse) were mounted. For each mouse, 300-900 cells were counted for each immunohistochemical marker analysis. The density of cells was determined by dividing the total number of cells quantified for each lineage by the total volume of the imaged frames (mm3). In all experiments, “n” refers to the number of mice. In every experiment, “n” equals at least 3 mice per group; the exact “n” for each experiment is defined in the figure legends.
For electron microscopy (EM) quantification of g-ratio and myelinated axon density, 6000X images were obtained. G-ratios were measured by dividing the axonal diameter by the diameter of the entire fiber (diameter of axon/diameter of axon + myelin sheath) using ImageJ software. For myelinated axon density, the number of axons surrounded by a myelin sheath were counted and normalized to tissue area. 29 images were scored for each mouse. Approximately 400-700 axons were scored for each mouse.
For COVID patients with and without reported symptoms of “brain fog”, a multiple linear regression with least squares method was performed for a variety of clinical characteristics against CCL11 levels. Subsequent comparisons were made with t-tests and 2-way ANOVA.
All statistics were performed using Prism Software (Graphpad). For all analyses involving comparisons of one or two variables, 1-way or 2-way ANOVAs, respectively, were used with Tukey’s multiple comparisons post hoc corrections used to assess main group differences. For analyses involving only two groups, unpaired two-tailed Student’s t tests were used. For measurements of individual cytokine levels, unpaired two-tailed Student’s t tests were used without correction for multiple comparisons. Shapiro-Wilk test was used to determine normality for all datasets; all datasets were parametric. Outliers were identified using ROUT method with Q=1%. A level of p < 0.05 was used to designate significant differences.
For single-cell RNA sequencing, all analysis, quantification, and statistical testing was completed using R (v4.1.1). Seurat (v4.1.0) (Hao et al., 2021) was used for preprocessing, dimensionality reduction, clustering, and differential expression testing. Cell Ranger count matrices for each sample were merged, and low-quality cells with fewer than 200 detected genes or greater than 10% mitochondrial mapping UMIs and putative doublets with greater than 5000 detected genes were removed. UMI count data was then normalized, such that the counts per each gene per cell were divided by the total UMIs per cell, multiplied by a scale factor of 10,000, and natural-log transformed (referred to here as “log-normalized”). Variance stabilizing transformation was performed, and the top 2000 variable genes were identified, followed by PCA. UMAP was conducted using the first 20 principal components, and graph-based clustering was used to identify clusters with a resolution parameter of 0.8. All following differential expression testing was performed using Wilcoxon rank-sum tests with Bonferroni corrections for multiple comparisons. Adjusted p-values < 0.05 with accompanying average log2-fold changes greater than 0.25 in magnitude were considered statistically significant. Differential expression testing was first performed to identify cell types based on canonical marker gene expression, and clusters of the same cell type were merged (Figures S5D and S5E). To identify microglia and remove contaminating cell types, the macrophages/microglia cluster was then subsetted, the top 3000 variable genes in the remaining cells shared across samples were calculated, and the normalized expression data was scaled across cells for each gene, regressing technical variables of genes detected, UMI number, and percentage of mitochondrial mapping UMIs against each feature. Harmony (v0.1.0) (Korsunsky et al., 2019) was used to integrate the data across samples. UMAP was then performed using the top 20 Harmony-adjusted principal components, followed by clustering with a resolution parameter of 0.1. Differential expression testing was again performed, and cell type labels were assigned based on expression of canonical markers (Figures S5F and S5G). A small number of putative microglia or doublets clustered together with the erythrocyte and pericyte clusters, resulting in expression of some microglia markers like Tmem119 and P2ry12 in these clusters (Figure S5 G)–these few cells were excluded from downstream analysis as outliers. The microglia were then subsetted, new top 2000 variable features were calculated, expression data was scaled as above, and PCA was performed followed by integration with Harmony. UMAP was performed using the top 20 Harmony-adjusted principal components, and clustering was conducted with a resolution of 1.9, yielding three small, highly distinct clusters (labeled ATM-like, Chemokine, & Interferon-responsive) and several smaller cluster with few defining marker genes which mixed at different resolution parameters and were thus merged. The merged cluster was labeled Homeostatic due to its higher expression of typical microglia marker genes and lower expression of inflammation-related genes. Differential expression testing across clusters and between microglia within each cluster from SARS-CoV-2 infected versus control samples was performed as described above. Differential abundance testing was performed for each cluster across mild COVID and control samples using propeller (part of the speckle v0.0.3 package) (https://doi.org/10.1101/2021.11.28.470236), conducting moderated t-tests using logit-transformed cluster proportions with the Benjamini and Hochberg procedure for FDR control (significance threshold: FDR < 0.05). Enrichment of gene ontology (GO) biological process terms in cluster differentially expressed genes was tested using a hypergeometric test in the clusterProfiler package (v4.0.5) (Wu et al., 2021) with the Benjamini and Hochberg procedure. Terms enriched with an adjusted p-value < 0.01 were considered significant. The simplifyEnrichment package (v1.2.0) (https://doi.org/10.1101/2020.10.27.312116) was used to cluster enriched GO terms based on semantic similarity, and clusters were then manually summarized.
To compare SARS-CoV-2 related microglia transcriptional changes to previously defined microglia states, we obtained differential expression test results from previous studies using supplemental data or through request. DAM data was obtained from Keren-Shaul et al. (2017; Table S3). A Seurat object containing the 10X expression data from Safaiyan et al. (2021) was obtained by request to the authors, and differential expression testing between the WAM cluster and other microglia was performed as above. LDAM data was obtained from Marschallinger et al. (2020; Table S3), and lysolecithin (LPC) count matrices from Hammond et al. (2019) were obtained from the Gene Expression Omnibus (GEO) under accession number GSE121654. From the LPC data, cells expressing fewer than 650 unique genes were removed following the authors’ methods, and differential expression testing as above was performed between microglia from LPC-injected samples and saline-injected samples. Genes were considered overlapping between two gene signatures if the gene was upregulated or downregulated in both sets with an adjusted p-value less than 0.05 and an average log2-fold change greater in magnitude than 0.25. Intersections were plotted using the UpsetR package (v1.4.0) (Conway et al., 2017)
Acknowledgments
The authors would like to thank members of the Yale Center for Genomic Analysis (YCGA), especially Guilin Wang and Jack Wheeler for help with 10X sequencing, the Human Immune Monitoring Core at Stanford, Erin Gibson for input on the manuscript, and Yoon Seok Kim for help with figure formatting. The graphical abstract was created with BioRender.com. This work was supported by grants from the National Institute of Neurological Disorders and Stroke (R01NS092597 to M.M., NS003130 and NS003157 to A.N., K23NS109284 to M.M.H.), NIH Director’s pioneer award (DP1NS111132 to M.M., supplement DP1NS111132-S1 to A.F.-C.), NIH National Eye Institute (R01EY033353 to S.A.L.), National Institute of Allergy and Infectious Diseases (R01AI157488 to A.I.), FDA Office of Women’s Health Research Centers of Excellence in Regulatory Science and Innovation (CERSI) (to A.I.), Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (to M.M.), Cancer Research UK (to M.M.), Waxman Family Research Fund (to M.M. and A.C.G.), Fast Grant from Emergent Ventures at the Mercatus Center (to A.I.), RTW Foundation (D.P.) MD Anderson Neurodegenerative Disease Consortium (to S.A.L.), the Cure Alzheimer’s Fund (to S.A.L.), the Howard Hughes Medical Institute Collaborative COVID-19 Initiative (to A.I.), and the Howard Hughes Medical Institute (to A.I. and M.M.).
Author contributions
Conceptualization, methodology, validation, and visualization were performed by M.M., A.I., A.F.-C., P.L., A.C.G., and E.S.; formal analysis by A.F.-C., A.C.G., M.R.O., K.S., R.M., M.B.K., S.L., R.F., M.M.H., H.V., and M.M.; resources provided by M.M. and A.I.; investigation was performed by A.F.-C., P.L., A.C.G., E.S., M.-H.L., J.W., S.D., K.N., B.Y., K.R.T., L.A.-A., K.M., L.N., P.J.W., D.C.-E., A.M.S.T., J.R.G., J.K., T.T., J.S., B.I., C.L., T.M., M.A.P.-H., A.T., R.J.H., L.T., J.T.-M., E.B., A.K., D.M.C., M.Q., H.V., M.M.H., D.P.P., R.F., D.P., and A.N.; writing – original draft by M.M.; writing – review & editing by A.F.-C., P.L., A.C.G., E.S., S.L., A.I., and M.M.; data curation by E.S., M.R.O., S.L.; supervised by M.M., A.I., H.V., S.L., D.P., and A.N.; funding acquisition by M.M., A.I., and D.P.
Declaration of interests
A.I. served as a consultant for Spring Discovery, Boehringer Ingelheim (Ingelheim, Germany), and Adaptive Biotechnologies. M.M. serves in the scientific advisory board of Cygnal Therapeutics. S.A.L. sits on the scientific advisory board and has a financial interest in AstronauTx Ltd. The opinions and assertions expressed herein are those of the authors and do not reflect the official policy or position of the Uniformed Services University of the Health Sciences or the Department of Defense.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. We actively worked to promote gender balance in our reference list.
Published: June 13, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.06.008.
Supplemental information
Values represent mean median fluorescence intensity (MFI) per group ± SEM from Luminex mouse cytokine assay. p values represent comparisons between control and mild COVID at each time point for each individual cytokine and were measured from unpaired, two-tailed t test.
For all CSF cytokine assays, n = 7 mice/group. For serum cytokine assays, n = 7 mice/group with the exception of
GMCSF, 7 DPI n = 6 control, n = 7 mild COVID
CXCL2, 7 DPI n = 6 control, n = 7 mild COVID
CXCL5, 7 DPI n = 7 control, n = 6 mild COVID
CCL5, 7 WPI n = 7 control, n = 6 mild COVID
IL18, 7 WPI n = 5 control, n = 7 mild COVID
CCXL10, 7 WPI n = 6 control, n = 6 mild COVID
TNFα, 7 WPI n = 5 control, n = 6 mild COVID
VEGF, 7 WPI n = 7 control, n = 6 mild COVID.
Abbreviations: PMI, postmortem interval; CMV, cytomegalovirus; COPD, chronic obstructive pulmonary disease; DAD, diffuse alveolar damage; HTN, hypertension; CAD, coronary artery disease; MI, myocardial infarction; HLD, hyperlipidemia; CABG, coronary artery bypass grafting; CHF, congestive heart failure; DM2, diabetes mellitus type 2; PTSD, post-traumatic stress disorder; BPH, benign prostatic hypertrophy; CKD, chronic kidney disease.
Values represent mean median fluorescence intensity (MFI) per group ± SEM from Luminex mouse cytokine assay. p values represent comparisons between control and influenza at each timepoint for each individual cytokine and were measured from unpaired, two-tailed t test. For all CSF cytokine assays, n = 5 mice/group. For serum cytokine assays, n = 5 mice/group with the exception of
IL18, 7 DPI n = 4 control, n = 5 influenza
IL6, 7 WPI n = 5 control, n = 4 influenza
IFNγ, 7 WPI n = 5 control, n = 4 influenza
CXCL5, 7 WPI n = 4 control, n = 5 influenza.
References
- Acharya M.M., Green K.N., Allen B.D., Najafi A.R., Syage A., Minasyan H., Le M.T., Kawashita T., Giedzinski E., Parihar V.K., et al. Elimination of microglia improves cognitive function following cranial irradiation. Sci. Rep. 2016;6:31545. doi: 10.1038/srep31545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Errico D., Aguilar H., Kitzig F., Brckalo T., Sayós J., López-Botet M. IREM-1 is a novel inhibitory receptor expressed by myeloid cells. Eur. J. Immunol. 2004;34:3690–3701. doi: 10.1002/eji.200425433. [DOI] [PubMed] [Google Scholar]
- Antonelli M., Penfold R.S., Merino J., Sudre C.H., Molteni E., Berry S., Canas L.S., Graham M.S., Klaser K., Modat M., et al. Risk factors and disease profile of post-vaccination SARS-CoV-2 infection in UK users of the COVID Symptom Study app: a prospective, community-based, nested, case-control study. Lancet Infect. Dis. 2022;22:43–55. doi: 10.1016/S1473-3099(21)00460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub R., Ruddy R.M., Cox E., Oyefiade A., Derkach D., Laughlin S., Ades-Aron B., Shirzadi Z., Fieremans E., MacIntosh B.J., et al. Assessment of cognitive and neural recovery in survivors of pediatric brain tumors in a pilot clinical trial using metformin. Nat. Med. 2020;26:1285–1294. doi: 10.1038/s41591-020-0985-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J.H., Lin J.J., Doernberg M., Stone K., Navis A., Festa J.R., Wisnivesky J.P. Assessment of cognitive function in patients After COVID-19 infection. JAMA Netw. Open. 2021;4:e2130645. doi: 10.1001/jamanetworkopen.2021.30645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.L., Bennett F.C., Liddelow S.A., Ajami B., Zamanian J.L., Fernhoff N.B., Mulinyawe S.B., Bohlen C.J., Adil A., Tucker A., et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA. 2016;113:E1738–E1746. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Wang K., Yu J., Howard D., French L., Chen Z., Wen C., Xu Z. The spatial and cell-type distribution of SARS-CoV-2 receptor ACE2 in the human and mouse brains. Front. Neurol. 2021;11:573095. doi: 10.3389/fneur.2020.573095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland C.D., Choi M., Romberg C., Clemenson G.D., Jr., Fragniere A., Tyers P., Jessberger S., Saksida L.M., Barker R.A., Gage F.H., Bussey T.J. A functional role for adult hippocampal neurogenesis in spatial pattern separation. Science. 2009;325:210–213. doi: 10.1126/science.1173215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis H.E., Assaf G.S., McCorkell L., Wei H., Low R.J., Re'em Y., Redfield S., Austin J.P., Akrami A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalmedicine. 2021;38:101019. doi: 10.1016/j.eclinm.2021.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douaud G., Lee S., Alfaro-Almagro F., Arthofer C., Wang C., McCarthy P., Lange F., Andersson J.L.R., Griffanti L., Duff E., et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature. 2022;604:697–707. doi: 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fünfschilling U., Supplie L.M., Mahad D., Boretius S., Saab A.S., Edgar J., Brinkmann B.G., Kassmann C.M., Tzvetanova I.D., Möbius W., et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 2012;485:517–521. doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty A.C., Gibson E.M., Ghanem R.A., Greene J.J., Ocampo A., Goldstein A.K., Ni L., Yang T., Marton R.M., Paşca S.P., et al. Loss of adaptive myelination contributes to methotrexate chemotherapy-related cognitive impairment. Neuron. 2019;103:250–265.e8. doi: 10.1016/j.neuron.2019.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson E.M., Monje M. Microglia in cancer therapy-related cognitive impairment. Trends Neurosci. 2021;44:441–451. doi: 10.1016/j.tins.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson E.M., Nagaraja S., Ocampo A., Tam L.T., Wood L.S., Pallegar P.N., Greene J.J., Geraghty A.C., Goldstein A.K., Ni L., et al. Methotrexate chemotherapy induces persistent tri-glial dysregulation that underlies chemotherapy-related cognitive impairment. Cell. 2019;176:43–55.e13. doi: 10.1016/j.cell.2018.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson E.M., Purger D., Mount C.W., Goldstein A.K., Lin G.L., Wood L.S., Inema I., Miller S.E., Bieri G., Zuchero J.B., et al. Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science. 2014;344:1252304. doi: 10.1126/science.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Hübschmann D. simplifyEnrichment: an R/Bioconductor package for Clustering and Visualizing Functional Enrichment Results. bioRxiv. 2021 doi: 10.1101/2020.10.27.312116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan K.A., Weigel M.K., Prakash P., Wijewardhane P.R., Hasel P., Rufen-Blanchette U., Münch A.E., Blum J.A., Fine J., Neal M.C., et al. Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature. 2021;599:102–107. doi: 10.1038/s41586-021-03960-y. [DOI] [PubMed] [Google Scholar]
- Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J., et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasel P., Rose I.V.L., Sadick J.S., Kim R.D., Liddelow S.A. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 2021;24:1475–1487. doi: 10.1038/s41593-021-00905-6. [DOI] [PubMed] [Google Scholar]
- Hinkle J.J., Olschowka J.A., Love T.M., Williams J.P., O'Banion M.K. Cranial irradiation mediated spine loss is sex-specific and complement receptor-3 dependent in male mice. Sci. Rep. 2019;9:18899. doi: 10.1038/s41598-019-55366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S., Wilk E., Michaelsen-Preusse K., Gerhauser I., Baumgärtner W., Geffers R., Schughart K., Korte M. Long-term neuroinflammation induced by influenza A virus infection and the impact on hippocampal neuron morphology and function. J. Neurosci. 2018;38:3060–3080. doi: 10.1523/JNEUROSCI.1740-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa K., Isobe M., Matsukawa T., Ito S., Maehara A., Takahashi M., Yamanishi Y., Kaitani A., Oki T., Okumura K., et al. Sphingomyelin and ceramide are physiological ligands for human LMIR3/CD300f, inhibiting FcepsilonRI-mediated mast cell activation. J. Allergy Clin. Immunol. 2014;133:270. doi: 10.1016/j.jaci.2013.08.008. 3.e1. [DOI] [PubMed] [Google Scholar]
- Jurgens H.A., Amancherla K., Johnson R.W. Influenza infection induces neuroinflammation, alters hippocampal neuron morphology, and impairs cognition in adult mice. J. Neurosci. 2012;32:3958–3968. doi: 10.1523/JNEUROSCI.6389-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Keswani T., Roland J., Herbert F., Delcroix-Genete D., Bauderlique-Le Roy H., Gaayeb L., Cazenave P.A., Pied S. Expression of CD300lf by microglia contributes to resistance to cerebral malaria by impeding the neuroinflammation. Genes Immun. 2020;21:45–62. doi: 10.1038/s41435-019-0085-9. [DOI] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Perl D.P., Nair G., Li W., Maric D., Murray H., Dodd S.J., Koretsky A.P., Watts J.A., Cheung V., et al. Microvascular injury in the brains of patients with Covid-19. N. Engl. J. Med. 2021;384:481–483. doi: 10.1056/NEJMc2033369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.S., Peterson T.C., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Wong P., Klein J., Castro T.B.R., Silva J., Sundaram M., Ellingson M.K., Mao T., Oh J.E., Israelow B., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschallinger J., Iram T., Zardeneta M., Lee S.E., Lehallier B., Haney M.S., Pluvinage J.V., Mathur V., Hahn O., Morgens D.W., et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020;23:194–208. doi: 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh S.E., Walker A.J., Kamath T., Dissing-Olesen L., Hammond T.R., de Soysa T.Y., Young A.M.H., Murphy S., Abdulraouf A., Nadaf N., et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci. 2022;25:306–316. doi: 10.1038/s41593-022-01022-8. [DOI] [PubMed] [Google Scholar]
- Mazza M.G., Palladini M., De Lorenzo R., Magnaghi C., Poletti S., Furlan R., Ciceri F., COVID-19 BioB Outpatient Clinic Study group. Rovere-Querini P., Benedetti F. Persistent psychopathology and neurocognitive impairment in COVID-19 survivors: Effect of inflammatory biomarkers at three-month follow-up. Brain Behav. Immun. 2021;94:138–147. doi: 10.1016/j.bbi.2021.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron V.E., Boyd A., Zhao J.W., Yuen T.J., Ruckh J.M., Shadrach J.L., van Wijngaarden P., Wagers A.J., Williams A., Franklin R.J.M., ffrench-Constant C. M2 microglia and macrophages drive oligodendrocyte differentiation during CNS remyelination. Nat. Neurosci. 2013;16:1211–1218. doi: 10.1038/nn.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje M.L., Mizumatsu S., Fike J.R., Palmer T.D. Irradiation induces neural precursor-cell dysfunction. Nat. Med. 2002;8:955–962. doi: 10.1038/nm749nm749. [DOI] [PubMed] [Google Scholar]
- Monje M.L., Toda H., Palmer T.D. Inflammatory blockade restores adult hippocampal neurogenesis. Science. 2003;302:1760–1765. doi: 10.1126/science.1088417. [DOI] [PubMed] [Google Scholar]
- Monje M.L., Vogel H., Masek M., Ligon K.L., Fisher P.G., Palmer T.D. Impaired human hippocampal neurogenesis after treatment for central nervous system malignancies. Ann. Neurol. 2007;62:515–520. doi: 10.1002/ana.21214. [DOI] [PubMed] [Google Scholar]
- Nasserie T., Hittle M., Goodman S.N. Assessment of the frequency and variety of persistent symptoms Among patients With COVID-19: a systematic review. JAMA Netw. Open. 2021;4:e2111417. doi: 10.1001/jamanetworkopen.2021.11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath A. Long-haul COVID. Neurology. 2020;95:559–560. doi: 10.1212/WNL.0000000000010640. [DOI] [PubMed] [Google Scholar]
- Nath A., Smith B. Neurological issues during COVID-19: an overview. Neurosci. Lett. 2021;742:135533. doi: 10.1016/j.neulet.2020.135533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noori R., Park D., Griffiths J.D., Bells S., Frankland P.W., Mabbott D., Lefebvre J. Activity-dependent myelination: a glial mechanism of oscillatory self-organization in large-scale brain networks. Proc. Natl. Acad. Sci. USA. 2020;117:13227–13237. doi: 10.1073/pnas.1916646117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajevic S., Basser P.J., Fields R.D. Role of myelin plasticity in oscillations and synchrony of neuronal activity. Neuroscience. 2014;276:135–147. doi: 10.1016/j.neuroscience.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandharipande P.P., Girard T.D., Jackson J.C., Morandi A., Thompson J.L., Pun B.T., Brummel N.E., Hughes C.G., Vasilevskis E.E., Shintani A.K., et al. Long-term cognitive impairment after critical illness. N. Engl. J. Med. 2013;369:1306–1316. doi: 10.1056/NEJMoa1301372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B., Sim C.B., Porrello E., Hewitt A.W., Powell J., Oshlack A. propeller: testing for differences in cell type proportions in single cell data. bioRxiv. 2021 doi: 10.1101/2021.11.28.470236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi A.I., Baskett W.I., Huang W., Naqvi S.H., Shyu C.R. New-onset dementia among survivors of pneumonia associated With severe acute respiratory syndrome coronavirus 2 infection. Open Forum Infect. Dis. 2022;9:ofac115. doi: 10.1093/ofid/ofac115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remsik J., Wilcox J.A., Babady N.E., McMillen T.A., Vachha B.A., Halpern N.A., Dhawan V., Rosenblum M., Iacobuzio-Donahue C.A., Avila E.K., et al. Inflammatory leptomeningeal cytokines mediate COVID-19 neurologic symptoms in cancer patients. Cancer Cell. 2021;39:276–283.e3. doi: 10.1016/j.ccell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadick J.S., O’Dea M.R., Hasel P., Dykstra T., Faustin A., Liddelow S.A. Astrocytes and oligodendrocytes undergo subtype-specific transcriptional changes in Alzheimer’s disease. Neuron. 2022;110:1788–1805.e10. doi: 10.1016/j.neuron.2022.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaiyan S., Besson-Girard S., Kaya T., Cantuti-Castelvetri L., Liu L., Ji H., Schifferer M., Gouna G., Usifo F., Kannaiyan N., et al. White matter aging drives microglial diversity. Neuron. 2021;109:1100–1117.e10. doi: 10.1016/j.neuron.2021.01.027. [DOI] [PubMed] [Google Scholar]
- Seeßle J., Waterboer T., Hippchen T., Simon J., Kirchner M., Lim A., Müller B., Merle U. Persistent symptoms in adult patients 1 year After coronavirus disease 2019 (COVID-19): a prospective cohort study. Clin. Infect. Dis. 2022;74:1191–1198. doi: 10.1093/cid/ciab611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.S., Koles Z.J. Myelinated nerve fibers: computed effect of myelin thickness on conduction velocity. Am. J. Physiol. 1970;219:1256–1258. doi: 10.1152/ajplegacy.1970.219.5.1256. [DOI] [PubMed] [Google Scholar]
- Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., Weizman O.E., Liu F., Dai Y., et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021:218. doi: 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steadman P.E., Xia F., Ahmed M., Mocle A.J., Penning A.R.A., Geraghty A.C., Steenland H.W., Monje M., Josselyn S.A., Frankland P.W. Disruption of oligodendrogenesis impairs memory consolidation in adult mice. Neuron. 2020;105:150–164.e6. doi: 10.1016/j.neuron.2019.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudre C.H., Murray B., Varsavsky T., Graham M.S., Penfold R.S., Bowyer R.C., Pujol J.C., Klaser K., Antonelli M., Canas L.S., et al. Attributes and predictors of long COVID. Nat. Med. 2021;27:626–631. doi: 10.1038/s41591-021-01292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabacof L., Tosto-Mancuso J., Wood J., Cortes M., Kontorovich A., McCarthy D., Rizk D., Rozanski G., Breyman E., Nasr L., et al. Post-acute COVID-19 syndrome negatively impacts physical function, cognitive function, health-related quality of life, and participation. Am. J. Phys. Med. Rehabil. 2022;101:48–52. doi: 10.1097/PHM.0000000000001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasek M.J., Garber C., Dorsey D., Durrant D.M., Bollman B., Soung A., Yu J., Perez-Torres C., Frouin A., Wilton D.K., et al. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 2016;534:538–543. doi: 10.1038/nature18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda S.A., Luo J., Mosher K.I., Zou B., Britschgi M., Bieri G., Stan T.M., Fainberg N., Ding Z., Eggel A., et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman S.G. Determinants of conduction velocity in myelinated nerve fibers. Muscle Nerve. 1980;3:141–150. doi: 10.1002/mus.880030207. [DOI] [PubMed] [Google Scholar]
- Wefel J.S., Kesler S.R., Noll K.R., Schagen S.B. Clinical characteristics, pathophysiology, and management of noncentral nervous system cancer-related cognitive impairment in adults. CA Cancer J. Clin. 2015;65:123–138. doi: 10.3322/caac.21258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A.C., Kern F., Losada P.M., Agam M.R., Maat C.A., Schmartz G.P., Fehlmann T., Stein J.A., Schaum N., Lee D.P., et al. Dysregulation of brain and choroid plexus cell types in severe COVID-19. Nature. 2021;595:565–571. doi: 10.1038/s41586-021-03710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.L., Zou Y., He W., Gage F.H., Evans R.M. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–1007. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G., et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Values represent mean median fluorescence intensity (MFI) per group ± SEM from Luminex mouse cytokine assay. p values represent comparisons between control and mild COVID at each time point for each individual cytokine and were measured from unpaired, two-tailed t test.
For all CSF cytokine assays, n = 7 mice/group. For serum cytokine assays, n = 7 mice/group with the exception of
GMCSF, 7 DPI n = 6 control, n = 7 mild COVID
CXCL2, 7 DPI n = 6 control, n = 7 mild COVID
CXCL5, 7 DPI n = 7 control, n = 6 mild COVID
CCL5, 7 WPI n = 7 control, n = 6 mild COVID
IL18, 7 WPI n = 5 control, n = 7 mild COVID
CCXL10, 7 WPI n = 6 control, n = 6 mild COVID
TNFα, 7 WPI n = 5 control, n = 6 mild COVID
VEGF, 7 WPI n = 7 control, n = 6 mild COVID.
Abbreviations: PMI, postmortem interval; CMV, cytomegalovirus; COPD, chronic obstructive pulmonary disease; DAD, diffuse alveolar damage; HTN, hypertension; CAD, coronary artery disease; MI, myocardial infarction; HLD, hyperlipidemia; CABG, coronary artery bypass grafting; CHF, congestive heart failure; DM2, diabetes mellitus type 2; PTSD, post-traumatic stress disorder; BPH, benign prostatic hypertrophy; CKD, chronic kidney disease.
Values represent mean median fluorescence intensity (MFI) per group ± SEM from Luminex mouse cytokine assay. p values represent comparisons between control and influenza at each timepoint for each individual cytokine and were measured from unpaired, two-tailed t test. For all CSF cytokine assays, n = 5 mice/group. For serum cytokine assays, n = 5 mice/group with the exception of
IL18, 7 DPI n = 4 control, n = 5 influenza
IL6, 7 WPI n = 5 control, n = 4 influenza
IFNγ, 7 WPI n = 5 control, n = 4 influenza
CXCL5, 7 WPI n = 4 control, n = 5 influenza.
Data Availability Statement
-
•
Single cell RNA sequencing data are publicly available on NCBI Sequencing Read Archive (SRA): BioProject ID PRJNA833593 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA833593/). Additional supplemental items, including the source data for all figures in this paper (Data S1) are available on Mendeley Data: https://data.mendeley.com/datasets/jw25y27vrm/3. Microscopy images reported in this paper will be shared by the lead contact upon request.
-
•
All original code is available on GitHub: https://github.com/liddelowlab/Mild_COVID_Microglia-Cell_2022.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.