Abstract
Cardiolipin (CL) has been shown to play a crucial role in regulating the function of proteins in the inner mitochondrial membrane. As the most abundant protein of the inner mitochondrial membrane, the ADP/ATP carrier (AAC) has long been the model of choice to study CL-protein interactions, and specifically bound CLs have been identified in a variety of crystal structures of AAC. However, how CL binding affects the structural dynamics of AAC in atomic detail remains largely elusive. Here we compared all-atom molecular dynamics simulations on bovine AAC1 in lipid bilayers with and without CLs. Our results show that on the current microsecond simulation time scale: 1) CL binding does not significantly affect overall stability of the carrier or structural symmetry at the matrix-gate level; 2) pocket volumes of the carrier and interactions involved in the matrix-gate network become more heterogeneous in parallel simulations with membranes containing CLs; 3) CL binding consistently strengthens backbone hydrogen bonds within helix H2 near the matrix side; and 4) CLs play a consistent stabilizing role on the domain 1-2 interface through binding with the R30:R71:R151 stacking structure and fixing the M2 loop in a defined conformation. CL is necessary for the formation of this stacking structure, and this structure in turn forms a very stable CL binding site. Such a delicate equilibrium suggests the strictly conserved R30:R71:R151stacking structure of AACs could function as a switch under regulation of CLs. Taken together, these results shed new light on the CL-mediated modulation of AAC function.
Supplementary key words: cardiolipin, phospholipid, mitochondrial ADP/ATP translocase, solute carrier family 25, SLC25, mitochondrial carriers, molecular dynamics simulation, arginine stacking, matrix-gate network, protein–lipid interactions
Abbreviations: AAC, ADP/ATP carrier; CATR, carboxyatractyloside; CL, cardiolipin; IMM, the inner mitochondrial membrane; MCF, the mitochondrial carrier family; MD, molecular dynamics; OXPHOS, Oxidative phosphorylation; POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; RMSF, root mean square fluctuations; TM, transmembrane
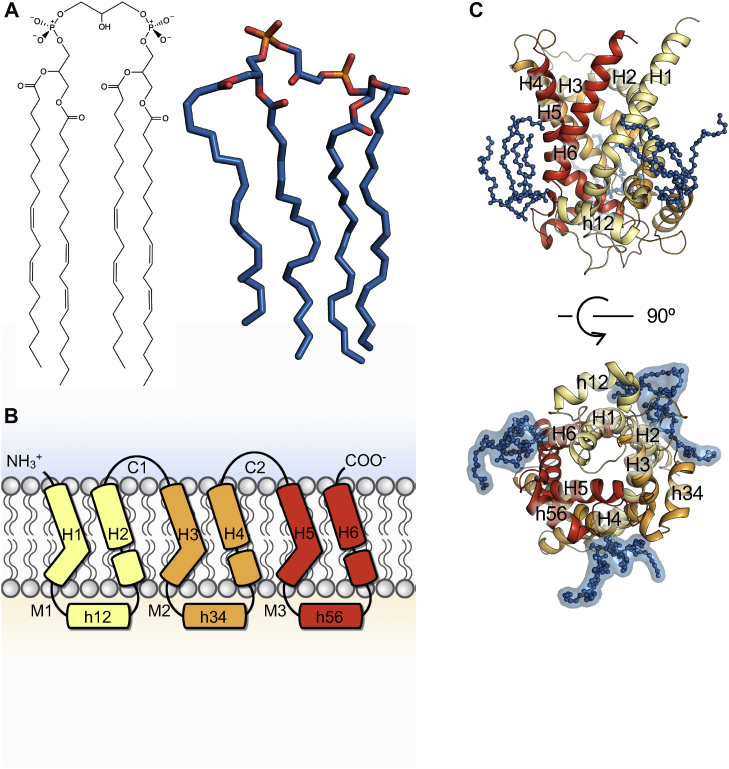
The inner mitochondrial membrane (IMM) exhibits a variety of unique properties: forming cristae leads to very large surface area; proteins are densely packed within it; and the membrane contains featured phospholipids, cardiolipins (CLs). IMM is crowded with membrane proteins involved in energy production and substrate transportation, including oxidative phosphorylation (OXPHOS) complexes and mitochondrial carriers. Some studies estimate the protein/lipid ratio reaches to as high as 3:1 (1, 2, 3), in contrast to roughly 1:1 in cell membranes (4). Because IMM is densely packed with OXPHOS complexes, there is little potential to improve OXPHOS capacity through increasing density of OXPHOS proteins. One important strategy is to increase the surface area of IMM through forming highly folded cristae structure. It is reported that CLs might play an important role on cristae biogenesis of mitochondria (5). CLs are unique lipids preserved during evolution from aerobic bacteria to mitochondria, and in mitochondria, CLs are mainly distributed within the inner leaflet of IMM where they are generated (6). CLs roughly account for 10%–20% of total phospholipids in IMM (7, 8). The structure of CL is featured with a small hydroxyl head group and four hydrophobic acyl tails (Fig. 1A), and it usually carries two negative net charges in physiological conditions. Its special conical shape makes CLs have potential to relax curvature strain in cristae. Moreover, CLs have been reported to stabilize and regulate the function of a bunch of mitochondrial proteins such as cytochrome c, ADP/ATP carrier (AAC), and ATPase (9, 10, 11, 12, 13, 14, 15).
Fig. 1.
Structures of cardiolipin and the ADP/ATP carrier. A: Chemical and tertiary structure of CL. B: The tripartite secondary structure of AAC. Domains are shown in distinct colors with domain 1 in yellow, domain 2 in orange, and domain 3 in red. C: Side view (top) and matrix view (bottom) of the tertiary structure of AAC with three specifically bound CLs. CLs are highlighted in blue stick-ball model. AAC, ADP/ATP carrier; CL, cardiolipin.
AAC is the most abundant protein in IMM and plays a central role in OXPHOS. It is responsible for importing ADP into the mitochondrial matrix and exporting ATP out of the mitochondria for living cells in a 1:1 exchange. The exchange of ADP and ATP across the IMM is completed through AAC alternating from the cytosol-open (c-) or ADP-waiting conformation to the matrix-open (m-) or ATP-waiting conformation via the transition state (16). Highly specific inhibitors including atractyloside and carboxyatractyloside (CATR) help lock AAC at c-state (17, 18, 19), while bongkrekic acid helps fix AAC at m-state (17, 19, 20). Benefitting from the high expression levels and the presence of state-specific inhibitors, AAC has long been paradigm of the big mitochondrial carrier family (MCF) (21, 22), and this family functions to mediate a variety of metabolites across IMM. Currently 53 MCF members have been identified in human genome, and they form the biggest solute carrier (SLC) subfamily which is also referred to as SLC25. Like other members of MCF, AAC is featured with three homologous repeats (Fig. 1B), each containing about 100 amino acids with highly conserved MCF motif Px[DE]xx[KR]xRxQ-(matrix loop and matrix helix)-[DE]Gxxxx[YWF][KR]G near the matrix side (23, 24, 25, 26, 27, 28, 29), with the cysteine-containing matrix short helices hosting the sequence motif -50-QYKGxxDCxRK-60 (sequence numbering referred to the bovine AAC1 sequence) (28).
Although CL is not absolutely required for AAC function (14), evidences have shown that CL can effectively stabilize AAC in purification and reconstitution (18, 20, 30), act as a transport activator (31, 32), affect conformational preference of AAC (15) and regulate the c/m conformational transition (30, 33). Tight binding between AAC and CL was first identified using 31P-NMR in 1985 (34) and was further confirmed by crystal structures of AAC in both c- and m-states (18, 20, 30, 35). In these structures, six transmembrane (TM) helices form a bundle surrounding a cone-shaped cavity opening toward the intermembrane space in c-state and toward the matrix in m-state. Each odd-numbered helix is connected to the following even-numbered helix at the matrix side through a flexible loop, a short amphipathic helix and a linker helix, which forms the basic structural scaffold for one repeat domain of AAC (Fig. 1B, C). In c-state structure, three repeated homologous domains of AAC show three-fold pseudosymmetry, and at each domain–domain interface, there is one specifically bound CL (Fig. 1C). Based on the static crystal structures, it was proposed that CLs bind with the exposed positively charged amide groups and helix dipoles at the N-terminal ends of even-numbered helices and short matrix helices (30). However, molecular dynamics (MD) simulations have shown that positively charged and polar residues also play an important role in binding with CLs (36, 37), and this result is in agreement with previous experiments (11, 38, 39). The difference from crystal structures and MD simulations highlights the importance of introducing structural dynamics in exploring lipid–protein interactions, which is also supported by studies from other groups (10, 40, 41, 42, 43, 44, 45). Due to varied distributions of the positively charged and polar residues in the three specific CL binding sites, our previous work reported that the three sites are highly asymmetric, and the asymmetric CL binding behavior at the three sites is quite consistent with the asymmetry observed in the matrix side of the m-state structure (20). However, how CL binding affects the structural dynamics of AAC in atomic detail remains largely elusive.
In the current work, three 3-μs all-atom MD simulations on c-state apo AAC with CLs (CL-1, 2, 3) are compared to the three 3-μs simulations without CLs (PC-1, 2, 3). We analyzed the impact of CLs on both macroscopic parameters such as pocket volume of AAC and microscopic parameters such as interactions among MCF motif residues. Significantly, we have revealed a strictly conserved stacking structure R30:R71:R151 at the domain 1-2 interface, and we speculate that this stacking structure functions as a structural switch under regulation of CLs.
Materials and methods
Initial preparation of the simulation systems
The starting protein coordinates were obtained from the CATR-inhibited crystal structure of AAC (PDB entry: 1okc) (18). The missing residues in the crystal structure of AAC were added using iTasser (46). The termini residues of AAC were manipulated into a charged state by protonating the N terminus and deprotonating the C terminus. A pure 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane and a mixed membrane composed of POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamine, and CL were built with the Membrane Builder modules of VMD (47) and CHARMM-GUI (48, 49, 50, 51), respectively. To build the AAC-POPC system, a hole was made within the POPC lipid bilayer, and the carrier alone was embedded. To build the AAC-CL system, the carrier bound with three CLs was inserted manually into the mixed bilayer with the same method. Then, the system was solvated with TIP3P (52) water molecules, and 100 mmol/L Na+ and Cl− were added to the system in addition to the neutralized chloride ions. The AAC-POPC system contains 70,769 atoms, including one AAC molecule, 219 POPC lipids, 12,226 water molecules, 23 Na+, and 42 Cl-. The AAC-CL system contains 82,971 molecules, including one AAC molecule, 110 POPCs, 88 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylethanolamines, 22 CLs, 15,740 water molecules, 53 Na+, and 28 Cl-. The detailed steps for systems setup were reported in our previous papers (29, 36).
MD simulations
MD simulations were performed using GROMACS 4.5.5 package (53) with the CMAP modified (54) CHARMM36 force field (55). The four-round energy minimizations were performed to remove unfavorable contacts and prevent the AAC-lipid system from collapsing at the subsequent MD simulations. Positional restraints were sequentially applied on all heavy atoms, main-chain atoms, and Cα atoms of AAC using a force constant of 1000 kcal/mol/Å2, and the last round was minimization without any positional restraint. Following energy minimizations, a series of equilibration steps were carried out for each system. In the first step, the system temperature was heated from 50 K to 310 K with positional restraints applied on heavy atoms of the protein, and the time step was set to 1 fs. In the subsequent three steps, the lipid bilayer and the solvent were fully equilibrated with positional restraints on all heavy atoms, main-chain atoms, and Cα atoms of AAC, respectively, and the time step was increased to 2 fs. After equilibration, three independent 3-μs production simulations were run in parallel for each system. The coordinates of each system were saved every 10 ps. The system temperature and pressure were constantly maintained (310 K, 1 bar) using the v-rescale method (56) and the Berendsen barostat algorithm (57), respectively. Other parameters and more detailed steps could be found in our previous papers (29, 36). The pdb files for the initial protein structure and structures after equilibrations are provided as a supplementary compressed file (pdb_structures.zip).
Data analyses and visualization
The programs in GROMACS 4.5.5 were utilized for most trajectory analyses. Specifically, root mean square deviation, root mean square fluctuations (RMSF), and spatial density function were calculated with the g_rms, g_rmsf, and g_spatial programs, respectively. Principal component analysis was calculated using g_covar and g_anaeig programs in Gromacs (53). Occupancies of salt bridges and H-bonds (hydrogen bonds) were all calculated based on the last 2 μs of each trajectory, with a distance cut-off of 0.33 nm. Time evolutions of the pocket volumes of AAC along the simulation trajectory were calculated with Epock (58).Trajectories were displayed with VMD (47), and structural graphics were prepared with PyMOL (59) and UCSF Chimera (60). The statistical graphics were plotted with R version 4.0.2 (61) and R studio IDE (62). The sequence logos were generated with TEXshade (63).
Results
Effect of CL on structural dynamics of c-state AAC in general
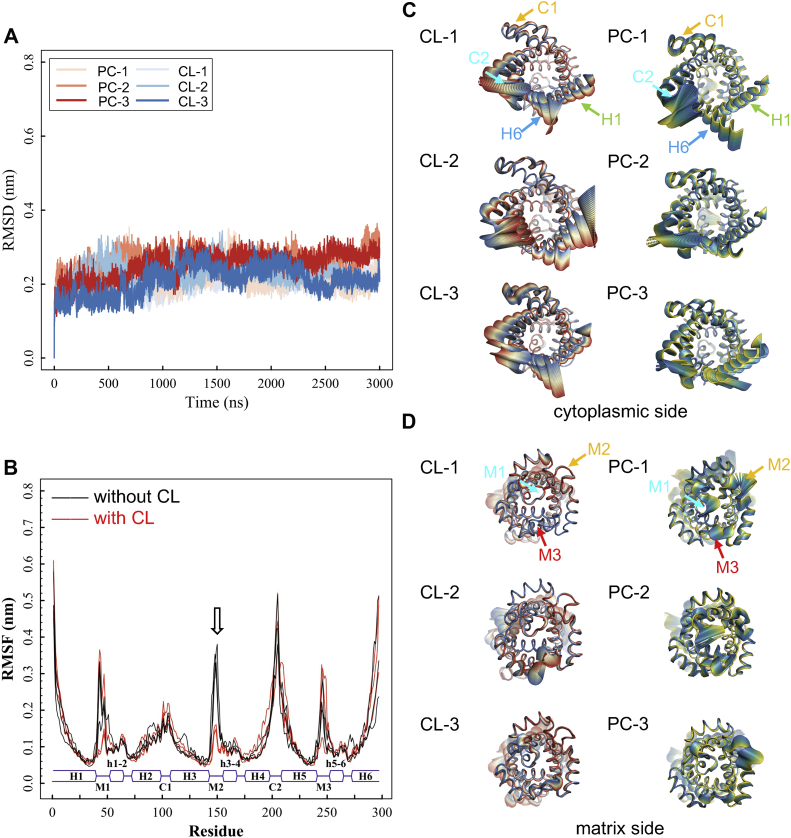
Structures of apo c-state AAC were well maintained in all six simulations with or without CLs in the lipid bilayer. Root mean square deviations of all Cα atoms of AAC during the simulations do not show obvious differences in the two conditions (Fig. 2A), indicating that the overall stability of AAC is not significantly affected by CL binding. Then we did RMSF calculations to further analyze the effect of CL binding on the structural flexibility of different regions in AAC. Of special interest, the M2 loop exhibits extreme stability in all three trajectories with CLs, while in all the other three trajectories without CLs the M2 loop becomes highly dynamic (Figs. 2B, D and S1). This suggests that CL dramatically stabilizes the M2 loop, but for the rest regions of the carrier, CL binding does not show obvious impact. In both conditions, the cytoplasmic sides of TM helices show much higher RMSF values than the matrix side, suggesting the cytoplasmic sides are more flexible (Fig. 2B). This is consistent with the fact that c-state structure closes at the matrix side and opens wide toward the cytoplasmic side. Moreover, flexibility at the cytoplasmic side is not symmetrically distributed among different domains. More specifically, the C2 loop and the cytoplasmic ends of H1 and H6 exhibit the highest RMSF values, while the values of the C1 loop is much lower than that of the C2 loop (Fig. 2B) which is consistent with previous simulations on shorter timescale (64). Principal component analysis also shows that major movements of AAC are observed in the region around the C2 loop and the cytoplasmic sides of H1 and H6, while the region around the C1 loop including cytoplasmic ends of H2 and H3 are relatively stable in both conditions with and without CLs (Fig. 2C, D). Our recent work had identified that the second basic patch (K91K95R187), N115 and the tyrosine ladder (Y186Y190Y194) including F191 form the specific ADP binding site (65), and ADP binding to this region was also reported by other groups (16, 66, 67). Thermodynamic stability of this region below the C1 loop could be crucial for efficient substrate recognition.
Fig. 2.
Effect of CL on the structural dynamics of AAC in general. A: Time evolutions of the root mean square deviation (RMSD) values of AAC Cα atoms during 3- s simulations in absence (red lines) and in presence (blue lines) of CLs. B: Time evolutions of root mean square fluctuations (RMSF) values of AAC backbone atoms in presence (red lines) and in absence of CLs (black lines). C–D: Principal component analysis on AAC trajectories. The motion of AAC C atoms is projected along the first eigenvector, with amplitude of the motion following the color scale from red to blue in simulations with CLs and from yellow to blue in simulations without CLs. AAC, ADP/ATP carrier; CL, cardiolipin.
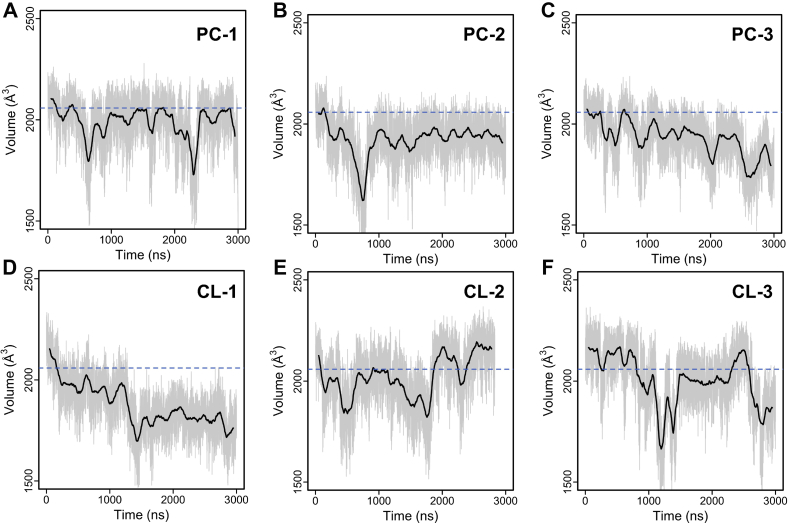
Impact of CLs on the pocket volume of c-state AAC
All released crystal structures of c-state AAC were cocrystallized with the inhibitor CATR (18, 30, 35). CATR is much bigger than the natural substrate ADP, and therefore it was proposed that the CATR-bound AAC could adopt an extreme configuration and open wider than the apo c-state which is the start of the translocation cycle (17). Our calculations show that in PC-1, PC-2, and CL-1, the pocket volume decreases first and then remains relatively stable till the end of the simulations (Fig. 3A, B and D). The significant decrease of the pocket volume at the beginning hundreds of nanoseconds indicates that the crystal conformation of AAC with CATR removed is actually unstable in the lipid bilayer. Similar results were also obtained in previous MD simulations on much shorter time scale (64). In PC-3, the pocket volume of AAC shows a decreasing trend throughout the simulation (Fig. 3C), and we reported similar results with an alternative method in our previous work (65). In CL-2 and CL-3, the pocket volumes fluctuate heavily during the 3-μs simulations. This result indicates that the cytoplasmic side of c-state AAC could be more flexible in presence of CL. However, we can not exclude the possibility that MD simulations with CL may need much longer time to get more converged results.
Fig. 3.
Time evolutions of pocket volume of c-state apo AAC in simulations without (A–C) and with (D–F) CLs. The smoothed average is shown in black lines, and the blue dashed lines highlight the volume calculated for the crystal structure of AAC. AAC, ADP/ATP carrier; CL, cardiolipin.
CL stabilizes the M2 loop through the conserved R30: R79: R151 stacking structure
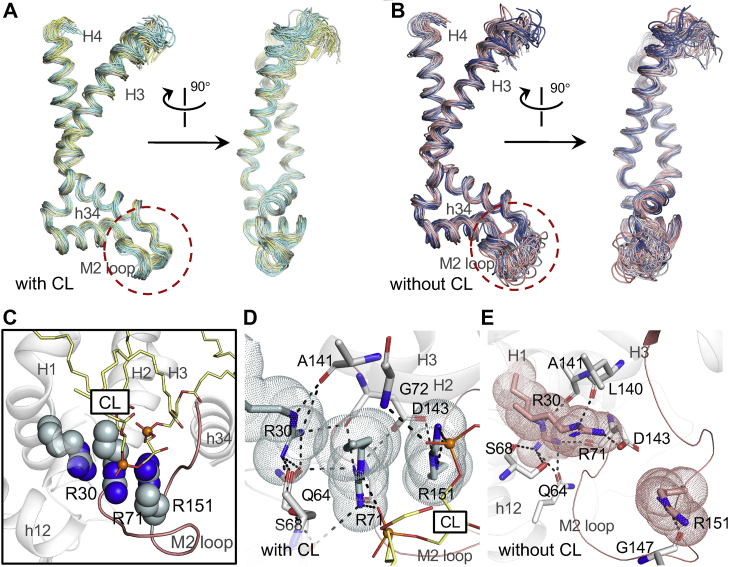
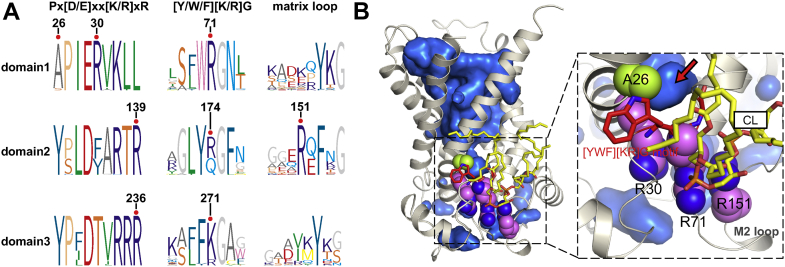
Although the three parallel trajectories in presence of CL show heterogeneity based on the above pocket volume analysis (Fig. 3B), they consistently show the stabilized M2 loop (Fig. 2A). The stabilizing effect of CL on the M2 loop is also illustrated through superposed snapshots taken from the trajectories with and without CLs (Fig. 4A, B). The M2 loop connects the matrix end of H3 with the N terminus of the matrix helix h34 within domain 2. In the crystal structure of bovine AAC1, guanidinium group of R151 in the M2 loop stacks with guanidinium group of R71 of [YWF][KR]G motif in domain 1 and therefore attaches the M2 loop to domain 1. We observe that R71 also stacks with R30, the capping arginine of Px[DE]xx[KR]xR motif in domain 1. Different from the capping arginine residues R139 and R236 in the other two domains, R30 moves four residues ahead from its canonical position in sequence and therefore moves about one helical turn upward in structure (28, 29). This movement makes the guanidinium group of R30 much closer to that of R71, which facilitates their stacking interaction. In this way, R151, R71, and R30 form a nice stacking structure (Fig. 4C), and similar stacking structure is commonly found in high-quality structures in PDB database (68).
Fig. 4.
The featured stacking structure on the domain 1-2 interface. A–B: Superposed conformations of 50 snapshots of domain 2 of AAC with and without CLs are extracted from MD simulation per 60 ns. The M2 loop is highlighted by the circle (red dashed line). C: The featured R30:R71:R151 stacking structure. R30, R71 and R151 are shown in light blue spheres, and the bound CL is shown in yellow sticks. The M2 loop is highlighted in salmon. D: Electrostatic interactions that stabilize the R30:R71:R151 stacking structure in the simulations with CL. The interactions are shown in black dashed lines. E: Dissociation of the stacking structure in the simulations without CL. The electrostatic interactions with the dissociated R30, R70, and R151 residues are shown in black dashed lines. AAC, ADP/ATP carrier; CL, cardiolipin; MD, molecular dynamics.
The electrostatic repulsion between these negatively charged guanidinium groups is balanced by interactions of these stacking residues with environment. In the crystal structure, R151 is also stabilized by side chain of D143 and backbone carbonyl of F153. R71 binds with backbone carbonyl oxygen atoms of D143 and G145. R30 forms hydrogen bonds with side chain of S68 and Q64 and backbone carbonyl oxygen atoms of A141 and D143. However, these interactions are not strong enough, and the R30:R71:R151 stacking structure is not maintained in all the simulations without CLs (Fig. 4E), which leads to thermodynamic flexibility of the M2 loop. In contrast, in presence of CLs, the stacking structure is well maintained throughout the simulations, and the M2 loop is extremely stable (Fig. 4A, C and D).
In our previous work, the stacked R71 and R151 bind to CL with high occupancies, and they are critical residues of the specific CL binding site I at the domain 1-2 interface (36). Therefore, these simulation results suggest that CL is necessary for the formation of the R30:R71:R151 stacking structure, and this stacking structure in turn forms a very stable CL binding site. Such delicate equilibrium prompts us to propose that the strictly conserved R30:R71:R151 stacking motif could serve as a structural switch under regulation of CLs. The simulation results also demonstrate that through stabilizing the R30:R71:R151 stacking structure, CLs play a crucial role in fixing the M2 loop in a pretty defined conformation.
CL binding stabilizes the domain 1-2 interface
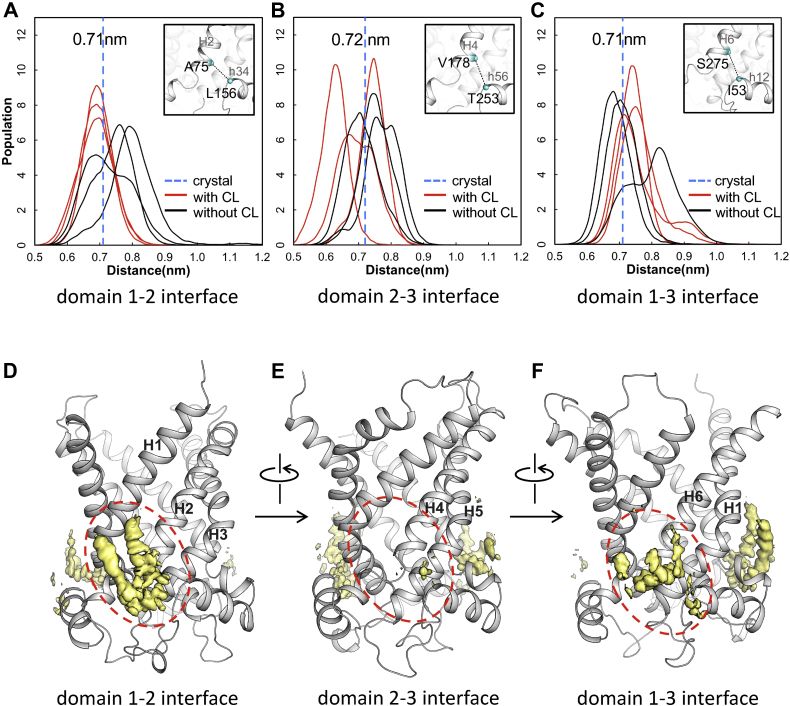
Specific CL binding sites are located at domain-domain interfaces, involving positively charged residues, N termini of even-numbered and short matrix helices (30, 36). To investigate the effect of CL binding on three domain-domain interfaces, we calculated distances between N terminus of even number helices and N terminus of matrix helices in the next domain with and without CLs. More specifically, minimum distances between C atoms of residues in triplet 75 (A75, V178, S275) and C atoms of corresponding residues in triplet 53 (L156, T253, I53) were calculated respectively for each simulation trajectory (Fig. 5A–C). Here, the triplet refers to three symmetric amino acids in the homologous tripartite structure, numbered according to the residue number in the first repeat of bovine AAC1 (28, 69). Frequency distributions of these distances suggest that CL binding consistently show a stabilization effect on the domain 1-2 interface (Fig. 5A). For the other two interfaces, big variations among different trajectories either with or without CLs indicate that CL binding does not have significant effect (Fig. 5B, C). The stabilization effect of CL binding on the domain 1-2 interface is also consistent with the spatial distribution function results in which higher density of CLs is observed at this interface compared to those at the other two interfaces (Fig. 5D–F). These results imply that CL is of special significance in stabilizing the domain 1-2 interface.
Fig. 5.
Impact of CL binding on domain-domain interfaces. A–C: Distribution of minimum distances between N terminus of even-numbered helices and N terminus of matrix helices in MD simulations with and without CLs. D–F: Isosurface of CLs density in the three CL binding sites of AAC. AAC, ADP/ATP carrier; CL, cardiolipin; MD, molecular dynamics.
Effect of CL binding on m-gate network
In the crystal structures of CATR-bound AAC, charged residues from the Px[DE]xx[KR] motifs form a symmetrical and cyclic interhelical salt bridge network on the matrix side (m-gate) (70, 71), and this network is proposed to be important to stabilize the c-state conformation (28). MD simulations from different groups consistently show that this network rearranges when the inhibitor CATR is removed (29, 64, 66, 67, 72). Based on simulations without CLs, we reported that after removing CATR, the charged residues from three Px[DE]xx[KR] motifs mainly form intrahelical salt bridges, and together with polar residues and positive residues K79, R235, and R279, form a broad asymmetric m-gate network (29). Here, we aim to evaluate how presence of CL may affect the salt bridges and H-bonds involved in the broad m-gate network.
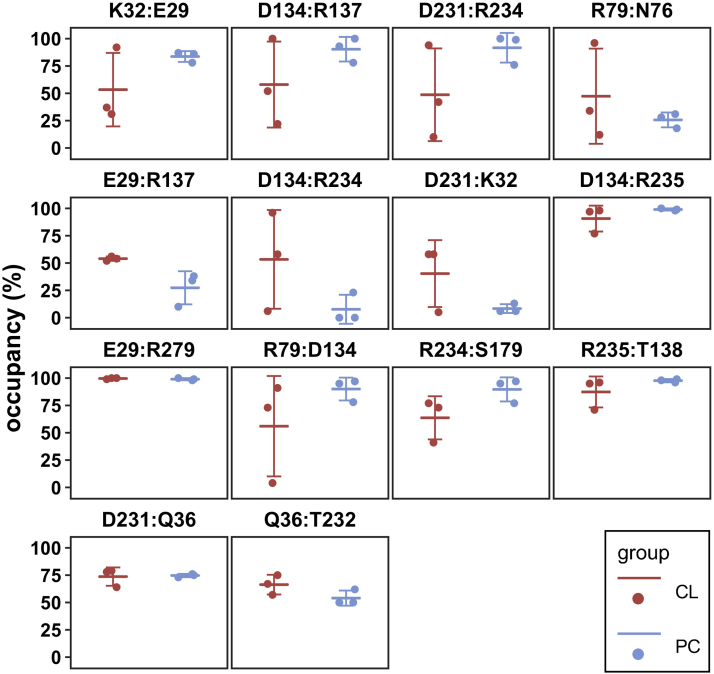
Calculations show that in contrast to the simulations without CL, three parallel simulations with CLs exhibit tremendous heterogeneity for a majority of interactions involved in the m-gate network (Figs. 6, S2 and S3, supplemental Table S1). For example, occupancies of the interhelical salt bridge D134:R234 in CL-1, CL-2, and CL-3 are 96%, 58%, and 6% respectively; for the intrahelical D231:R234, the occupancies are 10%, 42%, and 94% in CL-1, CL-2, and CL-3 respectively; and for D134:R79, the occupancies are 4%, 91%, and 73% in these three trajectories. The tremendous heterogeneity might reflect the increased flexibility of AAC in presence of CL. However, we can not exclude the possibility that the simulations with CLs may need longer time to get fully relaxed.
Fig. 6.
Effect of CL binding on the m-gate network. Scatter plots for the occupancies of the salt bridges and H-bonds involved in the broad m-gate network. Each data point of group CL (n=3) or group PC (n=3) is signified as a red or blue solid circle. Average occupancies are represented as longer horizontal lines, and the standard deviations are shown as shorter lines. CL, cardiolipin.
Worthy of mentioning, despite the heterogeneity for many m-gate interactions in simulations with CLs, the E29:R279 salt bridge is extremely stable with a high occupancy of almost 100% in each trajectory either with or without CL. This result is consistent with our previous finding that R279 has little impact on other interactions of the m-gate network (73). The stable E29:R279 salt bridge was also reported in previous simulations on c-state AAC (66, 72) and homologous SLC25A29 (D23:R257) (37), and the dissociation of E29:R279 is accompanied by the opening the matrix side during c- to m- state transition (74).
Effect of CL binding on other MCF structural elements
In addition to the m-gate, we also evaluate the effect of CL binding on interactions among other MCF structural elements including proline kink, capping arginine, [DE]G, and [YWF][KR]G motifs (supplemental Table S2 and supplemental Fig. S4). Results show that the electrostatic interactions between the [YWF][KR]G motif and Pro kink region are very strong and not affected by CL binding. The interactions between capping arginines and termini of even-numbered helices are different among different domains. R30 binds carbonyl oxygen of A141 with high occupancy in each trajectory, either with or without CLs. R139 binds carbonyl oxygen of M238 with lower occupancy in simulations without CLs than in simulations with CLs. One possible reason is that without CLs, the M2 loop is very flexible and therefore E152 may dynamically bind with R139 and deteriorate the R139:M238(O) interaction. Heterogeneity of the R236:Q36(O) interactions in parallel simulations either with or without CLs may just result from thermodynamic fluctuations of the carrier. The occupancies of the Q64:R30 interactions are quite different in parallel simulations with CLs, and based on our observations, it is mainly caused by different conformations of S68 which may bind with either G65 backbone, R30 or Q64. The M3 loop is intervened between D167 and R139, and based on our simulation trajectories, the interaction between R139 and the M3 loop is generally stronger than that between D167 and the M3 loop, and CL binding does not directly affect these interactions. The E264:R236 salt bridge is extremely stable and not affected by CL binding, and this salt bridge was proposed to be important in AAC transport (18). In the previous work, we have reported that the [YWF][KR]G motif lost its featured -turn structure in one of the three trajectories without presence of CLs (29). While this featured structure was perfectly maintained in all the three simulations with CLs, suggesting CLs help stabilize the -turn structure of the [YWF][KR]G motif (36). In fact, [YWF][KR]G motif forms the most critical part of the specific CL binding sites at domain-domain interfaces.
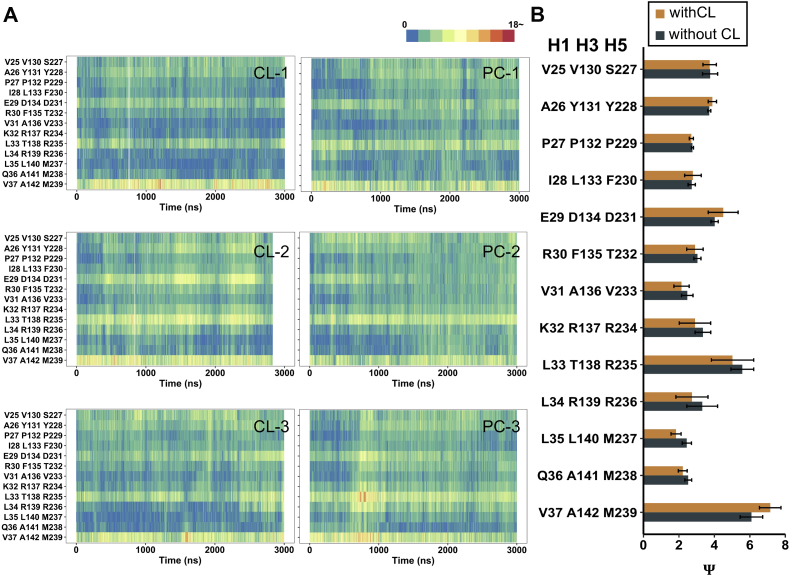
Effect of CL binding on structural symmetry of c-state AAC around m-gate level
Structural symmetry is of special significance in understanding transport mechanism of mitochondrial carriers (69). In a previous study, the metric ψ was introduced to evaluate the deviation degree of backbone structural symmetry of triplet from the ideal C3 symmetry (75). Specifically, ψ was calculated through averaging the deviations of the three internal angles of the triangle formed by three Cα atoms of a triplet from the internal angle of an equilateral triangle (60°). Higher ψ values suggest a higher deviation from C3 symmetry for the triplet residues. In that work, the authors reported that presence of CLs slightly improves the structural symmetry of the triplets in H2, H4, and H6 of m-state AAC around m-gate level (75). Following the same method, our recent work on wild-type and mutant c-state AAC showed that the R79A mutation drastically deteriorates structural symmetry of the triples in H1, H3, and H5 near the m-gate level (73). Here, we use this method to investigate effect of CL binding on the structural symmetry of c-state AAC around m-gate level. The heat maps of time evolutions of ψ values show no significant changes during each simulation, and no big differences were observed among different trajectories (Fig. 7A). In addition to the terminal triplet 37 (V37, A142, M239), the triplet 33 (L33, T138, R235) consistently show higher ψ values compared to other triplet planes in both conditions, highlighting the crucial role of the strictly conserved R235 in introducing structural asymmetry to c-state AAC near the m-gate level. To compare the differences in the two conditions in a quantitative way, we calculated the averaged ψ values over all three 3-μs trajectories with or without CLs. The results show that all triplets show no significant differences (P > 0.05) (Fig. 7B). These results suggest that presence of CL does not significantly affect structural symmetry of c-state AAC around the m-gate level on the current simulation time scale.
Fig. 7.
Structural symmetry of c-state AAC around the m-gate level in simulations with and without CLs. A: Time evolutions of ψ values in c-state AAC with (left panel) and without (right panel) CLs. Gradient colors from blue to red are used to indicate the deviation from three-fold symmetry of each triplet. B: The average ψ values calculated over trajectories with (orange) and without (black) CLs. The ψ values are presented as mean ± SEM. To evaluate the differences of ψ values in the two groups (with CLs and without CLs), a two independent-samples t test was performed, with a two-tailed P-value less than 0.05 considered to be significant. The P-values of all the triplets are bigger than 0.05 and therefore are not labeled in the figure. AAC, ADP/ATP carrier; CL, cardiolipin.
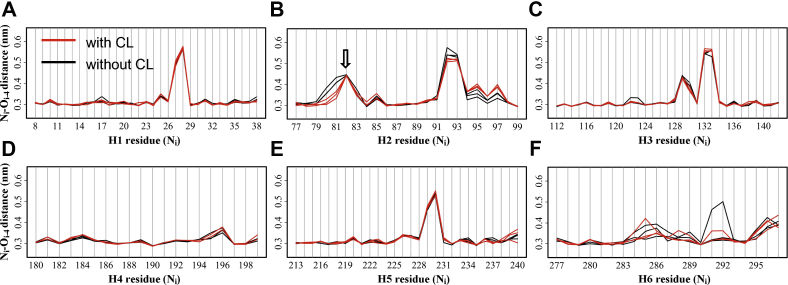
Effect of CL binding on backbone H-bonds within TM helices
Flexibility within the TM helices is of special significance for the normal function of membrane proteins. To evaluate the effect of CL binding on the flexibility of each TM helix, we calculated the average distance of each backbone H-bond (Ni-Oi-4) within the TM helices over the simulations with and without CLs. The increased Ni-Oi-4 distance is usually caused by structural kinks or increased flexibility within the TM helix. Results show that CL binding does not affect the average distances of backbone H-bonds within the odd-numbered helices H1, H3, and H5 (Fig. 8A, C and E), and within each of these helices, the conserved kink proline (P27, P132, P229) leads to a sharp peak for the Ni-Oi-4 distances in the region nearby (from triplet 26 to triplet 29). Compared to H1 and H5, H3 is different in that a secondary peak consistently appears around F129 in all the six trajectories (Fig. 8C), and this could be caused by the bent H2 that connects tightly with the H3 through the short C1 loop.
Fig. 8.
Effect of CL binding on backbone H-bonds within TM helices. (A–F) The average distance between the backbone N atom of residue i and the backbone O atom of neighboring fourth residue (i-4) within TM helices of AAC in simulations with (red lines) and without (black lines) CLs. AAC, ADP/ATP carrier; CL, cardiolipin; TM, transmembrane. The arrow in figure B highlights the only proline residue P82 within the transmembrane region besides the three conserved kink prolines.
The three even-numbered helices H2, H4, and H6 are quite different from each other in their backbone H-bonds and effect upon CL binding. H2 is special in its hydrophilic property and bent backbone (76). The bent backbone of H2 is demonstrated by two sharp peaks in the backbone Ni-Oi-4 distances in the region from R79 to Q84 and in the region from K91 to Y94 (Fig. 8B). Parallel simulations show consistent effect of CL binding on the backbone Ni-Oi-4 distances of H2 (Fig. 8B). CL binding consistently decreases the Ni-Oi-4 distances in the region from R79 to P82 (Fig. 8B), and the decreased amplitude is maintained relative stable throughout the simulations (supplemental Fig. S5). Although the average distances of F88(O)-D92(N) and A89(O)-93(N) are also decreased upon CL binding (Fig. 8B), time evolutions show that the decrease is discrete (supplemental Fig. S5), indicating that the decrease is caused by transient conformational changes within H2 backbone. Moreover, CL binding increases the average distances of K91(O)-D95(N) and K93(O)-97(N) (Fig. 8B), and the time evolutions also show that the increase is discrete (supplemental Fig. S5). These results indicate that CL binding stabilizes the matrix side of H2 but makes the cytosolic side of H2 more dynamic.
For H4, the low average Ni-Oi-4 distances throughout the TM helix indicates that H4 conserves its perfect α-helical structure during the simulations, and this structure is not affected by CL binding (Fig. 8D). For H6, whether in presence of CL or not, increased backbone Ni-Oi-4 distances are observed in many regions of H6, and parallel trajectories show tremendous heterogeneity (Fig. 8F). This indicates the dynamic property of H6 in both conditions. In fact, instability of H6 and its conformational changes during the simulations without CLs have been described in detail in our previous work (29).
Discussion
In the current work, we have investigated the effect of CL binding on the structure and dynamics of c-state AAC. Because of the dynamic property of AAC in apo state, parallel trajectories show considerable heterogeneity either with or without CLs, which makes it hard to get simple conclusions on the effect of CLs. However, we do observe that CLs play a consistent stabilizing role on the interface between domain 1 and domain 2. Our newly published results on wild-type and mutant AAC indicate the vulnerability of domain 1-2 interface near the matrix side without CLs (77). The current work shows that CLs could stabilize this interface through binding with the R30:R71:R151 stacking structure and therefore fix the M2 loop in a pretty defined conformation. Meanwhile, CLs stabilize the matrix part of the dynamic H2 and introduce more heterogeneity among parallel simulations for the m-gate network.
Previous studies have suggested that the M2 loop undergoes dramatic conformational changes during state transitions (77, 78, 79). Crystal structures of AAC in the two states also show significant difference in the conformations of the M2 loop. Our MD simulations on c-state apo bovine AAC1 in the current work have shown that CL plays a critical role in stabilizing the crystal conformation of the M2 loop through binding with the R30:R71:R151 stacking structure. Without CLs, the R30:R71:R151 stacking structure is disrupted, and the M2 loop loses its crystal conformation and becomes very flexible in the simulations. All the three Arg residues within the stacking structure are strictly conserved throughout different species of AAC (Fig. 9A), and they form a highly asymmetric structural element at the matrix side of the carrier. Among the capping arginine residues from the three homologous domains, R30 is the only one that moves one helical turn upward in structure (28, 29). It forms an extremely stable interdomain interaction with A141 but unstable intradomain interaction with Q64. Prior studies have noted the importance of these interactions. These interactions were reported to participate in protecting the dynamic interface between domains 1 and 2 (36) and closing the matrix side of AAC (28). R71 is the only strictly conserved middle residue of the three [YWF][KR]G motifs (Fig. 9A). The side chain of R71 is exposed to the lipid bilayer, while its backbone oxygen is oriented toward the interior of the AAC pocket. In all crystal structures of AAC, R71 binds with the guanidine group of the R137 through its backbone oxygen and is probably the only surface residue that has a chance to involve in the m-gate electrostatic network. In the three matrix loops, R151 is the only strictly conserved basic residue located just before the 50-QYKGxxDCxRK-60 motif (28) or [YF]xG motif (30) (Fig. 9A). At the matrix side, the three domains close tightly in the c-state AAC, while domain 1 separates from the other two domains in the m-state. This implies that the R30:R71:R151 stacking structure at domain 1-2 interface needs to dissociate and reform repetitively during state transitions.
Fig. 9.
The solvent exposed β-turn structure of the [YWF][KR]G motif in domain 1 is protected by the R30:R71:R151 stacking structure. A: Sequence logo presentations on the neighboring residues of A26, R30, R71, and R151 and their counterpart residues in homologous domains. Forty-three sequences of various AAC subtypes from totally 24 species were used for alignment. B: Structural illustration for the R30:R71:R151 stacking structure (magenta spheres) and β-turn structure of the [YWF][KR]G motif (shown in red sticks) that is exposed to the cavity solvent (highlighted with red arrow) due to the small residue A26 (green spheres). Solvent is shown in blue surface, and the stably bound CL is shown in yellow sticks. AAC, ADP/ATP carrier; CL, cardiolipin.
Previous analysis based on 70 thousand crystal structures suggests that stacking arginines are potentially controlled or switched by charged membrane lipids and may play an important role in protein function (68). CLs are featured negatively charged lipids within IMM, and in our previous work on c-state AAC, we highlighted the dynamic property of CLs and their potential involvement in adapting to the conformational changes during state transitions (36). Our results also show that in the presence of CLs, the R30:R71:R151 stacking structure is stably maintained throughout our simulations (Fig. 3D), and meanwhile, this stabilized stacking structure forms a good CLs binding site, and both R71 and R151 exhibit very high CLs binding occupancies in previous atomic and coarse grain MD simulations (36, 41, 42). Without presence of CLs, the stacking dissociates in all three trajectories (Fig. 3E). Therefore, based on the dynamic property of CLs binding and sensitivity of the R30:R71:R151 stacking structure to CLs, we infer that CLs play an important role in the dynamic dissociation and reformation of the stacking structure.
As described in previous work, the small alanine (A26) instead of a tyrosine appears before P27, and this makes the β-turn structure of the [YWF][KR]G motif in domain 1 faces directly to the cavity solvent (Fig. 9B) and therefore contributes to the vulnerability of the domain 1-2 interface. This conserved vulnerable interface is protected by the conserved R30:R71:R151 stacking structure that is under regulation of CLs (Fig. 9B), and this leads us to infer that CLs may play a regulation role on the function of the carrier through this stacking structure.
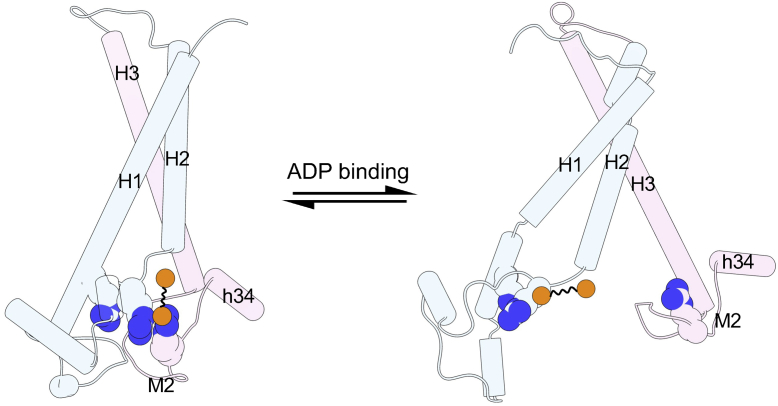
Here we propose a model to describe the relationship between CLs, R30:R71:R151 stacking structure and state transition (Fig. 10). In the ground c-state, the R30:R71:R151 stacking structure at the vulnerable domain 1-2 interface is stabilized by CLs binding. One typical binding mode is that one phosphate group of CL binds simultaneously with R71 and R151, with the other phosphate group binding preferentially with N terminus of H2, and the other typical binding mode is that one phosphate group binds with R71, and the other phosphate group binds with both R151 and N terminus of H2 (36). Upon ADP binding, the original CL binding mode with the R30:R71:R151 stacking structure can easily be disturbed. This will lead to the dissociation of the stacking structure, and further facilitates opening of the vulnerable domain 1-2 interface. Therefore, this model explains, at least partially, the activation role of CLs on the transport of AAC (11).
Fig. 10.
A model for how CLs regulate the c- to m- state transition of AAC through the featured arginine stacking structure. The linked orange circles represent the two phosphate groups of CLs. Residues of the R30:R71:R151 stacking structure are shown as spheres. Domain 1 and 2 are colored in light blue and pink, respectively. The N atoms in the side chain of stacking arginines are colored in dark blue. AAC, ADP/ATP carrier; CL, cardiolipin.
CL is also known for its stabilization role on the carrier purification with detergents (11). However, it was reported that the stabilization effect from the inhibitor CATR is much more significant, and little is known about the stabilization effect of CL on apo AAC (11). In physiological conditions, apo AAC predominantly adopts the ground c-state (17), accounting for 98% of total population at equilibrium based on NMR experiments (80). Well-tempered metadynamics simulations starting from c-state AAC embedded in the pure POPC lipid bilayer also showed that c-state is energetically more favorable than m-state (16). In fact, pure POPC lipid bilayer was widely used in previous all-atom MD simulations on AAC (16, 29, 64, 66, 67, 72, 74, 81, 82), and the c-state structure was well maintained in all these simulations. Nevertheless, a recent work demonstrated that in absence of CL, yeast AAC2 is mostly in the m-state (15), implying that only in presence of CLs c-state is more stable than m-state. In a previous model, the excess positive charges near the matrix side of AAC facilitate the opening of the m-gate when the bound CLs are sequestered by Ca2+ (17), which also implies that c-state becomes less stable without presence of CLs. Therefore, relationship between CL and AAC conformations seems rather complicated and merits further investigations. The current work is limited in that we only evaluate the effect of CL binding on structural dynamics of c-state AAC, and we believe that further work on m-state and state transition pathways combined with free energy calculations may help to better understand the regulation mechanism of CL on AAC conformations and functions.
Data availability
Data are available upon reasonable request to the corresponding author.
Supplemental data
This article contains supplemental data (29).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
MD simulations were carried out at National Supercomputer Center in Tianjin, and the calculations were performed on TianHe-1(A).
Author contribution
Q. Y. and S. Y. investigation; B. M., S. Y., and Q. Y. data curation; Q. Y. and S. Y. writing-original draft; X. C. conceptualization; X. C. supervision; X. C. funding acquisition; X. C. writing-review & editing.
Funding and additional information
The research was supported by the National Natural Science Foundation of China (Grant No. 32171241 to X. C.) and the Natural Science Foundation of Zhejiang Province (Grant No. LY18C050002 to X. C.).
Supplemental data
Reference
- 1.Hallermayer G., Neupert W. Lipid composition of mitochondrial outer and inner membranes of Neurospora crassa. Hoppe. Seylers. Z. Physiol. Chem. 1974;355:279–288. doi: 10.1515/bchm2.1974.355.1.279. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls D.G., Ferguson S.J. Gulf Professional Publishing; Houston: 2002. Bioenergetics 3. [Google Scholar]
- 3.Schenkel L.C., Bakovic M. Formation and regulation of mitochondrial membranes. Int. J. Cell Biol. 2014;2014:709828. doi: 10.1155/2014/709828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper G.M. 2nd Ed. Sinauer Associates; Sunderland: 2000. The Cell: A Molecular Approach. [Google Scholar]
- 5.Ikon N., Ryan R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017;1859:1156–1163. doi: 10.1016/j.bbamem.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baile M.G., Lu Y.-W., Claypool S.M. The topology and regulation of cardiolipin biosynthesis and remodeling in yeast. Chem. Phys. Lipids. 2014;179:25–31. doi: 10.1016/j.chemphyslip.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath S.E., Daum G. Lipids of mitochondria. Prog. Lipid. Res. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Dowhan W., Bogdanov M., Mileykovskaya E. In: Biochemistry of Lipids, Lipoproteins and Membranes. Sixth Edition. Ridgway N.D., McLeod R.S., editors. Elsevier; Boston: 2016. Functional Roles of Lipids in Membranes; pp. 1–40. Chapter 1. [Google Scholar]
- 9.Ren M., Phoon C.K.L., Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog. Lipid. Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Duncan A.L., Robinson A.J., Walker J.E. Cardiolipin binds selectively but transiently to conserved lysine residues in the rotor of metazoan ATP synthases. Proc. Natl. Acad. Sci. U. S. A. 2016;113:8687–8692. doi: 10.1073/pnas.1608396113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klingenberg M. Cardiolipin and mitochondrial carriers. Biochim. Biophys. Acta Biomembr. 2009;1788:2048–2058. doi: 10.1016/j.bbamem.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Schlame M., Rua D., Greenberg M.L. The biosynthesis and functional role of cardiolipin. Prog. Lipid. Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 13.Pfeiffer K., Gohil V., Stuart R.A., Hunte C., Brandt U., Greenberg M.L., et al. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 14.Claypool S.M. Cardiolipin, a critical determinant of mitochondrial carrier protein assembly and function. Biochim. Biophys. Acta Biomembr. 2009;1788:2059–2068. doi: 10.1016/j.bbamem.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senoo N., Kandasamy S., Ogunbona O.B., Baile M.G., Lu Y., Claypool S.M. Cardiolipin, conformation, and respiratory complex-dependent oligomerization of the major mitochondrial ADP/ATP carrier in yeast. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pietropaolo A., Pierri C.L., Palmieri F., Klingenberg M. The switching mechanism of the mitochondrial ADP/ATP carrier explored by free-energy landscapes. Biochim. Biophys. Acta Bioenerg. 2016;1857:772–781. doi: 10.1016/j.bbabio.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta Biomembr. 2008;1778:1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trézéguet V., Lauquin G.J., Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 19.Todisco S., Di Noia M.A., Onofrio A., Parisi G., Punzi G., Redavid G., et al. Identification of new highly selective inhibitors of the human ADP/ATP carriers by molecular docking and in vitro transport assays. Biochem. Pharmacol. 2016;100:112–132. doi: 10.1016/j.bcp.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Ruprecht J.J., King M.S., Zögg T., Aleksandrova A.A., Pardon E., Crichton P.G., et al. The Molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell. 2019;176:435–447.e15. doi: 10.1016/j.cell.2018.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boxer D.H., Feckl J., Klingenberg M. Identity between the major protein located at the outer face of the inner mitochondrial membrane and carboxyatractylate binding protein. FEBS Lett. 1977;73:43–46. [PubMed] [Google Scholar]
- 22.Palmieri F., Pierri C.L., De Grassi A., Nunes-Nesi A., Fernie A.R. Evolution, structure and function of mitochondrial carriers: a review with new insights. Plant J. 2011;66:161–181. doi: 10.1111/j.1365-313X.2011.04516.x. [DOI] [PubMed] [Google Scholar]
- 23.Saraste M., Walker J.E. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 1982;144:250–254. doi: 10.1016/0014-5793(82)80648-0. [DOI] [PubMed] [Google Scholar]
- 24.Nelson D.R., Felix C.M., Swanson J.M. Highly conserved charge-pair networks in the mitochondrial carrier family. J. Mol. Biol. 1998;277:285–308. doi: 10.1006/jmbi.1997.1594. [DOI] [PubMed] [Google Scholar]
- 25.Aquila H., Misra D., Eulitz M., Klingenberg M. Complete amino acid sequence of the ADP/ATP carrier from beef heart mitochondria. Hoppe. Seylers. Z. Physiol. Chem. 1982;363:345–349. [PubMed] [Google Scholar]
- 26.Walker J.E., Runswick M.J. The mitochondrial transport protein superfamily. J. Bioenerg. Biomembr. 1993;25:435–446. doi: 10.1007/BF01108401. [DOI] [PubMed] [Google Scholar]
- 27.Palmieri F. Mitochondrial carrier proteins. FEBS Lett. 1994;346:48–54. doi: 10.1016/0014-5793(94)00329-7. [DOI] [PubMed] [Google Scholar]
- 28.Pierri C.L., Palmieri F., De Grassi A. Single-nucleotide evolution quantifies the importance of each site along the structure of mitochondrial carriers. Cell Mol. Life Sci. 2014;71:349–364. doi: 10.1007/s00018-013-1389-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi Q., Li Q., Yao S., Chen Y., Guan M.-X., Cang X. Molecular dynamics simulations on apo ADP/ATP carrier shed new lights on the featured motif of the mitochondrial carriers. Mitochondrion. 2019;47:94–102. doi: 10.1016/j.mito.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Ruprecht J.J., Hellawell A.M., Harding M., Crichton P.G., McCoy A.J., Kunji E.R. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E426–434. doi: 10.1073/pnas.1320692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann B., Stöckl A., Schlame M., Beyer K., Klingenberg M. The reconstituted ADP/ATP carrier activity has an absolute requirement for cardiolipin as shown in cysteine mutants. J. Biol. Chem. 1994;269:1940–1944. [PubMed] [Google Scholar]
- 32.Jiang F., Ryan M.T., Schlame M., Zhao M., Gu Z., Klingenberg M., et al. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- 33.Beyer K., Nuscher B. Specific cardiolipin binding interferes with labeling of sulfhydryl residues in the adenosine diphosphate/adenosine triphosphate carrier protein from beef heart mitochondria. Biochemistry. 1996;35:15784–15790. doi: 10.1021/bi9610055. [DOI] [PubMed] [Google Scholar]
- 34.Beyer K., Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by phosphorus-31 nuclear magnetic resonance. Biochemistry. 1985;24:3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- 35.Nury H., Dahout-Gonzalez C., Trézéguet V., Lauquin G., Brandolin G., Pebay-Peyroula E. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 2005;579:6031–6036. doi: 10.1016/j.febslet.2005.09.061. [DOI] [PubMed] [Google Scholar]
- 36.Mao X., Yao S., Yi Q., Xu Z.-M., Cang X. Function-related asymmetry of the specific cardiolipin binding sites on the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta Biomembr. 2021;1863:183466. doi: 10.1016/j.bbamem.2020.183466. [DOI] [PubMed] [Google Scholar]
- 37.Pasquadibisceglie A., Polticelli F. Structural determinants of ligands recognition by the human mitochondrial basic amino acids transporter SLC25A29. Insights from molecular dynamics simulations of the c-state. Comput. Struct. Biotechnol. J. 2021;19:5600–5612. doi: 10.1016/j.csbj.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogner W., Aquila H., Klingenberg M. The transmembrane arrangement of the ADP/ATP carrier as elucidated by the lysine reagent pyridoxal 5-phosphate. Eur. J. Biochem. 1986;161:611–620. doi: 10.1111/j.1432-1033.1986.tb10485.x. [DOI] [PubMed] [Google Scholar]
- 39.Lin J.L.J. Characterization of the novel cardiolipin binding regions identified on the protease and lipid activated PKC-related kinase 1. Protein Sci. 2019;28:1473–1486. doi: 10.1002/pro.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller M.P., Jiang T., Sun C., Lihan M., Pant S., Mahinthichaichan P., et al. Characterization of Lipid–Protein Interactions and Lipid-Mediated Modulation of Membrane Protein Function through Molecular Simulation. Chem. Rev. 2019;119:6086–6161. doi: 10.1021/acs.chemrev.8b00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedger G., Rouse S.L., Domański J., Chavent M., Koldsø H., Sansom M.S.P. Lipid-loving ANTs: molecular simulations of cardiolipin interactions and the organization of the adenine nucleotide translocase in model mitochondrial membranes. Biochemistry. 2016;55:6238–6249. doi: 10.1021/acs.biochem.6b00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duncan A.L., Ruprecht J.J., Kunji E., Robinson A.J. Cardiolipin dynamics and binding to conserved residues in the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta Biomembr. 2018;1860:1035–1045. doi: 10.1016/j.bbamem.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cang X., Du Y., Mao Y., Wang Y., Yang H., Jiang H. Mapping the Functional binding sites of cholesterol in β2-adrenergic receptor by long-time molecular dynamics simulations. J. Phys. Chem. B. 2013;117:1085–1094. doi: 10.1021/jp3118192. [DOI] [PubMed] [Google Scholar]
- 44.Hanske J., Toffey J.R., Morenz A.M., Bonilla A.J., Schiavoni K.H., Pletneva E.V. Conformational properties of cardiolipin-bound cytochrome c. Proc. Natl. Acad. Sci. U. S. A. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cang X., Yang L., Yang J., Luo C., Zheng M., Yu K., et al. Cholesterol-β1AR interaction versus cholesterol-β2AR interaction. Proteins. 2014;82:760–770. doi: 10.1002/prot.24456. [DOI] [PubMed] [Google Scholar]
- 46.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2014;12:7. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Humphrey W., Dalke A., Schulten K. VMD – Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 49.Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Dávila-Contreras E.M., et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35:1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo S., Lim J.B., Klauda J.B., Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo S., Kim T., Im W. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 53.Hess B., Kutzner C., Van d.S.D., Lindahl E. GROMACS 4: a algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theor. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 54.Jr M.A., Feig M., Rd B.C. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 55.Huang J., Jr M.K.A. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:2384. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 57.Berendsen H.J.C., Postma J.P.M., Gunsteren W.F.V., Dinola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 58.Laurent B., Chavent M., Cragnolini T., Dahl A.C.E., Pasquali S., Derreumaux P., et al. Epock: rapid analysis of protein pocket dynamics. Bioinformatics. 2014;31:1478–1480. doi: 10.1093/bioinformatics/btu822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.
- 60.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 61.Core Team R. R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 62.RStudio Team . Integrated Development Environment for R. RStudio, Inc.; Boston, MA: 2016. RStudio. [Google Scholar]
- 63.Beitz E. TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinformatics. 2000;16:135–139. doi: 10.1093/bioinformatics/16.2.135. [DOI] [PubMed] [Google Scholar]
- 64.Falconi M., Chillemi G., Di Marino D., D'Annessa I., Morozzo della Rocca B., Palmieri L., et al. Structural dynamics of the mitochondrial ADP/ATP carrier revealed by molecular dynamics simulation studies. Proteins. 2006;65:681–691. doi: 10.1002/prot.21102. [DOI] [PubMed] [Google Scholar]
- 65.Yao S., Yi Q., Ma B., Mao X., Chen Y., Guan M.-X., et al. Mechanistic insights into multiple-step transport of mitochondrial ADP/ATP carrier. Comput. Struct. Biotechnol. J. 2022;20:1829–1840. doi: 10.1016/j.csbj.2022.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Tajkhorshid E. Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dehez F., Pebay-Peyroula E., Chipot C. Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J. Am. Chem. Soc. 2008;130:12725–12733. doi: 10.1021/ja8033087. [DOI] [PubMed] [Google Scholar]
- 68.Neves M.A.C., Yeager M., Abagyan R. Unusual arginine formations in protein function and assembly: rings, strings, and stacks. J. Phys. Chem. B. 2012;116:7006–7013. doi: 10.1021/jp3009699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson A.J., Overy C., Kunji E.R. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17766–17771. doi: 10.1073/pnas.0809580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palmieri F., Pierri C.L. Structure and function of mitochondrial carriers - role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 2010;584:1931–1939. doi: 10.1016/j.febslet.2009.10.063. [DOI] [PubMed] [Google Scholar]
- 71.Kunji E.R., Robinson A.J. Coupling of proton and substrate translocation in the transport cycle of mitochondrial carriers. Curr. Opin. Struct. Biol. 2010;20:440–447. doi: 10.1016/j.sbi.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 72.Johnston J.M., Khalid S., Sansom M.S. Conformational dynamics of the mitochondrial ADP/ATP carrier: a simulation study. Mol. Membr. Biol. 2008;25:506–517. doi: 10.1080/09687680802459271. [DOI] [PubMed] [Google Scholar]
- 73.Yao S., Ma B., Yi Q., Guan M.-X., Cang X. Investigating the Broad Matrix-Gate Network in the Mitochondrial ADP/ATP Carrier through Molecular Dynamics Simulations. Molecules. 2022;27:1071. doi: 10.3390/molecules27031071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tamura K., Hayashi S. Atomistic modeling of alternating access of a mitochondrial ADP/ATP membrane transporter with molecular simulations. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Montalvo-Acosta J.J., Kunji E.R.S., Ruprecht J.J., Dehez F., Chipot C. Structure, substrate binding, and symmetry of the mitochondrial ADP/ATP carrier in its matrix-open state. Biophys. J. 2021;120:5187–5195. doi: 10.1016/j.bpj.2021.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nury H., Dahout-Gonzalez C., Trezeguet V., Lauquin G.J., Brandolin G., Pebay-Peyroula E. Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu. Rev. Biochem. 2006;75:713–741. doi: 10.1146/annurev.biochem.75.103004.142747. [DOI] [PubMed] [Google Scholar]
- 77.Majima E., Koike H., Hong Y.M., Shinohara Y., Terada H. Characterization of cysteine residues of mitochondrial ADP/ATP carrier with the SH-reagents eosin 5-maleimide and N-ethylmaleimide. J. Biol. Chem. 1993;268:22181–22187. [PubMed] [Google Scholar]
- 78.Dahout-Gonzalez C., Ramus C., Dassa E.P., Dianoux A.C., Brandolin G. Conformation-dependent swinging of the matrix loop m2 of the mitochondrial Saccharomyces cerevisiae ADP/ATP carrier. Biochemistry. 2005;44:16310–16320. doi: 10.1021/bi0514820. [DOI] [PubMed] [Google Scholar]
- 79.Majima E., Shinohara Y., Yamaguchi N., Hong Y.-M., Terada H. Importance of loops of mitochondrial ADP/ATP carrier for its transport activity deduced from reactivities of its cysteine residues with the sulfhydryl reagent eosin-5-maleimide. Biochemistry. 1994;33:9530–9536. doi: 10.1021/bi00198a019. [DOI] [PubMed] [Google Scholar]
- 80.Sounier R., Bellot G., Chou J.J. Mapping conformational heterogeneity of mitochondrial nucleotide transporter in uninhibited states. Angew. Chem. Int. Ed. Engl. 2015;54:2436–2441. doi: 10.1002/anie.201408417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Di Marino D., Oteri F., Morozzo Della Rocca B., Chillemi G., Falconi M. ADP/ATP mitochondrial carrier MD simulations to shed light on the structural-dynamical events that, after an additional mutation, restore the function in a pathological single mutant. J. Struct. Biol. 2010;172:225–232. doi: 10.1016/j.jsb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 82.Mielke C., Lefort N., McLean C.G., Cordova J.M., Langlais P.R., Bordner A.J., et al. Adenine nucleotide translocase is acetylated in vivo in human muscle: Modeling predicts a decreased ADP affinity and altered control of oxidative phosphorylation. Biochemistry. 2014;53:3817–3829. doi: 10.1021/bi401651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request to the corresponding author.