Abstract
Supergenes are involved in adaptation in multiple organisms, but they are little known in humans. Genomic inversions are the most common mechanism of supergene generation and maintenance. Here, we review the information about two large inversions that are the best examples of potential human supergenes. In addition, we do an integrative analysis of the newest data to understand better their functional effects and underlying genetic changes. We have found that the highly divergent haplotypes of the 17q21.31 inversion of approximately 1.5 Mb have multiple phenotypic associations, with consistent effects in brain-related traits, red and white blood cells, lung function, male and female characteristics and disease risk. By combining gene expression and nucleotide variation data, we also analysed the molecular differences between haplotypes, including gene duplications, amino acid substitutions and regulatory changes, and identify CRHR1, KANLS1 and MAPT as good candidates to be responsible for these phenotypes. The situation is more complex for the 8p23.1 inversion, where there is no clear genetic differentiation. However, the inversion is associated with several related phenotypes and gene expression differences that could be linked to haplotypes specific of one orientation. Our work, therefore, contributes to the characterization of both exceptional variants and illustrates the important role of inversions.
This article is part of the theme issue ‘Genomic architecture of supergenes: causes and evolutionary consequences’.
Keywords: inversions, supergenes, humans, phenotypic traits, gene expression
1. Human supergenes and inversions
Supergenes are defined as clusters of tightly linked functional genetic elements spanning hundreds of kilobases that control complex balanced phenotypes and are inherited as a unit owing to reduced or absent recombination within them [1–4]. Since recombination reduction is essential, the most common mechanism of supergene formation are inversions [2,4], in which single crossovers between heterozygotes lead to unbalanced gametes [5,6]. This allows the generation of highly divergent haplotypes accumulating a large number of sequence differences, which can form coadapted gene complexes [5,6]. In fact, there are many examples of inversion supergenes involved in adaptation in multiple organisms, such as the polymorphic social behaviour of ants, the Batesian mimicry wing colour pattering in butterflies or mating morphs in sparrows and ruffs, among many others [7–11].
In humans, there are practically no described cases of supergenes so far. Recent studies have finally started to characterize in detail human inversion polymorphism, but most inversions are relatively small and their potential functional effects are limited to nearby genes [12–14]. In addition, bigger inversions tend to be mediated by large complex segmental duplications (SDs), which could result in high rates of inversion recurrence by non-allelic homologous recombination (NAHR) that complicate finding phenotypic associations [13,14]. Two good human supergene candidates are the large inversions in 17q21.31, which has already been proposed to act as a supergene [15,16], and 8p23.1 [17,18]. The two inversions are well known and have been linked to diverse effects [12,19,20], although it has not been possible to establish clearly the actual genes and molecular changes responsible for them.
Here, we investigate to what extent the two inversions are indeed supergenes. By integrating the published information and newest genomic data, including nucleotide variation, gene expression and genome-wide association studies (GWAS), we give a global overview of their genomic architecture and how they can cause their effects. Also, we discuss briefly other potential human inversion supergene candidates.
2. The 17q21.31 inversion
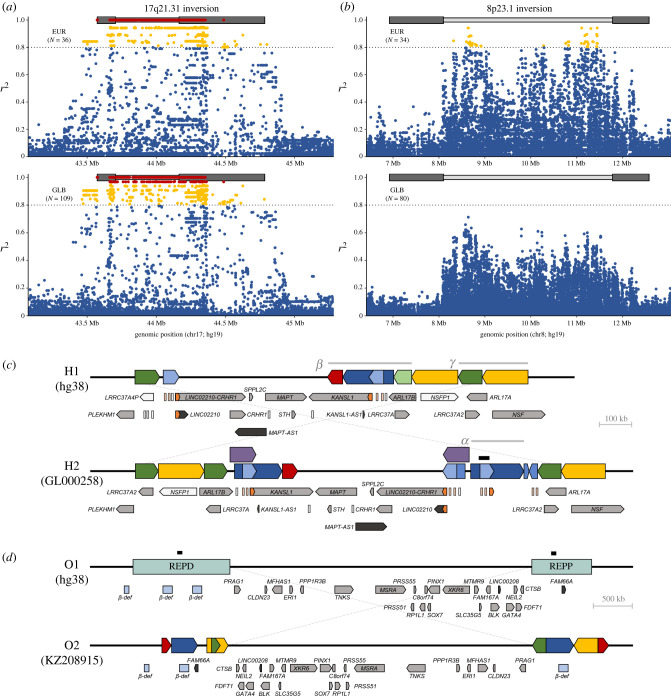
The 17q21.31 inversion spans approximately 900 kb flanked by variable complex SD blocks of 200–800 kb and it is located within a 1.5-Mb long Chr. 17 region of high linkage disequilibrium (LD), defining two divergent haplotypes (H1 and H2) with no recombination between them [15,21,22]. The inversion was initially genotyped in a limited number of diverse individuals by fluorescence in situ hybridizaton (FISH) [22,23] and more recently by droplet digital polymerase chain reaction [14]. Thus, early on it was established that H1 and H2 correspond to the reference and inverted orientation, respectively, which results in many completely linked tag single nucleotide polymorphisms (SNPs) (figure 1a; electronic supplementary material, table S1).
Figure 1.
Summary of the genomic structure of two potential human inversion supergenes. (a,b) Linkage disequilibrium (LD) distribution of the 17q21.31 (hg38, chr17: 45 495 836–46 707 123) (a) and 8p23.1 (hg38, chr8:7 064 966–12 716 088) (b) inversions in European (EUR) and global (GLB) populations. Each dot represents a polymorphic variant within the inversion ± 500 kb region. LD was calculated from 1000 Genome Project (1000GP) individuals in which the inversion has been genotyped experimentally (N) and is shown in different colours: r2 < 0.8 (blue), r2 = 0.8–0.95 (yellow) and r2 > 0.95 (red). The rectangles above the graphs summarize the inversion region (light grey) and the variable segmental duplication (SD) blocks at its breakpoints (dark grey). (c) Structure of the inversion 17q21.31 H1 and H2 haplotypes updated from Boettger et al. and Steinberg et al. [21,22]. Coloured blocks indicate repeated segments at the breakpoints, with partial copies from the same repeat in lighter colours. The copy number variable fragments corresponding to α, β and γ duplications are indicated above [21]. Arrows below each orientation represent protein-coding genes (grey), non-coding genes (black) and pseudogenes (white). Small orange boxes correspond to the duplication of the first KANSL1 exons and two pseudogenes. (d) Structure of the 8p23.1 inversion region according to Mohajeri et al. [24]. SD organization within the REPD and REPP repeat blocks in O1 (light green rectangles) is not indicated because it has not been completely resolved and it includes gaps (indicated as black bars) and known assembly errors. Owing to the size of the region, all protein-coding genes (grey arrows) and only two non-coding genes mentioned in the main text (black arrows) are shown, with β-defensin clusters pictured as light blue boxes.
Inversion imputation based on these tag SNPs showed a particular distribution pattern across the globe. H2 is low in Africa (less than 6%) and virtually absent in East Asian populations, while it forms a southwest/northeast cline in Europe, with highest frequencies in Mediterranean regions of southwest Asia and Europe (20–37%), high frequency in western, central, and southeast Europe (15–28%) and much lower frequency in eastern and northern Europe and on the Arabian Peninsula (5–10%) [22,25,26]. In addition, based on haplotype differentiation, the age of the inversion was estimated on 2–3 Myr [15,27] (although more recent estimates of approx. 100 000 years have been obtained using different methods [26]). According to primates, H2 is the ancestral orientation, although there could have been independent recurrent inversion events in human, chimpanzee and orangutan lineages [27]. Therefore, the derived H1 orientation probably increased in frequency to almost fixation in the human lineage, and a recent selective sweep of H2 haplotypes, possibly associated with higher fertility and recombination rate of H2 carrier females, resulted in the current distribution and lack of diversity of inverted chromosomes [15,27].
(a) . Inversion phenotypic effects
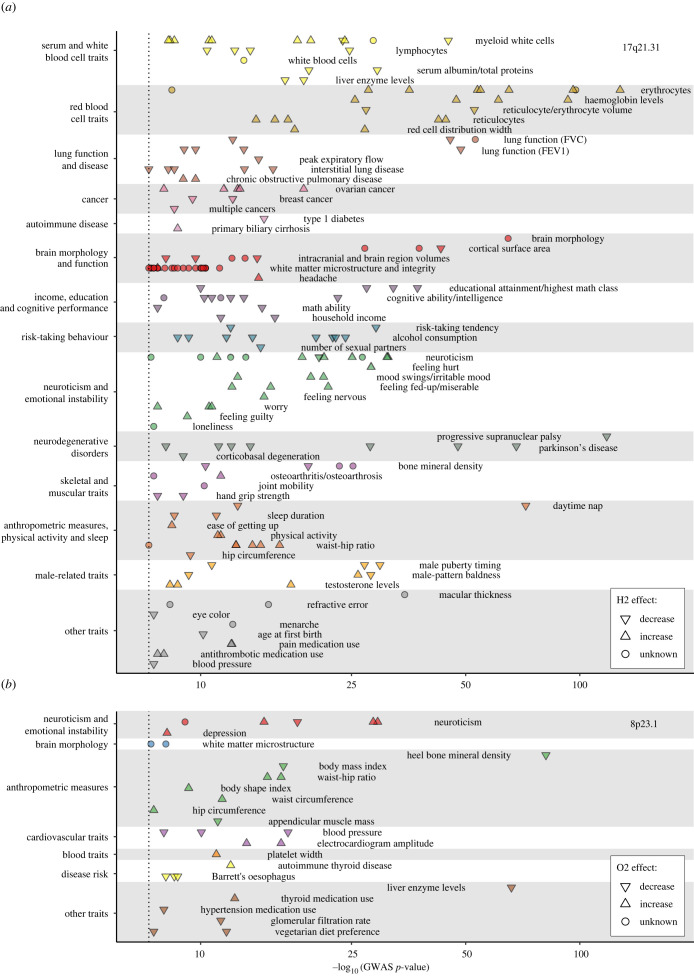
The 17q21.31 inversion stands out for its pleiotropic phenotypic effects and it has been involved in an increasing number of diverse traits [12,14,19,20,28,29]. To update and understand better these effects, we have done an exhaustive analysis and manual curation of the GWAS hits (p < 5 × 10−8) corresponding to all SNPs in high LD (r2 > 0.8) with the inversion listed in the GWAS Catalog [30] (summarized together with their supporting studies in the electronic supplementary material, table S2). This has allowed us to classify the 203 independent hits in 14 broad categories comprising 64 more or less different traits, showing quite consistent and related effects for the two orientations (figure 2a).
Figure 2.
Summary of phenotypic associations for 17q21.31 (a) and 8p23.1 (b) inversions. GWAS Catalog signals (p < 5 × 10–8, dotted line) in high LD (r2 ≥ 0.8) with the inversions in the closest studied population are shown grouped by categories of related traits with different colours. Upwards and downwards triangles illustrate direction of effect of the inverted allele (H2 or O2) and dots unknown direction. Complete data are described in the electronic supplementary material, tables S2 and S3. (Online version in colour.)
As shown in figure 2a, one of the first phenotypes the inverted haplotype has been associated with is a lower risk of several neurodegenerative disorders, including Parkinson's disease, progressive supranuclear palsy and corticobasal degeneration [12,20]. However, the H2 haplotype is also associated with very significant changes in cell blood composition (electronic supplementary material, table S2). These changes combine higher levels of mature and immature erythrocytes, haemoglobin and haematocrit (together with more variable and smaller cell volume), and a general increase of white blood cells, especially neutrophils and basophils, but not eosinophils and lymphocytes, that show lower counts. In addition, the inversion is linked to a reduction of several lung function parameters and to increased risk of chronic obstructive pulmonary disease, although it could protect from lung fibrosis (electronic supplementary material, table S2). It is also linked to opposite effects in ovarian (higher risk) and breast cancer (lower risk), which might be related to the different role of progesterone in these cancers, and in autoimmune diseases like type 1 diabetes (lower risk) and primary biliary cirrhosis (higher risk) (electronic supplementary material, table S2).
A large group of related phenotypes are those involved in brain morphology and different cognitive and behavioural traits (figure 2a). For example, H2 is associated with reduced intracranial volume and cortical surface area, plus additional changes in white matter microstructure and integrity (electronic supplementary material, table S2). Actually, a genetic correlation has been found between reduced brain surface and several of the inversion-associated traits, such as higher tendency to neuroticism and depressive behaviours, and a decrease of the highly interrelated variables cognitive ability, education and household income [31,32]. Similarly, apart from reduced sleep, the inversion appears to be associated with higher physical activity and less inclination towards risk-taking behaviours, like speeding, alcohol consumption and number of sexual partners (electronic supplementary material, table S2).
Finally, H2 is associated with several anthropometric, skeletal and muscular parameters that could be connected with the previous phenotypes: lower blood pressure; higher waist-hip ratio, which contrasts with the increased activity levels and could be caused by smaller hip circumference; and reduced hand grip strength and bone mineral density, combined with higher risk of osteoarthritis (figure 2a; electronic supplementary material, table S2). The inversion is also associated with alopecia, later puberty and higher testosterone levels in males (electronic supplementary material, table S2), although there is not an obvious relationship between them. In females, as already mentioned, the inversion was originally linked to higher fertility and higher recombination rates [15,33], which is confirmed here by a reduction in the age at first birth plus an association with menarche onset (electronic supplementary material, table S2). Other inversion associations involve eye phenotypes, including lighter colour, typical of Europeans where the inversion is most common (figure 2a; electronic supplementary material, table S2). Interestingly, the observed increase in medication use fits well with some other possible inversion effects, such as antithrombotic medication and higher number of red blood cells and pain medication and higher risk of headache and arthritis (electronic supplementary material, table S2).
Therefore, the number and diversity of inversion-associated traits make it clearly the best supergene example in humans. Moreover, our analysis suggests that H2 selection in Europeans could be more complex than initially thought, involving other traits besides fertility. In particular, there are several phenotypes potentially involved in immune response, a highly selected trait, and it has been recently found that the inversion protects against severe COVID-19 [29], which could be related to changes in immune cells, lung function or even red blood cell levels. Nevertheless, so far there is little information about the molecular causes behind these phenotypic differences.
(b) . Sequence differences between inversion haplotypes
Possible functional effects of 2568 variants that differentiate H1 and H2 haplotypes across diverse human populations (r2 > 0.95) were analysed using the Ensembl Variant Effect Predictor (VEP) [34] (electronic supplementary material, table S4). We found 17 SNPs that cause amino acid (aa) changes in five genes, with SPPL2C, MAPT and KANSL1 accumulating 7, 5 and 3 aa differences, respectively (table 1). Of those, two changes in SPPL2C and KANSL1 and one in MAPT have combined annotation dependent depletion (CADD) scores greater than 20, indicating that they are among the 1% most deleterious substitutions in the human genome (electronic supplementary material, table S4). Most of these variants affect the proteins produced by several alternatively spliced isoforms of the genes. There are also two SNPs in positions adjacent to splicing signals in the non-coding RNA LINC02210, which could affect the splicing of four different transcripts (table 1). In addition, 60 SNPs are located within sequences annotated as promoters in the Ensembl Regulatory Build [35], affecting up to 12 promoters of nine different genes (table 1). For example, a putative promoter region of the shorter KANSL1 transcripts and/or the KANSL1-AS1 long non-coding RNA (lncRNA) (ENSR00000095038), which is highly conserved in vertebrates, includes four nucleotide changes with CADD scores greater than 17. Finally, we detected many other SNPs that could affect gene regulation by altering untranslated regions (UTRs) or other types of regulatory elements, like enhancers or CTCF-binding sites (table 1; electronic supplementary material, table S4).
Table 1.
Potential functional effects of SNP differences in high LD (r2 > 0.95) with 17q21.31 H1 and H2 haplotypes according to VEP analysis. (The combined annotation dependent depletion (CADD) value scores the predicted deleteriousness of variants by integrating multiple annotations, including conservation and functional information. Variants with scores greater than 20 are predicted to be the 1% most deleterious substitutions in the human genome. n.a., not applicable.)
| SNP effect | no. SNPs | affected elements | no. genes | genes |
|---|---|---|---|---|
| amino acid change (CADD > 20) | 5 | n.a. | 3 | SPPL2C, MAPT, KANSL1 |
| amino acid change (CADD < 20) | 12 | n.a. | 5 | CRHR1, SPPL2C, MAPT, STH, KANSL1 |
| synonymous change | 16 | n.a. | 4 | CRHR1, SPPL2C, MAPT, KANSL1 |
| UTRs | 58 | n.a. | 3 | CRHR1, MAPT, KANSL1 |
| splicing signals | 2 | n.a. | 1 | LINC02210 |
| promoters | 60 | 12 | 9 | DND1P1, RPS26P8, LINC02210, CRHR1, MAPT, KANSL1, KANSL1-AS1, DND1P2, RP11-359G18.1 |
| transcription factor binding sites | 25 | 43 | n.a. | n.a. |
| enhancers | 76 | 24 | n.a. | n.a. |
| CTCF binding sites | 126 | 49 | n.a. | n.a. |
Apart from small sequence changes, the 17q21.31 inversion breakpoint regions (BP1 and BP2) are characterized by a considerable amount of structural diversity [21,22]. Within H1 chromosomes BP2, there are two copy number variable regions: duplication β (approx. 267 kb, 1–3 copies), containing two protein-coding genes (ARL17A and LRRC37A2), the first exons of KANSL1, KANSL1-AS and two pseudogenes (defective copies of MAPK8IP1 and DND1); and duplication γ (approx. 218 kb, 1–4 copies), which includes ARL17B and LRRC37A and part of NSF (pseudogene NSFP1) (figure 1c). Both duplications were generated independently from a haplotype with a single copy of each sequence (H1.β1.γ1), with the most common H1 haplotypes in human populations being H1.β1.γ1, H1.β2.γ1, H1.β1.γ2 and H1.β1.γ3 [21,22].
The inversion originated probably in an H1.β1.γ2 haplotype by NAHR between 59 kb SDs containing copies of genes ARL17 and LRRC37 in opposite orientation (figure 1c) [27]. However, inverted chromosomes have at least 32 kb extra sequence that creates the approximately 85 kb H2-specific inverted SDs at both breakpoints (figure 1c). Moreover, the most common H2 haplotype (H2.α2.γ2) contains an additional insertion in BP2 of around 200 kb, formed probably by a complex rearrangement of a complete 154 kb α duplication and two 12.3- and 28.4 kb α fragments (although a gap in the only available sequence complicates the analysis) [21,22]. Other H2 haplotypes with a deletion of the γ duplication in BP1 also exist at low frequencies [21,22]. Therefore, H2.α2.γ2 gene content has three main differences from haplotype H1.β1.γ2: (i) two functional LRRC37 copies (BP2) and one pseudogene (BP1) in H1 versus three functional copies in H2; (ii) an extra copy of the first three exons and an alternative first exon of KANSL1 and their respective promoters, which could generate novel transcripts in H2; and (iii) up to four copies of DND1 and MAPK8IP1 pseudogenes in H2 versus only two copies of each in H1 (figure 1c).
Besides these large changes at the breakpoints, we looked at the LD of the inversion with other structural variants (SVs) identified in the 1000GP [36,37] (electronic supplementary material, table S5). A 1383 bp SVA element insertion and two deletions (323 and 314 bp long) in different KANSL1 introns were associated with H1/H2 haplotypes (r2 > 0.8). In addition, a 238 bp MAPT intronic deletion that had been proposed to map to H2 haplotypes [15] showed a r2 = 0.85, which could be caused by errors in deletion genotype imputation.
(c) . 17q21.31 Haplotypes and gene expression
We identified gene expression differences associated with the 17q21.31 inversion and the β and γ duplications in multiple tissues and cell lines using Geuvadis and GTEx RNA-Seq data [38,39] (electronic supplementary material, table S6). Similar to previous results obtained from GTEx expression quantitative trait loci (eQTLs) information [14,29], the inversion acts as lead eQTL in at least one tissue of 29 genes located both within the inversion and in the nearby flanking regions (figure 3). Of those, 13 are protein-coding genes, nine non-coding RNAs and seven pseudogenes. Remarkably, the genes flanking BP1 tend to be downregulated in H2, whereas most of those within the inverted region and around BP2 are upregulated (figure 3). Actually, the inversion is associated with an alteration of cis-regulatory domains in the region, defined by correlated histone modifications, that mirrors the changes in expression patterns [40]. However, the consequences of this alteration are not known, and sequence differences between H1 and H2 could also be responsible. For example, in H2, missing pseudogene LRRC37A4P is consistently downregulated (figure 1c), whereas LRRC37A2 is upregulated in almost all tissues (figure 3), which could be explained by the presence of three functional LRRC37 copies. Similarly, the DND1 and MAPK8IP1 pseudogenes that have additional copies in H2 show widespread increased expression (which contrasts with the lack of expression differences associated with the H1 β-duplication, containing extra copies of these pseudogenes as well; see below).
Figure 3.
Tissue-specific association of gene expression with the inversion and β and γ duplications in the 17q21.31 region. Variant cis-eQTL effects were estimated by testing associations between genotypes and gene expression measures from multiple GTEx tissues and Geuvadis lymphoblastoid cell lines (LCLs) [38,39] (see the electronic supplementary material, methods). eQTL effect size and direction associated with the presence of the inversion or duplication is illustrated by the colour gradient and the p-value by the dot size, with black squares indicating when the variants are lead eQTLs or in high LD (r² ≥ 0.95) with top variants in the corresponding dataset (electronic supplementary material, table S6). Genes are ordered according to their genomic position. Figure is an updated version of that in Degenhardt et al. [29].
With respect to tissue distribution, many changes in gene expression accumulate in different parts of the brain, especially in the cerebellum (figure 3), a structure linked to some of the symptoms of the neurodegenerative diseases against which the inversion has protective effects [12,20]. In fact, the genes in the 17q21.31 inversion region show highest expression in only a few tissues [29,39]: cerebellum, pituitary, lung, testis and female reproductive organs for protein-coding genes and non-coding RNAs; and testis, followed by cerebellum and female reproductive organs, for pseudogenes. Thus, expression differences associated with both haplotypes (figure 3) are expected to have more functional consequences in these tissues, which could explain the phenotypic changes related to them.
The β and γ duplications were associated with expression differences of nine and four genes, respectively (figure 3). In this case, lower imputation accuracies complicate assessing if they are the causal variant, but the duplications were the lead eQTLs of at least three (β) and two (γ) genes. Moreover, most of these genes are upregulated and map inside the duplicated segments. Exceptions are the partially duplicated NSF gene, which is downregulated in lymphoblastoid cell lines (LCLs) with duplication γ, and duplication β pseudogenes MAPK8IP1P1 and DND1P2, which do not show higher expression with more β copies. Some of the genes that could be affected by the β and γ duplications also show expression changes between H1 and H2 haplotypes. However, only the KANSL1 expression change can be explained by the β duplication, although the inversion has a much stronger effect in just certain tissues (figure 3). Therefore, the observed expression changes appear to be more associated with the two inversion haplotypes than with the presence of either duplication.
(d) . Candidate integrative analysis
As usually happens with supergenes, the huge number of variants in high or almost complete LD complicates narrowing down the individual genetic changes responsible for the different phenotypes. Nevertheless, based on the sequence and expression changes observed and functional information of the genes, there are several good candidates to mediate the 17q21.31 inversion phenotypic associations.
One of the best candidates is CRHR1, which encodes a receptor involved in corticotropin-releasing hormone (CRH) family signalling. This protein is a major regulator of the hypothalamic-pituitary-adrenal (HPA) axis and activates transduction pathways that regulate diverse physiological processes in which the inversion could have an effect, such as stress, the immune system, mood and emotions, reproduction, sleep and obesity [41]. Importantly, the CRHR1 receptor is also involved in promoting embryo implantation [42] and modulates myometrial contractility [41], which could be related to the inversion effect on fertility. Therefore, the inversion probably increases CRHR1 activity and HPA axis signalling. According to our analysis (figure 3), H2 is associated with upregulation of CHRH1 in several tissues but not in brain, where the gene is most highly expressed (although a limited previous study using microarrays found increased CHRH1 expression in the cerebellum [43]). However, the inversion is lead eQTL for LINC02210 upregulation in almost all tissues. This lncRNA forms a fusion transcript with CRHR1 and could be driving a higher expression of some CRHR1 isoforms (figure 1c), which is consistent with changes in LINC02210 splicing patterns between the two haplotypes in GTEx data [29].
Another good candidate is KANSL1, which produces a histone acetylation regulatory subunit that could be related to transcription regulation and chromatin modification. KANSL1 pathogenic mutations in heterozygosis cause developmental psychomotor delay and mild/modest intellectual disability characteristic of Koolen-de Vries syndrome [44,45]. Moreover, the KANSL1 protein is involved in microtubule organization during mitosis [46]. The H2 haplotype is associated with higher KANSL1 expression just in muscle and cultured LCLs and fibroblasts, whereas the overlapping KANSL-AS1 transcript (figure 1c) is upregulated in all tissues (figure 3). These expression differences could be in part owing to the nucleotide changes accumulated in the joint promoter of the shorter KANSL1 isoforms and KANSL1-AS1. In addition, different KANSL1 coding and non-coding isoforms show inversion-associated expression changes in other conditions, such as Parkinson disease patients [47] or monocytes under immune stimuli [29], emphasizing the potential important functional consequences of this gene in many tissues and phenotypes.
The gene MAPT has been long implicated in neurodegenerative diseases caused by aggregation of the encoded microtubule-associated protein tau, which promotes assembly and stabilization of cytoskeletal microtubules [48]. It is expressed predominantly in the central nervous system, where there are six MAPT transcripts produced by alternative splicing of exons 2, 3 and 10 [49]. Alternative splicing of exon 10 results in MAPT protein isoforms with three (3R) or four (4R) repeat microtubule-interacting domains, and mutations disrupting the normal exon 10 splicing balance cause disease [49]. We have seen that MAPT expression changes are limited to H2 upregulation in the oesophagus, testis and lung (figure 3). Previous microarray expression data found increased MAPT expression in H1 chromosomes in frontal cortex and cerebellum [43,50], but it has been suggested that this difference was probably caused by a technical artefact [51]. However, reporter assays indicated that transcriptional activity of H1 MAPT promoter was higher than H2 [52]. In addition, several studies have reported MAPT splicing differences between H1/H2 haplotypes, such as higher expression of the isoform containing exon 10 in H1 [53,54] or the one containing exon 3 in H2 [51,55]. These differences are also supported by the association of the inversion with multiple splicing QTLs (sQTLs) in GTEx in several brain areas [29]. Remarkably, between H1 and H2 sequences, there are 42 SNPs plus the 238 bp deletion within the intron preceding exon 10 that could be affecting the splicing of this critical exon. Two additional MAPT SNPs linked to H1/H2 haplotypes produce aa substitutions with high CADD scores: rs17651549 (CADD = 26.9) and rs10445337 (CADD = 18.9). The most deleterious substitutes arginine (polar) by tryptophan (hydrophobic) in a position conserved in all mammals, although both changes occur in exons included in MAPT isoforms expressed in the peripheral nervous system and their consequences are not known [49]. Thus, the molecular mechanisms involved in neurodegenerative disease risk are not yet clear, but both MAPT increased expression and preferential inclusion of exon 10 could play a role.
One last candidate that accumulates many changes between H1 and H2 haplotypes is SPPL2C, which has seven aa substitutions with CADD scores ranging from 0.001 to 23.3. This gene encodes a signal peptide peptidase of 684 aa that is expressed predominantly in testis and could be related to male germ cell development and fertility [56], although it is upregulated in H2 in cerebellum (figure 3). In particular, H2 SPPL2c has a proline for arginine substitution in position 461 that might be important for protein conformation according to the predicted interaction with other positions. So, in summary, different type of molecular changes in diverse genes could contribute to the widespread phenotypic effects of the 17q21.31 inversion.
3. The 8p23.1 inversion
The other well-characterized human inversion is that in 8p23.1, which spans a 4.2 Mb region on Chr. 8 [17,18,24]. This inversion was mediated by NAHR between large blocks of high-identity SDs spanning 0.8–1.0 Mb, that are typically known as REPD and REPP [18,24,57] (figure 1d). The complexity of the breakpoints makes studying of the 8p23.1 inversion very challenging and the reference (O1) or inverted (O2) orientation status has been determined experimentally for just around 200 individuals by FISH [17,18,23]. This has shown that the genetic structure is very different from the 17q21.31 inversion, without two clearly differentiated haplotypes and relatively low LD with other variants in most populations (figure 1b). The only exception is Europeans, that have several highly linked SNPs close to each breakpoint (r2 = 0.94) (electronic supplementary material, table S1). Such LD pattern is consistent with double crossovers homogenizing the central region of the inversion, as happens in large Drosophila inversions, and a small degree of recurrence [18,20,23]. Actually, it has already been proposed that the inversion has occurred independently in humans and the chimpanzee-bonobo lineage from the ancestral O2 orientation [18,23].
Nevertheless, several computational algorithms based on genetic differentiation have made it possible to impute 8p23.1 inversion genotypes quite reliably, especially in European individuals [18,58,59]. Although these estimates have limitations, they have shown that the inversion follows a clinal distribution in which the geographical distance from Ethiopia correlates negatively with O2 frequency, with the highest frequency in African populations (60–80%), followed by Europeans (50–60%) and East Asians (10–20%) [18]. Therefore, 8p23.1 geographical distribution agrees with O2 ancestral status and human population expansion out of Africa, suggesting very weak or neutral selective pressure.
(a) . Inversion phenotypic effects
The imputation methods have also allowed linking the 8p23.1 inversion to diverse phenotypic effects, although much fewer than in 17q21.31. It is well known that the O1 orientation is associated with a higher risk for systemic lupus erythematosus (SLE) [18,60,61]. Moreover, the presence of the inversion has been associated with neuroticism [62] and several risky behaviour traits [63]. However, it has been related to a reduction of different metabolic risk factors, such as hypertension, obesity and the co-occurrence of diabetes, hypertension or asthma with obesity, and to an increase of triglyceride levels [28,58].
Our extended analysis of the most recent GWAS Catalog data with highly linked SNPs (r2 > 0.8) in Europeans (figure 2b) replicates several of these inversion associations, including increased risk of neuroticism and depression, together with brain microstructure changes, or protection against hypertension and changes in other cardiovascular traits, like platelet width and electrocardiogram. In the case of obesity, we also find a lower body mass index with the inversion, but this is accompanied by an increase in diverse anthropometric measures typically related to obesity, such as both waist and hip circumference, waist-hip ratio or body shape index, and a reduction in muscle mass and bone mineral density (electronic supplementary material, table S3). This suggests that the role of the inversion in obesity and body plan might be complex. In addition, we have found new interesting inversion phenotypic associations, like protection against Barrett's oesophagus inflammation, increased risk of autoimmune thyroid disease, and lower liver enzyme levels, glomerular filtration rate and vegetarian diet preference (electronic supplementary material, table S3). Nevertheless, owing to the low LD with SNPs in many human populations, the phenotype analysis is limited and many effects are probably missed. Thus, more studies that determine directly inversion orientation in different GWAS datasets are needed.
(b) . 8p23.1 Inversion sequence and expression differences
To determine the potential molecular mechanisms responsible for 8p23.1 inversion phenotypes, we also checked the associated sequence and gene expression changes. As mentioned above, this inversion does not have a clear haplotype structure with a large number of tag SNPs in high LD (figure 1b). In 1000GP Europeans, only nine SNPs have a r2 > 0.9 with the inversion and none of them affects the protein sequence or alters 5′ or 3′ UTRs (electronic supplementary material, table S4). CADD scores associated with these changes are all below 10 except for rs13255193 (CADD = 10.25). Furthermore, only rs560544 locates within a CTCF binding site and affects four putative transcription factor binding sites.
On the other hand, the exact structure of the complex SD blocks flanking the inversion is still not fully resolved [24,57]. These SD blocks contain large β-defensin gene clusters that show considerable variation between individuals [57]. A complete characterization of a 6.3 Mb alternate reference assembly of the inverted sequence has shown 20 structural differences ranging from 1 to 357 kb with respect to the non-inverted reference assembly, the largest of which is another polymorphic inversion [24]. Moreover, the amount of copy number variation in the breakpoint regions between humans could be as much as 1.2 Mb [24]. However, it has not yet been possible to do similar detailed analysis of the SVs in multiple individuals and their association with inversion status. Currently, there are not any known SVs from the 1000GP data [36,37] in high LD with the inversion, although this analysis probably misses most of the variation in the duplicated regions.
With regard to gene expression, previous studies have suggested that the 8p23.1 inversion influences the expression of more than 20 genes [18,28,62,64]. Apart from one spanning multiple tissues [28], most analysis were done in blood-derived cells, and consistent expression differences were found for BLK, NEIL2, MSRA, CTSB, FDFT1, MFHAS1, MTMR9, FAM167A and PPP1R3B. To test this in more detail, we imputed 8p23.1 inversion genotypes and analysed expression levels in Geuvadis LCLs [38] and GTEx tissues [39] (see the electronic supplementary material, methods). According to our strict criteria, the inversion is lead eQTL only of non-coding transcript RP11-148O21.4 in cultured fibroblasts and is highly linked (r2 = 0.81) to the lead variant for RP11-419I17.1 in the oesophagus (electronic supplementary material, table S6). Our analysis also confirmed the gene expression changes previously associated with the inversion, but other SNPs in lower LD were apparently the lead variant. Thus, as already suggested [18], the inversion regulatory effects could be primarily mediated by certain haplotypes specific of one orientation, although given the potential errors during imputation, the inversion could still be responsible of part of the expression variation.
(c) . Candidate integrative analysis
In this case, the lack of sequence differences between the two orientations probably limits the number of gene expression changes directly linked to the inversion, which together with its large gene content and the complexity of the breakpoint regions, make it difficult to identify the molecular mechanisms behind inversion phenotypic effects. In fact, variation in β-defensin content has been involved in different immune-related phenotypes, such as psoriasis and Crohn's disease [65], although how this variation is associated with the inversion has not been determined yet.
Aside from the β-defensins clusters, one of the main candidates for the phenotypic differences is BLK, which encodes a tyrosine kinase. Higher autoimmune disease risk in O1 orientation is apparently related to a common 16-kb O1 haplotype containing several risk alleles for SLE and rheumatoid arthritis [18,60,61]. These risk alleles are located in the bi-directional promoter region of BLK and FAM167A (figure 1d) and are associated, respectively, with decreased or increased expression of each gene [18,66]. The BLK overlapping transcript RP11-148O21.4 also shows lower expression levels in SLE [67], and its upregulation in fibroblasts with O2 fits well with the protective effect of the inversion. Furthermore, there are independent SLE GWAS signals close to other genes showing expression differences between orientations, such as CTSB, MFHAS1, PRAG1 and CLDN23 [61], and SLE risk of this locus could be related to the action of many genes.
Another inversion phenotypic association that could involve BLK is Barrett's oesophagus [68]. Two variants in high LD with the inversion (r2 = 0.89) located in the promoter region and first intron of lncRNA LINC00208, just downstream of BLK (figure 1d), have been associated with O2 protection against Barrett's oesophagus [68]. These variants are also associated with downregulation of lncRNA RP11-419I17.1 specifically in the oesophagus (electronic supplementary material, table S6). However, there is no more information about the possible consequences of this change. Similarly, it has been proposed that BLK, FDFT1 and FAM66A could be involved in diabetes [28]. Finally, PPP1R3B has been linked with serum lipid levels [18], but in most analysis no significant expression changes between inversion orientations have been found.
4. Are there other human inversion supergenes?
In the last few years, extended accurate genotyping in multiple individuals of a larger number of inversions has shown that a few other of the bigger inversions affect diverse functional and phenotypic traits [13,14,69]. An example is HsInv0786, a 171 kb inversion on Chr. 16, which has been associated with different phenotypes, including obesity and co-occurrence of obesity with asthma or hypertension, intelligence, type 1 diabetes, red blood cell count and pediatric autoimmune diseases [14,28,70]. This inversion has also been involved in expression changes of several genes, such as upregulation of SULT1A2 or TUFM, an elongation factor required for mitochondrial protein synthesis and potentially involved in energy metabolism [14,28,70]. However, in most cases, the relatively low LD with flanking SNPs precludes determining reliably the effects of these inversions in available GWAS and functional datasets, as happens for HsInv0290, a low-frequency Chr. 7 inversion of approximately 0.74 Mb [14]. Therefore, future large-scale genotyping of these and other inversions will help us understand better the role of inversion supergenes in the human genome. Interestingly, despite the added difficulty of recurrence, the lack of complete association between inversions and SNPs gives the opportunity to solve the long-debated question if their effects are mainly caused by the inversion itself or the combination of SNPs contained within [16].
Another important question is how inversion supergenes are maintained, given their expected negative fertility costs. Accordingly, we should assume that these costs were outweighed by some type of favourable selection. Thus, it is not surprising that common large inversions accumulate many different effects that probably were selected at some point during human evolution, although now they may increase our disease risk, as could be the case for obesity or autoimmune diseases.
Acknowledgements
We would like to thank the core members of the Severe COVID-19 GWAS Group for extensive discussions about the possible effects of the 17q21.31 inversion candidate genes and S. McCarroll for providing the 17q21.31 genotypes reference panel.
Data accessibility
The data are provided in the electronic supplementary material [71].
Authors' contributions
E.C.: formal analysis, writing—original draft; M.P.: formal analysis, writing—original draft; I.Y.: formal analysis, writing—original draft; J.L.-J.: formal analysis; M.C.: conceptualization, formal analysis, supervision, writing—original draft.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported by research grant nos. BFU2016-77244-R and PID2019-107836RB-I00 funded by the Agencia Estatal de Investigación (AEI, Spain) and the European Regional Development Fund (FEDER, EU) and 2017-SGR-1379 from the Generalitat de Catalunya (Spain) to M.C. E.C. and I.Y. were supported by AEI (Spain) Predoctoral Contracts BES-2017-082018 and PRE-2020-092440.
References
- 1.Thompson MJ, Jiggins CD. 2014. Supergenes and their role in evolution. Heredity 113, 1-8. ( 10.1038/hdy.2014.20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black D, Shuker DM. 2019. Supergenes. Curr. Biol. 29, R615-R617. ( 10.1016/J.CUB.2019.05.024) [DOI] [PubMed] [Google Scholar]
- 3.Rubenstein DR, et al. 2019. Coevolution of genome architecture and social behavior. Trends Ecol. Evol. 34, 844-855. ( 10.1016/J.TREE.2019.04.011) [DOI] [PubMed] [Google Scholar]
- 4.Gutiérrez-Valencia J, Hughes PW, Berdan EL, Slotte T. 2021. The genomic architecture and evolutionary fates of supergenes. Genome Biol. Evol. 13, evab057. ( 10.1093/GBE/EVAB057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39, 21-42. ( 10.1146/annurev.ecolsys.39.110707.173532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8, e1000501. ( 10.1371/journal.pbio.1000501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wellenreuther M, Bernatchez L. 2018. Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 33, 427-440. ( 10.1016/j.tree.2018.04.002) [DOI] [PubMed] [Google Scholar]
- 8.Jay P, Leroy M, Le Poul Y, Whibley A, Arias M, Chouteau M, Joron M. 2022. Association mapping of colour variations in a butterfly provides evidences that a supergene locks together a cluster of adaptive loci. Phil. Trans. R. Soc. B 377, 20210193. ( 10.1098/rstb.2021.0193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kay T, Helleu Q, Keller L. 2022. Iterative evolution of supergene-based social polymorphism in ants. Phil. Trans. R. Soc. B 377, 20210196. ( 10.1098/rstb.2021.0196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim K-W, De-Kayne R, Gordon IJ, Saitoti K, Martins DJ, Ffrench-Constant R, Martin S. 2022. Stepwise evolution of a butterfly supergene via duplication and inversion. Phil. Trans. R. Soc. B 377, 20210207. ( 10.1098/rstb.2021.0207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenløkk KSR, Saitou M, Rud-Johansen L, Nome T, Moser M, Árnyasi M, Kent MP, Barson N, Lien S. 2022. The emergence of supergenes from inversions in Atlantic salmon. Phil. Trans. R. Soc. B 377, 20210195. ( 10.1098/rstb.2021.0195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puig M, Casillas S, Villatoro S, Cáceres M. 2015. Human inversions and their functional consequences. Brief. Funct. Genomics 14, 369-379. ( 10.1093/bfgp/elv020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giner-Delgado C, et al. 2019. Evolutionary and functional impact of common polymorphic inversions in the human genome. Nat. Commun. 10, 4222. ( 10.1038/s41467-019-12173-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puig M, et al. 2020. Determining the impact of uncharacterized inversions in the human genome by droplet digital PCR. Genome Res. 30, 724-735. ( 10.1101/766915) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefansson H, et al. 2005. A common inversion under selection in Europeans. Nat. Genet. 37, 129-137. ( 10.1038/ng1508) [DOI] [PubMed] [Google Scholar]
- 16.Villoutreix R, Ayala D, Joron M, Gompert Z, Feder JL, Nosil P. 2021. Inversion breakpoints and the evolution of supergenes. Mol. Ecol. 30, 2738-2755. ( 10.1111/MEC.15907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giglio S, et al. 2002. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 71, 276-285. ( 10.1086/341610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salm MPA, et al. 2012. The origin, global distribution, and functional impact of the human 8p23 inversion polymorphism. Genome Res. 22, 1144-1153. ( 10.1101/gr.126037.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feuk L. 2010. Inversion variants in the human genome: role in disease and genome architecture. Genome Med. 2, 11. ( 10.1186/gm132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alves JM, Lopes AM, Chikhi L, Amorim A. 2012. On the structural plasticity of the human genome: chromosomal inversions revisited. Curr. Genomics 13, 623-632. ( 10.2174/138920212803759703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. 2012. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat. Genet. 44, 881-885. ( 10.1038/ng.2334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinberg KM, et al. 2012. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat. Genet. 44, 872-880. ( 10.1038/ng.2335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antonacci F, Kidd JM, Marques-Bonet T, Ventura M, Siswara P, Jiang Z, Eichler EE. 2009. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet. 18, 2555-2566. ( 10.1093/hmg/ddp187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohajeri K, et al. 2016. Interchromosomal core duplicons drive both evolutionary instability and disease susceptibility of the Chromosome 8p23.1 region. Genome Res. 26, 1453-1467. ( 10.1101/GR.211284.116/-/DC1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alves JM, et al. 2015. Reassessing the evolutionary history of the 17q21 inversion polymorphism. Genome Biol. Evol. 7, 3239-3248. ( 10.1093/gbe/evv214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donnelly MP, et al. 2010. The distribution and most recent common ancestor of the 17q21 inversion in humans. Am. J. Hum. Genet. 86, 161-171. ( 10.1016/j.ajhg.2010.01.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zody MC, et al. 2008. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 40, 1076-1083. ( 10.1038/ng.193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González JR, et al. 2020. Polymorphic inversions underlie the shared genetic susceptibility of obesity-related diseases. Am. J. Hum. Genet. 106, 846-858. ( 10.1016/J.AJHG.2020.04.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degenhardt F, et al. 2021. New susceptibility loci for severe COVID-19 by detailed GWAS analysis in European populations. medRxiv 2021.07.21.21260624. ( 10.1101/2021.07.21.21260624) [DOI]
- 30.MacArthur J, et al. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896-D901. ( 10.1093/nar/gkw1133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams HHH, et al. 2016. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat. Neurosci. 19, 1569-1582. ( 10.1038/NN.4398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grasby KL, et al. 2020. The genetic architecture of the human cerebral cortex. Science 367, eaay6690. ( 10.1126/SCIENCE.AAY6690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhury R, Bois PRJ, Feingold E, Sherman SL, Cheung VG. 2009. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 5, e1000648. ( 10.1371/JOURNAL.PGEN.1000648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F. 2016. The ensembl variant effect predictor. Genome Biol. 17, 1-14. ( 10.1186/s13059-016-0974-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. 2015. The ensembl regulatory build. Genome Biol. 16, 1-8. ( 10.1186/S13059-015-0621-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526, 68-74. ( 10.1038/nature15393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sudmant PH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75-81. ( 10.1038/nature15394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lappalainen T, et al. 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506-511. ( 10.1038/nature12531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550, 204-213. ( 10.1038/nature24277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delaneau O, et al. 2019. Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science 364, eaat8266. ( 10.1126/science.aat8266) [DOI] [PubMed] [Google Scholar]
- 41.Hillhouse EW, Grammatopoulos DK. 2006. The molecular mechanisms underlying the regulation of the biological activity of corticotropin-releasing hormone receptors: implications for physiology and pathophysiology. Endocr. Rev. 27, 260-286. ( 10.1210/ER.2005-0034) [DOI] [PubMed] [Google Scholar]
- 42.Makrigiannakis A, Zoumakis E, Kalantaridou S, Coutifaris C, Margioris AN, Coukos G, Rice KC, Gravanis A, Chrousos GP. 2001. Corticotropin-releasing hormone promotes blastocyst implantation and early maternal tolerance. Nat. Immunol. 2, 1018-1024. ( 10.1038/ni719) [DOI] [PubMed] [Google Scholar]
- 43.de Jong S, Chepelev I, Janson E, Strengman E, van den Berg LH, Veldink JH, Ophoff RA. 2012. Common inversion polymorphism at 17q21.31 affects expression of multiple genes in tissue-specific manner. BMC Genomics 13, 458. ( 10.1186/1471-2164-13-458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farnè M, Bernardini L, Capalbo A, Cavarretta G, Torres B, Sanchini M, Fini S, Ferlini A, Bigoni S. 2021. Koolen-de Vries syndrome in a 63-year-old woman: report of the oldest patient and a review of the adult phenotype. Am. J. Med. Genet. A 188, 692-707. ( 10.1002/AJMG.A.62536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dingemans AJM, Stremmelaar DE, van der Donk R, Vissers LELM, Koolen DA, Rump P, Hehir-Kwa JY, de Vries BBA. 2021. Quantitative facial phenotyping for Koolen-de Vries and 22q11.2 deletion syndrome. Eur. J. Hum. Genet. 29, 1418-1423. ( 10.1038/S41431-021-00824-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meunier S, Shvedunova M, Van Nguyen N, Avila L, Vernos I, Akhtar A.. 2015. An epigenetic regulator emerges as microtubule minus-end binding and stabilizing factor in mitosis. Nat. Commun. 6, 1-10. ( 10.1038/ncomms8889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tobin JE, et al. 2008. Haplotypes and gene expression implicate the MAPT region for Parkinson disease: the GenePD Study. Neurology 71, 28-34. ( 10.1212/01.wnl.0000304051.01650.23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb A, Miller B, Bonasera S, Boxer A, Karydas A, Wilhelmsen KC. 2008. Role of the tau gene region chromosome inversion in progressive supranuclear palsy, corticobasal degeneration, and related disorders. Arch. Neurol. 65, 1473-1478. ( 10.1001/archneur.65.11.1473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rademakers R, Cruts M, Van Broeckhoven C.. 2004. The role of tau (MAPT) in frontotemporal dementia and related tauopathies. Hum. Mutat. 24, 277-295. ( 10.1002/HUMU.20086) [DOI] [PubMed] [Google Scholar]
- 50.Myers AJ, et al. 2007. A survey of genetic human cortical gene expression. Nat. Genet. 39, 1494-1499. ( 10.1038/ng.2007.16) [DOI] [PubMed] [Google Scholar]
- 51.Trabzuni D, et al. 2012. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum. Mol. Genet. 21, 4094-4103. ( 10.1093/HMG/DDS238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwok JBJ, Teber ET, Loy C, Hallupp M, Nicholson G, Mellick GD, Buchanan DD, Silburn PA, Schofield PR. 2004. Tau haplotypes regulate transcription and are associated with Parkinson's disease. Ann. Neurol. 55, 329-334. ( 10.1002/ANA.10826) [DOI] [PubMed] [Google Scholar]
- 53.Myers AJ, et al. 2007. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol. Dis. 25, 561-570. ( 10.1016/J.NBD.2006.10.018) [DOI] [PubMed] [Google Scholar]
- 54.Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R. 2006. Haplotype-specific expression of exon 10 at the human MAPT locus. Hum. Mol. Genet. 15, 3529-3537. ( 10.1093/HMG/DDL429) [DOI] [PubMed] [Google Scholar]
- 55.Caffrey TM, Joachim C, Wade-Martins R. 2008. Haplotype-specific expression of the N-terminal exons 2 and 3 at the human MAPT locus. Neurobiol. Aging 29, 1923–1929. ( 10.1016/J.NEUROBIOLAGING.2007.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niemeyer J, et al. 2019. The intramembrane protease SPPL2c promotes male germ cell development by cleaving phospholamban. EMBO Rep. 20, e46449. ( 10.15252/EMBR.201846449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hollox EJ, Barber JC, Brookes AJ, Armour JA. 2008. Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 18, 1686-1697. ( 10.1101/gr.080945.108) [DOI] [PubMed] [Google Scholar]
- 58.Cáceres A, González JR. 2015. Following the footprints of polymorphic inversions on SNP data: from detection to association tests. Nucleic Acids Res. 43, e53. ( 10.1093/nar/gkv073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruiz-Arenas C, Cáceres A, López-Sánchez M, Tolosana I, Pérez-Jurado L, González JR. 2019. scoreInvHap: inversion genotyping for genome-wide association studies. PLoS Genet. 15, e1008203. ( 10.1371/journal.pgen.1008203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Namjou B, et al. 2014. The effect of inversion at 8p23 on BLK association with Lupus in Caucasian population. PLoS ONE 9, e115614. ( 10.1371/journal.pone.0115614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demirci YF, et al. 2017. Multiple signals at the extended 8p23 locus are associated with susceptibility to systemic lupus erythematosus. J. Med. Genet. 54, 381-389. ( 10.1136/JMEDGENET-2016-104247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okbay A, et al. 2016. Genetic variants associated with subjective well-being, depressive symptoms and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624-633. ( 10.1038/ng.3552) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karlsson LR, et al. 2019. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat. Genet. 51, 245-257. ( 10.1038/s41588-018-0309-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bosch N, Morell M, Ponsa I, Mercader JM, Armengol L, Estivill X. 2009. Nucleotide, cytogenetic and expression impact of the human chromosome 8p23.1 inversion polymorphism. PLoS ONE 4, e8269. ( 10.1371/journal.pone.0008269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantsilieris S, White SJ. 2013. Correlating multiallelic copy number polymorphisms with disease susceptibility. Hum. Mutat. 34, 1-13. ( 10.1002/HUMU.22172) [DOI] [PubMed] [Google Scholar]
- 66.Hom G, et al. 2008. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 358, 900-909. ( 10.1056/NEJMoa0707865) [DOI] [PubMed] [Google Scholar]
- 67.Hauberg ME, et al. 2017. Large-scale identification of common trait and disease variants affecting gene expression. Am. J. Hum. Genet. 100, 885–894. ( 10.1016/J.AJHG.2017.04.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gharahkhani P, et al. 2016. Genome-wide association studies in oesophageal adenocarcinoma and Barrett's oesophagus: a large-scale meta-analysis. Lancet. Oncol. 17, 1363-1373. ( 10.1016/S1470-2045(16)30240-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lerga-Jaso J. 2019. Integrative analysis of the functional consequences of inversions in the human genome . Barcelona, Spain: Universitat Autònoma de Barcelona. See http://hdl.handle.net/10803/669344. [Google Scholar]
- 70.González JR, et al. 2014. A common 16p11.2 inversion underlies the joint susceptibility to asthma and obesity. Am. J. Hum. Genet. 94, 361-372. ( 10.1016/j.ajhg.2014.01.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campoy E, Puig M, Yakymenko I, Lerga-Jaso J, Cáceres M. 2022. Genomic architecture and functional effects of potential human inversion supergenes. FigShare. ( 10.6084/m9.figshare.c.5986112) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Campoy E, Puig M, Yakymenko I, Lerga-Jaso J, Cáceres M. 2022. Genomic architecture and functional effects of potential human inversion supergenes. FigShare. ( 10.6084/m9.figshare.c.5986112) [DOI] [PMC free article] [PubMed]
Data Availability Statement
The data are provided in the electronic supplementary material [71].