Abstract
Protein disulfide isomerase (PDI) is important in assisting the folding and maturation of secretory proteins in eukaryotes. A gene, pdiA, encoding PDIA was previously isolated from Aspergillus niger, and we report its functional characterization here. Functional analysis of PDIA showed that it catalyzes the refolding of denatured and reduced RNase A. pdiA also complemented PDI function in a Saccharomyces cerevisiae Δpdi1 mutant in a yeast-based killer toxin assay. Levels of pdiA mRNA and PDIA protein were raised by the accumulation of unfolded proteins in the endoplasmic reticulum. This response of pdiA mRNA levels was slower and lower in magnitude than that of A. niger bipA, suggesting that the induction of pdiA is not part of the primary stress response. An increased level of pdiA transcripts was also observed in two A. niger strains overproducing a heterologous protein, hen egg white lysozyme (HEWL). Although overexpression of PDI has been successful in increasing yields of some heterologous proteins in S. cerevisiae, overexpression of PDIA did not increase secreted yields of HEWL in A. niger, suggesting that PDIA itself is not limiting for secretion of this protein. Downregulation of pdiA by antisense mRNA reduced the levels of microsomal PDIA activity by up to 50%, lowered the level of PDIA as judged by Western blots, and lowered the secreted levels of glucoamylase by 60 to 70%.
Filamentous fungi have a potential for the commercial production of heterologous proteins, although their production levels are usually much lower than those of homologous proteins. For example, production levels of ca. 40 g/liter are obtained for cellulase produced by Trichoderma reesei (8), while the production of heterologous proteins is usually on the order of milligrams per liter (39). For some heterologous proteins, the transit of newly synthesized protein from the fungal endoplasmic reticulum (ER) limits secretion (13).
One of the most important processes occurring in the ER is the controlled folding and assembly of newly synthesized proteins, a process mediated by ER-resident foldases and molecular chaperones. These ER proteins also associate with misfolded proteins and prevent their exit from the ER until they are correctly folded or are targeted for degradation by the cytoplasmic proteasomes (3, 19, 45). In normal cells, the concentrations of these foldases and chaperones are likely to be sufficient for proper folding and assembly of proteins destined for secretion. However, in expression systems where there is a greater flux of proteins being translocated into the ER, the folding, assembly, and secretion machinery may become saturated, leading to improperly folded structures or protein aggregates which are not secreted. The accumulation of improperly folded protein aggregates has been observed in Escherichia coli (as inclusion bodies), yeasts, mammalian cells, and insect cells as the result of overexpression of heterologous proteins (2, 12, 28). One approach to overcome the problem of protein aggregation is to overexpress chaperones or foldases in the ER. Two ER proteins, protein disulfide isomerase (PDI), a foldase, and BiP, a molecular chaperone, play important roles in the folding, assembly, and secretion of proteins in the ER. Overexpression of BiP and PDI in various expression systems including Saccharomyces cerevisiae, E. coli, and mammalian cells has improved the yields of some heterologous proteins (14, 16, 32).
PDI, a soluble protein resident in the ER, plays a key role in the folding and secretion of proteins (11). As a foldase, PDI catalyzes the formation or breakage of disulfide bonds, depending on the redox potential of the environment. PDI also catalyzes the rearrangement of preexisting disulfide bonds (11). We have previously isolated a gene, pdiA, from Aspergillus niger which encodes a putative PDI (30). We confirm here that pdiA encodes a functional PDI (PDIA), we examine the transcriptional response of pdiA to agents that perturb protein folding and secretion, and we also examine the impact of up- and downregulation of pdiA expression on the secretion of a homologous and a heterologous protein from A. niger. We also compare the transcriptional responses of pdiA with those of tigA, which encodes an ER-resident PDI family member in A. niger (18), and bipA, which encodes the major ER-resident chaperone in A. niger and A. awamori (15, 40).
MATERIALS AND METHODS
Strains, media, and culture conditions.
A. niger AB4.1 (41), which is a pyrG mutant, was used as the host for transformation and pdiA regulation studies. A. niger L11 and B1, which secrete different amounts of the heterologous protein hen egg white lysozyme (HEWL) (17), were used for pdiA regulation studies and as hosts for overexpressing pdiA. Inoculation and growth of A. niger strains at 25°C in ACM/N/P medium supplemented with 1% (wt/vol) filter-sterilized starch or xylose as carbon source was as previously reported (1). The final concentrations of inducing agents which were added 44 h after inoculation were as follows: dithiothreitol (DTT), 20 mM; Ca2+ ionophore A23187, 6 μg/ml. A. niger AB4.1 cultures were harvested at 20, 40, 60, 120, 180, and 240 min after treatment with DTT, while the Ca2+ ionophore A23187-treated cultures were harvested at 30, 60, 120, 180, and 240 min. A. niger SBD (25) overexpressing the homologous protein, glucoamylase, was used to compare microsomal free PDIA levels with those in A. niger B1 and L11. E. coli XL1-Blue (Stratagene) was used for plasmid propagation. S. cerevisiae strains were kindly provided by M. F. Tuite, University of Kent, Canterbury, England. The genotypes of strains were as follows: DNY5, MATa/α leu2/leu2,trp1/trp1 Δpdi1::HIS3/Δpdi1::HIS3(pVT-100U:KT) (pVT-100U:KT contains the killer toxin gene); SK14a, MATa ura3-52(pVT-100U:KT); S6, killer-sensitive strain, wild type. The strains were grown at 30°C. S. cerevisiae DNY5 was maintained in a vegetative state. S. cerevisiae DNY5 and SK14a were grown on SCD medium prepared as previously described (20). S. cerevisiae S6 was maintained on YEPD (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] glucose).
For many of the subsequent studies, mycelial cultures were examined for pdiA transcript levels and refolding activities either throughout growth or at 44 h because, at this time, cultures were still actively growing but would soon begin to decline in their rates of biomass increase. Levels of secreted protein were generally measured either during growth or at a fixed time point of 72 or 96 h. These time points reflect the approach to, or point of, maximum levels of protein secreted. Although protein secretion is a growth-linked function, the appearance of protein in culture filtrates is delayed relative to growth in batch cultures due to the kinetics of the secretory process and wall transit. Thus, the 44-h time point for measurement of pdiA transcript levels and refolding activities is relevant to the later times used for measured yields of secreted proteins.
Nucleic acid manipulations.
DNA manipulations were performed as described previously (36). DNA isolation, Southern blot analysis, RNA isolation, and Northern blot analysis were also performed as described previously (30). RNA from S. cerevisiae was isolated by using an RNeasy mini kit (Qiagen). Probes used for Northern analysis were as described for pdiA (30) and tigA (18). The probe for bipA was a gel-purified 445-bp PCR fragment corresponding to coordinates 712 to 1156 (40). To ensure equal loading between tracks on Northern gels and to afford comparison of transcript levels, all hybridization signals were standardized against an internal γ-actin control as described elsewhere (30). To compensate for differences in probe length, pdiA and bipA mRNA signals were also adjusted by using 831-bp tigA probe (18) as the standard (=1). Signals from the 32P-labeled probes were quantified by using a Fuji BAS-1500 phosphorimager.
Transformations of A. niger and S. cerevisiae were performed as described earlier (1, 9). PCR with 1-day-old mycelium was performed by using a method described by van Zeijl et al. (42). DNA sequencing was performed as previously described (30).
Reverse transcription (RT)-PCR from total RNA of starch (ACMS/N/P)- and xylose (ACMX/N/P)-grown A. niger antisense cultures was used to determine the presence of pdiA antisense transcripts. RT was performed as described previously (38). Primers CN63 (5′-CAC GAT TCT GTT TGC CTA GC-3′) (at coordinates 344 to 363 of pdiA) and DJ128 (5′-CCC ATC CTT TAA CTA TAG CG-3′), designed from the 3′ end of the glucoamylase gene 16 to 34 bp downstream of the stop codon, were used for PCR, and a PCR product of approximately 500 bp was expected.
Construction of Aspergillus expression vectors and strains.
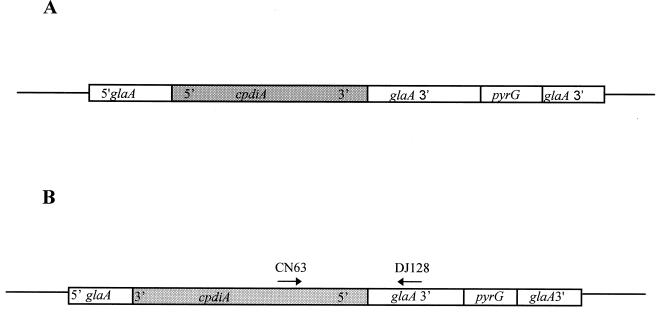
A. niger PO3 and PO13 were constructed by introducing plasmid pIGCPDI into A. niger AB4.1 (Table 1). pIGCPDI was constructed by inserting a 1.7-kb blunt-ended SalI/HpaI fragment of pdiA cDNA (cpdiA) into the HpaI site of pIGPG under the glucoamylase (glaA) promoter (Fig. 1A). The vector pIGPG, a pUC18-based plasmid, contains approximately 2 kb each of the 5′ (including the promoter sequences) and 3′ glucoamylase sequences and the A. niger pyrG gene (41).
TABLE 1.
A. niger transformants
| A. niger strain | Origin | Transforming plasmid | Plasmid origin | Promoter | Gene expressed | Copy no. |
|---|---|---|---|---|---|---|
| PO3 | AB4.1 | pIGCPDI | pIGPG | glaA | cpdiA | 1 |
| PO13 | AB4.1 | pIGCPDI | pIGPG | glaA | cpdiA | 2 |
| AS1.1 | AB4.1 | pAS | pIGPG | glaA | cpdiAa | 6–10 |
| AS2.2 | AB4.1 | pAS | pIGPG | glaA | cpdiAa | 4–6 |
| LO6 | L11 | pANpdiA | pAN7-Blue1 | pdiA | pdiA | 4–6 |
| LO9 | L11 | pANpdiA | pAN7-Blue1 | pdiA | pdiA | 2–4 |
| OB22 | B1 | pANpdiA | pAN7-Blue1 | pdiA | pdiA | 3–5 |
| OB38 | B1 | pANpdiA | pAN7-Blue1 | pdiA | pdiA | 3–5 |
Inserted in the antisense direction.
FIG. 1.
(A) pPIGCPDI was constructed from pIGPG. cpdiA was inserted at the HpaI site of pIGPG downstream of the starch inducible glaA promoter. (B) pAS was constructed from pIGPG. cpdiA was inserted in the antisense direction under control of the glaA promoter. Arrows associated with CN63 and DJ128 represent primers used for RT-PCR studies. The diagram is not to scale.
A. niger AS1.1 and AS2.2 were constructed by introducing pAS into A. niger AB4.1 (Table 1). pAS was constructed by inserting a blunt-ended SalI/HpaI fragment containing cpdiA into the HpaI site of pIGPG in the antisense direction under control of the glaA promoter (Fig. 1B). Transformants were selected by the ability to grow without uridine. A single spore from each transformant was plated on AMMN minimal agar (29) with 1% (wt/vol) xylose (AMMNX) or 1% (wt/vol) starch (AMMNS) as the sole carbon source.
A. niger strains OB22, OB38, LO6, and LO9 which overexpress pdiA were constructed by transforming pANpdiA into A. niger B1 and L11 (Table 1). pANpdiA was constructed by inserting a 4.7-kb blunt-ended ClaI/XbaI restriction fragment of pdiA, which contains the complete pdiA sequence including the promoter region, into a SmaI-digested vector, pAN7-BlueI. pAN7-BlueI is a vector derived from pAN7-1 (31) constructed by P. Bowyer (Long Ashton, United Kingdom). pAN7-BlueI contains the hygromycin selection marker, lacZ, and multiple cloning sites.
pdiA multicopy transformants of A. niger B1 and L11 were selected on AMMN plates containing 100 μg of hygromycin per ml. These transformants were further selected on AMMN slopes with 150 and 200 μg of hygromycin per ml (31). Those A. niger LO transformants (derived from L11) and A. niger OB transformants (derived from B1) which grew in AMMN with 200 μg of hygromycin per ml were selected for Northern analysis by using the pdiA probe (30).
Construction of yeast expression vector and transformation of S. cerevisiae strain DNY5.
The unwanted ATG in the polylinker of pYX243 (LEU2 selectable marker) (R&D Systems), a 2μm plasmid, was removed by NcoI restriction and mung bean nuclease (Promega) digestion to produce flush ends before religation with T4 ligase (Promega), creating pYX243ΔNcoI. A 1.7-kb SmaI/SalI fragment containing cpdiA (30) was cloned into SmaI/SalI-cut pYX243 ΔNcoI. The resulting plasmid, pYX:cpdiA, was transformed into S. cerevisiae DNY5. The transformants were selected on SCD plates without leucine. Transformant DNY5:cpdiA was selected for further analysis.
Microsomal preparation from A. niger.
The method for preparing microsomes from A. niger was adapted from a protocol kindly provided by G. Wallis (University of Nottingham, Nottingham, United Kingdom). A total of 2 to 3 g (wet weight) of A. niger mycelium was harvested and freeze ground in liquid nitrogen by using a mortar and pestle with 0.5 g of acid-washed sand. The freeze-ground mycelium was resuspended in 15 ml of TAS (20 mM Tris-acetate, pH 7.4; 0.25 M sucrose) containing 1 COMPLETE protease inhibitor cocktail tablet (Boehringer) per 50 ml of TAS and 1 μl of β-mercaptoethanol per ml and then centrifuged at 5,000 × g for 20 min at 4°C to remove the mycelial debris. The resulting supernatant was then subjected to ultracentrifugation at 120,000 × g for 90 min at 4°C. The pellet was resuspended in 200 μl of cold TAS buffer (without β-mercaptoethanol and protease inhibitors but containing 10 mM MgCl2). The protein concentration of the microsomes was determined by using a DC Protein Assay kit (Bio-Rad). In all microsome preparations, complete loss of pyruvate kinase activity (35) (a cytoplasmic enzyme marker) was confirmed.
RNase refolding assay.
An RNase A refolding assay adapted from Lyles and Gilbert (26) and Lee et al. (24) was used to estimate PDI activity in A. niger microsomes. Reduced and denatured RNase A (bovine pancreas type IIIA) (Sigma) was obtained as described elsewhere (26). Immediately before use, the reduced and denatured RNase A was separated from the DTT and guanidine hydrochloride by using a PD10 column (Pharmacia) which was equilibrated with 0.1% acetic acid. The concentration of denatured RNase and the number of thiol groups per reduced RNase molecule were determined as described earlier (4). Refolding activity was corrected against uncatalyzed reactions (i.e., controls without a microsomal fraction added). An average of three readings was taken for each reaction. One unit of refolding activity was defined as the amount of enzyme increasing RNase A activity by one unit. Refolding specific activity is defined as the units of refolding activity per milligram of microsomal protein. The average specific activity ± the standard error of the mean (SEM) presented is derived from three individual cultures. The refolding activities represent the levels available to assist RNase refolding; i.e., they are a measure of free, rather than total, refolding activity.
Yeast killer toxin assay.
The yeast killer toxin assay used was a modified form of those of Woods and Bevan (46) and Dunn et al. (7). Cells from the killer-sensitive S. cerevisiae strain, S6, were grown in YEPD at 30°C. A top agar killer medium lawn (0.5% [wt/vol] Bacto Yeast Extract, 0.5% [wt/vol] Bacto Peptone, 0.05 M citric acid, 0.1 M disodium-hydrogen phosphate buffer [pH 6.5], 1.5% [wt/vol] agar, and 4% [wt/vol] filter-sterilized galactose or glucose) was made by mixing 5 × 106 S6 cells/ml of medium with filter-sterilized methylene blue (final concentration, 0.003%) and overlaying it onto a killer medium plate. S. cerevisiae DNY5:cpdiA, DNY5, and SK14a were grown in minimal media with plasmid selection for 24 h at 30°C to late log phase. The cells were pelleted and resuspended in liquid killer medium (containing 4% filter-sterilized galactose or glucose) for the induction of cpdiA. Induction was performed at 25°C with moderate agitation at 120 rpm for a further 24 h. These cultures were tested for the ability to kill S. cerevisiae S6 cells by spotting 5 × 104 or 5 × 106 cells onto blank antibiotic disks (Oxoid) on killer medium plates containing a newly poured overlay of S. cerevisiae S6 cells in the top agar. The plates were left at 25°C for 3 to 4 days before the lysis zones were measured.
Analytical methods.
Dry weight was determined by filtering 100-ml A. niger culture samples through Miracloth. The resulting mycelium was squeezed, blotted dry, and dried in a freeze-dryer for 2 days. The amount of glucoamylase secreted into the supernatant by A. niger was determined as previously described (27). Assay of lysozyme activity from culture supernatants was performed as previously described (17). For Western analysis, 10 μg of microsomal protein was loaded onto precast 4 to 12% sodium dodecyl sulfate gels (Novex) and immunoblotted by using anti-PDIA (a gift from Danisco) as previously described (1). Bands were scanned, and those migrating at the predicted size for PDIA (56.3 kDa) were quantified by using Whole Band Analyser software (BioImage) on a SunSparc workstation. Additional bands observed in the PDIA standard lanes are likely to represent PDIA degradation products.
RESULTS
A. niger pdiA restores PDI function in Δpdi1 S. cerevisiae DNY5.
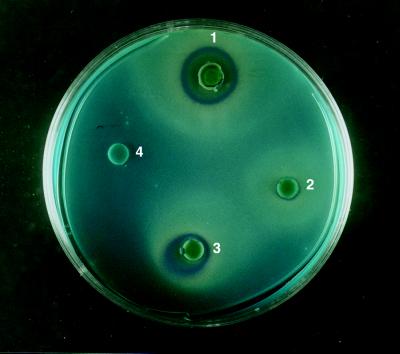
S. cerevisiae DNY5 contains a plasmid encoding the yeast killer toxin protein but is unable to secrete this disulfide-bridged protein due to disruption of the S. cerevisiae pdi1 gene (7). Killer toxin secretion was restored by the transformation of S. cerevisiae DNY5 with pYX:cpdiA, indicating that PDI-like functions are encoded by pdiA (Fig. 2). Evidence of killer toxin activity was determined against a lawn of sensitive S. cerevisiae S6 cells. Transparent halos surrounded by a blue ring of S. cerevisiae S6 lawn cells were apparent around the discs which had been spotted with galactose-induced S. cerevisiae DNY5:cpdiA at two different concentrations of cells (Fig. 2). A larger halo ca. 2 cm in diameter was observed around disks spotted with 5 × 104 S. cerevisiae SK14a cells containing the native pdi1. In contrast, a halo of 0.7 cm was observed in the presence of the same amount of S. cerevisiae DNY5:cpdiA cells. When 5 × 106 cells of DNY5:cpdiA were spotted onto the disks, a halo 1.2 to 1.4 cm in diameter was observed. No halos were seen around the untransformed S. cerevisiae DNY5 control. Similarly, no halo was observed with DNY5:cpdiA cells grown on glucose (data not shown). The smaller halo observed in S. cerevisiae DNY5:cpdiA could result from insufficient PDI being available for efficient secretion of the killer toxin. The killer toxin assay demonstrates that PDIA, encoded by multiple copies of pdiA, can functionally replace PDI in Δpdi1 strains of S. cerevisiae.
FIG. 2.
Complementation of S. cerevisiae Δpdi1 by the A. niger pdiA gene in a yeast killer toxin assay. Evidence of killer toxin secretion was determined by a transparent halo surrounded by a blue ring: 1, S. cerevisiae SK14a, 5 × 104 cells; 2, S. cerevisiae DNY5:cpdiA, 5 × 104 cells; 3, S. cerevisiae DNY5:cpdiA, 5 × 106 cells; and 4, S. cerevisiae DNY5, 5 × 104 cells.
PDIA refolds reduced and denatured RNase A.
The presence of microsomal extracts from A. niger AB4.1 catalyzed the refolding of denatured RNase A 10 to 12 times faster than the natural rate (data not shown). In the absence of a glutathione redox buffer, there was no increase in RNase A activity, and refolding activity was limited solely to the microsomal fraction. Thus, proteins present in the microsomes catalyze the refolding of denatured RNase A in the presence of glutathione redox buffers.
Transformant A. niger PO3 contained one copy of pIGCPDI and PO13 contained two copies of pIGCPDI. Since these additional copies of pdiA cDNA are under the control of the glaA promoter, they were analyzed for the level of pdiA transcripts in starch-containing (inducing) and xylose-containing (noninducing) media. Starch-grown A. niger PO3 and PO13 cultures showed a 2- to 3-fold increase in pdiA transcript levels (data not shown) and an approximately 1.7- to 2-fold increase in refolding activity compared to the xylose-grown cultures (2.6 ± 0.1 versus 1.4 ± 0.1 U/mg of microsomal protein). The level of refolding activity in the xylose-grown strains was the same as that in the untransformed AB4.1 strain grown on either starch or xylose (data not shown). That this increase in refolding activity in the PO3 and PO13 strains is induced by starch strongly suggests that PDIA functions as a true foldase in vivo.
Transcriptional responses of pdiA, bipA, and tigA to DTT and the Ca2+ ionophore A23187.
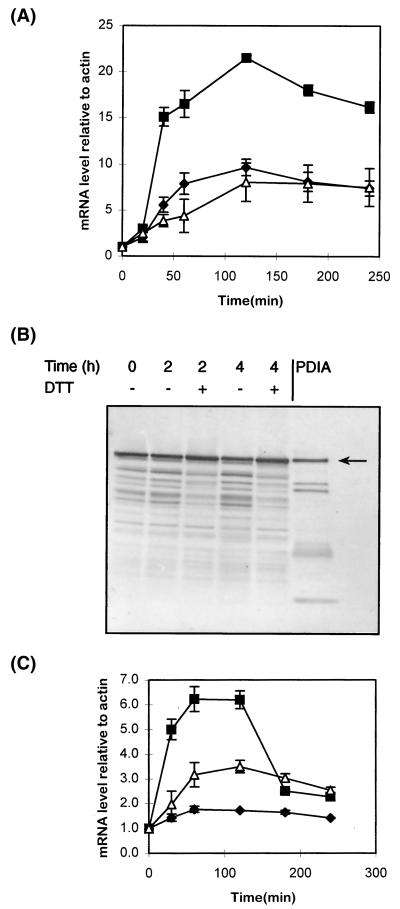
The effects of agents known to perturb ER function and induce the expression of genes governed by the unfolded protein response were examined. Exposure of A. niger AB4.1 cultures to 20 mM DTT raised the measured pdiA mRNA level by 5- to 6-fold in 40 min and by 10-fold within 2 h (Fig. 3A). The level of tigA mRNA showed a similar response: a fourfold induction was observed after 40 min, which rose to eightfold after 2 h. The response of bipA mRNA was higher than that of either pdiA or tigA, showing an increase of 15-fold after 40 min of exposure to DTT and a 20-fold increase within 2 h.
FIG. 3.
(A) Levels of pdiA (⧫), bipA (■), and tigA (▵) transcripts relative to actin mRNA at various time points after the addition of DTT. Values represent the average ± the SEM (bars) from triplicate experiments. (B) Western blots, probed for PDIA, of microsomal preparations from A. niger AB4.1 cultures at 0, 120, and 240 min after addition of DTT relative to untreated (water) controls. The arrow indicates the predicted size of PDIA. (C) Levels of pdiA (⧫), bipA (■), and tigA (▵) transcripts at various time points after the addition of the Ca2+ ionophore A23187. Values represent the average ± the SEM (bars) from triplicate experiments.
Western analysis of the microsomal preparations of A. niger AB4.1 treated with DTT showed that, after 4 h, PDIA protein levels were ca. 30% higher than in non-DTT-treated controls (Fig. 3B).
A. niger AB4.1 cultures were also treated with the calcium ionophore A23187, which affects the permeability of cellular membranes to Ca2+ and depletes the intracellular stores (6). Analysis by Northern blots indicated that pdiA transcripts showed a slight increase after 1 to 2 h compared to the control cultures (Fig. 3C). In contrast, tigA mRNA levels increased by 3.5-fold and bipA levels showed a 6.3-fold increase after 1 to 2 h. The levels of bipA mRNA decreased strongly after 2 h.
Levels of pdiA, bipA, and tigA transcripts in A. niger strains overproducing heterologous proteins.
pdiA, bipA, and tigA transcript levels were investigated in two strains of A. niger, B1 and L11 (17), which overexpress HEWL at different levels in the inducing medium ACMS/N/P. Strains B1 and L11 express similar levels of HEWL mRNA, but strain B1 secretes higher HEWL levels than does strain L11 (17).
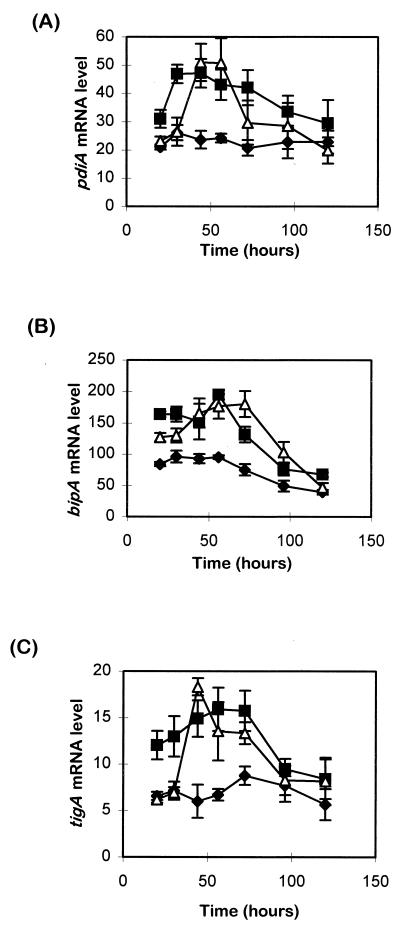
Transcriptional analysis of all three ER-specific genes showed that mRNA levels were higher in the HEWL-secreting A. niger B1 and L11 strains than in the parent strain AB4.1 (Fig. 4). Relative to the untransformed parent, peak levels of pdiA, tigA, and bipA transcripts are evident in both heterologous protein overexpressing strains, i.e., B1 and L11.
FIG. 4.
Time course showing levels of (A) pdiA, (B) bipA, and (C) tigA transcripts in A. niger strains AB4.1 (⧫), B1 (■), and L11 (▵). Transcript levels were standardized as a percentage against actin mRNA levels. Values represent the average ± the SEM (bars) from triplicate experiments.
Comparing the relative abundance of the three transcript species, in A. niger AB4.1 at 20 and 30 h, the bipA transcript levels were three- to fourfold more abundant than those of pdiA, while the tigA transcript level appeared to be three- to fourfold less than that of pdiA (Fig. 4). In B1, a twofold increase in the bipA transcript level was observed from 20 h, while an approximately twofold increase of pdiA and tigA transcript levels was detected from 30 to 60 h (Fig. 4). In L11, a 2- to 3-fold increase of pdiA and tigA mRNA levels was detected at 44 h, and the bipA transcript levels showed a 1.8-fold induction from 30 to 70 h. Taken together, these data suggest that pdiA, bipA, and tigA are responsive to the overexpression of heterologous proteins.
PDIA levels decrease in A. niger strains producing heterologous proteins.
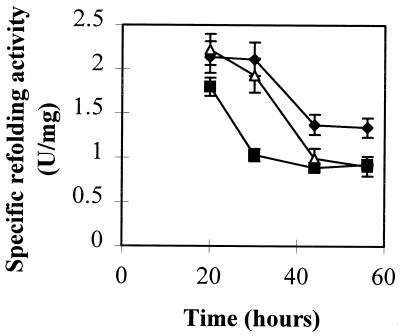
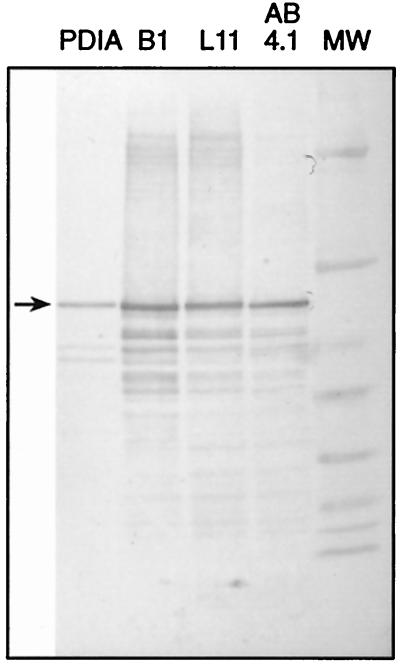
To further investigate the role of PDIA in A. niger strains B1 and L11 during the production of heterologous proteins in A. niger, the levels of PDIA in the microsomal preparations were estimated by a refolding assay and by Western blotting. In A. niger AB4.1, there was an approximately 40% decrease of refolding specific activity over 20 to 56 h (Fig. 5, top panel). In B1 and L11, a more rapid and significant decrease in the amount of refolding specific activity was observed over this period. These results suggest that PDIA associates with secretory proteins during transit through the ER, thereby decreasing the amount of PDIA detected by assay of refolding activity. PDIA protein levels in microsomes prepared from 44-h cultures of AB4.1, B1, and L11 were compared by Western blotting (Fig. 5, bottom panel). B1 samples showed a ca. 30% increase in PDIA levels over AB4.1, while L11 PDIA levels were comparable to those found in AB4.1. These data suggest an association between PDIA and secretory proteins in L11 and B1 which renders a fraction of PDIA unavailable in the refolding assay.
FIG. 5.
(Top) Levels of specific refolding activity (units per milligram of microsomal proteins) in microsomal preparations of A. niger AB4.1 (⧫), B1 (■), and L11 (▵) from 20- to 56-h cultures. (Bottom) Western blot of microsomal extracts from 44-h cultures of A. niger AB4.1, L11, and B1 probed with antiserum to PDI. The arrow indicates the predicted size of PDIA. MW, prestained broad range marker (New England Biolabs). Sizes are (from top to bottom) 83, 62, 47.5, 32.5, 25, 16.5, and 6.5 kDa.
To distinguish whether it is the increased flux of secreted protein or, alternatively, the particular protein which causes the decrease in refolding specific activity, microsomal preparations from 20- and 44-h cultures of A. niger SBD (25) were assayed for refolding activity. This strain secretes levels (ca. 200 mg/liter, mainly glucoamylase) of total protein similar to those secreted by strain B1 in this medium. The amount of refolding specific activity in A. niger SBD closely mimicked that observed for A. niger AB4.1 (data not shown), suggesting that overexpression of glucoamylase did not sequester PDIA to the same extent as lysozyme.
Overexpression of PDIA does not increase secreted yields of proteins.
Northern analysis on 44-h cultures of A. niger LO6, LO9, OB22, and OB38 (constructs in Table 1) showed that pdiA transcript levels increased two- to fourfold, although refolding specific activity from microsomal fractions indicated only a slight increase in the amount of PDIA (data not shown). To ensure that overexpression of PDI did not saturate the ER retention system, causing normal ER-resident proteins in the ER to be secreted, refolding assays were also performed on the supernatants and cytosolic fractions of A. niger LO6, LO9, OB22, and OB38 cultures. Refolding activity was not detected in either the cytoplasmic fractions or culture supernatants.
The effects of overexpression of pdiA on the secreted yields of both a homologous protein, glucoamylase, and a heterologous protein, HEWL, were determined. The amounts of glucoamylase secreted by A. niger L11, LO6, and LO9 did not show any detectable difference at 96 h. Similarly, no effect of pdiA overexpression on the secreted levels of HEWL in these strains was detected. A similar pattern was observed with the B1-derived strains.
Downregulation of PDIA activity in A. niger AS1.1 and AS2.2.
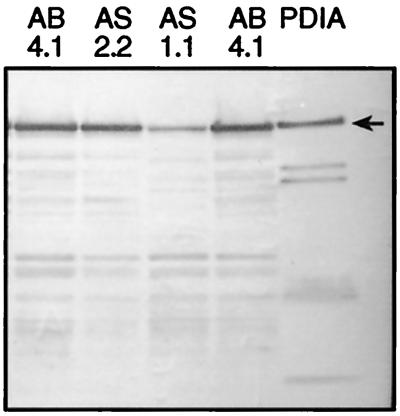
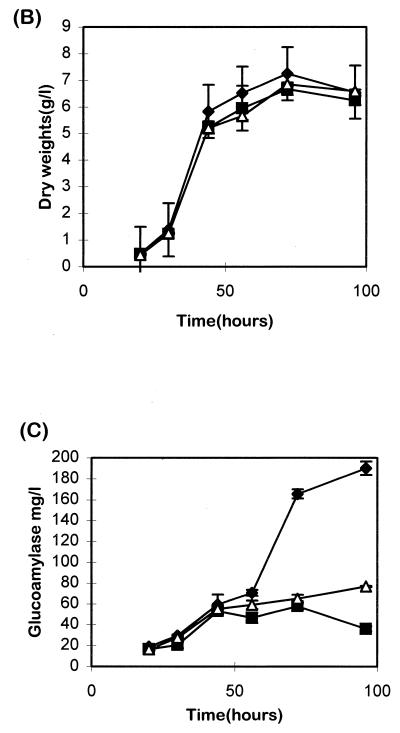
When A. niger AS1.1 and AS2.2 were grown in ACMS/N/P for induction of the glaA promoter driving expression of the antisense pdiA, AS1.1 showed a 45 to 50% decrease in refolding specific activity, and AS2.2 showed a 23 to 30% decrease compared to A. niger AB4.1 (Table 2). Western analysis (Fig. 6A [top panel]) showed reductions in PDIA levels in AS1.1 and AS2.2 of ca. 60 and 5%, respectively, compared to levels in AB4.1. Since PDI in S. cerevisiae is essential for sporulation (10, 22), the effects of downregulating pdiA in A. niger on its ability to sporulate and its growth rate were determined. Both A. niger AS1.1 and AS2.2 were able to sporulate efficiently on either xylose or starch plates. Dry weight determinations of A. niger AS1.1 and AS2.2 grown in ACMS/N/P showed no significant decrease in growth rates and final yields compared to AB4.1 (Fig. 6B). The effects of pdiA downregulation on the secretion of glucoamylase were also examined. Glucoamylase assays showed a significant drop in levels of glucoamylase secreted from 56 h in the ACMS/N/P-grown AS1.1 and AS2.2 cultures (Fig. 6C). At 72 h, AS1.1 showed a 60 to 70% decrease in glucoamylase, and AS2.2 showed a 55 to 65% decrease (Table 2). These results show that PDI plays a role in the secretion of glucoamylase.
TABLE 2.
Comparison of the number of copies of pAS, the specific refolding activities, and the levels glucoamylase secreted by A. niger AB4.1, AS1.1, and AS2.2 grown in ACMS/N/Pa
| A. niger strain | No. of copies of pAS | Specific refolding activity at 44 h (U/mg) | Level of glucoamylase at 72 h (mg/liter) |
|---|---|---|---|
| AB4.1 | 0 | 1.4 ± 0.1 | 166 ± 7 |
| AS1.1 | 6–10 | 0.7 ± 0.1 | 58 ± 6 |
| AS2.2 | 4–6 | 1.0 ± 0.1 | 65 ± 6 |
Refolding activities are presented in units per milligram of microsomal protein. Levels of glucoamylase secreted were assayed from the supernatant of 72-h cultures grown in ACMS/N/P. Values represent the average ± the SEM from triplicate experiments.
FIG. 6.
(A [top panel]) Western blot of microsomal extracts from A. niger AB4.1, AS1.1, and AS2.2 probed with antiserum to PDIA. The arrow indicates the predicted size of PDIA. (B) Dry weights of A. niger AB4.1 (⧫), AS1.1 (■), and AS2.2 (▵) in ACMS/N/P-grown cultures. Values represent the average ± the SEM (bars) from triplicate experiments. (C) Amount of glucoamylase secreted by A. niger AB4.1, AS1.1, and AS2.2 in ACMS/N/P-grown cultures. Values represent the average ± the SEM (bars) of triplicate experiments.
DISCUSSION
Proteins secreted from eukaryotes must be correctly folded, or aggregates may be formed and the unfolded proteins will then be targeted for degradation. Disulfide bond formation plays a critical role in the folding of many proteins in the ER. Since many recombinant proteins of interest contain disulfide bonds, formation of disulfide bonds to obtain protein with a native fold will probably be particularly important in obtaining high yields.
We previously described the gene, pdiA, from A. niger AB4.1 (30), and here we describe the regulation and function of the encoded protein. Although the A. niger ER contains foldases other than PDIA (18), the starch-inducible increase in refolding activity in A. niger PO3 and PO13, which contain extra copies of pdiA under the control of the glaA promoter, strongly suggests that the pdiA gene encodes a functional foldase. Functional analysis of the pdiA gene product was also confirmed by complementation of an S. cerevisiae Δpdi1 mutant. Attempts to delete pdiA in A. niger by gene replacement were unsuccessful.
The transcriptional regulation of pdiA was studied under conditions which perturb the ER through agents which cause the accumulation of misfolded proteins. The transcriptional response of pdiA was compared with that of two genes, bipA and tigA, which encode the ER-specific chaperone BiPA and the PDI family protein TIGA. DTT is a strong reducing agent which disrupts the highly oxidizing environment of the ER and prevents the formation of disulfide bonds. DTT may also chemically reduce disulfide bridges, thus causing accumulation of unfolded proteins in the ER. Treatment of A. niger AB4.1 with DTT produced a significant increase in the transcript level of pdiA and a moderate increase in PDIA protein. However, the change in pdiA mRNA levels is both smaller and delayed relative to that in bipA mRNA levels. This supports the suggestion of Dorner et al. (5) that PDI is not part of the primary cellular response to the accumulation of unfolded proteins in the ER.
The ER is the major intracellular reservoir of Ca2+ ions (21). The Ca2+ ionophore A23187 affects the permeability of cellular membranes to Ca2+ ions, causing their leakage from the ER and subsequent malfolding of secretory proteins (6). Transcript levels from GRP78 are increased in some mammalian systems by 20- to 30-fold when treated with the Ca2+ ionophore A23187 (23). Data obtained with A. niger indicated that only a small increase in pdiA transcripts occurred in the presence of A23187, a finding similar to that obtained with CHO cells (5), whereas tigA and bipA showed a more significant increase. This indicates that pdiA is less sensitive to decreases in Ca2+ concentration in the ER than bipA or tigA, suggesting that the binding affinities of the three ER proteins to Ca2+ ions may vary. This finding is particularly interesting in comparing TIGA with PDIA because both proteins are members of a clearly related thioredoxin-like family and the distinction between their respective roles in vivo is not clear.
In A. niger B1 and L11 strains overexpressing HEWL, transcript levels from pdiA, bipA, and tigA increased in comparison to those in the parent strain, AB4.1. The time of this increase corresponded to the time at which HEWL was detected in the supernatant, suggesting that transcription of pdiA, bipA, and tigA is induced by the accumulation of newly synthesized, unfolded nascent polypeptides in the ER. Refolding activity assays in A. niger B1 and L11 showed a decrease in the amount of refolding specific activity in the ER compared to the parent strain A. niger AB4.1, which does not produce HEWL. A similar finding was reported in S. cerevisiae when overexpression of heterologous proteins decreased extractable BiP and PDI levels (34). The lower refolding specific activities in HEWL-producing transformants is in contrast to the higher levels of pdiA mRNA in these strains. Also, total PDIA protein levels were judged by Western blotting to be higher in strain B1 and at least as high in L11 as in the parent strain AB4.1. As the refolding activity is probably a measure of free PDIA levels, these data suggest that a fraction of the PDIA in HEWL-producing transformants is bound to HEWL and thus inactive in the refolding assay.
Multiple copies of pdiA were introduced into two strains of A. niger, L11 and B1, which naturally secrete different amounts of HEWL. The resulting transformants were analyzed for secreted yields of HEWL and glucoamylase. Although increases in both pdiA transcript levels and refolding activity were observed, the amounts of HEWL and glucoamylase secreted by the transformants were similar to those of the single-copy pdiA A. niger L11 and B1 strains. In addition, the levels of refolding specific activity obtained in the pdiA-overexpressing L11 and B1 strains did not reach the levels found in AB4.1. This is probably due to the effective association of PDIA with HEWL during the folding process within the ER. Refolding assays on the supernatants and cytosolic fractions of selected A. niger LO and OB strains showed that PDIA was not present in the cytosolic fraction or culture media, indicating that the ER-Golgi-HDEL retention or retrieval system was not saturated. We conclude that the amount of folding activity due to PDIA is not a limiting factor in the production of HEWL or the secretion of glucoamylase. It may require the production of proteins with a higher degree of disulfide bonding before PDIA becomes limiting and overproduction of PDIA is needed.
Overexpression of PDI has been successful in increasing the yields of some heterologous proteins from S. cerevisiae and E. coli but not others (16, 33, 37). Coexpression of human PDI in E. coli strains producing antibody Fab fragments only increased secreted yields of one Fab variant, which was more susceptible to incorrect disulfide formation but not the other (16), suggesting that proteins which are susceptible to the formation of kinetically trapped intermediates may require higher concentrations of PDI, and overexpression of PDI will then ease this bottleneck.
Downregulating pdiA was achieved by expressing an antisense pdiA gene under the starch-inducible glaA promoter. Analysis of two antisense transformants revealed a decrease in refolding activity (by up to 50%) and of levels of glucoamylase secreted (by up to 70%) compared to those of A. niger AB4.1. The antisense strains contain up to 11 copies of the glaA promoter driving expression of the single copy glaA gene and the antisense pdiA genes. This number of glaA control sequences is lower than the number (ca. 20) shown to titrate transcription factors controlling this promoter (43, 44). The antisense pdiA results indicate that PDIA plays an important role in the secretion of glucoamylase and also suggest that PDIA levels are optimized to the levels of major extracellular proteins secreted by A. niger.
In conclusion, the gene pdiA isolated from A. niger encodes a PDIA protein which has disulfide isomerase activity and which is involved in the secretion of proteins. pdiA is induced by the accumulation of unfolded proteins in the ER and overproduction of heterologous proteins. Although overexpression of pdiA did not increase the secreted yields of the model protein HEWL in A. niger, this study has demonstrated the function of PDIA in A. niger and shown the importance of refolding activity in the secretion of a heterologous and a homologous protein. As part of the strategy of manipulating the secretory pathway of A. niger, the pdiA gene will play a key part in improving heterologous protein production by filamentous fungi.
ACKNOWLEDGMENTS
We gratefully acknowledge funding for this work from the EU Biotechnology Programme (Eurofung grant BIO4CT96-0535) and the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Archer D B, Jeenes D J, MacKenzie D A, Brightwell G, Lambert N, Lowe G, Radford S E, Dobson C M. Hen egg white lysozyme expressed in, and secreted from, Aspergillus niger is correctly processed and folded. Bio/Technology. 1990;8:741–745. doi: 10.1038/nbt0890-741. [DOI] [PubMed] [Google Scholar]
- 2.Betenbaugh M J, Ailor E, Whiteley E, Hinderlitre P, Hsu T-A. Chaperone and foldase coexpression in the baculovirus-insect cell expression system. Cytotechnology. 1996;20:149–159. doi: 10.1007/BF00350396. [DOI] [PubMed] [Google Scholar]
- 3.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 4.Creighton T E. Disulphide bonds. In: Pain R H, editor. Protein folding. Oxford, England: IRL Press; 1994. pp. 154–163. [Google Scholar]
- 5.Dorner A J, Wasley L C, Raney P, Haugejorden S, Green M, Kaufman R J. The stress response in Chinese hamster ovary cells. J Biol Chem. 1990;265:22029–22034. [PubMed] [Google Scholar]
- 6.Drummond I A S, Lee A S, Resendez E, Jr, Steinhard R A. Depletion of intracellular calcium stores by calcium ionophore A23187 induces the gene for glucose-regulated proteins in hamster fibroblasts. J Biol Chem. 1987;262:12801–12805. [PubMed] [Google Scholar]
- 7.Dunn A, Luz J M, Natalia D, Gamble J A, Freedman R B, Tuite M F. Protein disulphide isomerase (PDI) is required for the secretion of a native disulphide-bonded protein from Saccharomyces cerevisiae. Biochem Soc Trans. 1995;23:78S. doi: 10.1042/bst023078s. [DOI] [PubMed] [Google Scholar]
- 8.Durand H, Clanet M, Tiraby G. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microb Technol. 1988;10:341–346. [Google Scholar]
- 9.Elbe R. A simple and efficient procedure for transformation of yeasts. Bio/Techniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 10.Farquhar R, Honey N, Murant S J, Bossier P, Schultz L. Protein disulfide isomerase is essential for viability in Saccharomyces cerevisiae. Gene. 1991;108:81–89. doi: 10.1016/0378-1119(91)90490-3. [DOI] [PubMed] [Google Scholar]
- 11.Freedman R B, Hirst T R, Tuite M F. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 12.Georgiou G, Valax P. Expression of correctly folded proteins in Escherichia coli. Curr Opin Biotechnol. 1996;7:190–197. doi: 10.1016/s0958-1669(96)80012-7. [DOI] [PubMed] [Google Scholar]
- 13.Gouka R J, Punt P J, Hessing J G M, van den Hondel C A M J J. Analysis of heterologous protein production in defined recombinant Aspergillus awamori strains. Appl Environ Microbiol. 1996;62:1951–1957. doi: 10.1128/aem.62.6.1951-1957.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmsen M M, Bruyne M I, Raue H A, Maat J. Overexpression of binding protein and disruption of the PMR1 gene synergistically stimulate secretion of bovine prochymosin but not plant thaumatin in yeast. Appl Microbiol Biotechnol. 1996;46:365–370. doi: 10.1007/BF00166231. [DOI] [PubMed] [Google Scholar]
- 15.Hijarrubia M J, Casqueiro J, Gutierrez S, Fernandez F J, Martin J F. Characterization of the bip gene of Aspergillus awamori with an HDEL retention signal homologous to the mammalian BiP involved in polypeptide secretion. Curr Genet. 1997;32:139–146. doi: 10.1007/s002940050258. [DOI] [PubMed] [Google Scholar]
- 16.Humphreys D P, Weir N, Lawson A, Mountain A, Lund P A. Co-expression of human protein disulphide isomerase (PDI) can increase the yield of an antibody Fab′ fragment expressed in Escherichia coli. FEBS Lett. 1996;380:194–197. doi: 10.1016/0014-5793(96)00028-2. [DOI] [PubMed] [Google Scholar]
- 17.Jeenes D J, MacKenzie D A, Archer D B. Transcriptional and post-transcriptional events affect the production of secreted hen egg white lysozyme by Aspergillus niger. Transgenic Res. 1994;3:297–303. doi: 10.1007/BF01973589. [DOI] [PubMed] [Google Scholar]
- 18.Jeenes D J, Pfaller R, Archer D B. Isolation and characterization of a stress inducible PDI-family gene from Aspergillus niger. Gene. 1997;193:151–156. doi: 10.1016/s0378-1119(97)00098-x. [DOI] [PubMed] [Google Scholar]
- 19.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 21.Koch G, Smith M, Macer D, Webster P, Mortara R. Endoplasmic reticulum contains a common, abundant calcium-binding glycoprotein, endoplasmin. J Cell Sci. 1986;86:217–232. doi: 10.1242/jcs.86.1.217. [DOI] [PubMed] [Google Scholar]
- 22.Laboissiere M C A, Sturley S L, Raines R T. Protein disulfide isomerase in spore germination and cell division. Biol Chem. 1997;378:431–437. doi: 10.1515/bchm.1997.378.5.431. [DOI] [PubMed] [Google Scholar]
- 23.Lee A S. Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem Sci. 1987;12:20–23. [Google Scholar]
- 24.Lee B R, Yamada O, Kitamoto K, Takahashi K. Cloning, characterization and overexpression of a gene (pdiA) encoding protein disulfide isomerase of Aspergillus oryzae. J Ferment Bioeng. 1996;82:538–543. [Google Scholar]
- 25.Le Gal-Coëffet M F, Jacks A J, Sorimachi K, Williamson M P, Williamson G, Archer D B. Expression in Aspergillus niger of the starch-binding domain of glucoamylase. Comparison with the proteolytically produced starch-binding domain. Eur J Biochem. 1995;233:561–567. doi: 10.1111/j.1432-1033.1995.561_2.x. [DOI] [PubMed] [Google Scholar]
- 26.Lyles M M, Gilbert H F. Catalysis of the oxidative folding of ribonuclease A by protein disulphide isomerase: pre-steady-state kinetics and the utilization of the oxidizing equivalents of the isomerase. Biochemistry. 1991;30:619–625. doi: 10.1021/bi00217a005. [DOI] [PubMed] [Google Scholar]
- 27.MacKenzie D A, Gendron L C G, Jeenes D J, Archer D B. Physiological optimization of secreted protein production by Aspergillus niger. Enzyme Microb Technol. 1994;16:276–279. doi: 10.1016/0141-0229(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 28.Marquardt T, Helenius A. Misfolding and aggregation of newly synthesized protein in the endoplasmic reticulum. J Cell Biol. 1992;117:505–513. doi: 10.1083/jcb.117.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrice J, MacKenzie D A, Parr A J, Archer D B. Isolation and characterisation of the acetyl-CoA carboxylase gene from Aspergillus nidulans. Curr Genet. 1998;34:379–385. doi: 10.1007/s002940050410. [DOI] [PubMed] [Google Scholar]
- 30.Ngiam C, Jeenes D J, Archer D B. Isolation and characterisation of a gene encoding protein disulphide isomerase, pdiA, from Aspergillus niger. Curr Genet. 1997;31:133–138. doi: 10.1007/s002940050187. [DOI] [PubMed] [Google Scholar]
- 31.Punt P J, Oliver R P, Dingemanse M A, Pouwels P H, van den Hondel C A M J J. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 32.Punt P J, van Gemeren I A, Drint-Kuijvenhoven J, Hessing J G M, van Muijlwijk-Harteveld G M, Beijersbergen A, Verrips C T, van den Hondel C A M J J. Analysis of the role of the major ER chaperone-encoding gene bipA in the secretion of homologous and heterologous proteins in black aspergilli. Appl Microbiol Biotechnol. 1998;50:447–454. doi: 10.1007/s002530051319. [DOI] [PubMed] [Google Scholar]
- 33.Robinson A S, Hines V, Wittrup K D. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Bio/Technology. 1994;12:381–384. doi: 10.1038/nbt0494-381. [DOI] [PubMed] [Google Scholar]
- 34.Robinson A S, Wittrup K D. Constitutive overexpression of secreted heterologous proteins decreases extractable BiP and protein disulfide isomerase levels in Saccharomyces cerevisiae. Biotechnol Prog. 1995;11:171–177. doi: 10.1021/bp00032a009. [DOI] [PubMed] [Google Scholar]
- 35.Ruijter G J G, Panneman H, Visser J. Overexpression of phosphofructokinase and pyruvate kinase in citric acid-producing Aspergillus niger. Biochim Biophys Acta. 1997;1334:317–326. doi: 10.1016/s0304-4165(96)00110-9. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schultz L D, Markus H Z, Hofmann K J, Montgomery D L, Dunwiddie C T. Using molecular genetics to improve the production of recombinant proteins by the yeast Saccharomyces cerevisiae. Ann N Y Acad Sci. 1994;721:148–157. doi: 10.1111/j.1749-6632.1994.tb47387.x. [DOI] [PubMed] [Google Scholar]
- 38.Straffon M J, Hynes M J, Davis M A. Characterization of the ugatA gene of Ustilago maydis, isolated by homology to the gatA gene of Aspergillus nidulans. Curr Genet. 1996;29:360–369. doi: 10.1007/BF02208617. [DOI] [PubMed] [Google Scholar]
- 39.van den Hondel C A M J J, Punt P J, van Gorcom R F M. Heterologous gene expression in fungi. In: Bennett J W, Lasure L L, editors. More genetic manipulations of filamentous fungi. Orlando, Fla: Academic Press; 1991. pp. 396–428. [Google Scholar]
- 40.van Gemeren I A, Punt P J, Drint-Kuyvenhoven A, Broekhuijsen M P, van't Hoog A, Beijersbergen A, Verrips C T, van den Hondel C A M J J. The ER chaperone encoding bipA of black aspergilli is induced by heat shock and unfolded proteins. Gene. 1997;198:43–52. doi: 10.1016/s0378-1119(97)00290-4. [DOI] [PubMed] [Google Scholar]
- 41.van Hartingsveldt W, Mattern I E, van Zeijl C M, Pouwels P H, van den Hondel C A M J J. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet. 1987;206:71–75. doi: 10.1007/BF00326538. [DOI] [PubMed] [Google Scholar]
- 42.van Zeijl C M J, van de Kamp E H M, Punt P J, Selten G C M, Hauer B, van Gorcom R F M, van den Hondel C A M J J. An improved colony-PCR method for filamentous fungi for amplification of PCR-fragments of several kilobases. J Biotechnol. 1998;59:407–418. doi: 10.1016/s0168-1656(97)00170-3. [DOI] [PubMed] [Google Scholar]
- 43.Verdoes J C, Punt P J, Schrickx J M, van Verseveld H W, Stouthamer A H, van den Hondel C A M J J. Glucoamylase overexpression in Aspergillus niger: molecular genetic analysis of strains containing multiple copies of the glaA gene. Transgenic Res. 1993;2:84–92. doi: 10.1007/BF01969381. [DOI] [PubMed] [Google Scholar]
- 44.Verdoes J C, Punt P J, Stouthamer A H, van den Hondel C A M J J. The effects of multiple copies of the upstream region on expression of the Aspergillus niger glucoamylase-encoding gene. Gene. 1994;145:179–187. doi: 10.1016/0378-1119(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 45.Ward L C, Omura S, Kopito R R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 46.Woods D R, Bevan E A. Studies on the nature of the killer factor produced by Saccharomyces cerevisiae. J Gen Microbiol. 1968;51:115–125. doi: 10.1099/00221287-51-1-115. [DOI] [PubMed] [Google Scholar]