Abstract
Actinin alpha 4 (ACTN4) is expressed in the kidney podocytes. ACTN4 gene methylation in patients with diabetic nephropathy (DN) remains high. Underlying mechanism of epigallocatechin-3-gallate (EGCG) inducing ACTN4 demethylation, and its inhibitory effect on DN renal fibrosis remains unclear.
Methods:
Human podocyte cell line, HPC, was treated with high glucose to establish model of DN. The levels of cytokines, vascular endothelial growth factor (VEGF) and interleukin (IL)-8, and fibrosis markers, alpha smooth muscle actin (α-SMA) and fibronectin (FN), were determined using enzyme-linked immunosorbent assay. HPC cells were treated with EGCG, and cell viability was determined by MTT assay, ACTN4 gene methylation was analyzed by MSP. mRNA and protein expression levels were measured using RT-qPCR and Western blotting, respectively.
Results:
Actinin alpha 4 gene promoter was hypermethylated in the high glucose-treated groups. EGCG reversed the hypermethylated status of ACTN4, along with the upregulation of ACTN4 levels and downregulation of DNA methyltransferase 1 (DNMT1), NF-κB p65, p-NF-κB p65, IκB-α, VEGF, IL-8, α-SMA, and FN levels (P<.05).
Conclusion:
Epigallocatechin-3-gallate reduced hypermethylation of ACTN4 in HPC cells by downregulating DNMT1 expression and restoring ACTN4 expression, contributing to the upregulation of the NF-KB p65, p-NF-KB p65, IKB-α, VEGF, IL-8, α-SMA, and FN levels (P<.05).
Keywords: epigallocatechin gallate; actinin alpha 4; methylation; NF-KB signaling pathway,; diabetic nephropathy
Introduction
Diabetic nephropathy (DN) is a common refractory complication that increases the mortality of patients with diabetes. 1 Early morphological abnormalities are characterized by glomerular hypertrophy, thickening of the glomerular basement membrane, and production of mesangial extracellular matrix, which develops into glomerulosclerosis and tubulointerstitial fibrosis, eventually leading to kidney failure.2,3 Therefore, determining the key mechanism of DN development and developing an effective treatment method for delaying or halting DN progression is crucial. Recently, mounting evidence has demonstrated that the mechanism of DN is related to genetic abnormalities and epigenetic changes. DNA methylation is one of the most common epigenetic changes, and numerous studies have shown that gene hypermethylation promotes the occurrence of DN.4-7 The number of differentially methylated regions (DMRs) in DN is significantly higher than that in diabetes without nephropathy. 8 Therefore, reducing the hypermethylation status represents a novel approach for DN treatment.
The current management for reducing the hypermethylation status focuses on DNA methyltransferase (DNMT) inhibitors. DNMTs play an important role in DNA methylation, while DNMT inhibitors reduce hypermethylation and restore the expression of silenced genes. 9 However, the available DNMT inhibitors, such as 5-Aza-2′-deoxycytidine, lack target specificity, have high side effects, and are toxic to healthy cells. 10 Compared with these synthetic inhibitors, natural products exhibit low toxicity and fewer side effects and are cost-effective, emerging as targeted epigenetic modulators. 11 Green tea is a natural drink that is popular worldwide. The major chemical component of green tea, epigallocatechin-3-gallate (EGCG), can inhibit various inflammatory, toxic, proliferative, and angiogenic effects.12,13 EGCG forms hydrogen bonds with the GLU1265, Arg1310, Arg1311, and Lys1482 amino acid residues in the catalytic site of DNMT, which directly inhibits the function of DNMT and reduces the hypermethylation status of genes. 14
Actinin alpha 4 (ACTN4) belongs to the actin family and is highly expressed in podocytes and the vascular wall of the kidney. 15 Mutations in human ACTN4 gene cause autosomal dominant focal segmental glomerulosclerosis. 16 A high glucose environment can decrease the expression of ACTN4 and lead to the development of proteinuria. 17 Furthermore, the expression levels of ACTN4 in patients with DN were significantly lower than those in healthy individuals. 18 The methylation levels of ACTN4 were higher in patients with DN than in those without DN; in particular, DMR in the promoter region of ACTN4 exhibited high methylation status in patients with DN, indicating that differential methylation of ACTN4 is involved in the occurrence and development of DN. 5 In this study, we constructed a DN cell model to investigate whether EGCG could induce the demethylation of ACTN4 and inhibit renal fibrosis in DN, and to determine the underlying molecular mechanisms.
Methods
Cell Culture
The human podocyte cell line, HPC, was obtained from the National Collection of Authenticated Cell Cultures. Cells were maintained in 1640 medium (Cat. No. 11875101; Gibco, USA) containing 10% fetal bovine serum (Cat. No. 10439016; Life Technologies/Thermo Fisher Scientific; USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Cat. No. 15140122; Gibco, USA) in 5% CO2 at 37°C in a humid environment.
Construction of a Cell Model of DN
HPC cells were cultured in 5.5 and 30 mmol/L glucose medium, and when the cell growth density reached 80–90%, the cells incubated with high glucose were used to establish a cell model of DN. The construction of the cell model was evaluated by determining the levels of vascular endothelial growth factor (VEGF), interleukin (IL)-8, alpha smooth muscle actin (α-SMA), and fibronectin (FN) using enzyme-linked immunosorbent assay (ELISA). The cells were then treated with EGCG at different concentrations (25, 50, 75, 100, and 150 μg/mL) and the levels of VEGF, IL-8, α-SMA, and FN were determined using ELISA.
Cell Viability
HPC cells at a density of 5 x 104 cells were seeded into a 96-well plate and treated with EGCG at various concentrations (25, 50, 75, 100, and 150 μg/mL) for 24 h. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide solution (20 μL/well) was used to incubate the cells at 37°C for 2° h. Then, the supernatant was aspirated after centrifugation at 1000 rpm for 10 min and washed once with phosphate-buffered saline, and 150 μL dimethyl sulfoxide was added to dissolve the formazan crystals. The optical density value was measured at a wavelength of 570 nm using a microplate reader (Bio-Rad, Hercules, CA, USA), which was used to calculate the relative cell viability.
Isolation of DNA
Total genomic DNA was isolated using a TIANamp Genomic DNA Kit (Cat. No. DP304; TIANGEN, China), according to the manufacturer’s protocol, to determine the promoter methylation profile of ACTN4. The purity and amount of the extracted genomic DNA were evaluated using a Nanodrop microspectrophotometer (Thermo Fisher, USA).
Actinin Alpha 4 Promoter Methylation
Methylation-specific polymerase chain reaction (MSP) was performed to analyze the methylation profile of the promoter regions of ACTN4 in HPC cells. MethPrimer program (http://www.urogene.org/methprimer/index1.html) was used to analyze the presence of CpG islands in the promoter regions of ACTN4 and to design the primers for MSP. Nucleotide sequences of ACTN4 gene promoter regions were obtained from the National Center for Biotechnology Information database. The sequences of methyl-specific primers of ACTN4 were: F: 5′-AGTTTGGTTAATATGGTGAAATTTC-3′ and R: 5′-CAAACTAAAATACAATAACGCGAT-3′
The sequences of the non-methyl-specific primers of ACTN4 were: F: 5′-TTTGGTTAATATGGTGAAATTTTGT-3′ and R: 5′-TCTCCCAAACTAAAATACAATAACACA-3′. EZ-96 DNA methylation gold (Cat. No. ZD5023; Zymo Research, USA) was used to bisulfite-treat the isolated genomic DNA. After sodium bisulfite conversion, the parameters of amplification of PCR reaction were applied as follows: preliminary denaturation for 3 min at 95°C, 35 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 55°C, extension for 30 s at 72°C, and final extension for 4 min at 72°C. A 1.5% agarose gel was prepared and PCR products were detected via electrophoresis.
Reverse Transcription-Quantitative PCR
Various concentrations of EGCG (25, 50, 75, 100, and 150 μg/mL) were used to treat the HPC cells for 48 h. TRIzol Reagent (Cat. No. 15596026; Thermo Fisher Scientific) was used to isolate the nuclear and cytoplasmic RNA, which was then reverse transcribed into cDNA using the iScript cDNA synthesis kit (Cat. No. 170889017; Bio-Rad, Carlsbad, CA, USA), according to the manufacturer’s protocol. SYBR premix Ex Taq (Cat. No. RR820A; Takara) was used to perform quantitative PCR on an ABI 7900HT real-time PCR system. The mRNA of β-actin was used as the internal control for normalization. Primer 5 software (Premier; USA) was used to design the primers. The primers used for PCR are listed in Table 1. The relative RNA expression was determined using the 2−ΔΔCt method.
Table 1.
The Primers Used in RT-qPCR.
| Gene | Up-stream primer | Down-stream primer |
|---|---|---|
| ACTN4 | AGCAAATGCAGGAGTTCCGA | GCCAAGGGAGTCTGTTCAGG |
| DNMT1 | AAGAATGGTGTTGTCTACCGAC | CATCCAGGTTGCTCCCCTTGGATGG |
| NF-κB p65 | CATGCCAGTGAGAATGTATGCCAT | ACGCAGGAGACGGAAGAATAAAT |
| IκBα | TGACCATGGAAGTGATTGGTCAG | GATCACAGCCAAGTGGAGTGGA |
| VEGF | TGGCAAAACTGCACCTTCACA | TGGCAAAACTGCACCTTCACA |
| IL-8 | CCAGGAAGAAACCACCGGAAG | TGGCAAAACTGCACCTTCACA |
| α-SMA | TCTTGACCTGGCTGGACG | TCTCCTTGATGTCTCGCACA |
| FN | GCCATTTGCTCCTGCA | CAATTGGGCAATTAACATTA |
| IL-10 | CACCTACTTCCCAGCCAACC | TCAGCAGAGACTCACTCA |
| BMP-7 | AGACGCCAAAGAACCAAGAG | GCTGTCGTCGAAGTAGAGGA |
| β-actin | TGGCACCCAGCACAATGAA | CTAAGTCATAGTCCGCCTAGAAGCA |
Western Blotting
Cells were harvested, washed with PBS, and dissolved in the radioimmunoprecipitation assay buffer (Cat. No. P0013C; Beyotime, China), and bicinchoninic acid protein assays were performed to measure the protein concentrations. Total cellular protein was separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were then transferred from the gel onto polyvinylidene fluoride (PVDF) membranes using an electroblotting apparatus. PVDF membranes were then incubated with Tris-buffered saline with Tween (TBST) containing 5% skim milk at 25°C for 1 h. The membranes were incubated with primary antibodies at 4°C overnight. The primary antibodies used in this study were mouse monoclonal anti-ACTN4 (Cat. No. sc-390205; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-DNMT1 (Cat. No. sc-271729; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-nuclear factor (NF)-κB p65 (Cat. No. sc-8008; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-p-NF-κB p65 (Cat. No. sc-13579; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-inhibitor κB (IκB)-α (Cat. No. sc-1643; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-VEGF (Cat. No. sc-57496; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-IL-8 (Cat. No. sc-376750; Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-α-SMA (Cat. No. sc-53142; Santa Cruz Biotechnology), and mouse monoclonal anti-FN (Cat. No. sc-8422; Santa Cruz, USA) antibodies. Membranes were incubated with primary antibodies overnight at 4°C and then washed with TBST, followed by incubation with a secondary antibody at room temperature for 2 h. ECL detection reagent (Cat. No. 32106; Thermo Fisher Scientific, USA) was used to visualize the protein bands. Image Lab software 1.8.0 (Bio-Rad, USA) was used for densitometric quantification.
Statistical Analysis
All experiments were repeated at least three times, and data are presented as the mean ± standard deviation. A group t-test was used for two-sample mean comparison, while differences among multiple groups were compared using one-way analysis of variance. SPSS 17.0 software was used to perform all statistical analyses. P < .05 was considered to be statistically significant (*P < .05).
Results
Secreted Levels of VEGF and IL-8 in HPC Cells Under High Glucose Exposure and EGCG Treatment
The secreted levels of serum fibrosis markers, α-SMA and FN, and inflammatory cytokines, VEGF, and IL-8, were significantly increased in the high glucose-treated HPC cells (HG group) compared to those in the normal glucose-treated HPC cells (NG group), suggesting that the cell model of DN was successfully constructed. The levels of α-SMA, FN, IL-8, and VEGF were found to be significantly decreased in the HG group after EGCG treatment (Table 2).
Table 2.
The Secretion Levels (ng/L) of VEGF and IL-8 in the HK-2 Cells (Mean ± SD. n = 3).
| α-SMA | FN | VEGF | IL-8 | |
|---|---|---|---|---|
| NG | 534 ± 46 | 3865 ± 103 | 110 ± 10 | 6046 ± 184 |
| HG | 1747 ± 120 | 6762 ± 214 | 478 ± 16 | 12,975 ± 593 |
| EGCG (25 μg/mL) | 1567 ± 88 | 6409 ± 180 | 404 ± 16 | 11,246 ± 594 |
| EGCG (50 μg/mL) | 1324 ± 120 | 5961 ± 259 | 370 ± 13 | 9887 ± 403 |
| EGCG (75 μg/mL) | 1038 ± 74 | 5604 ± 186 | 326 ± 25 | 8787 ± 426 |
| EGCG (100 μg/mL) | 884 ± 80 | 4978 ± 211 | 264 ± 19 | 7695 ± 455 |
| EGCG (150 μg/mL) | 664 ± 78 | 4527 ± 163 | 162 ± 25 | 6596 ± 401 |
Treatment with EGCG Influences HPC Cell Viability
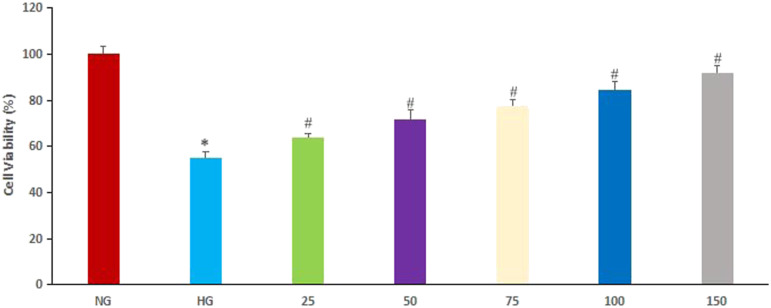
As shown in Figure 1, cell viability in the HG group was lower than that in the NG group. However, various concentrations of EGCG increased the cell viability of HPC cells in the HG group in a concentration-dependent manner compared to the cell viability of HPC cells treated with HG but without EGCG (P < .05).
Figure 1.
Effect of epigallocatechin-3-gallate (EGCG) on cell viability. Normal glucose (NG), cells treated with 5.5 mmol/L glucose; High glucose (HG), cells treated with 30 mmol/L glucose, 25: 25 μg/mL EGCG, 50: 50 μg/mL EGCG, 75: 75 μg/mL EGCG, 100: 100 μg/mL EGCG, 150: 150 μg/mL EGCG. (*P < .05, #P < .01 vs NG group).
Methylation of the ACTN4 Gene Promoter
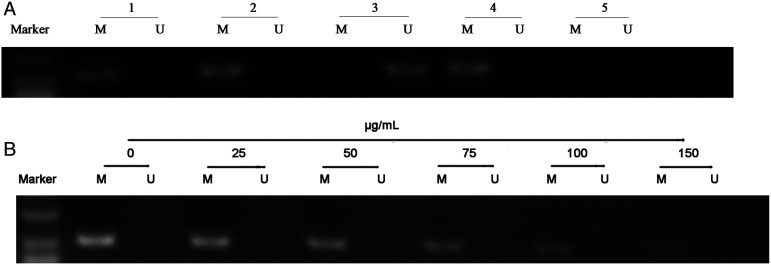
Genomic DNA was extracted from HG and NG groups using a methylation kit. PCR amplification was performed using the extracted genomic DNA as templates, and electrophoresis was performed to analyze the ACTN4 gene promoter. We observed significantly high ACTN4 promoter CpG methylation in the cell model of DN in the NG group (Figure 2A). To further explore the possibility that EGCG could reverse the methylation of the ACTN4 gene promoter in the DN cell model, we tested the methylation status of ACTN4 in the HG and NG groups. As shown in Figure 2B, EGCG reversed the hypermethylated status of ACTN4 in HPC cells of HG group in a dose-dependent manner. The gray scan results of PCR are shown in Table 3.
Figure 2.
Methylation status of actinin alpha 4 (ACTN4) in HPC cells and ACTN4 gene demethylation induced by EGCG. M: Methylation-specific PCR. U: Unmethylation-specific PCR. (A): Methylation status of ACTN4 in HPC cells. Line 1: Methylation status of ACTN4 in HPC without HG treatment. Line 2: Methylation status of ACTN4 in the diabetic nephropathy (DN) cell model. Line 3: Unmethylated positive control. Line 4: Methylated positive control. Line 5: Negative Control. (B): ACTN4 demethylation induced by EGCG.
Table 3.
Gray Scan Results of MSP (Mean ± SD. n = 3).
| Methylation-specific | Unmethylation-specific | |
|---|---|---|
| Line1 (Figure 2A) | 16,697.7 ± 156.3 | 0 |
| Line2 (Figure 2A) | 15,420 ± 381.1 | 0 |
| Line3 (Figure 2A) | 0 | 14,552.7 ± 125.2 |
| Line4 (Figure 2A) | 16,687.7 ± 82.6 | 0 |
| HG (Figure 2B) | 18,593.3 ± 491.9 | 0 |
| EGCG (25 μg/mL) (Figure 2B) | 17,425.3 ± 256.3 | 0 |
| EGCG (50 μg/mL) (Figure 2B) | 16,411.7 ± 266.6 | 0 |
| EGCG (75 μg/mL) (Figure 2B) | 15,268.7 ± 215.8 | 0 |
| EGCG (100 μg/mL) (Figure 2B) | 14,346.3 ± 269.6 | 0 |
| EGCG (150 μg/mL) (Figure 2B) | 13,726.7 ± 395.2 | 0 |
mRNA Expression Levels of ACTN4, DNMT1, NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, FN, IL-10, and Bone Morphogenetic Protein (BMP)-7
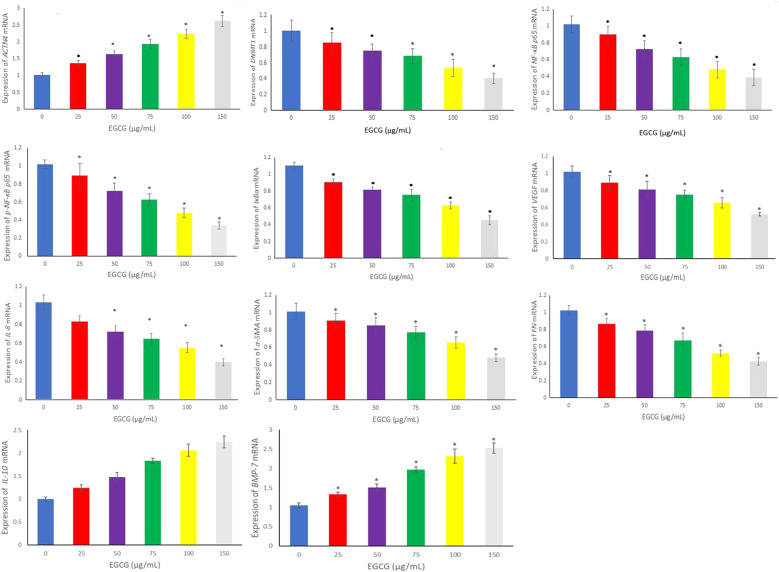
To assess ACTN4 mRNA expression in the cell model of DN, we extracted total RNA and used quantitative fluorescence PCR to detect the expression of ACTN4 mRNA. The results showed ACTN4 mRNA expression was decreased in the cell model of DN (.673 ± .086), but not in 5.5 mmol/L glucose-treated HPC cells (1.021 ± .056). These results suggest that ACTN4 is commonly expressed in the DN cell model and is inversely associated with promoter region methylation. To further explore the possibility that EGCG influences the expression of ACTN4 mRNA and its underlying mechanism, we used various concentrations of EGCG to analyze the mRNA expression levels of ACTN4, DNMT1, NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, FN, IL-10, and BMP-7. The results showed that the mRNA expression levels of ACTN4, IL-10, and BMP-7 were significantly higher, while those of DNMT1, NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, and FN were downregulated in the DN cell model compared with the NG group (P < .05, Figure 3).
Figure 3.
mRNA expression levels of ACTN4, DNA methyltransferase 1 (DNMT1), nuclear factor (NF)-κB p65, p-NF-κB p65, inhibitor κB (IκB)-α, vascular endothelial growth factor (VEGF), interleukin (IL)-8, alpha smooth muscle actin (α-SMA), fibronectin (FN), IL-10, and bone morphogenetic protein (BMP-7) were determined by RT-qPCR in the cell model of DN treated with EGCG (*P < .05 vs Control group, n = 3).
Protein Expression Detected via Western Blotting
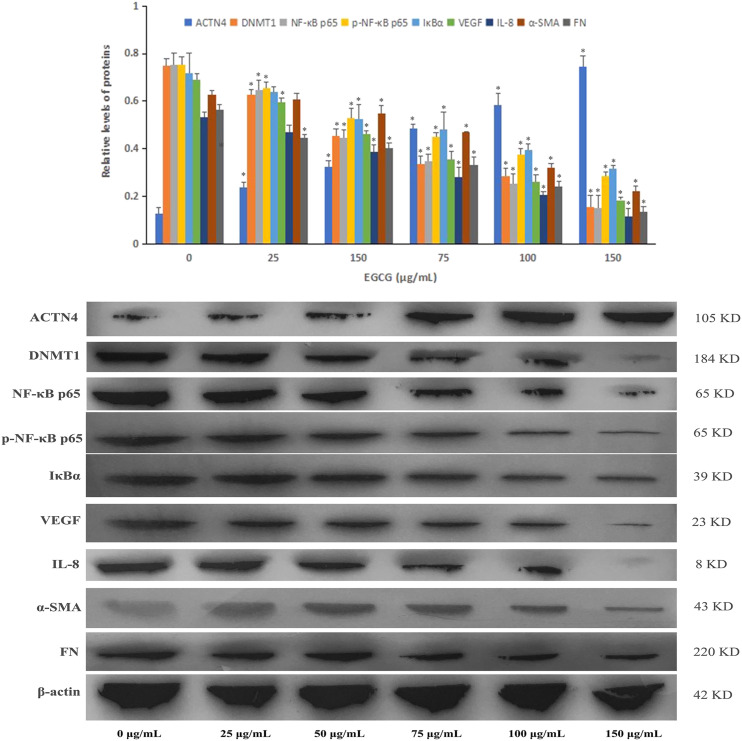
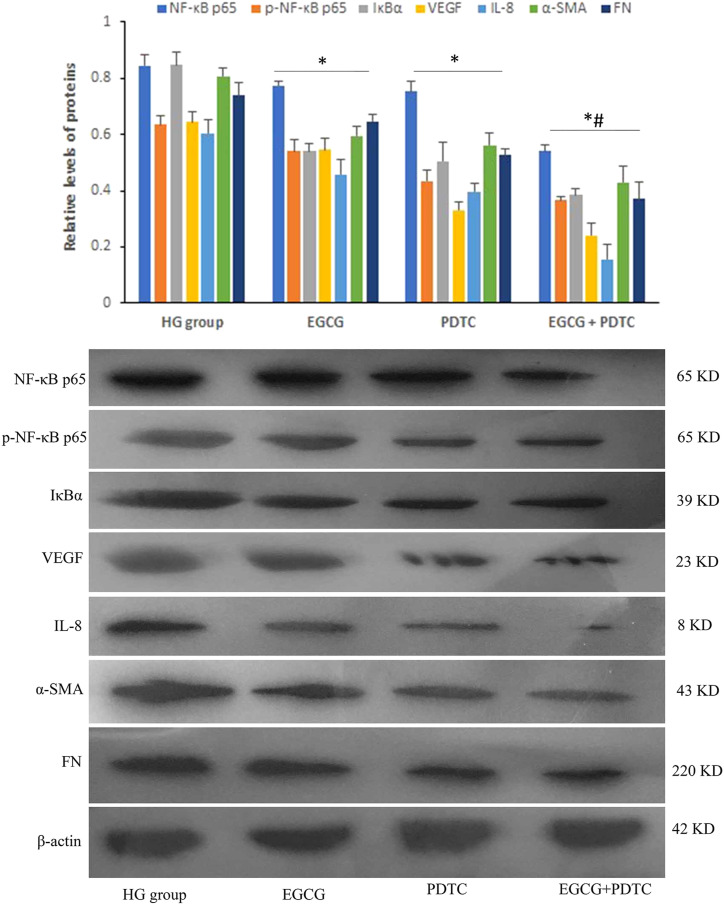
Western blotting was performed to determine the protein expression in HPC cells treated with or without various concentrations of EGCG. Protein levels of ACTN4 were increased with EGCG treatment in the DN cell model compared with the NG group at 150 μg/mL EGCG (Figure 4). In contrast, DNMT1, NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, and FN protein levels were moderately decreased in the DN cell model treated with EGCG, and the protein expression levels were downregulated in a dose-dependent manner (P < .05). After HPC cells were treated with 20 μmol/L ammonium 1-pyrrolidinedithiocarbamate (PDTC), an inhibitor of NF-κB, the protein expression levels of NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, and FN were decreased in the cell model of DN, and the combination of EGCG and PDTC exhibited a significant reduction in protein expression compared with PDTC treatment alone (Figure 5).
Figure 4.
Protein expression levels of ACTN4, DNMT1, NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, and FN were determined by Western blotting in the cell model of DN treated with EGCG (*P < .05 vs Control group, n = 3).
Figure 5.
Protein expression levels of NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, and FN were determined by Western blotting in the cells treated with EGCG and pyrrolidinedithiocarbamate (PDTC) (*P < .05 vs HG group, n = 3; #P < .05 vs PDTC group).
Discussion
Diabetic nephropathy is a fatal complication of diabetes and the main cause of end-stage renal disease (ESRD). 19 DN is one of the most challenging health problems worldwide. Although the therapeutic strategies for DN have remarkably improved in recent years, nearly half of the patients inevitably develop ESRD. 20 Renal fibrosis is a common pathway of ESRD in the vast majority of chronic kidney diseases. Various factors, such as hypertension, inflammation, hyperglycemia, hyperlipidemia, and drug damage, cause renal fibrosis. 21 However, the molecular mechanisms underlying high glucose-induced renal fibrosis remain unclear. Therefore, studying this mechanism is expected to provide a theoretical basis for identifying effective treatment targets for DN.
Epigenetic regulation plays a crucial role in DN occurrence. 5 As one of the most important epigenetic modifications, DNA methylation contributes to the pathogenesis of renal fibrosis and causes DN via phenotypic alterations and modifications in functional gene expression. 6 DNA methylation alteration induced by high glucose levels plays an important role in the pathogenesis of DN by altering peripheral leukocyte DNA or salivary cell DNA in patients with DN. 7 DNA methylation refers to a chemical reaction in which the cytosine (C) in CpG dinucleotides is methylated into 5-methylated cytosine (5−MC) with methyl groups from S-adenosine methionine via DNMTs. 10 The methylation status of eukaryotic DNA is related to gene transcription regulation. During DN formation, DNA methylation patterns, including partial hypermethylation of genes, change significantly. 7 A study revealed that the methylation status of 6246 genes in patients with DN was significantly increased compared to that in diabetic patients without DN, while the methylation status of 3680 genes in DN was higher than that in control groups. 22 The overall CpG methylation level in DN patients was 72.62%, which was higher than that in diabetic patients without DN and the control group, and Maghbooli et al 23 obtained similar results. Dayeh et al demonstrated that the ATP binding cassette subfamily G member 1 (ABCG1) gene methylation was associated with a 9% increased risk of future diabetes morbidity, and silencing the expression of ABCG1 via hypermethylation caused an accumulation of cholesterol, leading to atherosclerosis, glucose-stimulated insulin secretion impairment, and inflammation of islet beta cells. 24 Marumo et al also revealed that kinesin family member 20B, claudin 18, and solute carrier organic anion transporter family member 1A1 exhibited hypermethylation in DN mice compared to the control groups. 25
Actinin alpha 4 plays a crucial role in kidney diseases. 26 In several kidney diseases, including DN, the protein expression levels of ACTN4 are reduced in kidney tissues, which have been reported in some animal and clinical studies. ACTN4 downregulation is closely associated with the development of diabetes. 27 ACTN4 can alleviate inflammatory and fibrosis impairment in DN. 28 In this study, we obtained similar results; the protein expression of ACTN4 was decreased in the cell model of DN induced by high glucose conditions. Moreover, ACTN4 gene hypermethylation was observed in diabetic patients with DN and DMR in the promoter region was highly methylated, suggesting that ACTN4 aberrant methylation is associated with the pathogenesis and progression of DN. Therefore, reducing hypermethylation may be a new strategy for the treatment of DN.
Numerous studies have reported that 5-aza-2′-deoxycytidine reverses methylation and alleviates diabetic kidney inflammation in DN. 29 However, 5-aza-2′-deoxycytidine and other chemical compounds exhibit unpredictable side effects and cell toxicity. 10 The identification of new agents for reversing DNA methylation appears to be particularly important. Natural products that inhibit DNMT activity exhibit some advantages, such as minor side effects and low toxicity. Green tea exhibits both physiological and pharmacological effects. As the major polyphenol of green tea, EGCG is a potential demethylating agent that directly inhibits DNMT, demethylates CpG islands, and reactivates the methylation-silenced genes.12,13 In 2020, Yang et al 30 revealed that EGCG treatment significantly reduced the levels of Klotho promoter methylation under high glucose conditions. In addition, EGCG treatment exerts a protective effect on the Klotho protein by upregulating its expression. However, the mechanism by which EGCG exerts its protective effects remains unclear.
In our study, we established a cell model of DN and detected the secreted levels of serum fibrosis markers, α-SMA and FN, and inflammatory cytokines, VEGF, and IL-8, to evaluate the successful construction of the DN cell model. While the DN cell model was treated with EGCG, the levels of α-SMA, FN, IL-8, and VEGF were significantly decreased, indicating that EGCG plays a role in inhibiting fibrosis and inflammation in DN. In addition, EGCG increased the viability of HPC cells treated with high glucose, possibly by the alleviation of fibrosis and inflammation. ACTN4 is rich in CpG islands, indicating that the promoter of ACTN4 gene is closely related to DNA methylation. MSP assay revealed that the level of ACTN4 gene methylation in the DN cell model was higher than that in normal glucose-treated cells. In the DN cell model, the protein expression of ACTN4 was downregulated, while that of DNMT1 was upregulated. However, EGCG treatment reversed these effects, indicating that EGCG reduced the expression of DNMT1, thereby demethylating the ACTN4 gene promoter to reverse the protein expression of ACTN4. Similar to our results, Yang et al found that the protein expression levels of DNMT1 and DNMT3a were increased in diabetic kidney disease. 30 Moreover, Liu et al found that the protein expression of DNMT3b was increased in chronic diabetic kidney disease. 31
Renal inflammation and fibrosis are associated with DN pathogenesis and progression. The secretion of cytokines, VEGF, and IL-8, and the expression levels of fibrosis markers, α-SMA, and FN, were significantly increased in DN. 19 The glomeruli were hypertrophied and the mesangial matrix was broadened, eventually leading to ESRD due to the increase in α-SMA and FN levels. 20 In contrast, some anti-inflammatory and antifibrotic cytokines have been shown to alleviate the development and progression of DN. In Wang’s study, schisandrin C exerted protective effects against DN by increasing the expression of the anti-inflammatory cytokine, IL-10, in macrophages. 32 Meng et al revealed that lncRNA zinc finger E-box binding homeobox 1 antisense RNA 1 (ZEB1-AS1) inhibits high glucose-induced fibrogenesis by improving the expression level of BMP7 in DN, which provides a novel target for DN therapy. 33 In this study, we demonstrated that EGCG significantly reduced the expression levels of inflammatory and fibrotic markers, while increasing the expression levels of anti-inflammatory and anti-fibrotic cytokines by upregulating the protein expression of ACTN4, suggesting that the reversal of ACTN4 protein expression can alleviate the cell damage induced by high glucose treatment.
However, the molecular mechanism by which EGCG regulates ACTN4 expression via demethylation to alleviate high glucose-induced cell damage needs to be further explored. ACTN4 functions as a selective regulator of RelA/p65 which is subunit of the transcription factor, NF-kB, and P65 inhibits the proliferation of H1299 cells, while ACTN4 overexpression regulates this inhibitory effect via RelA/p65 interacting with nuclear ACTN4. 34 Huang indicated that ACTN4 overexpression affects the activation of P65 in osteosarcoma cells and ultimately regulates their invasion ability via the NF-κB pathway. 35 Thus, we presume that EGCG regulates ACTN4 expression via demethylation to alleviate high glucose-induced cell damage, which may be related to the regulation of the NF-KB signaling pathway. Therefore, we investigated the effects of EGCG on the mRNA and protein expression levels of the major components of the NF-KBB signaling pathway. The results showed that the mRNA and protein expression levels of NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, and FN with EGCG treatment were decreased in the DN cell model compared with those in the NG group. In addition, the NF-κB inhibitor, PDTC, decreased the expression levels of NF-κB p65, p-NF-κB p65, IκBα, VEGF, IL-8, α-SMA, and FN, suggesting that EGCG, similar to PDTC, inhibits renal inflammation and fibrosis in DN by regulating the NF-κB signaling pathway. Hence, these results show that EGCG induces the demethylation of ACTN4, suppressing ACTN4 expression, and inhibits renal fibrosis in DN via the NF-KB signaling pathway.
In summary, these results indicate that the methylation level of the ACTN4 gene promoter was increased by upregulating DNMT1 expression in the cell model of DN, along with low protein expression of ACTN4. The expression levels of fibrosis and inflammatory markers were also increased in the cell model of DN compared with those in the NG group. EGCG significantly increased the viability of high glucose-treated HPC cells. EGCG treatment induced demethylation and reduced promoter methylation of ACTN4 to reverse ACTN4 expression. Moreover, EGCG downregulated the mRNA and protein expression levels of NF-κB p65, p-NF-κB, p65, and IκBα, inhibited the expression levels of fibrosis and inflammatory markers, and increased the levels of anti-inflammatory and anti-fibrotic cytokines in the cell model of DN. Furthermore, EGCG showed similar inhibition as PDTC on renal inflammation and fibrosis in DN via the suppression of the NF-κB signaling pathway. Hence, this study indicated that EGCG exerts its protective effects via the demethylation of ACTN4 to reverse its expression and alleviate renal fibrosis in DN and that the NF-KB signaling pathway is involved in this process.
Conclusions
Collectively, we found that EGCG reduced the hypermethylated status of ACTN4 by downregulating DNMT1 expression and restoring ACTN4 expression, resulting in the alleviation of renal inflammation and fibrosis in DN via upregulation of the NF-KB signaling pathway.
Acknowledgments
We would like to thank Yu Meng and Ting Dong for their helpful comments and suggestions.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
M. W., C. H., and J. L. designed the study. Y. H., X. H., and S. S. performed the experiments. C. H., D. W., and J. L. analyzed the data. M. W. and C. H. prepared the article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by The Excellent Young Talents Fund Program of the Higher Education Institutions of Anhui Province (gxyq2017035, gxyq2020025), Independent Research Program of the province Key Laboratory of Active Biological Macromolecules of Anhui (LAB201808), and Key programs of Wannan Medical College (WK2021ZF19), National Innovation Training and Entrepreneurship Plan for College Students (202110368028, S202110368103), and The Young Backbone Talents Fund Project of Wannan Medical College (wyqnyx201906).
Availability of Data and Materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
ORCiD iD
Mingcai Wu https://orcid.org/0000-0002-7434-3669
References
- 1.Yang HS, Wang J, Zhang Z, et al. Sp1-induced lncRNA Rmrp promotes mesangial cell proliferation and fibrosis in diabetic nephropathy by modulating the miR-1a-3p/JunD pathway. Front Endocrinol. 2021;12:690784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam-Chung CE, Zavala NM, Ibarra-Salce R, et al. Association of estimated glucose disposal rate and chronic diabetic complications in patients with type 1 diabetes. Endocrinol Diabetes Metab. 2021;4:e00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu JY, Tang ZH, He YW, et al. l-3-n-Butylphthalide ameliorates diabetic nephropathy by ameliorating excessive fibrosis and podocyte apoptosis. Front Pharmacol. 2021;12:628950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang XH, Feng SY, Yu Y, Liang Z, et al. Study on the relationship between the methylation of the MMP-9 gene promoter region and diabetic nephropathy. Endokrynol Pol. 2018;69(3):269-275. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Natarajan R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol. 2019;15(6):327-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang XH, Cao RF, Yang Y, et al. A study on the correlation between MTHFR promoter methylation and diabetic nephropathy. Am J Transl Res. 2016;8(11):4960-4967. [PMC free article] [PubMed] [Google Scholar]
- 7.Coskun ZM, Ersoz M, Adas M, et al. Kruppel-like transcription factor-4 gene expression and DNA methylation status in type 2 diabetes and diabetic nephropathy patients. Arch Med Res. 2019;50(3):91-97. [DOI] [PubMed] [Google Scholar]
- 8.Chen DQ, Zhang AP, Fang M, et al. Increased methylation at differentially methylated region of GNAS in infants born to gestational diabetes. BMC Med Genet. 2014;15:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardenas H, Fang F, Jiang G, et al. Methylomic signatures of high grade serous ovarian cancer. Epigenetics. 2020;8:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunes SP, Henrique R, Jerónimo C, Paramio JM. DNA Methylation as a therapeutic target for bladder cancer. Cells. 2020;9(8):1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnekenburger M, Diederich M. Epigenetics offer new horizons for colorectal cancer prevention. Curr Colorectal Cancer Rep. 2012;8(1):66-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braicu C, Gherman CD, Irimie A, Berindan-Neagoe I. Epigallocatechin-3-gallate (EGCG) inhibits cell proliferation and migratory behavior of triple negative breast cancer cells. J Nanosci Nanotechnol. 2013;13(1):632-637. [DOI] [PubMed] [Google Scholar]
- 13.Gan RY, Li HB, Sui ZQ, Corke H. Absorption, metabolism, anti-cancer effect, and molecular targets of epigallocatechin gallate (EGCG): An updated review. Crit Rev Food Sci Nutr. 2018;58(6):924-941. [DOI] [PubMed] [Google Scholar]
- 14.Mereles D, Hunstein W. Epigallocatechin-3-gallate (EGCG) for clinical trials: more pitfalls than promises? Int J Mol Sci. 2011;12:5592-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto S, Tsuda H, Honda K, et al. ACTN4 gene amplification and actinin-4 protein overexpression drive tumour development and histological progression in a high-grade subset of ovarian clear-cell adenocarcinomas. Histopathology. 2012;60(7):1073-1083. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan JM, Kim SH, North KN, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24(3):251-256. [DOI] [PubMed] [Google Scholar]
- 17.Jefferson JA, Shankland SJ, Pichler RH. Proteinuria in diabetic kidney disease: a mechanistic viewpoint. Kidney Int. 2008;74(1):22-36. [DOI] [PubMed] [Google Scholar]
- 18.Ma FZ, Sun T, Wu MY, Wang W, Xu Z. Identification of key genes for diabetic kidney disease using biological informatics methods. Mol Med Rep. 2017;16(6):7931-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krolewski AS, Skupien J, Rossing P, Warram JH. Fast renal decline to end-stage renal disease: an unrecognized feature of nephropathy in diabetes. Kidney Int. 2017;91(6):1300-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alicic RZ, Rooney MT, Tuttle KR. Diabetic kidney disease: challenges, progress, and possibilities. Clin J Am Soc Nephrol. 2017;12(12):2032-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haneda M. Diabetic nephropathy. Nihon Rinsho. 2015;73(3):489-494. [PubMed] [Google Scholar]
- 22.Miao A, Lu J, Wang Y, et al. Identification of the aberrantly methylated differentially expressed genes in proliferative diabetic retinopathy. Exp Eye Res. 2020;199:108141. [DOI] [PubMed] [Google Scholar]
- 23.Maghbooli Z, Hossein-nezhad A, Larijani B, Amini M, Keshtkar A. Global DNA methylation as a possible biomarker for diabetic retinopathy. Diabetes Metab Res Rev. 2015;31(2):183-189. [DOI] [PubMed] [Google Scholar]
- 24.Dayeh T, Tuomi T, Almgren P, et al. DNA methylation of loci within ABCG1 and PHOSPHO1 in blood DNA is associated with future type 2 diabetes risk. Epigenetics. 2016;11(7):482-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marumo T, Yagi S, Kawarazaki W, et al. Diabetes induces aberrant DNA methylation in the proximal tubules of the kidney. J Am Soc Nephrol. 2015;26(10):2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng D, DuMontier C, Pollak MR. The role of alpha-actinin-4 in human kidney disease. Cell Biosci. 2015;18:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bostrom MA, Perlegas P, Lu L, et al. Relevance of the ACTN4 gene in African-Americans with non-diabetic end-stage renal disease. Am J Nephrol. 2012;36(3):252-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benoit G, Machuca E, Nevo F, Gribouval O, Lepage D, Antignac C. Analysis of recessive CD2AP and ACTN4 mutations in steroid-resistant nephrotic syndrome. Pediatr Nephrol. 2010;25(3):445-451. [DOI] [PubMed] [Google Scholar]
- 29.Fontecha-Barriuso M, Martin-Sanchez D, Ruiz-Andres O, et al. Targeting epigenetic DNA and histone modifications to treat kidney disease. Nephrol Dial Transplant. 2018;33(11):1875-1886. [DOI] [PubMed] [Google Scholar]
- 30.Yang K, Yang J, Bi X, et al. Serum klotho, cardiovascular events, and mortality in nondiabetic chronic kidney disease. Cardiorenal Med. 2020;10(3):175-187. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Sun LT, Fong P, et al. An association between overexpression of DNA methyltransferase 3B4 and clear cell renal cell carcinoma. Oncotarget. 2017;8(12):19712-19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Cui J, Liu M, Shao Y, Dong X. Schisandrin C attenuates renal damage in diabetic nephropathy by regulating macrophage polarization. Am J Transl Res. 2021;13(1):210-222. [PMC free article] [PubMed] [Google Scholar]
- 33.Meng Q, Zhai X, Yuan Y, Ji Q, Zhang P. lncRNA ZEB1-AS1 inhibits high glucose-induced EMT and fibrogenesis by regulating the miR-216a-5p/BMP7 axis in diabetic nephropathy. Braz J Med Biol Res. 2020;53(4):e9288. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Lomert E, Turoverova L, Kriger D, et al. Co-expression of RelA/p65 and ACTN4 induces apoptosis in non-small lung carcinoma cells. Cell Cycle. 2018;17(5):616-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang QS, Li XD, Huang Z, et al. ACTN4 promotes the proliferation, migration, metastasis of osteosarcoma and enhances its invasive ability through the NF-κB pathway. Pathol Oncol Res. 2020;26(2):893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.