Abstract
The endophytic lifestyle of Klebsiella pneumoniae is described, including the production of dinitrogenase reductase by bacteria residing in maize root tissue. The green fluorescent protein (GFP) was used to detect the colonization of maize by K. pneumoniae strains 2028 and 342. These strains were found to reside in intercortical layers of the stem and within the region of maturation in the root. The production of dinitrogenase reductase by GFP-tagged bacteria was visualized using immunolocalization. This activity was only apparent when bacteria were supplied with an exogenous carbon source. The results suggest that maize provides a suitable habitat for K. pneumoniae and that this species is capable of producing nitrogenase under the appropriate plant cultivation conditions.
Recent research on diazotroph-plant associations has revealed that unrelated groups of diazotrophs can inhabit plant tissue without causing disease (3, 11, 12, 13, 14). In the case of Gluconacetobacter diazotrophicus, a growth benefit is provided to sugarcane directly via nitrogenase activity (6, 20). Although this direct correlation is lacking with other associations, both Azospirillum brasilense and Azoarcus sp. abundantly colonize and express nif genes within plant tissue (2, 8, 9, 24). This suggests that there is potential for a plant growth benefit from bacterial nitrogen fixation if the appropriate bacterial and/or plant growth conditions are met (20).
Previous attempts to isolate diazotrophs from surface-sterilized maize have resulted in the recovery of Klebsiella pneumoniae from stem tissue (16). The strains grew well on sucrose (a potential carbon source produced by the plant), fixed nitrogen in pure culture, and were recovered from both Zea mays and Z. luxurians. The cumulative evidence suggested an endophytic lifestyle; however, this was not verified by microscopic analysis.
The green fluorescent protein (GFP) is an ideal marker for studying microbial colonization of plants (4, 7, 10, 21, 22, 23). This is an especially powerful tool when used with scanning confocal laser microscopy (SCLM), in part because the optical sectioning properties of this microscopy produce a three-dimensional image with minimal sample preparation. As a result, the integrity of the plant structures and the bacteria residing within them is preserved. In addition, this tool can be combined with immunofluorescence to colocalize dinitrogenase reductase (NifH) production by inoculated bacteria. In this work, the colonization pattern and expression of NifH by GFP-tagged K. pneumoniae associated with maize were determined.
MATERIALS AND METHODS
Bacterial isolations and characterization.
Strains of K. pneumoniae were isolated from the stems of Z. mays L. cv. Alaquat (field grown for 90 days) and cv. CIMMYT 342 (greenhouse grown for 50 days). The stems were surface sterilized in 1.3% sodium hypochlorite for 5 min and then washed five times with sterile distilled water. Surface sterility was confirmed when no bacterial growth was observed on R2A (Difco) after contact with the plant stem. After the stem had been crushed in sterile water using a mortar and pestle, the homogenized material was incubated at 28°C in semisolid AcD (5). After growth and gas production were apparent, several transfers to fresh AcD were made, followed by bacterial isolations on R2A (Difco). Biochemical tests were performed using the API 20E system for the identification of members of the family Enterobacteriaceae.
Amplification by PCR was done by using 16S rRNA gene (rDNA) primers (Escherichia coli numbering) 27f and 1492r (15). Purified DNA was used as the template in a 50-μl PCR mixture containing 1× PCR Buffer (Promega), 5 mM MgCl2, 0.5 μg of bovine serum albumin, 200 μM deoxynucleoside triphosphates, and 0.5 U of Taq polymerase. Thermal cycle conditions were 3 min of denaturation at 95°C; 30 cycles of 94°C for 20 s, 54°C for 20 s, and 72°C for 30 s; and then an extension at 72°C for 7 min. The PCR product was purified using a Qiagen PCR purification kit. ABI's cycle sequencing kit was used by the University of Wisconsin Biotechnology Center to bidirectionally sequence all strains using primers (E. coli numbering) 27f, 536f, 1115f, 343r, 907r, 1165r, and 1492r (15). Small-subunit rDNA sequences were assembled using Sequencher computer software, and sequence similarities were determined using BLAST Sequence Similarity Search (1).
GFP-tagged strains.
E. coli DH5α, containing a red-shifted GFP allele on plasmid pPROBEKT-kan, was provided by Steven Lindow at the University of California, Berkeley. Conjugal transfer of the plasmid into K. pneumoniae was accomplished using E. coli containing helper plasmid pRK2073. Selection of transconjugants was done on Luria-Bertani agar supplemented with ampicillin (25 μg/ml) and kanamycin (50 μg/ml).
Plant growth conditions and bacterial inoculations.
Z. mays L. cv. Mo17 was grown in either a sand-vermiculite soil or nutrient agar medium. The agar medium consisted of (in millimolar) 2.0 CaCl2 · 2H2O, 0.5 MgSO4 · 7H2O, 2.0 KCl, 0.4 KH2PO4, 0.065 FeSO4 · 7H2O, 2.3 H3BO3, 0.9 MnSO4 · H2O, 0.6 ZnSO4 · H2O, 0.1 NaMoO4 · H2O, 0.11 NiCl2 · 6H2O, 0.01 CoCl2 · 6H2O, and 0.15 CuSO4 · 5H2O plus 0.1% sucrose and 8 g of agar per liter, pH 6.5. For all immunostaining experiments, plants were grown in the nutrient agar medium, where root health and development could be monitored. Greenhouse lighting included a 13-h day cycle at 25°C and 18°C at night. Seeds were surface sterilized as described above and allowed to germinate from 3 days to 1 week prior to inoculation. Bacteria were scraped from the surface of Luria-Bertani agar and diluted in sterile water to provide an inoculum containing 104 bacteria/seed.
Immunostaining.
From 3 to 8 days after inoculation, duplicate samples of plant roots were removed and fixed for 30 min in 4% paraformaldehyde. Protein blocking was done with 20% goat serum for 90 min, followed by primary-antibody staining (1:100 dilution) overnight at room temperature. Roots were washed three times for 5 min each time with 10 mM NaCl–130 mM sodium phosphate buffer (pH 7.2) after both the primary- and secondary-antibody stainings. The primary antibody, raised in a rabbit against purified NifH from both Rhodospirillum rubrum and Azotobacter vinelandii, was obtained from Paul Ludden (University of Wisconsin-Madison). The secondary antibody was a rhodamine red-X conjugate (Jackson ImmunoResearch) used at a 1:250 dilution. Roots were stained with the secondary antibody for 2 h at room temperature and mounted using an antifade kit (Prolong; Molecular Probes).
SCLM.
SCLM was done on a Bio-Rad MRC-1000 SCLM system attached to a Nikon Diaphot 200 inverted microscope. GFP-labeled cells were excited with the 488-nm laser line, and rhodamine red-X was excited with the 568-nm laser line. Images from both channels were collected sequentially in a z series from 10 to 25 optical sections ranging from 0.5 to 2.0 μm in thickness.
RESULTS
Bacterial isolation and identification.
K. pneumoniae 2028 was isolated from Z. mays cv. Alaquat that was grown in a sandy soil at Hancock, Wis., without nitrogen fertilizer. The plant was harvested after 3 months of growth, and the lower portion of the stem was used for bacterial isolations. K. pneumoniae 342 was isolated from a greenhouse-grown, nitrogen-efficient CIMMYT 342 (International Maize and Wheat Improvement Center) variety of maize that was watered without nitrogen fertilizer. This plant was harvested after approximately 7 weeks of growth.
Sequence analysis of the 16S rDNAs from both strains revealed 99% similarity to K. pneumoniae. This designation was substantiated by biochemical tests (API 20E) designed for identification of Enterobacteriaceae. The two strains gave identical results when examined for colonization and NifH production on Z. mays roots.
Visualization of GFP-tagged strains in maize tissue.
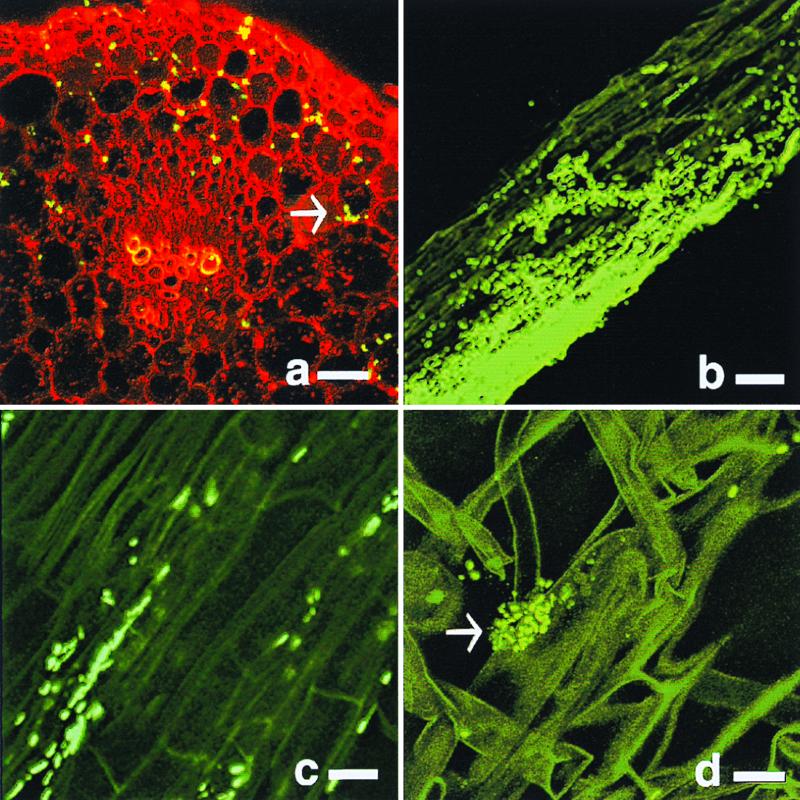
Bacteria were detected in both the plant roots and stems, although visualization of bacteria in the stem after approximately 2 weeks of growth was problematic due to plant autofluorescence. The stem cortex was colonized by bacteria typically in intercellular spaces, whereas colonization of the vascular bundle was only demonstrated by an occasional cell (Fig. 1a). Bacterial colonies on the roots were typically seen in the regions of elongation and more frequently in the root hair region of maturation (Fig. 1b to d). Within this region, bacteria colonized the epidermis and penetrated the surface to reside in or near the cortex region (Fig. 1c). The root hairs themselves were often colonized by single cells that occasionally formed bacterial colonies (Fig. 1d). Bacteria were rarely detected at the point of lateral root emergence or at the root cap.
FIG. 1.
Maize stem (a) and root (b to d) colonization by GFP-labeled K. pneumoniae strain 2028 (a and b) or 342 (c and d). Sections were visualized 8 (a) or 4 (b to d) days after inoculation. (a) Transverse section of a maize stem (red) colonized by K. pneumoniae (yellow-green). (b to d) Longitudinal section of a maize root (dark green) colonized by GFP-tagged cells (bright green). Arrows point to GFP-tagged cells. Bar, 20 (a and b) or 10 (c and d) μm.
Expression of NifH by endophytic bacteria.
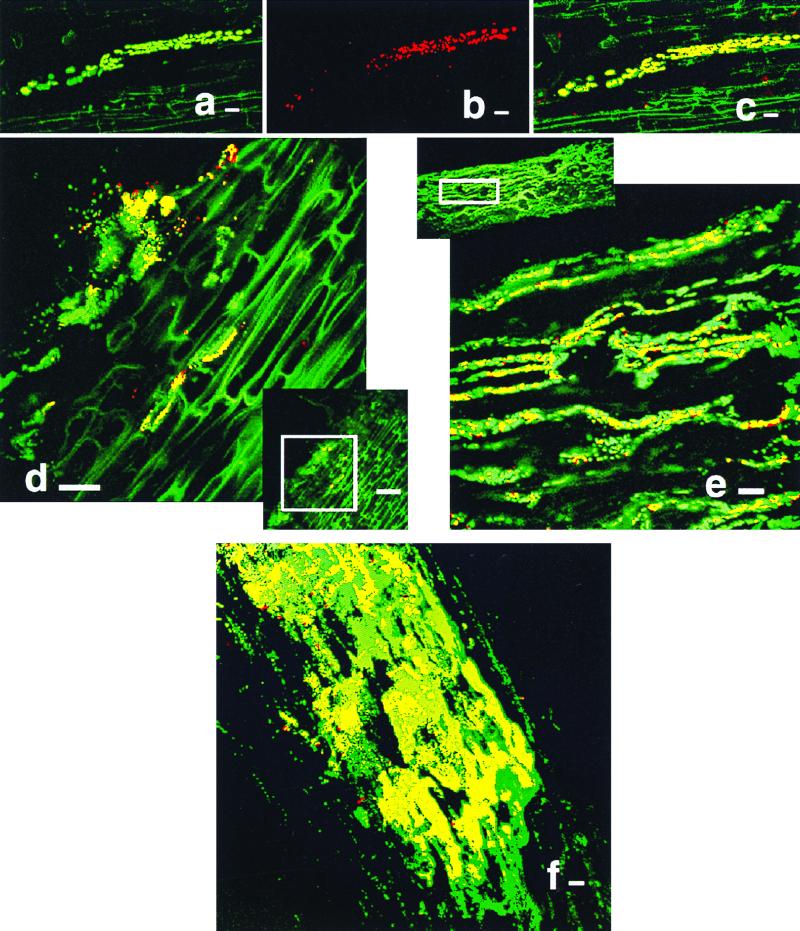
Patterns of colonization by GFP-tagged bacteria were easily monitored by their green fluorescence. Bacteria that expressed NifH emitted red light that appeared yellow when colocalization of both fluorophores was observed (Fig. 2a to f). Bacteria expressing NifH were generally attached to plant tissue with the long axis parallel to the long axis of the plant cell (Fig. 2a to e). NifH most often occurred in clusters of cells, and usually only a fraction of the colonizing bacteria within the same region showed production of NifH.
FIG. 2.
Immunolocalization of NifH produced by K. pneumoniae 2028 in maize roots. All images are longitudinal sections. (a to c) Series of images demonstrating fluorescence generated by green GFP-labeled cells (a), red NifH-producing cells (b), and a combination of the preceding images resulting in yellow cells (c). Immunolocalization of NifH at 3 (d), 4 (e), and 8 (f) days after inoculation. Bar, 10 (a to c), 20 (d, e, and f), or 50 (d offset) μm.
Preparation of the plant tissue for immunostaining involved minimal manipulation of plant tissue. Because of this, the natural colonization pattern and anti-NifH cross-reactivity of bacterial cells could be monitored without compromising the integrity of the plant structures. This worked especially well for the plant roots, where bacterial abundance on and within roots after immunostaining was significant. However, when this method was applied to plant stem sections, no GFP-labeled bacteria could be detected in this region, although visualization of bacteria in the stem prior to immunostaining was achieved. In this case, embedding the sample would probably improve the retention of bacteria within the plant tissue.
Attempts to immunostain root tissue from plants grown in a medium lacking a carbon substrate showed no anti-NifH cross-reactivity, even though bacteria substantially colonized these roots. After the addition of 0.1% sucrose to the plant growth medium, NifH production could be easily visualized. The correlation between external substrate addition and nitrogenase production corresponded to the results of Vande Broek et al. (24), who reported that carbon-limited A. brasilense growing on wheat roots could not be derepressed without the addition of an exogenous carbon source. With K. pneumoniae, cells inhabiting plant tissue at and near the agar-plant root interface produced NifH. These cells were in close proximity to the exogenously supplied carbon source, further supporting the hypothesis that a carbon supply is necessary for NifH production in this system.
To test the primary-antibody specificity, GFP-labeled cells including repressed K. pneumoniae and E. coli DH5α were carried through the immunostaining procedure. Preimmune serum was also used to detect potential specificity unrelated to NifH. For all of the controls tested, no cross-reactivity could be detected. Because GFP is a rather stable protein in bacterial cells, plant inoculations were run with E. coli in parallel to K. pneumoniae inoculations. E. coli was visualized on the plant surface, but the immunostaining yielded no antibody fluorescence (data not shown).
DISCUSSION
Colonization of maize by K. pneumoniae.
The use of GFP as a marker is a useful tool for studying plant-microbe interactions (4, 10, 21, 22, 23). This marker is well suited to studying the colonization patterns of bacteria within plants because there is no requirement for exogenous substrate or cofactors. In addition, GFP can be detected in single cells with no concern for its presence in the background. A potential disadvantage to using fluorophore markers is the deterioration in contrast generated by autofluorescing plant tissue. In this work, younger plants did not exhibit autofluorescence. However, this became problematic in root tissue as the plants matured.
The stability of the marker plasmid is also a cause for concern, but preliminary data shows that pPROBEKT-kan is stably maintained in the absence of selection pressure for at least 11 generations. In any case, because this work was qualitative in nature, any plasmid loss during the course of experimentation would not have significantly affected these results. Indeed, the abundance of GFP-tagged bacteria colonizing the plant tissue did not suggest significant marker loss.
Colonization events by diazotrophs have been studied in several grass species. Cells of A. brasilense demonstrate a characteristic pattern of wheat colonization that involves invasion of root hair cells (3) and colonization of the root tip (19). Herbaspirillum seropedicae colonizes the sugarcane root cortex intra- and intercellularly, as well as the xylem vessels (12). Azoarcus sp. colonizes rice and kallar grass in the root cortex, root cap, epidermis, exodermis, and xylem (11, 17). Pantoea agglomerans has been found in the intercellular spaces of wheat roots (18). The colonization of maize by K. pneumoniae was distinct in that it was typically found in the zone of root hair formation but only occasionally formed colonies on or in the root hairs. The pattern of stem colonization contrasted with root colonization in that the intercellular spaces of the stem cortex were inhabited by single cells or small clusters of cells (Fig. 1).
Expression of NifH by endophytic K. pneumoniae.
A fluorescent protein detection system, when used with SCLM, produced a useful tool for studying in planta activity of inoculated diazotrophs. SCLM is ideally suited to studies involving relatively thick tissues, where a three-dimensional analysis is desirable. Because the tissue is optically sectioned, there is no need for extensive tissue manipulation and the plant structure remains in tact. This advantage is readily apparent when evaluating the endophytic nature of bacterial cells.
The use of a plasmid marker in combination with direct protein detection minimized chromosomal manipulation and allowed direct detection of NifH production by the inoculant strain (Fig. 2). This is in contrast to other work that used a chromosomal lacZ-, gusA-, or GFP-nif fusion to monitor the nif transcript and not the protein product made by bacteria associating with plant roots (2, 9, 24). More recently, Egener et al. (8) used anti-NifH to observe cross-reactivity in rice plants inoculated with Azoarcus sp. strain BH72. The evidence for NifH production by inoculated Azoarcus sp. was indirect, since immunodetection of bacterial colonies and that of the iron protein of nitrogenase were sequential, not simultaneous, events.
Carbon limitation to NifH production by endophytic K. pneumoniae.
NifH production by K. pneumoniae in maize was carbon limited. The protein was not detected when the plants were cultivated without an external carbon source. A. brasilense reacted similarly to carbon amendment (24), although Arsene et al. (2) reported nif transcription in the absence of external carbon and Egener et al. (9) supplied only 5 mg of carbon per liter for nifH expression by Azoarcus sp. This level of carbon amendment might approach the lower threshold concentration needed for NifH expression, since expression and acetylene reduction by Azoarcus sp. were significantly augmented by medium supplementation with 400 mg of malic acid per liter (8). Although K. pneumoniae showed NifH production after amendment with a relatively high level (1 g/liter) of sucrose, the threshold level of carbon needed for NifH production was not determined.
In this work, cells repressed for nitrogenase activity were used to monitor subsequent NifH production by bacteria colonizing maize roots grown in a medium with an external carbon supply. Because the cells were repressed prior to inoculation, the production of NifH was unequivocally an in planta event. This suggests that nitrogenase activity is possible during natural colonization of maize if the appropriate plant and bacterial cultivation conditions are met, including the presence of a carbon supply sufficient for bacterial growth and nitrogenase production.
Pattern of NifH production.
NifH production by K. pneumoniae associating with maize roots was apparent with cells growing in colonies. In these areas, the pattern of production was patchy, possibly due to changes in suitable oxygen tension or the inability to sufficiently permeabilize some cells with the fixative in these areas. The latter explanation seems unlikely, since cells that appeared to be growing in the same plane showed differential antibody staining and immunostaining of control cells grown in an N-limited medium and derepressed for nitrogenase showed uniform fluorescence. Another reason for the differential staining could be that bacteria growing on the root surface are in direct contact with the exogenous carbon source, and once this resource is depleted, there is a minimal energy supply for NifH production and activity. This might also explain why cells growing in deeper layers of the plant tissue rarely produce detectable protein. Additional experimentation is needed to reveal whether inadequate oxygen tension or carbon limitation is responsible for the lack of NifH production by some cells.
In conclusion, K. pneumoniae is a common isolate from maize grown in temperate North America, where it exhibits an endophytic lifestyle, occupying both the stem and root tissue. The production of NifH by this species when associating with maize suggests that this may be a naturally occurring phenomenon if plant exudates or soil organic carbon meets the bacterial energy requirements for nitrogen fixation. Without a sufficient energy supply, nitrogen fixation may be prevented at the seedling stage. This limitation may not exist in sugarcane, where a plant growth benefit from nitrogen fixation has been observed (20). This work shows that K. pneumoniae cells can be repeatedly isolated from maize, that they can re-enter maize seedlings following inoculation, and that they can produce an important component of nitrogenase inside maize tissue following the addition of a carbon source.
ACKNOWLEDGMENTS
We thank Steven Lindow at the University of California, Berkeley, for supplying the GFP-containing plasmid; Paul Ludden for the NifH antibody; and Shawn Kaeppler for maize seeds.
This work was funded by University of Wisconsin-Madison College of Agriculture and Life Sciences Hatch project 5201.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsene F, Katupitiya S, Kennedy I R, Elmerich C. Use of lacZ fusions to study the expression of nif genes of Azospirillum brasilense in association with plants. Mol Plant-Microbe Interact. 1994;7:748–757. [Google Scholar]
- 3.Assmus B, Hutzler P, Kirchhof G, Amann R, Lawerence J R, Hartmann A. In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloemberg G V, O'Toole G A, Lugtenberg B J J, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burris R H. Comparative study of the response of Azotobacter vinelandii and Acetobacter diazotrophicus to changes in pH. Protoplasma. 1994;183:62–66. [Google Scholar]
- 6.Cavalcante V A, Dobereiner J. A new acid-tolerant nitrogen-fixing bacterium associated with sugar-cane. Plant Soil. 1988;108:23–31. [Google Scholar]
- 7.Chalfie M. Green fluorescent protein. Photochem Photobiol. 1994;62:651–656. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 8.Egener T, Hurek T, Reinhold-Hurek B. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol Plant-Microbe Interact. 1999;12:813–819. doi: 10.1094/MPMI.1998.11.1.71. [DOI] [PubMed] [Google Scholar]
- 9.Egener T, Hurek T, Reinhold-Hurek B. Use of green fluorescent protein to detect expression of nif genes of Azoarcus sp. BH72, a grass-associated diazotroph, on rice roots. Mol Plant-Microbe Interact. 1998;11:71–75. doi: 10.1094/MPMI.1998.11.1.71. [DOI] [PubMed] [Google Scholar]
- 10.Gage D J, Bobo T, Long S R. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa) J Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurek T, Reinhold-Hurek B, Van Montagu M, Kellenberger E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J Bacteriol. 1994;176:1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.James E K, Olivares F L. Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Crit Rev Plant Sci. 1998;17:77–119. [Google Scholar]
- 13.James E K, Olivares F L, Baldani J I, Dobereiner J. Herbaspirillum, an endophytic diazotroph colonizing vascular tissue in leaves of Sorghum bicolor L Moench. J Exp Bot. 1997;48:785–797. [Google Scholar]
- 14.James E K, Reis V M, Olivares F L, Baldani J I, Dobereiner J. Infection of sugarcane by the nitrogen-fixing bacterium Acetobacter diazotrophicus. J Exp Bot. 1994;45:757–766. [Google Scholar]
- 15.Lane D J. 16s/23s rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 115–175. [Google Scholar]
- 16.Palus J A, Borneman J, Ludden P W, Triplett E W. A diazotrophic bacterial endophyte isolated from stems of Zea mays L. and Zea luxurians Itlis and Doebley. Plant Soil. 1996;186:135–142. [Google Scholar]
- 17.Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;6:139–144. doi: 10.1016/s0966-842x(98)01229-3. [DOI] [PubMed] [Google Scholar]
- 18.Ruppel S, Hecht-Buchholz C, Remus R, Ortmann U, Schmelzer R. Settlement of the diazotrophic, phytoeffective bacterial strain Pantoea agglomerans on and within winter wheat: an investigation using ELISA and transmission electron microscopy. Plant Soil. 1992;145:261–273. [Google Scholar]
- 19.Schloter M, Hartmann A. Endophytic and surface colonization of wheat roots (Triticum aestivum) by different Azospirillum brasilense strains studied with strain-specific monoclonal antibodies. Symbiosis. 1998;25:159–179. [Google Scholar]
- 20.Sevilla M, De Oliveira A, Baldani I, Kennedy C. Contributions of the bacterial endophyte Acetobacter diazotrophicus to sugarcane nutrition: a preliminary study. Symbiosis. 1998;25:181–191. [Google Scholar]
- 21.Spellig T, Bottin A, Kahmann R. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet. 1996;252:503–509. doi: 10.1007/BF02172396. [DOI] [PubMed] [Google Scholar]
- 22.Tombolini R, van der Gaag D J, Gerhardson B, Jansson J K. Colonization pattern of the biocontrol strain Pseudomonas chloraphis MA 342 on barley seeds visualized by using green fluorescent protein. Appl Environ Microbiol. 1999;65:3674–3680. doi: 10.1128/aem.65.8.3674-3680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tombolini R, Unge A, Davey M E, de Bruijn F J, Jansson J K. Flow cytometric and microscopic analysis of GFP-tagged Pseudomonas fluorescens bacteria. FEMS Microbiol Ecol. 1997;22:17–28. [Google Scholar]
- 24.Vande Broek A, Michiels J, Van Gool A, Vanderleyden J. Spatial-temporal colonization patterns of Azospirillum brasilense on the wheat root surface and expression of the bacterial nifH gene during association. Mol Plant-Microbe Interact. 1993;5:592–600. [Google Scholar]