Abstract
Atopic dermatitis is a common inflammatory skin disease with a complex pathogenesis that includes imbalanced immune system signalling, impaired skin barrier and enhanced Staphylococcus aureus skin colonization. The skin bacterial communities are characterized by increasing abundance of S. aureus, leading to reduced diversity compared with the bacterial communities on healthy skin, and increasing disease severity. In contrast, fungal communities are richer and more diverse on the skin of patients with atopic dermatitis, although distribution of the most common species is similar in patients and controls. Filaggrin deficiency in atopic dermatitis skin might be related to the enhanced skin colonization by S. aureus. In addition, S. aureus expressing variant virulence factors have been shown to elicit atopic dermatitis-like phenotypes in mice, indicating that specific S. aureus strains can induce flare-ups. This review aims to provide an overview of the recent literature on the skin microbiome in atopic dermatitis.
Key words: atopic dermatitis, skin microbiome, Staphylococcus aureus, filaggrin
Atopic dermatitis (AD) is a common inflammatory skin disease that affects 10–20% of children and 2–10% of adults in developed countries (1, 2). The pathogenesis of the disease is complex and includes impaired skin barrier function and an imbalanced immune system with enhanced Th2, Th17, and Th22 signalling (3). Furthermore, patients with AD have an increased burden of Staphylococcus aureus skin colonization, which is associated with disease severity and exacerbation (4–8). Within recent years, where it has become possible to examine complete microbial communities using advanced DNA sequencing technologies, it is evident that cutaneous S. aureus is associated with decreased bacterial diversity on AD skin (8–13). The aim of this review is to provide an overview of the recent literature on the skin microbiome as well as microbe-host interactions in AD.
There is no single established accepted definition of the term “microbiome” in the scientific community. Often, this term is defined as the composition of all microbial genes in a community (14), but it has been argued that this definition rather describes the “metagenome” and that the word “microbiome” should be defined as all microorganisms in a habitat (the “microbiota”), their genomes, and the surrounding environmental conditions (15). In this review, the latter definition is used.
SIGNIFICANCE
Atopic dermatitis is a common skin disease characterized by dry and itchy skin with eczema flares. The disease is associated with changes in the skin microbiota, which constitutes all microorganisms present on the skin surface. The greatest difference is due to increased abundance of Staphylococcus aureus, a bacterium that can cause skin infections and probably contributes to aggravation of the disease. This review aims to provide an overview of recently published literature regarding changes in the skin microflora in atopic dermatitis and its association with disease severity and exacerbation.
Skin microorganisms can be identified using culture-based assays, and complete microbial communities can be examined by DNA sequencing (Box 1) followed by diversity and taxonomy analysis (Box 2). Recently, Grogan et al. (16) have summarized the techniques used for studying the skin microbiome, and the methods are therefore not described in detail in this review.
Box 1. Assays for examination of microbial organisms and communities.
Culture assays: Plating and culturing of samples in order to detect and isolate specific viable microorganisms of interest. Advantages of this method are the possibility to use isolated strains for additional analysis, e.g. testing for antimicrobial resistance and examine the gene content using molecular methods. Also, it is known that the detected microbes are alive and viable. Disadvantages are that it is difficult to detect microorganisms that do not grow easily under standard laboratory conditions and that it is not possible to examine the microbial community as a whole.
Whole genome sequencing: Sequencing genomes of specific microorganism of interest allows to examine the genetic content and thus properties of the single strain. This method is especially useful for comparing strains and their relatedness, e.g. to examine the similarity of strains isolated from distinct skin sites within individuals. A disadvantage of this method is that only selected strains are examined.
Targeted amplicon sequencing: Sequencing of selected variable regions of the 16S rRNA gene can be used to identify most bacteria in a sample, making it possible to examine the composition of the bacterial community. The transcriptional spacer regions ITS1/2 and variable regions of the 18S rRNA gene can be used to study eukaryotic microbial communities, such as fungal communities. An advantage of targeted amplicon sequencing is thus the ability to examine whole microbial communities, though this method has its limitations as it often is impossible to differentiate between related species. Another disadvantage of this method is that all available target DNA is sequenced, including DNA from microbial contaminants and human skin cells, which especially constitutes a problem for low biomass samples, such as skin samples.
Metagenomic shot-gun sequencing: Shot-gun sequencing is used to examine the complete genomes of the microbiota. This allows to detect all species constituting the microbiota at the strain level as well as examining specific properties of the community, e.g. metabolic pathway genes. A disadvantage of shot-gun sequencing is that deep-sequencing is required in order to obtain a high resolution making the method very expensive. As a consequence, most metagenomic studies are based on minor sample sets. Another disadvantage, which also applies to the amplicon DNA sequencing method, is the lack of discrimination between live and dead organisms.
Box 2. Diversity and taxonomy analysis.
-
Alpha-diversity: Diversity within samples.
Richness: the total number of species or unique sequences in a sample.
Shannon Index: a diversity measure that takes into account both species richness and evenness.
-
Beta-diversity: Diversity between samples.
-
Pairwise comparison of community structures can be measured using distance-based methods, e.g.:
Weighted UniFrac: Dissimilarity measure based on species presence/ absence data. Takes into account the phylogenetic relatedness of species.
Unweighted UniFrac: Dissimilarity measure based on the relative abundances of species. Takes into account the phylogenetic relatedness of species.
Jaccard: Dissimilarity measure based on species presence/absence data. Does not take into account the phylogenetic relatedness of species.
Bray-Curtis: Dissimilarity measure based on the relative abundances of species. Does not take into account the phylogenetic relatedness of species.
Ordination plots: Visualization of beta-diversity based on the pairwise distance measures. Samples with similar community structures are clustered together. The axes determine the degree of variance between samples.
Hierarchical clustering: Samples are clustered in a dendrogram based on the pairwise distance measures.
-
Taxonomy analysis: Analysis of species distributions in a community/sample.
HEALTHY SKIN MICROBIOME
The skin is an important first-line defence against pathogenic microbial invasion. Tight connections between corneocytes in the stratum corneum form a physical barrier, and antimicrobial peptides and lipids secreted from keratinocytes and glands provide a chemical barrier (17). In addition, commensal skin microorganisms can impede growth of pathogens, either directly by secreting antimicrobial molecules, or indirectly by occupying space and competing for nutritional resources (18, 19).
The skin microbiota consists of diverse organisms, including bacteria and fungi. In adults, the microbial community composition is rather stable over time, despite of the constant exposure to external microorganisms from other humans and the surrounding environment (20). However, the composition of the microbiota changes during puberty, with children having a more diverse microbiota compared with adults (21–23). Bacteria constitute the greatest proportion of the micro-biota, representing more than 70% of species in most skin areas (24). 16S rRNA gene sequence analysis has shown that Corynebacterium, Cutibacterium (formerly Propionibacterium (25)), Micrococcus, Staphylococcus, Streptococcus, Betaproteobacteria, and Gammaproteobacteria are common skin-colonizing bacteria (9, 21, 23, 24, 26). Microbial richness and Shannon-diversity are influenced by the microenvironmental conditions on the skin, including pH, moisture, sebum content, and topography (24, 26, 27). Sebaceous skin sites (e.g. facial areas and the upper part of the chest and back) are dominated by Cutibacterium acnes and are less diverse and rich compared with moist skin (e.g. nares, axillary vault, antecubital fossa, and popliteal fossa) and dry skin (e.g. volar forearm) (24, 26). C. acnes is also the most abundant species in dry skin, whereas no single bacterial species is over-represented in moist skin, although Corynebacterium spp. and Staphylococcus spp. relative abundances are greatest (24). Fungi constitute 1–5% of the skin microbiota (24), with Malassezia being the most common and abundant habitant (27, 28).
SKIN MICROBIOME IN ATOPIC DERMATITIS
AD is clinically characterized by red, dry, and itchy skin, with eczema flares and disease exacerbation. Interestingly, the clinical presentation of AD changes with age (29). Infants (<1 year) are primarily affected by acute lesions of the cheeks, scalp, neck, trunk, and extensor parts of the extremities. Children (2–12 years of age) are mostly affected by eczema at the antecubital and popliteal fossa, and adolescents and adults by chronic lesions comprising the head, neck, hands, and flexural areas (and sometimes widespread disease). Consequently, published studies on the skin microbiome in AD have focussed on distinct skin areas depending on the age group investigated. A major genetic risk factor of AD in Asian and Caucasian populations is loss-of-function mutations in the FLG gene encoding the skin protein filaggrin (30). Filaggrin is essential for the alignment of keratin in the corneocytes, and filaggrin breakdown products act as natural moisturizing factors (NMFs) important for proper skin hydration. Thus, filaggrin is important for maintaining a functional skin-barrier. Th2 and Th22 cytokines can down-regulate FLG expression, and thus lead to filaggrin deficiency in AD independently of loss-of-function mutations in FLG (3). Filaggrin deficiency and reduced levels of NMFs and free fatty acids, followed by an increase in skin pH, lead to an altered skin ecology in AD (31–34). Also, microbial communities are altered on AD skin compared with normal healthy skin, as described below.
Bacterial community on atopic dermatitis skin during infancy
As in healthy control skin, the skin microbial composition in AD differs between age groups, with distinct bacteria being over-represented at different ages (10, 21). Two case-control studies have compared the bacterial community composition on skin from infants with and without AD (35, 36). Zheng et al. (35) examined the bacterial community composition in perioral skin in infants with clinical signs of AD at the sample site and in age-matched healthy controls. The microbial diversity was lower on AD skin compared with healthy control skin, with the largest difference observed between patients with severe AD and healthy controls. Streptococcus was the most common bacterial genus at the perioral skin, with mean relative abundances exceeding 40% in both healthy control skin and lesional skin from patients with mild/ moderate AD. However, in the severe AD patient group, relative abundances of Streptococcus spp. were significantly reduced and replaced with Staphylococcus spp, primarily S. aureus. In contrast, bacterial communities on skin from the cheeks, nose tip, antecubital fossa, and popliteal fossa were generally similar in infants with or without AD (36). Importantly, the AD group consisted of infants who not necessarily had developed AD or had active disease at the sampling time-points, which very well can influence the results. S. aureus was not identified in any of the skin samples (36), despite the fact that S. aureus is frequently detected on antecubital and popliteal fossa skin regions in older children and adults with AD (8, 10). Although this could indicate that S. aureus colonization at the antecubital and popliteal fossa is not an essential marker for AD during disease development in the first year of life, culture-based analysis has indicated the opposite (37). Thus, Meylan et al. (37) found that frequencies of S. aureus colonization at axillary and antecubital fossa skin were significantly higher at the time of diagnosis among infants and toddlers (0–2 years of age) developing AD compared with non-AD age-matched controls. However, frequencies of S. aureus colonization were less than 15% and thus remarkably lower compared with the prevalence in older children and adults with AD (6). In addition, S. aureus colonization of the anterior nares, which is a major habitat for S. aureus in both healthy and AD individuals (6, 38), was not considered to be a risk factor for AD development among infants with familial predisposition (39). Thus, a possible role of cutaneous S. aureus colonization during development of AD still needs further investigation.
Bacterial community on atopic dermatitis skin during childhood and adulthood
Several studies have shown that bacterial communities on skin of children and adults with established AD are less diverse, and are dominated by increased proportions of S. aureus compared with communities on healthy skin (8–13, 21, 35). Quantification of cutaneous S. aureus abundances has shown that the proportional increase of S. aureus is due to a significant greater absolute abundance of S. aureus on AD lesional and non-lesional skin compared with healthy control skin (40–42). No difference in absolute abundances of the 3 common skin bacteria Corynebacterium, Cutibacterium, and Streptococcus were observed between AD and healthy control skin in children (40). Thus, the increased relative abundance of S. aureus on AD skin is probably not due to decreased colonization with these bacteria, but mainly a result of an enhanced burden of S. aureus on the skin. This could also explain that the total bacterial load is significantly greater on AD skin compared with healthy control skin (Fig. 1) (40, 43).
Fig. 1.
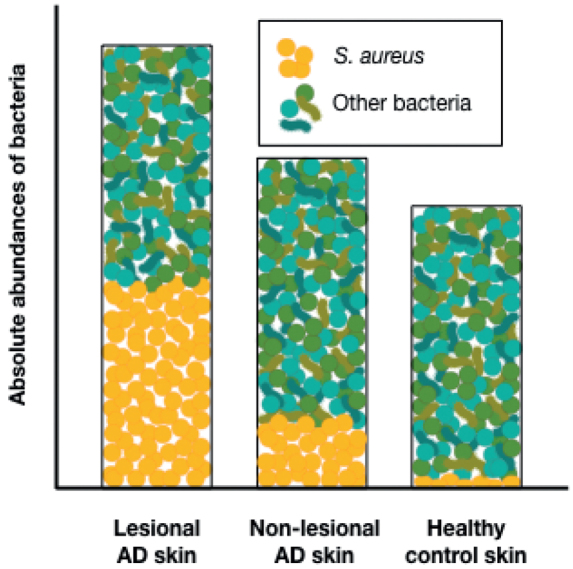
Absolute abundances of bacteria in atopic dermatitis (AD) skin and healthy skin. Bacterial densities are significantly greater on AD lesional skin compared with healthy control skin, which mainly is due to significantly increased abundances of S. aureus in AD lesional skin. S. aureus absolute abundances are also increased in AD non-lesional skin, but not as much as in lesional skin.
Kong and colleagues (8) were the first to examine the temporal bacterial variation on antecubital and popliteal fossa skin in children during and after an AD flare episode (8, 12). Bacterial diversity was significantly reduced during flares at both skin sites, compared with baseline and post-flare samples. The decreased diversity during AD flares was associated with increased relative abundances of S. aureus, which exceeded 40% in many of the samples. The increase in relative abundances of S. aureus was accompanied by a decrease in the relative abundance of Streptococcus salivarius (8, 44), a commensal bacteria of the oral cavity, intestines and skin that has been shown to possess anti-inflammatory potentials in vitro (45). In addition, higher proportions of S. salivarius contributed to greater bacterial diversity on the skin of the cheek, volar- and dorsal forearm in healthy infants with a family history of atopic diseases and thus at higher risk of developing AD (44). The proportional abundances of S. aureus decreased significantly at the post-flare sample time-point, but were still slightly higher compared with S. aureus proportional abundances in the healthy control samples (8, 12). Yet, no significant difference in alpha-diversity was observed between baseline, post-flare, and healthy control skin, which could indicate that skin bacterial diversity is only reduced during flare-ups. Several studies have compared alpha-diversity on lesional and non-lesional AD skin, but with different conclusions. Three studies found that the bacterial diversity was lower on lesional skin compared with nonlesional skin (13, 21, 46), whereas 2 other studies found that the diversity was equally reduced on affected and un-affected AD skin compared with healthy control skin (9, 10). Neither age nor sampling sites can explain the conflicting results. These studies might indicate that it is not the eczema itself that drives the changes in diversity, but other AD-related factors in or on the skin that not only are associated with lesional skin areas (e.g. S. aureus colonization, lipid composition or pH).
Clausen et al. (9) discovered that the bacterial community composition (beta-diversity) varied significantly between lesional and non-lesional skin areas in adult AD patients, with the greatest variance being due to a different distribution of Staphylococcal species. One-third of the lesional skin samples were dominated by S. aureus (relative abundances greater than 50%), whereas only a few non-lesional skin samples were characterized by high proportions of S. aureus. Instead, coagulase-negative staphylococcal species (CoNS), such as S. epidermidis and S. hominis, dominated the bacterial community on non-lesional skin in a majority of patients. In accordance, Baurecht et al. have shown that relative abundances of CoNS are reduced and S. aureus abundances increased in acute and chronic lesional skin compared with nonlesional AD skin (46). Though the identified CoNS are common colonizers of moist skin, their abundances were lower on healthy control skin compared with non-lesional AD skin at the antecubital fossa (9, 46). It could thus be hypothesized that the skin ecology in AD supports enhanced staphylococcal growth on both lesional and non-lesional skin, and that changes in the distribution among the staphylococcal species towards greater abundances of S. aureus can contribute to the development of eczema locally on the skin. However, no changes in the proportion of either S. epidermidis, S. hominis or S. capitis at the antecubital- and popliteal fossa during or after a flare-up episode was identified among paediatric AD patients (12), suggesting that a potential role for the CoNS spp. in AD still needs to be clarified. Furthermore, species level analysis might not be sufficient, as distinct strains within a species can have distinct phenotypes. For example, Nakatsuji et al. have shown that CoNS strains isolated from the skin of healthy individuals more often are capable of killing S. aureus compared with CoNS strains isolated from AD skin (42). Also, colonization of specific subspecies of S. aureus seems to be favoured in AD, as S. aureus clonal complex 1 (CC1) strains are more often detected on skin and in nares from AD patients compared with healthy controls (47). The increased prevalence of CC1 S. aureus colonization might be due to intrinsic factors in AD, e.g. CC1 S. aureus colonization have been associated with carriage of loss-of-function mutations in FLG (7), or due to extrinsic factors, such as treatment practice leading to selection of antibiotic resistance (48, 49).
Eukaryotic microbial community on atopic dermatitis skin
Few studies have examined the eukaryotic microbial community on AD skin, and only in adults and Asian populations (50–52). Focus has been on fungal communities, which was found to be richer and more diverse on AD lesional skin compared with healthy control skin (50, 51). Malassezia, especially M. globosa and M. restricta, was the dominant fungus in both AD lesional skin and healthy control skin (50, 51). An increase in the proportional abundance of M. dermatitis and M. sympodialis was identified on the skin of individuals with a history of AD (no active disease) compared with individuals without AD (52). However, no differences in the proportions of these 2 species were found between lesional skin of AD patients with active disease and healthy control skin (51, 53). Another fungal species, Candida albicans, was found to be over-represented on AD lesional skin on the cheeks (presence in 100% of samples), compared with healthy control skin from the same area (presence in 10% of samples) (51). The literature regarding skin eukaryotic microbial communities in AD is limited, and thus, additional studies with more attendees are needed in order to validate the presented results.
Atopic dermatitis disease severity is associated with changes in skin microbial communities
Significant differences in alpha- and beta-diversity across AD severity scores have been identified in both lesional and non-lesional skin sites, with patients with more severe disease having the lowest bacterial diversity on the skin (9, 10, 13, 54). Brandwein et al. (10) found that the bacterial community composition in antecubital- and popliteal fossa in patients with mild/moderate AD was more similar to the community composition of healthy control skin than to skin areas in patients with severe AD, regardless of whether samples were collected from lesional or non-lesional skin. This finding supports the hypothesis that the AD phenotype, such as an overall impaired skin barrier and skin inflammation, has a widespread effect on the skin microbial community and not only on lesional skin areas.
S. aureus skin and nasal colonization is significantly more prevalent among patients with more severe disease (6, 7, 55, 56), and increased relative abundances of S. aureus, at least at the antecubital fossa, have been associated with increasing AD severity scores (10–13). However, conflicting results regarding total S. aureus densities on skin in relation to AD severity have been published. Thus, S. aureus absolute abundances have been associated with increasing severity scores among adult patients (13, 57), whereas no association was detected in a paediatric AD population (54).
Studies investigating eukaryotic microbial communities on AD skin are sparse, but one study implies that there is an association between AD severity scores and the fungal community on skin, as beta-diversity analysis showed distinct community compositions in samples from patients with severe AD compared with samples from those with mild/moderate AD (51).
MICROBE-HOST INTERACTIONS IN ATOPIC DERMATITIS
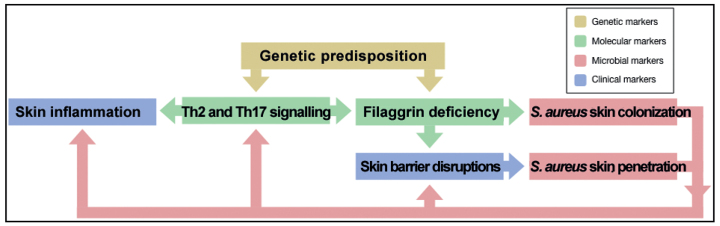
Microbiome studies have made it evident that microbial communities on AD skin differ from those of healthy skin, and that the greatest difference is due to an over-representation and greater abundance of S. aureus on AD skin. What are the mechanisms behind these differences? Functional analysis studies suggest that the AD phenotype, including impaired skin barrier function, increased pH, and skin inflammation, can promote changes in the skin microbial communities (43, 58, 59). Moreover, S. aureus can induce skin inflammation and aggravate AD (12, 60–62). Thus, a vicious circle might exist, with filaggrin deficiency in skin leading to enhanced colonization of S. aureus, which through the expression of virulence factors then can induce skin inflammation and contribute to further skin barrier impairment, and, in turn, can facilitate the maintenance of an imbalanced skin microbial community (Fig. 2). The mechanism behind these connections is elaborated below.
Fig. 2.
Proposed connections between human factors involved in AD pathogenesis and S. aureus colonization and virulence.
Atopic dermatitis pathogenesis facilitates changes in skin microbial communities
In AD, loss-of-function mutations in the FLG gene have been associated with changes in the overall bacterial community composition on non-lesional AD skin (9, 46), as well as with an increased risk of S. aureus colonization on lesional skin and in anterior nares (7). These studies indicate that filaggrin can influence bacterial growth and colonization on the skin. In accordance, presence of the filaggrin breakdown products urocanic acid (UCA) and pyrrolidone carboxylic acid (PCA), which contribute to skin acidification, have been shown to reduce S. aureus growth in vitro (58). More neutral pH, reflecting the skin pH in AD, has been associated with increased expression of S. aureus genes involved in colonization, including the gene encoding clumping factor B, which mediates adherence to keratinocytes (58, 59). Thus, increased S. aureus colonization among AD patients with FLG loss-of-function mutations (7) might be due to changes in skin pH caused by UCA and PCA deficiency. In addition to increased S. aureus adherence in epidermis, filaggrin deficiency is also associated with enhanced migration of S. aureus into the dermis skin layer. Nakatsuji et al. (43) showed that skin barrier impairment in mice, induced by genetic predisposition (FLG loss-of-function mutations) and physical skin disruptions (tape stripping), led to enhanced penetration of S. aureus into the dermis where it could activate the host immune system. In humans, the absolute abundance of S. aureus was significantly greater in dermis of AD lesional skin compared with healthy control skin, indicating that S. aureus can migrate more easily into the deeper skin layers of patients with AD with a disrupted skin barrier (43). Disrupted AD skin is also more permeable to allergens, which can trigger type I allergic responses in sensitized individuals. To corroborate this, patients with AD are also more often hypersensitive to a wide range of microbial allergens, including allergens from S. aureus and the skin colonizing fungal species Malassezia furfur and Candida albicans, compared with the general population (63–65).
AD skin might not only be more susceptible to S. aureus colonization, but also more vulnerable to S. aureus virulence. Alpha-haemolysin (also known as alpha-toxin), a virulence factor secreted by S. aureus, has thus been shown to adhere more easily to keratinocytes in AD skin compared with keratinocytes in healthy skin (66, 67). Alpha-haemolysin adheres to sphingomyelin lipids in the membranes of keratinocytes, leading to cell lysis and contribution to skin barrier disruptions (68). The density of sphingomyelin lipids, and thus the amount of free adherence sites for alpha-haemolysin, is regulated by the enzyme acid sphingomyelinase. Filaggrin deficiency as well as Th2 cytokines promote down-regulation of acid sphingomyelinase, thus enhancing alpha-haemolysin binding efficiency (66, 67). Thus, filaggrin deficiency in AD probably both favours S. aureus colonization and enhanced S. aureus mediated cytotoxicity and immune activation (Table I).
Table I.
The effect of filaggrin deficiency on S. aureus skin colonization and virulence
| Effect of filaggrin deficiency | ||
|---|---|---|
| Primary outcomes | Secondary outcomes | Refs |
| Increased skin pH | Enhanced S. aureus growth and colonization | (58, 59) |
| Impaired skin barrier | Enhanced S. aureus migration through the epidermal barrier and into dermis | (43) |
| Increased density of sphingomyelin lipids in keratinocyte membranes | Enhanced binding of alpha-haemolysin (cytotoxic S. aureus virulence factor) | (67) |
S. aureus as an inducer of clinical atopic dermatitis
Byrd et al. (12) have shown that S. aureus isolated from AD skin, but not S. aureus from normal healthy skin, was able to induce skin inflammation in wild-type mice with no genetic predisposition. Skin inflammation, assessed by epidermal thickening and cutaneous infiltration of immune cells, including Th2 and Th17 cells, was more pronounced in mice inoculated with S. aureus from patients with more severe AD. This study highly suggests that certain strains of S. aureus are able to elicit lesions similar to those observed in AD. The detected effect of S. aureus is probably mediated by the production of virulence factors, such as phenol-soluble modulins (PSM) and enterotoxins.
Several studies indicate that S. aureus induced skin inflammation and barrier disruption in mice are dependent on secretion of PSM-alpha, which promotes interleukin (IL)-17A mediated pro-inflammatory responses in vitro (human keratinocytes) and in vivo (mice) (60, 69, 70). Another PSM, known as delta-toxin, was also able to mediate S. aureus induced skin inflammation in mice, an effect that probably is mediated by delta-toxin induced mast cell degranulation, IgE production and enhanced IL-4 expression (62, 71). Interestingly, PSM-alpha transcripts are significantly more abundant in S. aureus isolated from AD skin compared with those from S. aureus isolated from healthy control skin (60), and delta-toxin production has been found to be considerably higher among S. aureus from lesional skin compared with nonlesional skin on patients with AD (62). These findings might explain why S. aureus strains isolated from AD lesional skin were better at eliciting skin inflammation compared with S. aureus from healthy skin (12).
S. aureus enterotoxins have also been proposed to be important mediators of S. aureus induced skin inflammation. Thus, topical application of staphylococcal enterotoxin B (SEB) to skin have been shown to cause erythema and epidermal thickening in both healthy volunteers and patients with AD (72), an effect which likely is mediated by enhanced T-cell signalling (72, 73). Studies indicates that S. aureus from AD skin more often carry genes encoding enterotoxins (sea, seb, sec, and sed) and more often produce these toxins compared with S. aureus isolated from non-AD individuals (73, 74). Furthermore, carriage of enterotoxin producing S. aureus has been associated with increased AD severity (assessed by SCORAD) (73, 75).
In one study, alpha-haemolysin was also found to be produced more frequently by AD S. aureus (91% of isolates) compared with production rates among S. aureus from healthy volunteers (33% of isolates) (61), which in combination with AD genetic predisposition for enhanced binding efficiency of the toxin (66, 67) could make alpha-haemolysin a potent inducer of skin barrier disruptions in AD (61). However, two other studies found lower proportions of S. aureus producing alpha-haemolysin on AD skin (30–63% of isolates) (76, 77) and a third study reported an alpha-haemolysin gene (hla) expression frequency of 59% among S. aureus nasal isolates from healthy carriers (78), highlighting that population-based differences and use of distinct assays can influence the results. Thus, future studies need to elucidate whether alpha-haemolysin, and other S. aureus toxins, is upregulated in S. aureus colonizing AD skin.
The above-mentioned studies support the hypothesis that S. aureus virulence is a major driver of AD disease exacerbation and might even be a direct cause of flare-ups. In order to cause disease, S. aureus must first colonize the skin. S. aureus isolated from AD skin has an enhanced binding activity of clumping factor B, leading to increased adhering to corneocytes, compared with S. aureus from healthy skin (79). In addition, CC1 S. aureus, which is a dominant clone in AD (7, 79, 80), had a slightly higher binding affinity compared with other S. aureus lineages (79). Thus, the increased prevalence of S. aureus skin colonization in AD might both be due to host factors and S. aureus factors (58, 59, 79). A summary of the described S. aureus virulence factors shown to be involved in AD is given in Table II.
Table II.
Virulence factors upregulated in S. aureus isolated from atopic dermatitis skin compared with 5. aureus from healthy control skin
| Virulence factors | Clinical outcomes | Mediators | Refs |
|---|---|---|---|
| PSM-alpha | Skin inflammation | IL-17A signalling | (60, 70) |
| Skin barrier disruptions | Protease activity | ||
| Delta-toxin | Skin inflammation | IL-4 signalling | (62, 71) |
| Mast cell degranulation IgE release | |||
| IgE release | |||
| Alpha-haemolysin | Skin barrier disruptions | Keratinocyte lysis | (61) |
| Enterotoxin B | Skin inflammation | T-cell signalling | (72) |
| Clumping factor B | S. aureus colonization | Cell adherence | (79) |
PSM: phenol-soluble modulin; IL: interleukin.
EFFECT OF ATOPIC DERMATITIS TREATMENTS ON SKIN MICROBIAL COMMUNITIES
Topical application of corticosteroid (glucocorticoids) based creams is a common treatment of AD lesions. Prospective studies examining the effect of topical corticosteroid treatment on skin microbial communities in AD, have shown that 4–6 weeks of treatment led to significant increases in bacterial Shannon-diversity and richness (40, 81), whereas 7–10 days of treatment had no influence on alpha-diversity, though an clinical improvement was observed (35). Thus, a possible effect of topical corticosteroid on skin microbial communities is dependent of several weeks of continuous treatment. Comparative studies also imply that topical corticosteroid treatments have an effect on the skin microbial community, as AD patients undergoing topical corticosteroid treatments prior to sample collections often have a more diverse bacterial population with lower relative abundances of S. aureus compared with non-treated patients (8, 9, 81). This effect might be due to direct inhibition of S. aureus as well as to a general improvement on skin conditions due to the anti-inflammatory properties of corticosteroids (82).
A keystone treatment practice in AD is application of emollients and moisturizers, which restore skin barrier integrity and prevents flare-ups. Despite extensive use, little is known about what effect this treatment approach has on skin microbial communities, but one study indicates that emollient application leads to decreased proportions of Staphylococcus spp. on AD lesional skin (83). Although it indeed would be interesting to examine the long-term effect of emollient usage on the skin micro-biome, it might be challenging and ethically unjustifiable to set up such study, as it would include an AD patient group that will be denied treatment with emollients and moisturizers for a longer period.
One study has examined the effect of dupilumab treatment, an anti-inflammatory systemic therapy offered to adults with severe and chronic AD, on the skin bacterial community (13). Sixteen weeks of treatment led to increased alpha-diversity and a decrease in relative and absolute S. aureus abundances on lesional as well as non-lesional AD skin. However, this effect was lost 18 weeks after treatment termination. Dupilumab inhibits IL-4/IL-13 signalling, and the study thus shows that reduction of Th2-mediated signalling may influence S. aureus skin colonization.
Another common treatment practice, at least in some countries, is topical application of fusidic acid, which is a narrow-spectrum antibiotic used against S. aureus. Unfortunately, bacterial growth of other common bacterial species on skin, including CoNS, are also inhibited by fusidic acid (84), and recent studies have shown a high prevalence of fusidic acid resistant S. aureus on AD skin and nares (48, 49), signifying that alternative treatment regimens are needed for the control of S. aureus colonization. Future treatment approaches could include S. aureus anti-virulence therapy (71) or application of commensal skin bacteria with anti-S. aureus properties (42). Oral administered antibiotics might also impact the cutaneous bacterial community composition and select for antibiotic resistance among skin bacteria (85–87).
CONCLUSION
Multiple studies have shown that increased abundance of S. aureus and loss of bacterial diversity on skin are associated with disease severity and flares in children and adults with AD. The enhanced burden of S. aureus skin colonization is probably facilitated by AD-related changes in the skin, including reduced levels of filaggrin and NMFs leading to increased skin pH and skin barrier impairment. In addition, deficiency of commensal bacterial strains with S. aureus inhibitory properties may contribute to the increased density of S. aureus on AD skin. Functional assays indicate that cutaneous S. aureus can exacerbate AD by expressing virulence factors that can induce skin inflammation and skin barrier disruption. Thus, changes in the composition of the skin bacterial community may be an important inducer of the clinical manifestations in AD patients with established disease. Whether bacterial community dysbiosis is also considered to be present prior to AD development is still unclear, and needs further investigation. Increasing knowledge regarding S. aureus as a potent promoter of AD exacerbation, has highlighted the skin microbial community as a potential target for future treatment strategies, and is a research field of great interest. Future studies are needed to explore the potentials, efficiency and safety of these novel anti-bacterial treatment approaches.
Footnotes
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Henriksen L, Simonsen J, Haerskjold A, Linder M, Kieler H, Thomsen SF, et al. Incidence rates of atopic dermatitis, asthma, and allergic rhinoconjunctivitis in Danish and Swedish children. J Allergy Clin Immunol 2015; 136: 360–366.e362. [DOI] [PubMed] [Google Scholar]
- 2.Carroll CL, Balkrishnan R, Feldman SR, Fleischer AB, Jr., Manuel JC. The burden of atopic dermatitis: impact on the patient, family, and society. Pediatr Dermatol 2005; 22: 192–199. [DOI] [PubMed] [Google Scholar]
- 3.Leung DY, Guttman-Yassky E. Deciphering the complexities of atopic dermatitis: shifting paradigms in treatment ap proaches. J Allergy Clin Immunol 2014; 134: 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol 1974; 90: 525–530. [DOI] [PubMed] [Google Scholar]
- 5.Williams JV, Vowels BR, Honig PJ, Leyden JJ. S. aureus isolation from the lesions, the hands, and the anterior nares of patients with atopic dermatitis. Pediatr Dermatol 1998; 15: 194–198. [DOI] [PubMed] [Google Scholar]
- 6.Totte JE, van der Feltz WT, Hennekam M, van Belkum A, van Zuuren EJ, Pasmans SG. Prevalence and odds of Staphylococcus aureus carriage in atopic dermatitis: a systematic review and meta-analysis. Br J Dermatol 2016; 175: 687–695. [DOI] [PubMed] [Google Scholar]
- 7.Clausen ML, Edslev SM, Andersen PS, Clemmensen K, Krogfelt KA, Agner T. Staphylococcus aureus colonization in atopic eczema and its association with filaggrin gene mutations. Br J Dermatol 2017; 177: 1394–1400. [DOI] [PubMed] [Google Scholar]
- 8.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res 2012; 22: 850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clausen ML, Agner T, Lilje B, Edslev SM, Johannesen TB, Andersen PS. Association of disease severity with skin micro-biome and filaggrin gene mutations in adult atopic dermatitis. JAMA Dermatol 2018; 154: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandwein M, Fuks G, Israel A, Sabbah F, Hodak E, Szitenberg A, et al. Skin microbiome compositional changes in atopic dermatitis patients accompany dead sea climatotherapy. Photochem Photobiol 2019; 95: 1446–1453. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Xu X, Wen H, Wang Z, Ding C, Liu X, et al. Inverse association between the skin and oral microbiota in atopic dermatitis. J Invest Dermatol 2019; 139: 1779–1787.e12. [DOI] [PubMed] [Google Scholar]
- 12.Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Conlan S, et al. Staphylococcus aureus and Staphylococcus epider midis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med 2017; 9. pii: eaal4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callewaert C, Nakatsuji T, Knight R, Kosciolek T, Vrbanac A, Kotol P, et al. IL-4Ralpha blockade by dupilumab decreases staphylococcus aureus colonization and increases microbial diversity in atopic dermatitis. J Invest Dermatol 2020; 140: 191–202.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrd AL, Belkaid Y, Segre JA. The human skin microbiome. Nat Rev Microbiol 2018; 16: 143–155. [DOI] [PubMed] [Google Scholar]
- 15.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome 2015; 3: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grogan MD, Bartow-McKenney C, Flowers L, Knight SAB, Uberoi A, Grice EA. Research techniques made simple: profiling the skin microbiota. J Invest Dermatol 2019; 139: 747–752.e741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandner JM. Importance of tight junctions in relation to skin barrier function. Curr Probl Dermatol 2016; 49: 27–37. [DOI] [PubMed] [Google Scholar]
- 18.Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol 2013; 25: 370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science 2012; 337: 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh J, Byrd AL, Park M, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell 2016; 165: 854–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi B, Bangayan NJ, Curd E, Taylor PA, Gallo RL, Leung DYM, et al. The skin microbiome is different in pediatric versus adult atopic dermatitis. J Allergy Clin Immunol 2016; 138: 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI, et al. Diverse human skin fungal communities in children converge in adulthood. J Invest Dermatol 2016; 136: 2356–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh J, Conlan S, Polley EC, Segre JA, Kong HH. Shifts in human skin and nares microbiota of healthy children and adults. Genome Med 2012; 4: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Segre JA. Biogeography and individuality shape function in the human skin metagenome. Nature 2014; 514: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scholz CF, Kilian M. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol 2016; 66: 4422–4432. [DOI] [PubMed] [Google Scholar]
- 26.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science 2009; 324: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 2013; 498: 367–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol 2010; 48: 3575–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bieber T, D’Erme AM, Akdis CA, Traidl-Hoffmann C, Lauener R, Schappi G, et al. Clinical phenotypes and endophenotypes of atopic dermatitis: where are we, and where should we go? J Allergy Clin Immunol 2017; 139: S58–s64. [DOI] [PubMed] [Google Scholar]
- 30.Cabanillas B, Novak N. Atopic dermatitis and filaggrin. Curr Ppin Immunol 2016; 42: 1–8. [DOI] [PubMed] [Google Scholar]
- 31.Jakasa I, Koster ES, Calkoen F, McLean WH, Campbell LE, Bos JD, et al. Skin barrier function in healthy subjects and patients with atopic dermatitis in relation to filaggrin loss-offunction mutations. J Invest Dermatol 2011; 131: 540–542. [DOI] [PubMed] [Google Scholar]
- 32.Seidenari S, Giusti G. Objective assessment of the skin of children affected by atopic dermatitis: a study of pH, capa citance and TEWL in eczematous and clinically uninvolved skin. Acta Derm Venereol 1995; 75: 429–433. [DOI] [PubMed] [Google Scholar]
- 33.Kezic S, Kemperman PM, Koster ES, de Jongh CM, Thio HB, Campbell LE, et al. Loss-of-function mutations in the filaggrin gene lead to reduced level of natural moisturizing factor in the stratum corneum. J Invest Dermatol 2008; 128: 2117–2119. [DOI] [PubMed] [Google Scholar]
- 34.van Smeden J, Bouwstra JA. Stratum corneum lipids: their role for the skin barrier function in healthy subjects and atopic dermatitis patients. Curr Probl Dermatol 2016; 49: 8–26. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Y, Wang Q, Ma L, Chen Y, Gao Y, Zhang G, et al. Alterations in the skin microbiome are associated with disease severity and treatment in the perioral zone of the skin of infants with atopic dermatitis. Eur J Clin Microbiol Infect Dis 2019; 38: 1677–1685. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WHI, Murray D, et al. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol 2017; 139: 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meylan P, Lang C, Mermoud S, Johannsen A, Norrenberg S, Hohl D, et al. Skin colonization by staphylococcus aureus precedes the clinical diagnosis of atopic dermatitis in infancy. J Invest Dermatol 2017; 137: 2497–2504. [DOI] [PubMed] [Google Scholar]
- 38.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mecha nisms, and associated risks. Clin Microbiol Rev 1997; 10: 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skov L, Halkjaer LB, Agner T, Frimodt-Moller N, Jarlov JO, Bisgaard H. Neonatal colonization with Staphylococcus aureus is not associated with development of atopic dermatitis. Br J Dermatol 2009; 160: 1286–1291. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez ME, Schaffer JV, Orlow SJ, Gao Z, Li H, Alekseyenko AV, et al. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. J Am Acad Dermatol 2016; 75: 481–493.e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tauber M, Balica S, Hsu CY, Jean-Decoster C, Lauze C, Redoules D, et al. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. J Allergy Clin Immunol 2016; 137: 1272–1274. e1273. [DOI] [PubMed] [Google Scholar]
- 42.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med 2017; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, et al. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol 2016; 136: 2192–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glatz M, Jo JH, Kennedy EA, Polley EC, Segre JA, Simpson EL, et al. Emollient use alters skin barrier and microbes in infants at risk for developing atopic dermatitis. PloS One 2018; 13: e0192443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaci G, Goudercourt D, Dennin V, Pot B, Dore J, Ehrlich SD, et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol 2014; 80: 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baurecht H, Ruhlemann MC, Rodriguez E, Thielking F, Harder I, Erkens AS, et al. Epidermal lipid composition, barrier integrity, and eczematous inflammation are associated with skin microbiome configuration. J Allergy Clin Immunol 2018; 141: 1668–1676. e1616. [DOI] [PubMed] [Google Scholar]
- 47.Iwamoto K, Moriwaki M, Miyake R, Hide M. Staphylococcus aureus in atopic dermatitis: Strain-specific cell wall proteins and skin immunity. Allergol Int 2019; 68: 309–315. [DOI] [PubMed] [Google Scholar]
- 48.Edslev SM, Clausen ML, Agner T, Stegger M, Andersen PS. Genomic analysis reveals different mechanisms of fusidic acid resistance in Staphylococcus aureus from Danish atopic dermatitis patients. J Antimicrob Chemother 2018; 73: 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harkins CP, McAleer MA, Bennett D, McHugh M, Fleury OM, Pettigrew KA, et al. The widespread use of topical antimicrobials enriches for resistance in Staphylococcus aureus isolated from patients with atopic dermatitis. Br J Dermatol 2018; 179: 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han SH, Cheon HI, Hur MS, Kim MJ, Jung WH, Lee YW, et al. Analysis of the skin mycobiome in adult patients with atopic dermatitis. Exp Dermatol 2018; 27: 366–373. [DOI] [PubMed] [Google Scholar]
- 51.Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A, Sugita T. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol 2011; 55: 625–632. [DOI] [PubMed] [Google Scholar]
- 52.Chng KR, Tay AS, Li C, Ng AH, Wang J, Suri BK, et al. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nat Microbiol 2016; 1: 16106. [DOI] [PubMed] [Google Scholar]
- 53.Amoako DG, Bester LA, Somboro AM, Baijnath S, Govind CN, Essack SY. Plasmid-mediated resistance and virulence mechanisms in the private health sector in KwaZulu-Natal, South Africa: an investigation of methicillin resistant Staphylococcus aureus (MRSA) clinical isolates collected during a three month period. Int J Infect Dis 2016; 46: 38–41. [DOI] [PubMed] [Google Scholar]
- 54.Totte JEE, Pardo LM, Fieten KB, Vos MC, van den Broek TJ, Schuren FHJ, et al. The nasal and skin microbiome are associated with disease severity in pediatric atopic dermatitis. Br J Dermatol 2019; 181: 796–804. [DOI] [PubMed] [Google Scholar]
- 55.Hill SE, Yung A, Rademaker M. Prevalence of Staphylococcus aureus and antibiotic resistance in children with atopic dermatitis: a New Zealand experience. Australas J Dermatol 2011; 52: 27–31. [DOI] [PubMed] [Google Scholar]
- 56.Simpson EL, Villarreal M, Jepson B, Rafaels N, David G, Hanifin J, et al. Patients with atopic dermatitis colonized with staphylococcus aureus have a distinct phenotype and endotype. J Invest Dermatol 2018; 138: 2224–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guzik TJ, Bzowska M, Kasprowicz A, Czerniawska-Mysik G, Wojcik K, Szmyd D, et al. Persistent skin colonization with Staphylococcus aureus in atopic dermatitis: relationship to clinical and immunological parameters. Clin Exp Allergy 2005; 35: 448–455. [DOI] [PubMed] [Google Scholar]
- 58.Miajlovic H, Fallon PG, Irvine AD, Foster TJ. Effect of filaggrin breakdown products on growth of and protein expression by Staphylococcus aureus. J Allergy Clin Immunol 2010; 126: 1184–1190. e1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mempel M, Schmidt T, Weidinger S, Schnopp C, Foster T, Ring J, et al. Role of Staphylococcus aureus surface-associated proteins in the attachment to cultured HaCaT keratinocytes in a new adhesion assay. J Invest Dermatol 1998; 111: 452–456. [DOI] [PubMed] [Google Scholar]
- 60.Williams MR, Costa SK, Zaramela LS, Khalil S, Todd DA, Winter HL, et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci Transl Med 2019; 11; eaat8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong SW, Choi EB, Min TK, Kim JH, Kim MH, Jeon SG, et al. An important role of alpha-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PloS One 2014; 9: e100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Munoz-Planillo R, Hasegawa M, et al. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature 2013; 503: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol 2016; 55: 494–504. [DOI] [PubMed] [Google Scholar]
- 64.Faergemann J. Atopic dermatitis and fungi. Clin Microb Rev 2002; 15: 545–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jinnestal CL, Belfrage E, Back O, Schmidtchen A, Sonesson A. Skin barrier impairment correlates with cutaneous Staphylococcus aureus colonization and sensitization to skin-associated microbial antigens in adult patients with atopic dermatitis. Int J Dermatol 2014; 53: 27–33. [DOI] [PubMed] [Google Scholar]
- 66.Brauweiler AM, Goleva E, Leung DYM. Th2 cytokines increase Staphylococcus aureus alpha toxin-induced keratinocyte death through the signal transducer and activator of transcription 6 (STAT6). J Invest Dermatol 2014; 134: 2114–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brauweiler AM, Bin L, Kim BE, Oyoshi MK, Geha RS, Goleva E, et al. Filaggrin-dependent secretion of sphingomyelinase protects against staphylococcal alpha-toxin-induced keratinocyte death. J Allergy Clin Immunol 2013; 131: 421–427. e 421–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seiti Yamada Yoshikawa F, Feitosa de Lima J, Notomi Sato M, Alefe Leuzzi Ramos Y, Aoki V, Leao Orfali R. Exploring the role of staphylococcus aureus toxins in atopic dermatitis. Toxins 2019; 11. pii: E321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu H, Archer NK, Dillen CA, Wang Y, Ashbaugh AG, Ortines RV, et al. Staphylococcus aureus epicutaneous exposure drives skin inflammation via IL-36-mediated T cell responses. Cell Host Microbe 2017; 22: 653–666.e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakagawa S, Matsumoto M, Katayama Y, Oguma R, Wakabayashi S, Nygaard T, et al. Staphylococcus aureus virulent PSMalpha peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe 2017; 22: 667–677.e665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baldry M, Nakamura Y, Nakagawa S, Frees D, Matsue H, Nunez G, et al. Application of an AGR-specific antivirulence compound as therapy for staphylococcus aureus-induced inflammatory skin disease. J Infect Dis 2018; 218: 1009–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Skov L, Olsen JV, Giorno R, Schlievert PM, Baadsgaard O, Leung DY. Application of Staphylococcal enterotoxin B on normal and atopic skin induces up-regulation of T cells by a superantigen-mediated mechanism. J Allergy Clin Immunol 2000; 105: 820–826. [DOI] [PubMed] [Google Scholar]
- 73.Bunikowski R, Mielke ME, Skarabis H, Worm M, Anagnostopoulos I, Kolde G, et al. Evidence for a disease-promoting effect of Staphylococcus aureus-derived exotoxins in atopic dermatitis. J Allergy Clin Immunol 2000; 105: 814–819. [DOI] [PubMed] [Google Scholar]
- 74.Schlievert PM, Case LC, Strandberg KL, Abrams BB, Leung DY. Superantigen profile of Staphylococcus aureus isolates from patients with steroid-resistant atopic dermatitis. Clin Infect Dis 2008; 46: 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomi NS, Kranke B, Aberer E. Staphylococcal toxins in patients with psoriasis, atopic dermatitis, and erythroderma, and in healthy control subjects. J Am Acad Dermatol 2005; 53: 67–72. [DOI] [PubMed] [Google Scholar]
- 76.Breuer K, Wittmann M, Kempe K, Kapp A, Mai U, Dittrich-Breiholz O, et al. Alpha-toxin is produced by skin colonizing Staphylococcus aureus and induces a T helper type 1 response in atopic dermatitis. Clin Ext Allergy 2005; 35: 1088–1095. [DOI] [PubMed] [Google Scholar]
- 77.Wichmann K, Uter W, Weiss J, Breuer K, Heratizadeh A, Mai U, et al. Isolation of alpha-toxin-producing Staphylococcus aureus from the skin of highly sensitized adult patients with severe atopic dermatitis. Br J Dermatol 2009; 161: 300–305. [DOI] [PubMed] [Google Scholar]
- 78.Aarestrup FM, Larsen HD, Eriksen NH, Elsberg CS, Jensen NE. Frequency of alpha- and beta-haemolysin in Staphylococcus aureus of bovine and human origin. A comparison between pheno- and genotype and variation in phenotypic expression. APMIS 1999; 107: 425–430. [PubMed] [Google Scholar]
- 79.Fleury OM, McAleer MA, Feuillie C, Formosa-Dague C, San-severe E, Bennett DE, et al. Clumping factor B promotes adherence of staphylococcus aureus to corneocytes in atopic dermatitis. Infect Immun 2017; 85. pii: e00994–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harkins CP, Pettigrew KA, Oravcova K, Gardner J, Hearn RMR, Rice D, et al. The microevolution and epidemiology of staphylococcus aureus colonization during atopic eczema disease flare. J Invest Dermatol 2018; 138: 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kwon S, Choi JY, Shin JW, Huh CH, Park KC, Du MH, et al. Changes in lesional and non-lesional skin microbiome during treatment of atopic dermatitis. Acta Derm Venereol 2019; 99: 284–290. [DOI] [PubMed] [Google Scholar]
- 82.Goggin R, Jardeleza C, Wormald PJ, Vreugde S. Corticosteroids directly reduce Staphylococcus aureus biofilm growth: an in vitro study. Laryngoscope 2014; 124: 602–607. [DOI] [PubMed] [Google Scholar]
- 83.Seite S, Bieber T. Barrier function and microbiotic dysbiosis in atopic dermatitis. Clin Cosmet Investig Dermatol 2015; 8: 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castanheira M, Watters AA, Mendes RE, Farrell DJ, Jones RN. Occurrence and molecular characterization of fusidic acid resistance mechanisms among Staphylococcus spp. from European countries (2008). J Antimicrob Chemother 2010; 65: 1353–1358. [DOI] [PubMed] [Google Scholar]
- 85.Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol 2014; 15: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kelhala HL, Aho VTE, Fyhrquist N, Pereira PAB, Kubin ME, Paulin L, et al. Isotretinoin and lymecycline treatments modify the skin microbiota in acne. Exp Dermatol 2018; 27: 30–36. [DOI] [PubMed] [Google Scholar]
- 87.Xu H, Li H. Acne, the Skin Microbiome, and Antibiotic Treatment. Am J Clin Dermatol 2019; 20: 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]