Abstract
Background
Dendritic cells (DCs) are professional antigen presenting cells that initiate immune defense to pathogens and tumor cells. Human tumors contain only few DCs that mostly display a non-activated phenotype. Hence, activation of tumor-associated DCs may improve efficacy of cancer immunotherapies. Toll-like receptor (TLR) agonists and interferons are known to promote DC maturation. However, it is unclear if DCs in human tumors respond to activation signals and which stimuli induce the optimal activation of human tumor DCs.
Methods
We first screened combinations of TLR agonists, a STING agonist and interferons (IFNs) for their ability to activate human conventional DCs (cDCs). Two combinations: TL8-506 (a TLR8 agonist)+IFN-γ and TL8-506+Poly(I:C) (a TLR3 agonist) were studied in more detail. cDC1s and cDC2s derived from cord blood stem cells, blood or patient tumor samples were stimulated with either TL8-506+IFN-γ or TL8-506+Poly(I:C). Different activation markers were analyzed by ELISA, flow cytometry, NanoString nCounter Technology or single-cell RNA-sequencing. T cell activation and migration assays were performed to assess functional consequences of cDC activation.
Results
We show that TL8-506 synergized with IFN-γ or Poly(I:C) to induce high expression of different chemokines and cytokines including interleukin (IL)-12p70 in human cord blood and blood cDC subsets in a combination-specific manner. Importantly, both combinations induced the activation of cDC subsets in patient tumor samples ex vivo. The expression of immunostimulatory genes important for anticancer responses including CD40, IFNB1, IFNL1, IL12A and IL12B were upregulated on stimulation. Furthermore, chemokines associated with CD8+ T cell recruitment were induced in tumor-derived cDCs in response to TL8-506 combinations. In vitro activation and migration assays confirmed that stimulated cDCs induce T cell activation and migration.
Conclusions
Our data suggest that cord blood-derived and blood-derived cDCs are a good surrogate to study treatment responses in human tumor cDCs. While most cDCs in human tumors display a non-activated phenotype, TL8-506 combinations drive human tumor cDCs towards an immunostimulatory phenotype associated with Th1 responses on stimulation. Hence, TL8-506-based combinations may be promising candidates to initiate or boost antitumor responses in patients with cancer.
Keywords: immunotherapy; dendritic cells; interferon inducers; drug therapy, combination; immunity, innate
WHAT IS ALREADY KNOWN ON THIS TOPIC
In human tumors, the majority of conventional dendritic cells shows an inactivated phenotype. It is unclear if they can be converted into activated and immunostimulatory dendritic cells.
WHAT THIS STUDY ADDS
Our study shows that patient tumor-derived conventional dendritic cells from different cancer indications can be activated by Toll-like receptor 8 agonist combinations to express chemokines and proinflammatory cytokines including interleukin-12 and type I interferons.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE AND/OR POLICY
Toll-like receptor 8 agonist-based combinations may enhance antitumor immunity in patients with cancer by targeting and activating conventional dendritic cells in the tumor microenvironment.
Background
Dendritic cells (DCs) are specialized antigen presenting cells, which are essential for the initiation and orchestration of the adaptive immunity against pathogens or tumor cells.1 2 DCs are crucial for priming of tumor antigen-specific T cells in the tumor draining lymph nodes.3 In addition, DCs support T cell effector functions in the tumor microenvironment.4 5 However, the generation of immunostimulatory DCs depends on appropriate activation cues counterbalancing potential suppressive factors in the tumor microenvironment.6 7 Recent single-cell RNA-sequencing (scRNA-seq) data confirmed that only a small population of DCs in human tumors expresses maturation markers such as CCR7 and LAMP3, whereas most DCs display a non-activated phenotype.7 8
DCs in human tumors consist of conventional DC (cDC)1 and cDC2 subsets.7 8 cDC1s are present in blood and tissues in low numbers. They are identified by the expression of CLEC9A, an endocytic receptor for actin filaments exposed on dead cells that favors cross-presentation of cell-associated antigens.9 Therefore, cDC1s can efficiently cross-present endocytosed tumor antigens to CD8+ T cells.10 The more abundant and heterogeneous cDC2s express CD1c and CLEC10A, markers that are shared to some extent with other immune cell types such as monocytes or macrophages.11 cDC2s are thought to mainly activate CD4+ helper T cells because of their high expression of major histocompatibility complex (MHC)-II pathway-associated genes and co-localization with CD4+ T cells in secondary lymphoid organs.12 13 However, some studies have challenged these strict functional separations and highlighted the importance of cDC1–cDC2 crosstalk for effective antitumor immunity.14 Human cDC2s, when properly activated, can produce high amounts of interleukin (IL)-12p70 and efficiently cross-present viral antigens to CD8+ T cells.15 In this context, it is interesting that the phenotypes of cDC1 and cDC2 converge on activation and display a similar gene expression profile.7 8
Harnessing the potential of DCs for cancer immunotherapy has gained interest since several studies have found a correlation of DC gene signatures in human tumors with improved response to immune checkpoint inhibitors and patient survival.16 17 Along the same lines, it was shown in preclinical models that DC activation promoted T cell infiltration into the tumor, restricted tumor growth and prolonged survival.18 19
Pattern recognition receptor (PRR) agonists and interferons (IFNs) can induce DC maturation and promote their immunostimulatory functions.20–22 Although a synergy between PRR agonists and IFNs concerning IL-12p70 expression and T cell activation was described,23 24 it is unclear which stimuli induce the optimal activation of cDCs in human tumors.
Toll-like receptor 8 (TLR8), a PRR, is expressed on both cDC subsets in humans.8 21 Although TLR8 agonists were tested in the past and are currently being evaluated for the treatment of different human cancer indications including head and neck or ovarian cancer in clinical trials (NCT03906526, NCT04460456, NCT02431559, NCT01836029), their effects on human tumor-derived cDC1s and cDC2s are unknown.
Here, we tested various combinations of TLR agonists, a STING agonist and IFNs for their ability to activate cDCs. TLR8 agonist TL8-506 synergized with TLR3 agonist Poly(I:C) or IFN-γ in the expression of combination-specific cytokines in human cord blood and blood cDC subsets. Importantly, TL8-506-based combinations activated both cDC1s and cDC2s derived from tumor explants of patients with cancer. To our knowledge this is the first study that shows the ex vivo activation of patient tumor-derived cDCs from different solid tumor indications and investigates the molecular alterations in activated human tumor cDCs. Our results suggest that targeting TL8-506-based combinations to tumor cDCs may open new opportunities to improve antitumor immunity in patients with cancer.
Materials and methods
Human samples
Human cord blood CD34+ stem cells were purchased from STEMCELL Technologies (#70008). Buffy coats were obtained from the Blood Donation Center Zurich (BASEC-Nr: 2020–01208). Frozen dissociated lung and ovarian tumor cells were purchased from Discovery Life Sciences, Huntsville, Alabama, USA. Melanoma and colorectal tumor samples were obtained from University Hospital of Zurich (BASEC-Nr.2017–00494) and Hirslanden Klinik Zurich (BASEC-Nr: 2016–02013). Declaration of informed consent was signed by all patients for all samples.
Isolation of peripheral blood mononuclear cells
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats using the SepMate PBMC Isolation Tubes (STEMCELL Technologies, #85450) according to the manufacturer’s instructions.
Enrichment of DCs from blood
DCs were enriched from freshly isolated PBMCs using the Miltenyi PanDC Enrichment Kit (#130-100-777) according to the manufacturer’s protocol and 2× enrichment was performed for all experiments.
Expansion of cord blood stem cells
1×106 CD34+ cord blood stem cells were thawed in a 37°C water bath and pipetted into 20 mL pre-warmed StemSpan SFEM medium (STEMCELL Technologies, #09650). Cells were centrifuged and resuspended into StemSpan medium containing 10% fetal bovine serum (FBS) (Sigma, #F4135). Cells were adjusted to 2.5×104 cells/mL, supplemented with 20 ng/mL huIL-3 (PeproTech, #200–03), 100 ng/mL huSCF (PeproTech, #300–07), 100 ng/mL huFLT3L (PeproTech, #300–19), 50 ng/mL huTPO (PeproTech, #300–18), plated at 0.5×104 cells per well in a 96-well U bottom plate and cultured at 37°C in a CO2 incubator. After 7 days of expansion, cells were harvested and frozen in 80% MEM-α (Gibco, #12 561–056), 10% DMSO (Sigma, #D2650), 10% FBS at 2.6×106 cells/mL. Cells were stored in liquid nitrogen.
Differentiation of DCs from cord blood stem cells
Murine stromal MS-5 cells were obtained from DMCZ (#ACC 441) and cultured in MS-5 medium (MEM-α, Gibco, #15 070–063, 10% FBS, 1% Pen/Strep, Gibco, #15 070–063, 2 mM Sodium Pyruvate, Gibco, #11 360–039). For in vitro cord blood DC differentiation, MS-5 cells were treated with 10 µg/mL Mitomycin C (Sigma, #M4287) for 3 hours at 37°C to stop cell proliferation. After washing, cells were detached by Trypsin-EDTA (Gibco, #25 300–054) and plated at 2.5×105 cells/mL in 100 µL volume per well in a 96-well F bottom plate. The day after, expanded CD34+ cord blood stem cells were added to the pre-treated MS-5 cells to start DC differentiation. 2.6×106 frozen cord blood stem cells were thawed and washed once. Cells were resuspended in 20 mL MS-5 medium supplemented with 5 ng/mL huGM-CSF (PeproTech, #300–03), 5 ng/mL huIL-4 (PeproTech, #200–04), 40 ng/mL huSCF and 200 ng/mL huFLT3L. 100 µL cord blood stem cells at 1.3×105 cells/mL were added to each well to the plated MS-5 cells in the 96-well F bottom plates and cultured at 37°C in a CO2 incubator. Cells were fed on day 6 with 50 µL/well MS-5 medium supplemented with 12.5 ng/mL huGM-CSF, 12.5 ng/mL huIL-4, 100 ng/mL huSCF and 500 ng/mL huFLT3L. In vitro differentiated cord blood DCs were harvested on day 12 or 13 after initiation of the culture.
Digestion of tumor tissue
Human tumor tissues were cut with a scalpel into small pieces at room temperature in the presence of 0.5 mL digestion mix (50% Accutase, Sigma, #A6964, 44% MACS Tissue Storage Solution, Miltenyi, #130-100-008, 1% BSA, Sigma, #A9576, 275 U/mL Collagenase IV, Worthington, LS004189, 10 U/mL DNase I Type 4, Sigma, #D5025, 471 U/mL Hyaluronidase, Sigma, #H6254). Pieces were further incubated with 10 mL digestion mix for 20–30 min at 37°C on a Miltenyi MACS rotator with medium speed. Cells were harvested through a 70 µm cell strainer on ice and remaining tumor pieces were carefully mashed through the cell strainer with the plunger of a 10 mL syringe. The tumor digestion solution was then centrifuged for 15 min at 300 × g, 4°C and the supernatant was carefully removed. Dissociated tumor cells were resuspended in cold RPMI medium (Gibco, #42 401–042) and counted. Cells were stored at 1–5×106 cells/mL in freezing medium (IBIDI, #89020) in liquid nitrogen.
Stimulation of DCs
Enriched blood DCs or sorted cord blood cDCs were resuspended at 1×106 cells/mL, PBMCs or tumor digests were resuspended at 5–10×106 cells/mL in DC medium (RPMI GlutaMAX, Gibco, #72 400–021, 1% human serum, Sigma, #H4522, 1% Pen/Strep). 1×105 cells (enriched blood DCs or sorted cord blood cDCs) or 0.5–1×106 cells (PBMCs or tumor digests) in 200 µL/well were treated with the indicated stimuli in a 96-well plate at 37°C in a CO2 incubator. For DC activation in the presence of tumor-conditioned medium, 1:1 diluted supernatant from a 6-hour culture of patient-derived tumor digest in DC medium or a 2-day culture of COR-L105 tumor cell line (ECACC, #92031918) in RPMI GlutaMAX, 10% FBS, 1% Pen/Strep was added to sorted cord blood cDCs during stimulation. The following concentrations were used: 10'000 U/mL huIFN-α (R&D, #11100–1), 10'000 U/mL huIFN-β (R&D, #8499-IF-010), 1 µg/mL huIFN-λ (R&D, #1598-IL-025), 50'000 U/mL huIFN-γ (PeproTech, #300–02), 1 µg/mL Pam3CSK4 (Invivogen, #vac-pms), 10 µg/mL Poly(I:C) (Invivogen, #vac-pic), 0.1 µg/mL LPS (Sigma, #L2880), 1 µM TL8-506 (Invivogen, #tlrl-tl8506), 10 µM CL075 (Invivogen, #tlrl-c75), 10 µM R848 (Invivogen, #vac-r848), 10 µg/mL ssRNA40 (Invivogen, #tlrl-lrna40), 10 µg/mL 2’3’-cGAM(PS)2(Rp/Sp) (Invivogen, #tlrl-nacga2srs).
Quantification of cytokines and chemokines in DC supernatant
1×105 enriched blood DCs or sorted cord blood cDCs were stimulated in 200 µL DC medium/well in a 96-well U bottom plate at 37°C in a CO2 incubator for 18 hours. Stimulated cells were centrifuged at 300 × g for 5 min. DC supernatants were collected and cytokine concentrations were determined using either Cisbio human cytokine HTRF kits (Cisbio, #62HIL12PEG) or customized ProcartaPlex Multiplex kits (Life Technologies).
Flow cytometry
0.5–1×106 PBMCs or tumor digests were stimulated in 200 µL DC medium/well in a 96-well V bottom plate at 37°C in a CO2 incubator for 3 hours in total, with GolgiPlug 1:1000 (BD, #555029) added after 1 hour stimulation. Stimulated cells were centrifuged at 300 × g for 5 min at room temperature. Cells were resuspended in 50 µL/well human BD Fc block 1:50 in eBioscience staining buffer (eBioscience, #0-4222-26) and incubated for 15 min at 4°C. After removal of human BD Fc block, cells were incubated with the indicated antibodies in Brilliant Stain Buffer (BD, #566349) for 20 min at 4°C. For intracellular staining, cells were washed, fixed and permeabilized using the Foxp3 staining buffer set (eBioscience, #00-5523-00). Antibodies for intracellular staining were incubated with cells for 20 min at 4°C. Cells were washed, resuspended in eBioscience staining buffer and acquired on the BD LSRFortessa or BD FACSymphony A5. Data were analyzed using FlowJo V.10. cDC1s were gated as CD45+, CD3−, CD56−, CD16−, CD14−, CD19−, CD68−, MHC-II+, CD11c+ and CLEC9A+ cells. cDC2s were gated as CD45+, CD3−, CD56−, CD16−, CD14−, CD19−, CD68−, MHC-II+, CD11c+ and CD1c+ cells. CXCL9, CXCL10, IFN-λ, TNF-α, CLEC9A and CD68 were stained intracellularly. Further details of the used antibodies are listed in online supplemental table 1.
jitc-2021-004268supp001.pdf (10.3MB, pdf)
Fluorescence-activated cell sorting of cord blood DCs
In vitro differentiated cord blood DCs were harvested on day 12 or 13 and filtered through a 40 µm cell strainer. Cells were centrifuged at 300 × g for 5 min and resuspended at 10×106 cells/mL in ice cold phosphate-buffered saline (PBS) containing 1:50 human BD Fc block (BD, #564220). Cells were incubated for 15 min on ice. Cells were centrifuged at 300 × g for 5 min, human BD Fc block was removed and cells were stained with antibodies in PBS for 20 min on ice. Cells were washed with ice cold PBS, passed through a 35 µm cell strainer and resuspended at 10×106 cells/mL in ice cold PBS for sorting on a BD FACSAria III. cDC1s were sorted as CD45+, CD14−, CLEC9A+ and CD141+ cells. cDC2s were sorted as CD45+, CD14−, CLEC9A−, CD141− and CD1c+ cells. Sorting speed was kept below 4000 events/s.
NanoString nCounter gene expression analysis
1×105 sorted cord blood cDCs were stimulated in 200 µL DC medium/well in a 96-well U bottom plate for 15 hours at 37°C in a CO2 incubator. Cells were collected, centrifuged at 300 × g for 5 min and washed 1× with PBS. 1×105 cells were lysed in 5 µL RLT buffer containing 0.142 M β-mercaptoethanol (Qiagen, #79216, Bio-Rad, #1610710) on ice and vortexed for 1 min. Cell lysates were snap frozen and stored at −80°C. 1.5 µl cell lysate was used for each NanoString reaction. Messenger RNA expression was detected using the human Myeloid Innate Immunity panel (NanoString, #115000171) according to the manufacturer’s instructions. In brief, samples were hybridized to the CodeSet probes for 24 hours at 65°C. Samples were then processed on NanoString nCounter and gene counts were normalized to the 40 reference genes provided by the panel.
Single-cell RNA-sequencing
Patient tumor digests were stimulated at 1×106 cells/well in 200 µL DC medium/well in a 96-well V bottom plate for 4 hours at 37°C in a CO2 incubator. Cells were harvested and incubated with 1:25 human BD Fc block for 10 min on ice. Human BD Fc block was removed and cells were stained with antibodies in PBS for 20 min on ice. Cells were washed, passed through a 35 µm cell strainer and resuspended in ice cold PBS for sorting. Live CD45+, CD3– single cells were sorted for scRNA-seq. 1×104 sorted cells per condition were loaded on the 10x Genomics Chromium Connect. The automated single cell library construction workflow was applied using Single Cell 3’ kit V.3.1 (#PN-1000128, #PN-1000127, #PN-1000213) purchased from 10x Genomics. Pooled libraries were sequenced at the Functional Genomics Center Zurich on the Illumina NextSeq 2000.
ScRNA-seq data analyses
Publicly available scRNA-seq data sets of different tissues (online supplemental table 2) were collected either as raw count matrices or fastq files. Internal and external fastq files were aligned and quantified using the Cell Ranger Single-Cell Software25 with default parameters against the GRCh38 human reference genome. The data were further pre-processed by following the standard workflow in Besca.26 For each data set, the genes showing highest variability using besca.st.highly_variable_genes function were selected. Principal component analysis with 50 components was performed and the first 50 components retained to build a nearest neighbor graph and to derive clusters using the Leiden community detection algorithm.27 To identify the cell type of the clusters returned by the Leiden algorithm, curated signatures and the sig-annot workflow available in Besca were used based on the expression of signature markers, applying the filter parameters in online supplemental table 3.26 Differential expression (DE) analysis between different cell groups was performed using a Wilcoxon rank-sum test and multiple hypothesis testing correction using the Benjamini-Hochberg procedure (function scanpy.tl.rank_genes_groups). Top 50 DE genes based on the highest log2FC change and adjusted p-value less than 0.05 are listed in online supplemental table 4. The Velocyto V.0.17.17 package28 was used to obtain spliced and unspliced read counts from the previously aligned scRNA-seq files from melanoma, lung and colon cancer samples and RNA velocity was calculated using the scvelo V.0.2.3 package.29 The detailed procedure for scRNA-seq data analyses is described in Supplementary Materials.
HEK-Blue reporter cell assay
HEK-Blue huTLR7 (Invivogen, #hkb-htlr7) and HEK-Blue huTLR8 (Invivogen, #hkb-htlr8) were maintained according to manufacturer’s recommendations. 4×104 HEK-Blue cells were treated with the indicated stimuli in 200 µL HEK-Blue Detection medium/well in a 96-well F bottom plate for 18 hours at 37°C in a CO2 incubator. Secreted embryonic alkaline phosphatase (SEAP) expression was measured using HEK-Blue Detection (Invivogen, #hb-det2) according to manufacturer’s instructions.
Activation of naive T cells by DCs
1×105 sorted cord blood cDCs were stimulated in 200 µL DC medium/well in a 96-well U bottom plate for 18 hours at 37°C in a CO2 incubator. Stimulated cDCs were then washed three times with DC medium to remove remaining stimuli. Naive T cells were isolated from PBMCs using the Miltenyi Naive Pan T Cell Isolation Kit (#130-097-095) according to the manufacturer’s protocol. 1×105 isolated naive T cells were co-cultured 5:1 with 2×104 stimulated cDCs for 4 days at 37°C in a CO2 incubator. Cytokine in cell culture supernatants was quantified using the Cisbio human IFN-γ HTRF kit (#62HIFNGPET) or customized ProcartaPlex Multiplex kits (Life Technologies).
DC migration assay
1×105 sorted cord blood cDCs were stimulated in 200 µL/well OPTI-MEM (Gibco, #31985062) in a 96-well U bottom plate for 18 hours at 37°C in a CO2 incubator and supernatant was collected. 100 µL of supernatant were diluted 1:1 with OPTI-MEM and plated in the lower compartment of a transwell plate. Transwell inlets with 5 µm pore size (Corning, #CLS3387-8EA) were used and 5×105 cells of cord blood DC culture at day 13 were added in 100 µL volume to the inlets. DCs were allowed to migrate for 3 hours and the sum of cord blood cDC1s and cDC2s migrated to the lower compartment was quantified by flow cytometry. Normalization for absolute counting of cells was performed using 123count eBeads (Invitrogen, #01-1234-42) according to the manufacturer’s instructions.
CD8+ T cell migration assay
T cell migration assays were performed in three-lane OrganoPlate (MIMETAS, #4004–400-B) using collagen as extracellular matrix barrier and human umbilical vein endothelial cells (HUVECs) (Lonza, #C2517AS) for vessel formation. Isolated CD8+ T cells (Miltenyi, #130-096-495) were activated by CD3/CD28 cross-linking (STEMCELL, #10971) and labeled with CMFDA (Life Technologies, #C7025). T cell migration towards supernatants of stimulated cord blood cDCs or recombinant cytokine dilutions was measured after 48 hours using the PerkinElmer Operetta High Content Imaging System. Quantification of the migrated T cells was done using ImageJ. The detailed experimental procedure is described in Supplementary Materials.
Statistics
For comparison of two groups, Student’s t-test was used. For comparison of multiple groups one-way analysis of variance was performed with Tukey’s multiple comparison correction. Statistical tests were performed using GraphPad Prism V.8. Error bars show SD if not otherwise specified.
Illustrations
The graphical abstract and illustrations were created with BioRender.com.
Results
TL8-506-based combinations synergize to induce the production of proinflammatory cytokines and chemokines by human cord blood DCs
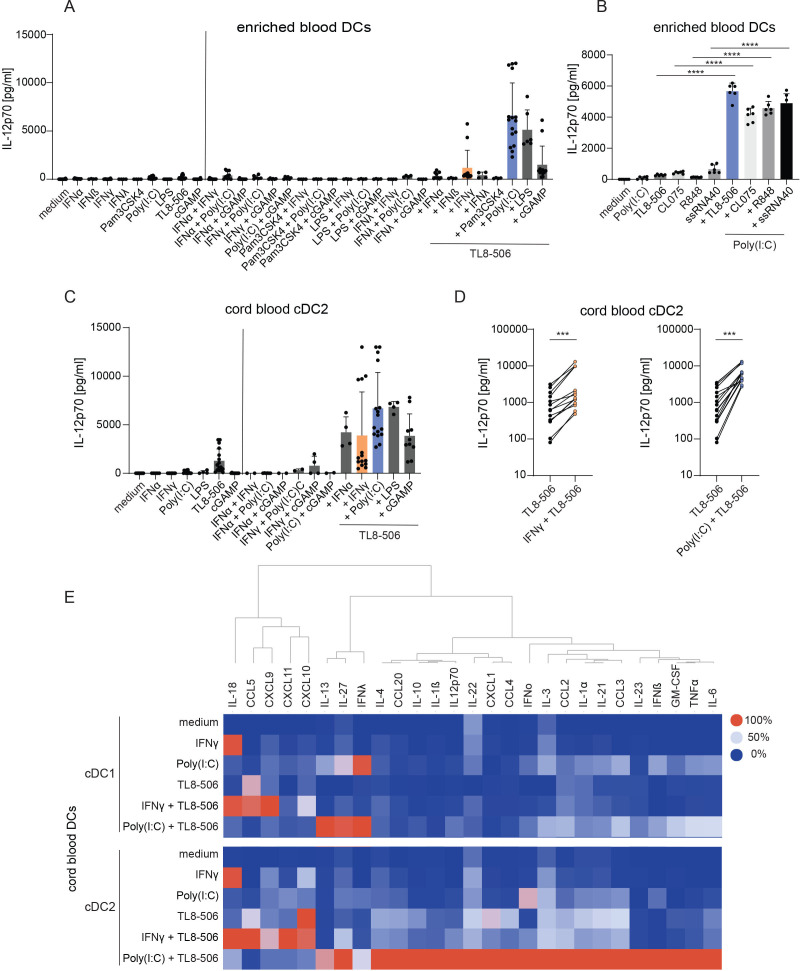
To identify stimuli for tumor DC activation, we screened combinations of TLR agonists, a STING agonist and IFNs, which could induce strong secretion of IL-12p70, an important mediator of DC functions for Th1 T cell responses.30 IL-12p70 supports the expansion of naive T cells during T cell priming and promotes IFN-γ release in T cells.31 32 Different TLR agonists, a STING agonist and IFNs were tested as single stimuli and in combinations in enriched human blood DC cultures. TL8-506, a TLR8-selective agonist, synergized with other stimuli to induce the highest concentrations of IL-12p70 in the supernatant among the tested combinations (figure 1A, online supplemental figure S1). Strong IL-12p70 secretion was observed for TL8-506+Poly(I:C) and this synergistic effect was not restricted to TL8-506, but detected for different TLR8 agonists in combination with Poly(I:C) (figure 1B).
Figure 1.
TL8-506-based combinations synergize in cytokine and chemokine secretion in cord blood DC subsets. (A, B) Enriched blood DCs were stimulated for 18 hours with the indicated stimuli. IL-12p70 concentrations were measured in the cell culture supernatant by ELISA. (A) IL-12p70 concentrations in enriched blood DC cultures on stimulation with combination of agonists, n=two–eight donors, from one to six independent experiments, mean+SD is shown. (B) IL-12p70 concentrations in enriched blood DC cultures on stimulation with combination of Poly(I:C) and different TLR8 agonists, n=4 donors, two independent experiments, representative data of two donors from one experiment, mean+SD is shown. One-way analysis of variance was used for statistical analysis, ****p≤0.0001. (C – E) Sorted cord blood DCs were stimulated for 18 hours with the indicated stimuli. Cytokine concentrations were determined in the cell culture supernatant by ELISA. (C) IL-12p70 concentrations in the supernatant of sorted cord blood cDC2s on stimulation with combination of agonists, n=two–seven batches of cord blood from mixed donors, from two to eight independent experiments, mean+SD is shown. (D) IL-12p70 concentrations in the supernatant of cord blood cDC2s treated with TL8-506 alone or in combination with IFN-γ or Poly(I:C), n= seven batches of cord blood from mixed donors, 14 donors in total, from seven independent experiments, paired Student’s t-test, ***p≤0.0002. (E) Cytokine concentrations in the supernatant of cord blood cDC1s and cDC2s treated with the indicated stimuli, n=three batches of cord blood from mixed donors, 6 donors in total, from three independent experiments, colors displaying the maximum (100%) to minimum (0%) mean cytokine concentrations per column. The following concentrations were used for DC stimulation: 10'000 U/mL huIFN-α, 10’000 U/mL huIFN-β, 1 µg/mL huIFN-λ, 50'000 U/mL huIFN-γ, 1 µg/mL Pam3CSK4, 10 µg/mL Poly(I:C), 0.1 µg/mL LPS, 1 µM TL8-506, 10 µM CL075, 10 µM R848, 10 µg/mL ssRNA40, 10 µg/mL 2’3’-cGAM(PS)2(Rp/Sp). cDC, conventional DC; DC, dendritic cell; IFN, interferon; IL, interleukin.
To study the effects of the combinations in specific cDC subsets and exclude the possibility that IL-12p70 was produced by other cells in the enriched blood DC fractions, we sorted cord blood stem cell-derived cDCs before stimulation with the different TL8-506 combinations. cDCs differentiated in vitro from cord blood stem cells can be generated in higher numbers and transcript expression analysis showed comparable overall gene expression to blood and tumor cDCs (online supplemental figure S2A, B). This assessment was based on an integrated collection of reanalyzed publicly available data that resulted in a reference map of cDCs across tissues and cancer indications (online supplemental table 2).7 8 33–43 We performed scRNA-seq of in vitro differentiated cord blood cDCs and blood cDCs, and compared their gene expression with those present in healthy tissues and different tumor indications of public scRNA-seq studies. Both cord blood cDC subsets expressed the cDC subset-specific markers, PRRs and IFNRs (online supplemental figure S2C, D), suggesting their relevance for our screening experiments. In accordance with the findings in enriched blood DC cultures, TL8-506 combinations induced the highest amount of IL-12p70 in supernatants of cord blood cDCs among the tested treatments (figure 1C). Synergy in IL-12p70 induction by IFN-γ+TL8-506 and Poly(I:C)+TL8-506 stimulation was consistently detected in all tested donors and experiments (figure 1D). We selected these two TL8-506 combinations to study in more detail, as IFN-γ, TLR3 agonists and TLR8 agonists are potentially clinically relevant molecules, which are currently being evaluated in clinical trials for cancer treatment (NCT02614456, NCT02834052, NCT01976585, NCT03732547).
Next, we analyzed the expression of chemokines and cytokines other than IL-12p70. As a single stimulus, each compound induced a specific cytokine profile and combinations selectively synergized in the expression of certain cytokines (figure 1E). TL8-506 in combination with Poly(I:C) induced the highest release of IL-12 family cytokines (IL-12p70, IL-23, IL-27), IFNs (IFN-β, IFN-λ), different proinflammatory cytokines (tumor necrosis factor (TNF)-α, IL-6, IL-1α, IL-1β) and chemokines (CCL3, CCL4, CCL20). IFN-γ+TL8-506 synergized in the induction of IFN-γ-inducible chemokines (CXCL9, CXCL10, CXCL11). Gene expression data further supported these combination-specific synergies (online supplemental figure S3A, B). For most cytokines including IL-12p70, secretion was stronger in cDC2s compared with cDC1s in response to treatment with TL8-506 combinations (figure 1E).
TL8-506-based combinations synergize to activate human blood DC subsets
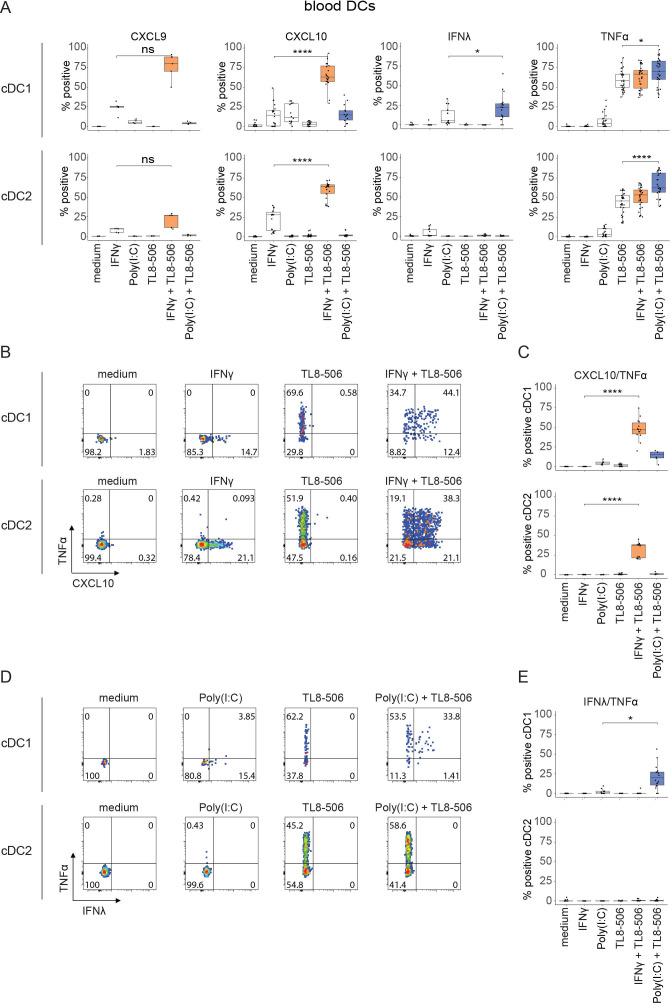
We next tested the effects of TL8-506-based combinations in cDC subsets derived from peripheral blood, a more physiologically relevant source of cDCs. As the numbers of blood cDCs were limiting, we focused on a selection of cytokines and chemokines that were synergistically induced by TL8-506-based combinations (figure 1E). Consistent with observations in cord blood cDCs, CXCL9 and CXCL10 expression was induced by IFN-γ and the percentage of CXCL9 or CXCL10 positive cDCs was further increased by the combination with TL8-506. Similarly, IFN-λ was induced by Poly(I:C) in cDC1s and its expression was further amplified by combination with TL8-506. TNF-α was induced by TL8-506 and combination with Poly(I:C) further enhanced this effect (figure 2A). Co-production of TNF-α and CXCL10 was only induced by combinatorial IFN-γ+TL8-506 stimulation (figure 2B, C) whereas the co-expression of TNF-α and IFN-λ was only triggered by combinatorial Poly(I:C)+TL8-506 treatment (figure 2D, E). Taken together, our data show that similar to cord blood cDCs, blood cDC1s and cDC2s are activated by TL8-506 combinations, which synergize in a stimulus-specific manner.
Figure 2.
TL8-506-based combinations synergize in the production of cytokines and chemokines in human blood DC subsets. Human peripheral blood mononuclear cells (PBMCs) were treated with the indicated stimuli for three hours. Intracellular cytokine staining was performed and analyzed by flow cytometry. (A) Quantification of cytokine producing blood cDCs, n=two–11 donors, from two to 16 independent experiments. (B) Flow cytometry plots of IFN-γ+TL8-506 and single stimuli treated blood cDCs showing the distribution of CXCL10 or/and TNF-α expressing cDCs. Percentage of positive cells are indicated in the quadrants. (C) Quantification of CXCL10/TNF-α double positive blood cDCs on stimulation with the indicated stimuli, n=four–eight donors, from four to eight independent experiments. (D) Flow cytometry plots of Poly(I:C)+TL8-506 and single stimuli treated blood cDCs showing the distribution of IFN-λ or/and TNF-α expressing cDCs. Percentage of positive cells are indicated in the quadrants. (E) Quantification of IFN-λ/TNF-α double positive blood cDCs on stimulation with the indicated stimuli, n=four–six donors, from four to six independent experiments. Statistics for A, C, E: paired Student’s t-test, *p≤0.03, ****p≤0.0001, ns, not significant. The following concentrations were used for DC stimulation: 50'000 U/mL huIFN-γ, 10 µg/mL Poly(I:C), 1 µM TL8-506. cDC, conventional DC; DC, dendritic cell; IFN, interferon; TNF, tumor necrosis factor.
TL8-506 combinations activate DCs in human tumors
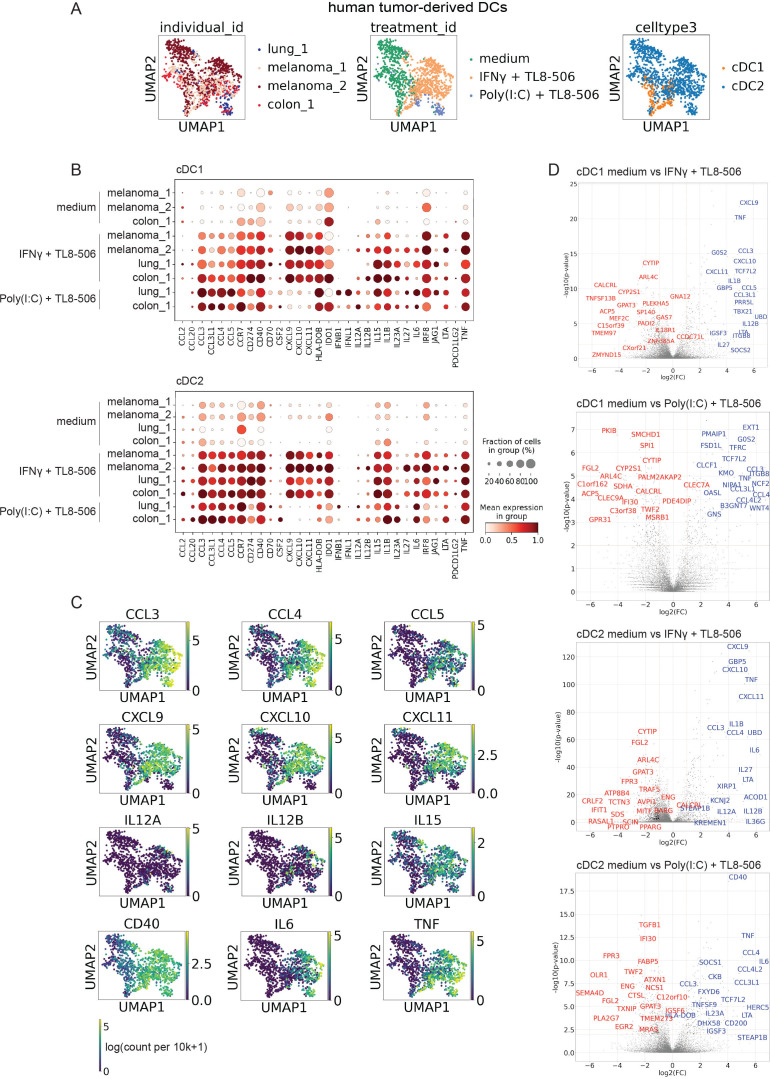
cDCs with an activated phenotype are scarce in different human tumor indications (online supplemental figure S4A, B), therefore we attempted to test whether the two selected TL8-506 combinations could activate cDCs in patient tumor explants ex vivo. To comprehensively analyze the effects of TL8-506 combinations on patient tumor-derived cDCs, we performed scRNA-seq of stimulated tumor cDCs derived from two melanoma samples of lymph node metastases, one colorectal cancer (CRC) sample of liver metastasis and one lung adenocarcinoma sample. Percentages of CD45+ cells in tested samples are shown in the supplementary files (online supplemental table 5). IFN-γ+TL8-506 and Poly(I:C)+TL8-506 stimulated cDCs clustered separately from untreated cDCs (figure 3A) and showed upregulation of many known DC activation markers (figure 3B). Important signals for T cell activation such as IL12A, IL12B, IL15, CD40 were induced by both TL8-506 combinations. In contrast, the expression of the CD8+ T cell recruiting chemokines CXCL9, CXCL10, CXCL11 and their co-expression with TNF was specific to the IFN-γ+TL8-506 combination (figure 3B, C). The expression of IFNB1 and IFNL1 was specific to the Poly(I:C)+TL8-506 combination, as previously observed for cord blood and blood-derived cDCs. Of note, both cDC1s and cDC2s showed comparable gene induction and increased IL12A and IL12B expression on stimulation with the two TL8-506 combinations. Differential expression (DE) analysis confirmed that CXCL9, CXCL10, CXCL11 or IFNB1 were among the DE genes specifically induced on IFN-γ+TL8-506 or Poly(I:C)+TL8-506 stimulation, respectively (figure 3D, online supplemental table 4). We have focused our analyses on cytokines and chemokines as readouts for cDC activation since we have observed strongest synergy in the induction of cytokines and chemokines by TL8-506 combinations compared with the expression of genes involved in co-stimulation and antigen presentation. However, TL8-506 combinations also upregulated other DC maturation markers including co-stimulatory molecules CD80, CD83, CD86 and components of the antigen presentation machinery such as TAP1, TAP2 and HLA-DOB (online supplemental figure S5A, B). The changes in gene expression in activated cDCs are likely regulated by both TLR and IFN signaling (online supplemental figure S6A, B). In summary, we demonstrate that tumor-derived cDCs from multiple indications respond to TL8-506 combinations and upregulate different relevant DC activation markers. In addition, the scRNA-seq analysis of treated tumor-derived cDCs revealed that the combination-specific induction of different cytokines and chemokines was remarkably conserved between cord blood, blood and patient tumor cDCs.
Figure 3.
TL8-506-based combinations activate human tumor-derived cDCs. Digested tumor samples from patients with melanoma, colorectal cancer and lung cancer were treated with 1 µM TL8−506+50’000 U/mL IFN-γ or 1 µM TL8−506+10 µg/mL Poly(I:C) for four hours. 10x Genomics scRNA-seq was performed on fluorescence-activated cell sorting (FACS) sorted CD45+, CD3– cells, n=four donors, from three independent experiments. (A) UMAP of 1264 cDCs profiled across all tumor samples with each cell color-coded for patient (left), treatment (middle) and cDC subset (right). (B) Fraction positive and mean expression of activation markers in cDC1s or cDC2s from IFN-γ+TL8-506 or Poly(I:C)+TL8-506 stimulated and control tumor samples. Mean expression was calculated across all the cells in the group and then scaled to a 0–1 range. (C) UMAP of tumor-derived cDCs showing expression values of different activation markers in log(count per 10k+1). (D) Volcano plots showing differentially expressed (DE) genes (blue: upregulated, red: downregulated) between stimulated and control treated cDCs from tumor samples. Top 20 DE genes were labeled based on p-values and fold changes, Wilcoxon rank-sum test. Top 50 DE genes ranked by log2 fold change (log2FC) are listed in online supplemental table 4. cDC, conventional DC; DC, dendritic cell; IFN, interferon; IL, interleukin; scRNA, single-cell RNA; TNF, tumor necrosis factor.
TL8-506 combinations induce a Th1-supportive gene signature in human tumor-derived DCs
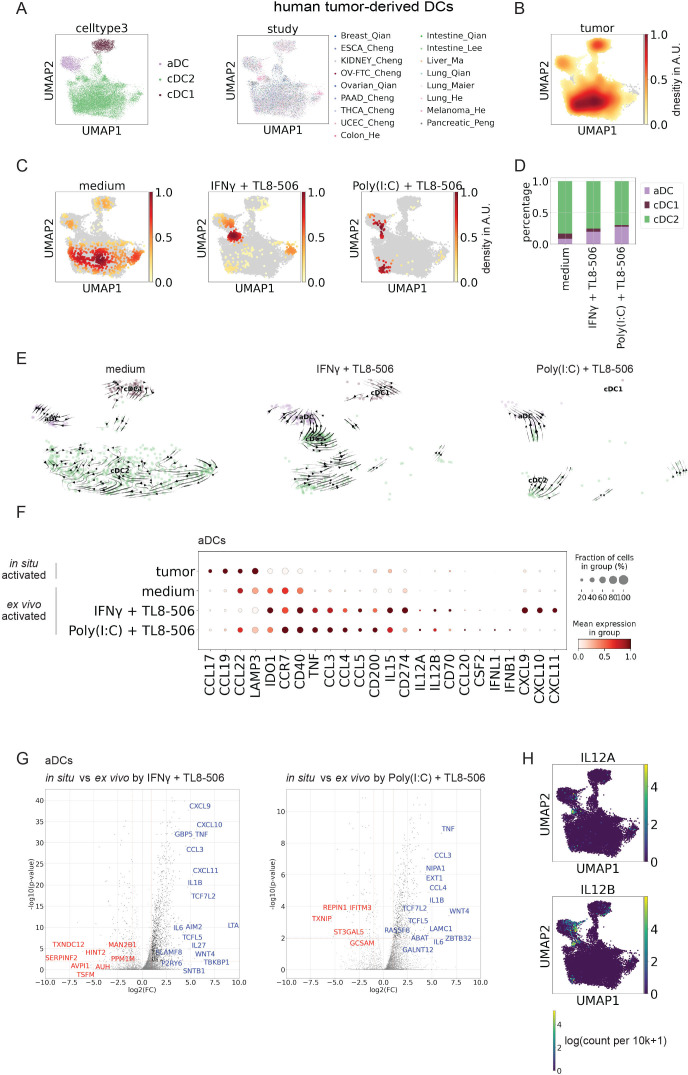
Since cDCs with an activated phenotype are present in different tumor indications (figure 4A, B, online supplemental figure S4A, B), we were interested in comparing the expression profile of treated tumor-derived cDCs to tumor cDCs activated in situ in human tumors. After four hours stimulation with TL8-506 combinations most treated cDCs clustered closely with the in situ activated DC population (figure 4C, D). Some of these treated cDCs fell into the in situ activated DC cluster and others were localized as an intermediate population between the cDC2 and the in situ activated DC cluster.
Figure 4.
Human tumor-derived cDCs show an immunostimulatory phenotype associated with Th1 responses on stimulation with TL8-506 combinations. (A) UMAP of 18'651 human tumor-derived cDC1s, cDC2s and activated DCs (aDCs) profiled across different single-cell RNA-sequencing studies with each cell color-coded for cDC subset (left) and study (right). (B) Gaussian kernel density plot of human tumor-derived cDCs from integrated public studies, AU, arbitrary units. (C) Gaussian kernel density plot of human tumor-derived cDCs on treatment with TL8-506 combinations; AU, arbitrary units. (D) Quantification of activated DCs in patient tumor digests on treatment with TL8-506 combinations. (E) Velocity analysis of human tumor cDCs on treatment with TL8-506 combinations. (F) Fraction positive and mean expression of activation markers in ex vivo activated tumor DCs on treatment with TL8-506 combinations or in situ activated tumor DCs from integrated public studies. Mean expression was calculated across all the cells in the group and then scaled to a 0–1 range. (G) Volcano plots showing differentially expressed (DE) genes (blue: upregulated, red: downregulated) between ex vivo activated tumor DCs on treatment with TL8-506 combinations and in situ activated tumor DCs from integrated public studies. Top 20 DE genes scored by p-values and fold changes are labeled, Wilcoxon rank-sum test. Top 50 DE genes ranked by log2 fold change (log2FC) are listed in online supplemental table 4. (H) UMAP plots showing expression values of IL12A and IL12B in tumor cDCs across different studies in log(count per 10k+1). cDC, conventional DC; DC, dendritic cell; IFN, interferon; IL, interleukin.
The fraction of unspliced versus spliced reads was recently employed to show a differentiation trajectory, referred to as a cell’s velocity.28 To estimate the likely directionality of transitions of stimulated tumor cDC subsets, we performed velocity analysis. Our results suggest that in response to TL8-506-based combinations activated tumor DCs are derived from immature cDCs (figure 4E). Compared with in situ activated cDCs present in cancer tissues, cDCs activated by TL8-506-based combinations showed increased expression of many DC activation markers including IL12A, IL12B, CD40, IFNB1 and IFNL1 (figure 4F, G), across the analyzed tumor indications (online supplemental figure S7A, B). In addition, tumor cDCs activated by TL8-506 combinations displayed higher expression of CXCL9, CXCL10, CXCL11 and CCL4, chemokines associated with CD8+ T cell recruitment.5 44 In contrast, cDCs activated in situ by the tumor microenvironment showed increased expression of CCL17 and CCL22, chemokines described to recruit CD4+ regulatory T cells.45 Consistent with earlier reports, the highest levels of IL12A and IL12B were observed in the activated DC cluster, validating our readout to screen and select the best cDC activating compounds (figure 4H). In conclusion, our data show that most tumor-derived cDCs can be activated by TL8-506-based combinations. Activated cDCs generated by ex vivo stimulation acquired a phenotype that is associated with a Th1 response, displaying increased expression of IL12A, IL12B, IFNB1, IFNL1, CD40 and CD8+ T cell chemokines when compared with cDCs activated in situ by the tumor microenvironment.
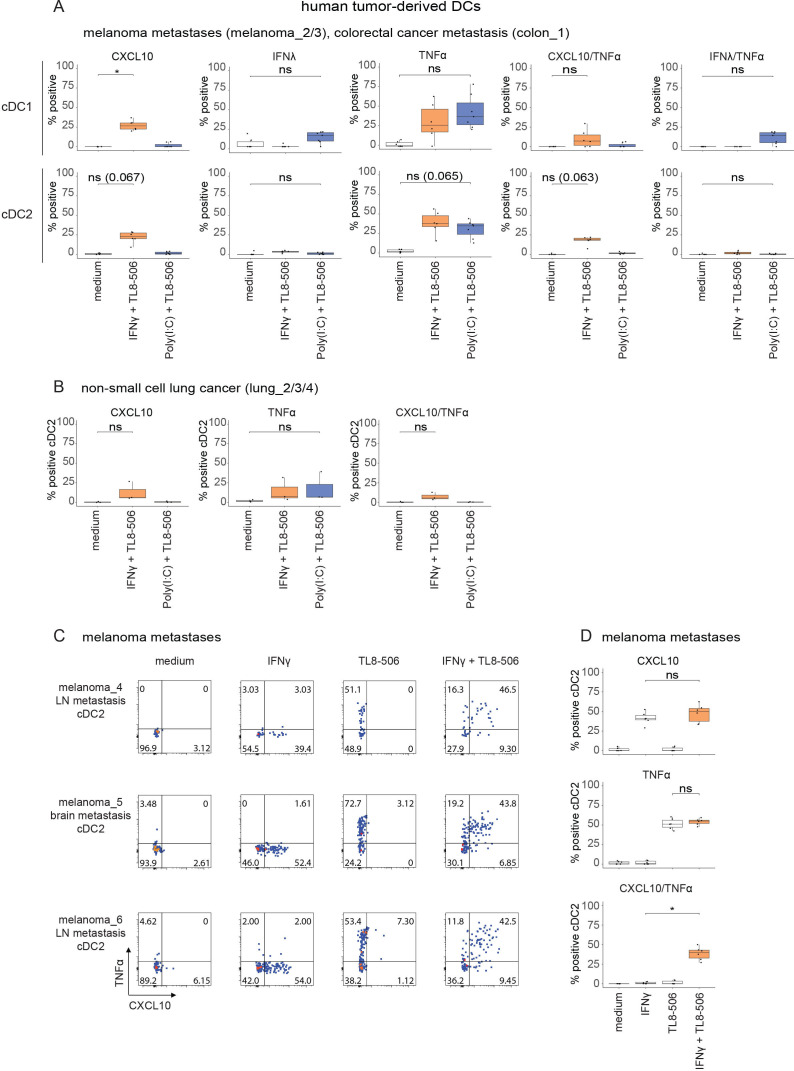
As transcript and protein levels do not necessarily correlate, we validated selected genes for which antibodies were available by intracellular flow cytometry. In agreement with our earlier observations (figure 2B, D), metastatic melanoma-derived, CRC-derived (figure 5A) and non-small cell lung cancer-derived (figure 5B) cDCs co-expressed intracellular TNF-α and CXCL10 proteins on IFN-γ+TL8-506 treatment. Treatment of tumor-derived cDC1s with Poly(I:C)+TL8-506 induced the co-expression of intracellular TNF-α and IFN-λ proteins, but not TNF-α and CXCL10 (figure 5A). The availability of sufficient patient-derived melanoma material allowed us to also test the individual effects of TL8-506 and IFN-γ on tumor-derived cDCs. Consistent with our observations in blood cDCs, TL8-506 or IFN-γ by themselves did not induce the co-expression of intracellular TNF-α and CXCL10 proteins in tumor-derived cDCs (figure 5C, D). In conclusion, our data confirm that TL8-506-based combinations induce the stimulus-specific protein expression of cytokines and chemokines in tumor-derived cDCs.
Figure 5.
TL8-506-based combinations induce stimulus-specific cytokine protein expression in human tumor-derived cDCs. Tumor digests from patients with melanoma, CRC and lung cancer were treated with 1 µM TL8−506+50’000 U/mL IFN-γ or 1 µM TL8-506+10 µg/mL Poly(I:C) for three hours. Intracellular cytokine staining was performed for CXCL10, IFN-λ and TNF-α. Protein expression in tumor-derived cDCs was analyzed by flow cytometry. (A) Quantification of indicated cytokine producing cDC1s and cDC2s from digested metastases of patients with melanoma and CRC on treatment with TL8-506 combinations, n=three donors, from three independent experiments. (B) Quantification of indicated cytokine expressing cDC2s in non-small cell lung cancer digests on treatment with TL8-506 combinations, n=three donors, from three independent experiments. (C) Flow cytometry plots of IFN-γ+TL8-506 or single agent treated cDC2s from digested melanoma lymph node (LN) and brain metastases showing the distribution of CXCL10 or/and TNF-α expressing cDC2s. Percentage of positive cells are indicated in the quadrants. (D) Quantification of CXCL10 or/and TNF-α expressing tumor-derived cDC2s from C, n=three donors, from three independent experiments. Statistics for A, B, D: Student’s paired t-test, *p≤0.03, ns, not significant. The following concentrations were used for DC stimulation: 50'000 U/mL huIFN-γ, 10 µg/mL Poly(I:C), 1 µM TL8-506. cDC, conventional DC; CRC, colorectal cancer; DC, dendritic cell; IFN, interferon; TNF, tumor necrosis factor.
DC activation by Poly(I:C)+TL8-506 results in enhanced T cell activation, DC and T cell recruitment
The previous results suggest that selected cytokines and chemokines are produced by tumor-derived cDCs in response to TL8-506-based combinations. To better understand the functional relevance of the factors secreted by tumor-derived cDCs on stimulation with TL8-506-based combinations, we performed co-culture and migration assays. We sorted cord blood cDCs as cDCs from patient tumor material were insufficient in numbers to perform functional assays and the response of cord blood cDCs to TL8-506 combinations was comparable to those of tumor-derived cDCs (figures 1E and 3B).
First, we assessed the effects of activated cDCs on Th1 differentiation using the production of IFN-γ as a proxy. Sorted cDCs were treated for 18 hours and the stimuli were removed by several washing steps. Activated cDCs were then co-cultured with allogeneic naive T cells and after four days the IFN-γ concentrations in the supernatant were measured. cDCs stimulated with Poly(I:C)+TL8-506 induced the highest IFN-γ concentrations among the tested stimuli with a 140-fold increase compared with unstimulated cDCs (figure 6A). In addition, we investigated the induction of other Th-associated cytokines in DC/T cell co-cultures such as IL-4 and IL-10 and did not observe their secretion (online supplemental figure S8).
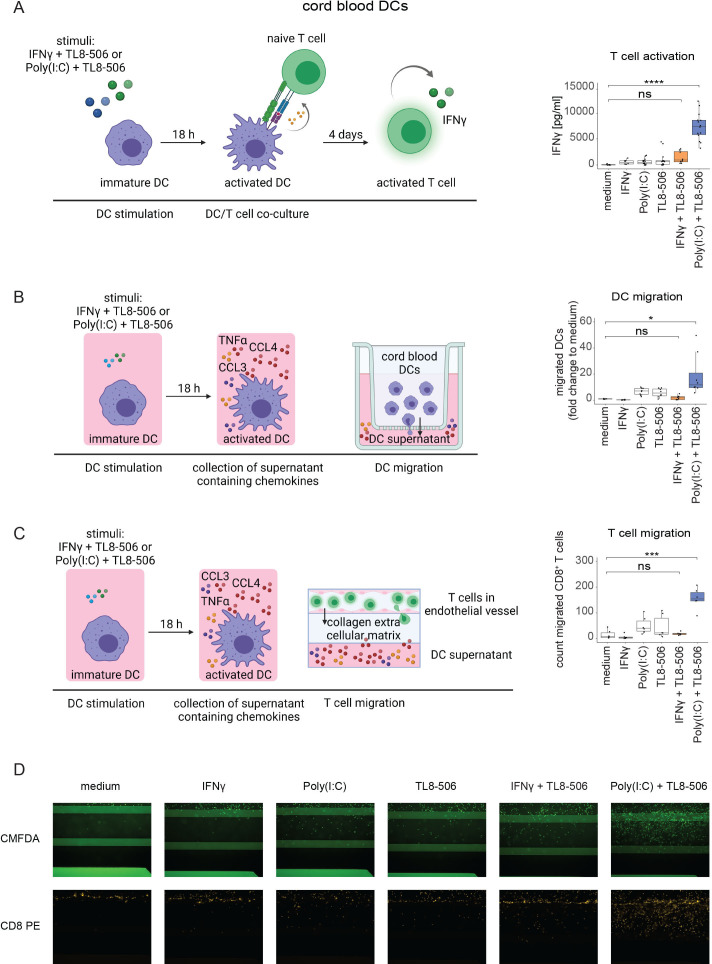
Figure 6.
Poly(I:C)+TL8-506 stimulated cord blood cDCs induce T cell activation, DC and T cell recruitment. The experimental set-up for the functional assays is shown on the left. (A) Sorted cord blood cDC2s were stimulated with the indicated compounds for 18 hours. Treated cDC2s were washed and co-cultured with allogeneic naive T cells for four days. IFN-γ concentrations in the supernatant were determined by ELISA as a readout for T cell activation. IFN-γ concentrations are shown on the right, n=three batches of cord blood from mixed donors, six T cell donors, from four independent experiments. (B) Sorted cord blood cDC2s were stimulated with the indicated compounds for 18 hours. The supernatants were collected and placed into the lower compartment of a transwell. Untreated cord blood DCs were added into the transwell insert and allowed to migrate for three hours. Fold change to medium of migrated cord blood cDC1s+cDC2s is depicted on the right, n=four batches of cord blood from mixed donors, from four independent experiments. (C) Sorted cord blood cDC2s were stimulated with the indicated compounds for 18 hours. The supernatants were collected and placed into the bottom channel of a three-dimensional tissue culture device. Activated CD8+ T cells were labeled with CMFDA and added to the top channel that was coated with an artificial endothelial vessel. Migrated T cells were detected after 48 hours by imaging of the collagen layer that separated the two channels. Cell counts of migrated CD8+ T cells are shown on the right, n=two batches of cord blood from mixed donors, three T cell donors, from three independent experiments. (D) Representative microscopy images of CD8+ T cell migration described in C using the Operetta system at 10x magnification. One-way analysis of variance was used for statistical analysis, *p≤0.03, ***p≤0.0002, ****p≤0.0001, ns, not significant. The following concentrations were used for DC stimulation: 50'000 U/mL huIFN-γ, 10 µg/mL Poly(I:C), 1 µM TL8-506. Illustrations were created with BioRender.com. cDC, conventional DC; DC, dendritic cell; IFN, interferon; TNF, tumor necrosis factor.
Next, we analyzed how the different chemokines induced by TL8-506-based combinations impact the recruitment of DCs. Supernatants of stimulated cDCs were placed in the lower compartment of a transwell. Untreated cord blood DCs were added into the transwell inserts and DC migration was measured after three hours by quantification of cDCs in the lower compartment by flow cytometry. Treatment of cDCs with Poly(I:C)+TL8-506 induced a 20-fold higher recruitment of cDCs compared with untreated cDCs (figure 6B). This was likely related to our observation that Poly(I:C)+TL8-506 synergized in the secretion of CCL3, CCL4 and CCL20, a set of chemokines described to promote DC migration.46 47 Since the supernatants also contained the TLR agonists, it cannot be excluded that the agonists have influenced the migratory capacity of DCs in the presence of cytokines and chemokines released by activated cDCs.
T cell migration was studied using a three-dimensional (3D) tissue culture platform in which activated CD8+ cells were allowed to transmigrate through an artificial endothelial vessel and collagen layer towards supernatants of stimulated cDCs. Migrated T cells were quantified after 48 hours by imaging of the collagen layer. Poly(I:C)+TL8-506 treatment of cDCs led to a ten-fold increase in CD8+ T cell recruitment when compared with untreated cDCs (figure 6C, D). In contrast, IFN-γ+TL8-506 failed to induce the recruitment of significant numbers of CD8+ T cells. IFN-γ+TL8-506 stimulation induced the strongest CXCL10 secretion while Poly(I:C)+TL8-506 treatment induced the highest CCL4 and TNF-α release in cDCs among the tested combinations (online supplemental figure S9A). CXCL10, CCL4 and TNF-α are all associated with CD8+ T cell chemotaxis.48–50 Hence, it is possible that the CCL4/TNF-α axis was more efficient in attracting CD8+ T cells in our 3D system. Blocking of CCL4 in cDC supernatants had no impact on CD8+ T cell migration suggesting that the recruitment depends on additional chemokines (online supplemental figure S9B). However, recombinant CCL4 and TNF-α at concentrations present in supernatants of Poly(I:C)+TL8-506 treated cDCs were sufficient to induce CD8+ T cell migration (online supplemental figure S9C). In summary, factors released by cDCs in response to Poly(I:C)+TL8-506 stimulation increased the recruitment of cDCs, CD8+ T cells and T cell activation. Hence, treatment of patients with cancer with TLR8 agonist-based combinations may help to inflame tumors, enhance tumor specific CD8+ T cell activities and boost anticancer immunity in patients.
Discussion
Most cDCs in human tumors show a non-activated phenotype.7 8 Two potential non-mutually exclusive explanations have been proposed in the literature: (1) Lack of cDC activating signals in the tumor; (2) the tumor microenvironment releases factors that preclude the differentiation of immunostimulatory cDCs. We stimulated human tumor cDCs derived from different cancer indications with TL8-506-based combinations, which induced the strongest activation of cord blood and blood cDCs among the tested treatments. We found that a majority of human tumor-derived cDCs were activated by TL8-506-based combinations ex vivo. Hence, our data indicate that most tumor-derived cDCs are responsive to activating signals. In accordance, transcriptome analysis revealed no major differences between cDCs present in healthy tissues and different cancer indications.
Low numbers of cDCs with an activated phenotype are observed in different human tumor indications. These in situ activated cDCs are characterized by the expression of CCL17, CCL19 and CCL22, chemokines associated with CD4+ regulatory T cell recruitment.44 45 They also show increased expression of both co-stimulatory and co-inhibitory molecules such as CD40 and CD274 when compared with non-activated cDCs.7 However, no or only very few transcripts for factors associated with antitumor responses were detected including IL12A, IL12B and IFNB1. Interestingly, human tumor cDCs activated by TL8-506 combinations showed increased expression of IL12A, IL12B, IFNB1, chemokines for CD8+ T cells including CXCL9, CXCL10 and CCL4 but decreased expression of CCL17, CCL19 and CCL22 when compared with in situ activated tumor cDCs. Functional assays using in vitro activated cord blood cDCs confirmed that cDC-derived factors mediated CD8+ T cell recruitment and activation. In summary, our data indicate that inadequate or insufficient levels of activating signals are present in the tumor to induce proinflammatory factors associated with Th1 responses in cDCs. Importantly, TL8-506-based combinations were sufficient to induce a Th1-like phenotype in tumor cDCs even though tumor cells or the tumor microenvironment might release suppressive factors that counteract DC activation (online supplemental figure S10A, B). Future experiments will be required to explore the effects of in situ and ex vivo activated tumor DCs on T cell activation and recruitment in more detail.
In patients, it is possible that additional cells contribute to the proinflammatory effects of TL8-506-based combinations. TLR3 and TLR8 are preferentially, but not exclusively expressed on cDCs while IFN-γ receptors (IFNGRs) are ubiquitously expressed.51 Macrophages express both TLR8 and IFNGRs.52 Tumor-derived macrophages upregulated CXCL10 and TNF-α expression on IFN-γ+TL8-506 treatment, but failed to induce IL12A, IL12B, IFNB1 or IFNL1 on stimulation with TL8-506 combinations (online supplemental figure S11A, B).
Different human cDC sources have been explored for drug screenings to predict treatment effects on human tumor cDCs due to their scarcity.53 54 We tested the usefulness of in vitro differentiated cord blood and blood cDCs that can be isolated in greater numbers to predict the response of tumor cDCs. Cord blood cDC1s and cDC2s formed well separated clusters in scRNA-seq analyses despite the fact that cord blood cDC1s expressed some cDC2-specific markers and vice versa. Using different readouts, we found that the response of in vitro differentiated cord blood and blood cDCs to different stimuli was strikingly similar compared with tumor-derived cDCs although the cultures contained other cell types that might have influenced the activation of cDCs (online supplemental figure S12A–C). In a stimulus-specific manner, IFN-γ induced CXCL10 production, Poly(I:C) induced IFN-λ expression and TL8-506 treatment resulted in TNF-α expression in cord blood, blood and tumor-derived cDCs. Surprisingly, only the stimulation with Poly(I:C)+TL8-506 but not IFN-γ+TL8-506 led to IFN-β expression in cord blood and tumor cDCs. However, it is possible that the experimental set-up influenced the IFN-β expression as the induction of IFN-β by TLR8 agonists in human DCs was inconsistent in previous studies.55 56 We conclude that in vitro differentiated cord blood and peripheral blood cDCs are useful surrogates to test the ability of drug candidates to activate human tumor-associated cDCs.
One focus of the study was also to compare treatment responses of human cDC1s and cDC2s. Although activated cDC1 and cDC2 clustered together and had similar overall gene expression profile, selected genes were differentially induced. As previously described in the literature, IFN-λ was selectively induced by Poly(I:C) in cDC1s.17 Poly(I:C) consistently showed stronger effects on cDC1s compared with cDC2s, which might be explained by the higher TLR3 expression in cDC1s.21 In contrast, we did not identify any cytokine that was selectively induced in cDC2s on stimulation with TL8-508 combinations. This highlights the importance of differential PRR expression in human cDCs and how it may guide the selection of appropriate agonists to target different cDC subsets.
TLR3 agonists, TLR8 agonists and IFN-γ are currently being tested for cancer treatment in different clinical trials.57 58 The results from past clinical trials have been rather disappointing showing no significant improvement in patient outcome, probably due to systemic toxicities and the lack of preferential accumulation in the tumor of untargeted molecules. cDC-targeted or/and tumor-targeted versions of current TLR3 and TLR8 agonists in development might improve treatment efficacy while reducing toxicity. To exploit synergistic effects in cDC activation, dual agonists can be designed to activate both TLR3 and TLR8 to increase their potency specifically in cDCs because they co-express TLR3 and TLR8. In addition, cDC activation in combination with radiotherapy or immune checkpoint inhibition has shown promising results in many preclinical models and may further improve treatment efficacy. Our study provides insights for the rational combination of cDC agonists to optimally activate human tumor cDCs and improve antitumor immunity in patients with cancer.
Acknowledgments
We thank Professor Christian Münz for the scientific inputs. We thank Thuy Trinh Nguyen for her support in T cell migration assays. Many thanks to Ramona Schlenker, Marisa Mariani and Emilio Yangueez for their support in single-cell RNA-sequencing. We thank Regula Buser for the fluorescent labeling of antibodies. We thank Said Aktas for his input for the NanoString and statistical analyses. We want to thank Eva Sum, Floriana Cremasco, Philipp Fröbel and Lucia Campos for the helpful discussions. Finally, we thank all our colleagues from Roche for their continuous support through all phases of this project.
Footnotes
Twitter: @bsoni08, @vandenBroek_Lab
Contributors: Concept and experimental design: MH, SC, IM, MvdB, SG. Acquisition of data: MH, YP, CW, LK. Data analysis and interpretation: MH, BS, PCS, MvdB, SG. Assay development: MH, TH, IM, CW, DDS. Provision of study materials: MPL, RD. Writing, review and/or revision of the manuscript: MH, BS, PCS, SC, IM, CT, MB, PU, MPL, MvdB, SG. Study supervision: MvdB, SG. Guarantor: SG.
Funding: This project was funded by Roche.
Competing interests: All authors, except MPL, RD and MvdB are employees of Roche.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Single-cell RNA-sequencing data are available in a public, open access repository. All other data relevant to the study are included in the article or uploaded as supplementary information. Single-cell RNA-sequencing data are stored at https://www.ebi.ac.uk/arrayexpress Single-cell RNA-seq of human tumor digests treated with Toll-like receptor agonists and interferons against untreated controls (E-MTAB-11734), Single-cell RNA-seq of human blood dendritic cells from buffy coats and in vitro differentiated dendritic cells from cord blood CD34+ stem cells (E-MTAB-11735) and at https://zenodo.org: Integration of single-cell RNA-sequencing data across tissues and cancer types towards immune cell characterization (https://doi.org/10.5281/zenodo.6514005).
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by Business Administration System for Ethics Committees BASEC-Nr: 2020-01208, BASEC-Nr: 2017-00494, and BASEC-Nr: 2016-02013. Participants gave informed consent to participate in the study before taking part.
References
- 1. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature 1998;392:245–52. 10.1038/32588 [DOI] [PubMed] [Google Scholar]
- 2. Hildner K, Edelson BT, Purtha WE, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science 2008;322:1097–100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roberts EW, Broz ML, Binnewies M, et al. Critical role for CD103(+)/CD141(+) dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in Melanoma. Cancer Cell 2016;30:324–36. 10.1016/j.ccell.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garris CS, Arlauckas SP, Kohler RH, et al. Successful anti-PD-1 cancer immunotherapy requires T Cell-Dendritic cell crosstalk involving the cytokines IFN-γ and IL-12. Immunity 2018;49:1148–61. 10.1016/j.immuni.2018.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chow MT, Ozga AJ, Servis RL, et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti-PD-1 therapy. Immunity 2019;50:1498. 10.1016/j.immuni.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tran Janco JM, Lamichhane P, Karyampudi L, et al. Tumor-Infiltrating dendritic cells in cancer pathogenesis. J Immunol 2015;194:2985–91. 10.4049/jimmunol.1403134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maier B, Leader AM, Chen ST, et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 2020;580:257–62. 10.1038/s41586-020-2134-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng S, Li Z, Gao R, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021;184:792–809. 10.1016/j.cell.2021.01.010 [DOI] [PubMed] [Google Scholar]
- 9. Sancho D, Joffre OP, Keller AM, et al. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature 2009;458:899–903. 10.1038/nature07750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canton J, Blees H, Henry CM, et al. The receptor DNGR-1 signals for phagosomal rupture to promote cross-presentation of dead-cell-associated antigens. Nat Immunol 2021;22:140–53. 10.1038/s41590-020-00824-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Villani A-C, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017;356:aah4573. 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dudziak D, Kamphorst AO, Heidkamp GF, et al. Differential antigen processing by dendritic cell subsets in vivo. Science 2007;315:107–11. 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- 13. Eisenbarth SC. Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol 2019;19:89–103. 10.1038/s41577-018-0088-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noubade R, Majri-Morrison S, Tarbell KV. Beyond cDC1: emerging roles of DC crosstalk in cancer immunity. Front Immunol 2019;10:1014. 10.3389/fimmu.2019.01014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nizzoli G, Krietsch J, Weick A, et al. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood 2013;122:932–42. 10.1182/blood-2013-04-495424 [DOI] [PubMed] [Google Scholar]
- 16. Truxova I, Kasikova L, Hensler M, et al. Mature dendritic cells correlate with favorable immune infiltrate and improved prognosis in ovarian carcinoma patients. J Immunother Cancer 2018;6:139. 10.1186/s40425-018-0446-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hubert M, et al. IFN-III is selectively produced by cdc1 and predicts good clinical outcome in human breast cancer. Cancer Immunol Res 2020;8:70. 10.1126/sciimmunol.aav3942 [DOI] [PubMed] [Google Scholar]
- 18. Oba T, Long MD, Keler T, et al. Overcoming primary and acquired resistance to anti-PD-L1 therapy by induction and activation of tumor-residing cDC1s. Nat Commun 2020;11:5415. 10.1038/s41467-020-19192-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roselli E, Araya P, Núñez NG, et al. TLR3 activation of intratumoral CD103+ dendritic cells modifies the tumor infiltrate conferring anti-tumor immunity. Front Immunol 2019;10:503. 10.3389/fimmu.2019.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gautier G, Humbert M, Deauvieau F, et al. A type I interferon autocrine-paracrine loop is involved in Toll-like receptor-induced interleukin-12p70 secretion by dendritic cells. J Exp Med 2005;201:1435–46. 10.1084/jem.20041964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hémont C, Neel A, Heslan M, et al. Human blood mDC subsets exhibit distinct TLR repertoire and responsiveness. J Leukoc Biol 2013;93:599–609. 10.1189/jlb.0912452 [DOI] [PubMed] [Google Scholar]
- 22. Kratky W, Reis e Sousa C, Oxenius A, et al. Direct activation of antigen-presenting cells is required for CD8+ T-cell priming and tumor vaccination. Proc Natl Acad Sci U S A 2011;108:17414–9. 10.1073/pnas.1108945108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Napolitani G, Rinaldi A, Bertoni F, et al. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat Immunol 2005;6:769–76. 10.1038/ni1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lövgren T, Sarhan D, Truxová I, et al. Enhanced stimulation of human tumor-specific T cells by dendritic cells matured in the presence of interferon-γ and multiple toll-like receptor agonists. Cancer Immunol Immunother 2017;66:1333–44. 10.1007/s00262-017-2029-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng GXY, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mädler SC, Julien-Laferriere A, Wyss L, et al. Besca, a single-cell transcriptomics analysis toolkit to accelerate translational research. NAR Genom Bioinform 2021;3:lqab102. 10.1093/nargab/lqab102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 2019;9:5233. 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature 2018;560:494–8. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergen V, Lange M, Peidli S, et al. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 2020;38:1408-1414. 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- 30. Vignali DAA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol 2012;13:722–8. 10.1038/ni.2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Valenzuela J, Schmidt C, Mescher M. The roles of IL-12 in providing a third signal for clonal expansion of naive CD8 T cells. J Immunol 2002;169:6842–9. 10.4049/jimmunol.169.12.6842 [DOI] [PubMed] [Google Scholar]
- 32. Lichtenegger FS, Mueller K, Otte B, et al. CD86 and IL-12p70 are key players for T helper 1 polarization and natural killer cell activation by Toll-like receptor-induced dendritic cells. PLoS One 2012;7:e44266. 10.1371/journal.pone.0044266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qian J, Olbrecht S, Boeckx B, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res 2020;30:745–62. 10.1038/s41422-020-0355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee H-O, Hong Y, Etlioglu HE, et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 2020;52:594–603. 10.1038/s41588-020-0636-z [DOI] [PubMed] [Google Scholar]
- 35. Ma L, Hernandez MO, Zhao Y, et al. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell 2019;36:418–30. 10.1016/j.ccell.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peng J, Sun B-F, Chen C-Y, et al. Author correction: single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:777. 10.1038/s41422-019-0212-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cillo AR, Kürten CHL, Tabib T, et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity 2020;52:183–99. 10.1016/j.immuni.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martin JC, Chang C, Boschetti G, et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 2019;178:1493–508. 10.1016/j.cell.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and Inter-cellular rewiring of the human colon during ulcerative colitis. Cell 2019;178:714–30. 10.1016/j.cell.2019.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramachandran P, Dobie R, Wilson-Kanamori JR, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019;575:512–8. 10.1038/s41586-019-1631-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raredon MSB, Adams TS, Suhail Y, et al. Single-cell connectomic analysis of adult mammalian lungs. Sci Adv 2019;5:eaaw3851. 10.1126/sciadv.aaw3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kotliarov Y, Sparks R, Martins AJ, et al. Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus. Nat Med 2020;26:618–29. 10.1038/s41591-020-0769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Madissoon E, Wilbrey-Clark A, Miragaia RJ, et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol 2019;21:1. 10.1186/s13059-019-1906-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hoch T, Schulz D, Eling N, et al. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci Immunol 2022;7:eabk1692. 10.1126/sciimmunol.abk1692 [DOI] [PubMed] [Google Scholar]
- 45. Rapp M, Wintergerst MWM, Kunz WG, et al. CCL22 controls immunity by promoting regulatory T cell communication with dendritic cells in lymph nodes. J Exp Med 2019;216:1170–81. 10.1084/jem.20170277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Williford J-M, Ishihara J, Ishihara A, et al. Recruitment of CD103+ dendritic cells via tumor-targeted chemokine delivery enhances efficacy of checkpoint inhibitor immunotherapy. Sci Adv 2019;5:eaay1357. 10.1126/sciadv.aay1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231–61. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- 48. Liu J, Li F, Ping Y, et al. Local production of the chemokines CCL5 and CXCL10 attracts CD8+ T lymphocytes into esophageal squamous cell carcinoma. Oncotarget 2015;6:24978–89. 10.18632/oncotarget.4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Castellino F, Huang AY, Altan-Bonnet G, et al. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 2006;440:890–5. 10.1038/nature04651 [DOI] [PubMed] [Google Scholar]
- 50. Jaczewska J, Abdulreda MH, Yau CY, et al. TNF-α and IFN-γ promote lymphocyte adhesion to endothelial junctional regions facilitating transendothelial migration. J Leukoc Biol 2014;95:265–74. 10.1189/jlb.0412205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol 2004;5:987–95. 10.1038/ni1112 [DOI] [PubMed] [Google Scholar]
- 52. Schroder K, Hertzog PJ, Ravasi T, et al. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 2004;75:163–89. 10.1189/jlb.0603252 [DOI] [PubMed] [Google Scholar]
- 53. Balan S, Ollion V, Colletti N, et al. Human XCR1+ dendritic cells derived in vitro from CD34+ progenitors closely resemble blood dendritic cells, including their adjuvant responsiveness, contrary to monocyte-derived dendritic cells. J Immunol 2014;193:1622–35. 10.4049/jimmunol.1401243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee J, Breton G, Oliveira TYK, et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med 2015;212:385–99. 10.1084/jem.20141442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mäkelä SM, Osterlund P, Julkunen I. TLR ligands induce synergistic interferon-β and interferon-λ1 gene expression in human monocyte-derived dendritic cells. Mol Immunol 2011;48:505–15. 10.1016/j.molimm.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 56. Larangé A, Antonios D, Pallardy M, et al. TLR7 and TLR8 agonists trigger different signaling pathways for human dendritic cell maturation. J Leukoc Biol 2009;85:673–83. 10.1189/jlb.0808504 [DOI] [PubMed] [Google Scholar]
- 57. Chiang CL-L, Kandalaft LE. In vivo cancer vaccination: which dendritic cells to target and how? Cancer Treat Rev 2018;71:88–101. 10.1016/j.ctrv.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ni L, Lu J. Interferon gamma in cancer immunotherapy. Cancer Med 2018;7:4509–16. 10.1002/cam4.1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-004268supp001.pdf (10.3MB, pdf)
Data Availability Statement
Single-cell RNA-sequencing data are available in a public, open access repository. All other data relevant to the study are included in the article or uploaded as supplementary information. Single-cell RNA-sequencing data are stored at https://www.ebi.ac.uk/arrayexpress Single-cell RNA-seq of human tumor digests treated with Toll-like receptor agonists and interferons against untreated controls (E-MTAB-11734), Single-cell RNA-seq of human blood dendritic cells from buffy coats and in vitro differentiated dendritic cells from cord blood CD34+ stem cells (E-MTAB-11735) and at https://zenodo.org: Integration of single-cell RNA-sequencing data across tissues and cancer types towards immune cell characterization (https://doi.org/10.5281/zenodo.6514005).