Abstract
The growth activity of Pseudomonas putida cells colonizing the rhizosphere of barley seedlings was estimated at the single-cell level by monitoring ribosomal contents and synthesis rates. Ribosomal synthesis was monitored by using a system comprising a fusion of the ribosomal Escherichia coli rrnBP1 promoter to a gene encoding an unstable variant of the green fluorescent protein (Gfp). Gfp expression in a P. putida strain carrying this system inserted into the chromosome was strongly dependent on the growth phase and growth rate of the strain, and cells growing exponentially at rates of ≥0.17 h−1 emitted growth rate-dependent green fluorescence detectable at the single-cell level. The single-cell ribosomal contents were very heterogeneous, as determined by quantitative hybridization with fluorescently labeled rRNA probes in P. putida cells extracted from the rhizosphere of 1-day-old barley seedlings grown under sterile conditions. After this, cells extracted from the root system had ribosomal contents similar to those found in starved cells. There was a significant decrease in the ribosomal content of P. putida cells when bacteria were introduced into nonsterile bulk or rhizosphere soil, and the Gfp monitoring system was not induced in cells extracted from either of the two soil systems. The monitoring system used permitted nondestructive in situ detection of fast-growing bacterial microcolonies on the sloughing root sheath cells of 1- and 2-day-old barley seedlings grown under sterile conditions, which demonstrated that it may be possible to use the unstable Gfp marker for studies of transient gene expression in plant-microbe systems.
The use of environmental biotechnology in agriculture is becoming an increasingly promising alternative to chemical control of plant diseases and pests. The current emphasis is on the development of microbial inoculants for biological control of pests (21, 42, 53), for promoting plant growth (32, 38), and more recently, for plant-assisted microbial elimination of pollutants (7, 18). In all of these cases and independent of the specific ecological conditions required by different agricultural practices, successful use of microbial inoculants in the field requires efficient colonization of the root system and a high level of competence of the introduced microbe for the desired biological property (e.g., production of antibiotics or plant growth-stimulating hormones or biosynthesis of catabolic enzymes for degradation of xenobiotic compounds) to be expressed. Hence, the effectiveness of an inoculant depends on the survival and physiological status of the inoculant cells in the rhizosphere and/or in the root tissues of the plant. However, little is known about the physiological status of bacterial cells in plant-microbe systems.
The rhizosphere is commonly perceived as a site where there are high levels of microbial activity and large numbers of bacteria. In general, the microbial densities in the rhizosphere, which are commonly measured by determining the number of CFU or total bacteria per gram of rhizosphere soil, are 2 to 3 orders of magnitude greater than the microbial densities in bulk soil (9). However, high densities do not necessarily indicate that the levels of activity of all of the microorganisms associated with the rhizosphere throughout the entire root system are high. In fact, the rhizosphere has been recognized as an oligotrophic environment which contains minute cells whose growth is limited by a lack of substrates (22). On the other hand, probably due to variations in nutrient availability, as well as differences in the compositions of plant-derived compounds, spatial heterogeneity and temporal heterogeneity in microbial activity have been observed in the rhizosphere (8, 17, 27, 35, 45, 51).
The methods that have been used to assess the physiological state of cells in the rhizosphere include indirect estimation of growth rates based on differences in the number of CFU or the total bacterial counts measured at various times (47) and estimation of bacterial activity by determining the rate of respiration (56) and the rate of DNA and/or protein synthesis (35, 51). To ascertain the in situ physiology of microbial cells in the rhizosphere, workers can use a number of methods based on cell extraction and subsequent microscopic observation; these methods include enumeration of cells having a measurable membrane potential by using redox dyes (27, 46) and assessment of variations in cell length by using fluorescent dyes in combination with image analysis (5). To differentiate introduced bacteria from indigenous soil bacteria in natural samples, very sensitive and specific molecular methods, such as methods involving fluorescent antibodies (26, 50) and rRNA probes (1, 3, 15, 25) directed toward the introduced strain, can be used to label the inoculated cells. Hybridization to whole cells in which fluorescently labeled rRNA probes and epifluorescence microscopy are coupled to digital image analysis can also be used to measure cell size and rRNA content (5, 23, 40, 43). In several bacteria the amount of rRNA in cells is correlated with growth or metabolic rate (4, 31, 49), and measurements of these parameters can therefore be used as indicators of metabolic activity.
Reporter gene technology is very useful in studies of the metabolic status of bacteria introduced into natural environments (44), and different markers have been used to identify cells and to assess physiological activities in the rhizosphere; these markers include lacZ (20) and luxAB (8, 10, 17, 19, 34, 45). More recently, the green fluorescent protein (Gfp) (11) has been used as a marker for in situ detection of bacterial cells in soil (54, 55) and in the rhizosphere (6). However, the great stability of Gfp makes it less valuable for studies of transient (real-time) gene expression. A novel Gfp-based reporter system, whose expression is cell growth regulated, has recently been developed (14, 52). This system, which includes a fusion of a mutant gfp gene encoding a short-half-life Gfp (2) to a ribosomal Escherichia coli promoter, has been used to study the in situ and in vivo activity of Pseudomonas putida cells introduced into complex environments, such as microbial biofilms (52).
In the present study, a monitoring system consisting of an unstable Gfp marker fused to a ribosomal promoter was used to analyze in situ the spatial and temporal heterogeneity of P. putida cells in the rhizosphere of young barley seedlings at the population and single-cell levels. Additionally, the ribosomal contents of bacteria extracted from this plant-microbial system under different conditions were estimated by quantitative hybridization with rRNA target probes and epifluorescence microscopy.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or characteristics | Reference |
|---|---|---|
| E. coli strains | ||
| HB101 | SmrrecA thi por leu hsdRM+ | 30 |
| MV1190λ-pir | Δ(lac proAB) Δ(srl-recA)306::Tn10 [F′ traD36 proAB lacIq Δ(lacZ)M15] thi supE, lysogenized with λ-pir phage | 28 |
| P. putida strains | ||
| JB156 | Nalr derivative of P. putida RI | 13 |
| CRR300 | P. putida JB156 × E. coli HB101(pRK600) × MV1190λ-pir(pJBA29), P. putida JB156 with mini-Tn5-Kmr-PA1/04/03::RBSII-gfpmut3*-T0-T1 randomly inserted into the chromosomea | This study |
| SM1700 | P. putida JB156 E. coli HB101(pRK600) × MV1190λ-pir(pSM1696), P. putida JB156 with mini-Tn5-Kmr-rrnBP1::gfp[AGA]-T0-T1 randomly inserted into the chromosomea | This study |
| Plasmids | ||
| pRK600 | Cmr ColE1oriV RP4oriT, helper plasmid in triparental matings | 30 |
| pJBA29 | Apr Kmr, delivery plasmid for mini-Tn5-Kmr-PA1/04/03::RBSII-gfpmut3*-T0-T1 derived from pUT-mini-Tn5-Kmr | 2 |
| pSM1696 | Apr Kmr, delivery plasmid for mini-Tn5-Kmr-rrnBP1::gfp[AGA]-T0-T1 derived from pUT-mini-Tn5-Kmr | This study |
Triparental matings were performed as described by Herrero et al. (28).
P. putida SM1700 carries a chromosomal insertion of gfp[AGA], which encodes an unstable Gfp (14), fused to the E. coli rrnB ribosomal P1 promoter. This strain was constructed as follows. First, the gfp[AGA] gene was constructed by adding a short peptide sequence, Ala-Gly-Ala, to the C-terminal end of intact Gfp as described by Andersen et al. (2) for similar mutant Gfp variants. This gfp allele was fused to the E. coli rrnB ribosomal P1 promoter, and the mini-Tn5 delivery vector pSM1696 was constructed as described by Sternberg et al. (52) for other gfp alleles. Finally, the mini-Tn5-Kmr transposon bearing the rrnBP1::gfp[AGA] fusion was transferred to the chromosome of P. putida JB156 by triparental mating as described by Herrero et al. (28). E. coli MV1190λ-pir(pSM1696) and HB101(pRK600) were used as donor and helper strains, respectively.
E. coli strains were grown in Luria-Bertani (LB) medium at 37°C. P. putida strains were grown at either 30 or 20°C in LB medium or FAB minimal medium [1 mM MgCl2, 0.1 mM CaCl2, 0.01 mM Fe-EDTA (catalog no. E6760; Sigma Chemical Co., St. Louis, Mo.), 0.15 mM (NH4)SO4, 0.33 mM Na2HPO4, 0.2 mM KH2PO4, 0.5 mM NaCl]. Unless indicated otherwise, minimal medium contained 10 mM sodium citrate as the only carbon source. Antibiotics were added at the following concentrations: chloroamphenicol, 30 μg/ml; kanamycin, 25 μg/ml; nalidixic acid, 50 μg/ml; and rifampin, 30 μg/ml.
Chemostat experiments.
To determine the specific activity of the rrnBP1 promoter fusion to gfp[AGA] at different growth rates, P. putida SM1700 cells were grown at 20°C in chemostats operated at four or five different dilution rates, as described by Sternberg et al. (52). Bacteria were grown to the stationary phase in FAB medium containing 10 mM sodium citrate as the only carbon source prior to inoculation. After inoculation, the chemostats were operated with the same medium for at least 60 h in order to allow establishment of the introduced strain at a stable population size. After this, three samples were taken from each chemostat at intervals of approximately 24 h to determine the optical density at 450 nm (OD450) of the culture, the fluorescence of the cells, and the ribosomal contents of the cells after hybridization with a fluorescently labeled rRNA probe.
Quantification of green fluorescence.
Emission of green fluorescence from P. putida SM1700 cells growing in batch or chemostat cultures was quantified as follows. Two-milliliter cell suspensions were harvested at various times by centrifugation, the cells were resuspended in the same volume of 0.9% NaCl, and green fluorescence was measured with a fluorometer (model RF-1501; Shimadzu, Tokyo, Japan) set at an excitation wavelength of 475 nm with emission detection at 515 nm. The specific fluorescence activities of the cells were expressed as the measured green fluorescence values (relative fluorescence units [RFU]) per turbidity (OD450) unit. The background green autofluorescence of P. putida cells was determined by measuring the fluorescence emitted by P. putida JB156 cells grown under the same culture conditions.
Seed sterilization and coating with bacteria.
Barley seeds (Hordeum vulgare var. Alexis) were surface sterilized by immersing them in 70% (vol/vol) ethanol and then treating them with 3% (vol/vol) hypochloride as described by Kragelund and Nybroe (33). Sterilization of seeds was verified by incubating 10 to 20 seeds on LB agar at 25°C for several days without any contamination appearing. Sterile seeds were pregerminated on moist filter paper for 24 h prior to inoculation.
Bacterial inocula for seed coating or soil inoculation were prepared as follows. A colony of the corresponding P. putida strain was inoculated into 10 ml of FAB minimal medium containing 10 mM sodium citrate as the only carbon source, and the culture was incubated at 20°C until the stationary phase. Cells were washed twice in M8 buffer (22 mM Na2HPO4 · 2H2O, 22 mM KH2PO4, 100 mM NaCl) prior to use.
Surface-sterilized seeds were coated with bacteria by soaking them for 30 min at 20°C in a 10-ml bacterial inoculum preparation adjusted turbidometrically to the appropriate cell density. The nonadhering liquid was decanted, and the seeds were air dried for 10 min in a petri dish. Bacteria were washed from three seeds into 3 ml of 0.9% NaCl and serially diluted onto selective media to determine the number of bacteria applied to the seeds.
Plant germination and growth conditions.
Seeds were germinated and plants were grown in nonsterile soil in 50-ml plastic tubes filled with 50 g of a 1:1 mixture of sand and sandy loam soil. The soil moisture content was adjusted to 15% (wt/wt) with sterile water. The soil used has been described previously (33). Coated seeds were sown about 1 cm below the soil surface; the seeds were germinated and the plants were grown at room temperature (18 to 20°C) with the natural daily light period (approximately 11 h of light and 13 h of darkness). To avoid soil drying, tubes were placed in a beaker containing moist filter paper, and the whole system was placed in a transparent plastic bag. Soil was inoculated with approximately 107 CFU/g of soil by using bacterial inocula prepared as described above.
Barley seeds were germinated and barley seedlings were grown under sterile conditions in M8 minimal medium plates (M9 minimal medium [48] containing no carbon or nitrogen source) containing 1% agar. No carbon source or nitrogen source was added to the plates. The plates were incubated in the dark at 20°C for 3 days.
Recovery of bacteria from soil and rhizosphere samples.
Bacteria were extracted from soil samples essentially as described by Unge et al. (55). All of the solutions were precooled on ice before they were used, and samples were kept on ice during the extraction procedure. Soil samples (500 mg) were placed into microcentrifuge tubes containing 800 μl of cold distilled water and approximately 16 mg of acid-washed polyvinylpolypyrrolidone prepared as described by Holben et al. (29). After the preparations were vortexed for 3 min, the large soil particles settled in each tube, and the upper phase (700 μl) was transferred to a new tube, serially diluted, and plated onto selective media in order to determine the number of bacteria as described below. The remaining soil suspension was centrifuged for 4 min at 4°C and 200 × g, and the supernatant was carefully transferred to a microcentrifuge tube containing 700 μl of Nycodenz (Nycomed Pharma, Oslo, Norway) with a density of 1.3 g/ml. The discontinuous Nycodenz-cell suspension gradient was centrifuged for 30 min at 12,000 × g and 4°C, and then the upper 500 μl of the gradient was discarded and the lower 500 μl, which contained the soil bacterial fraction, was transferred to a new microcentrifuge tube containing 1.5 ml of phosphate-buffered saline. To visualize Gfp-tagged cells, 1 ml of the bacterial suspension was diluted with 9 ml of 0.9% NaCl, and the cells were concentrated by filtration through 25-mm-diameter black polycarbonate membrane filters (pore size, 0.2 μm; Osmonics, Vista, Calif.). Cell fixation, hybridization, and visualization of Gfp-fluorescent or hybridized cells were performed as described below.
Bacteria were recovered from rhizosphere samples as follows. After different periods of plant growth, seedlings were removed from the soil system or the agar plates, and the roots were separated from the seeds. When the plants were harvested from the soil system, the roots and the soil adhering to the roots (the rhizosphere) were placed into 10-ml plastic tubes containing 5 ml of cold 0.9% sodium chloride and 16 mg of polyvinylpolypyrrolidone. Bacteria were separated from the rhizosphere soil suspensions as described above.
Bacteria were extracted from the root systems of agar-grown seedlings by placing the roots into 2 ml of 0.9% cold sodium chloride and vortexing the preparation for 3 min. The bacteria were counted on LB medium plates containing the necessary antibiotics for selection of the corresponding strain. As a sterility control, bacteria were also counted on LB medium plates, and we observed no difference in colony numbers between the two media for any of the strains tested.
Competitiveness of P. putida CRR300 and SM1700 in the rhizosphere soil.
Sterile barley seeds were coated with approximately 104 CFU of P. putida JB156 and the same number of either P. putida CRR300 or P. putida SM1700 CFU. Coated seeds were sown in nonsterile soil, and the number of CFU per gram of rhizosphere soil was estimated by dilution plating performed with selective media. P. putida CRR300 and SM1700 were counted on FAB minimal medium containing 10 mM sodium benzoate as the only carbon source (FAB-benzoate medium) supplemented with 50 μg of nalidixic acid per ml and 25 μg of kanamycin per ml. The size of the P. putida JB156 population was determined by counting the number of non-green-fluorescent colonies able to grow on FAB-benzoate medium containing 50 μg of nalidixic acid per ml. No indigenous soil bacteria were able to grow on the two media used. The parental and Gfp derivative strains were equally competitive in the barley rhizosphere. One day after planting, the three strains were established at a density of about 106 to 107 CFU/g of rhizosphere soil. The number of bacteria in the rhizosphere declined to about 105 CFU/g of rhizosphere soil after 7 days and then remained relatively constant (data not shown).
Whole-cell hybridization.
Cells extracted from soil or the rhizosphere, as well as cells obtained from batch or chemostat cultures, were fixed in 3% paraformaldehyde as described previously (43). Fixed cells were stored at −20°C in storage buffer (50% ethanol, 10 mM Tris [pH 7.5], 0.1% Nonidet P-40) until they were used. 16S rRNA hybridization of fixed cells was performed as described by Poulsen et al. (43) by using probe PP986, an oligonucleotide probe specific for P. putida subgroup A, labeled with the incarbocyanine fluorescent dye CY3 (41).
Epifluorescence microscopy.
An Axioplan epifluorescence microscope (Carl Zeiss) was used to visualize the hybridized cells. The microscope was equipped with a 100-W mercury lamp, and no. 10 (Carl Zeiss) and XF40 (Omega Optical, Brattlebror, Vt.) filter sets were used to visualize Gfp and CY3, respectively. A slow-scan charge-coupled device camera (model CH250; Photometrics, Tucson, Ariz.) equipped with a type KAF 1400 chip (pixel size, 6.8 by 6.8 μm) was used to capture digitized images (PMIS). The camera was operated at −40°C, and the chip was read out in 12 bits (4,096 intensity levels) at a rate of 200 kHz. To quantify fluorescence, the PMIS images were analyzed by using the Cellstat program (40). A DOS-based 486 computer was used as a controller for the charge-coupled device camera, and a SUN IPX computer was used to run Cellstat.
Nondestructive system for in situ visualization of bacterial cells on the root surface.
A nondestructive system for studying rhizosphere colonization of young barley seedlings by Gfp-tagged cells was developed. Rhizosphere chambers (length, 55 mm; width, 20 mm; depth, 3 mm) were constructed by sticking a silicone rubber gasket on top of a microscope slide and mounting a coverslip (24 by 60 mm) on top of the silicone layer. The chambers were filled with vermiculite soaked with 500 μl of water. A barley seed coated with bacteria was placed about 5 mm below the soil or vermiculite surface, and the whole system was placed inside a 50-ml centrifuge tube. To avoid drying, moist filter paper was placed inside each tube. To ensure that the root system was localized in the vicinity of the coverslip, the tubes containing the growth chambers were placed on a rack at an angle of approximately 45°. Seeds were allowed to germinate in the dark at 20°C for 3 days. At different times the Gfp-tagged cells colonizing the roots were visualized by scanning confocal laser microscopy (SCLM).
SCLM.
Microscopic observations were made and images of rhizosphere bound cells were acquired with a confocal microscope (model TCS4D; Leica Lasertechnik GmbH, Heidelberg, Germany) equipped with detectors and filter sets that simultaneously monitored Gfp and red fluorescence. The red autofluorescence exhibited by the root material was used to visualize the root surface. Simulated fluorescence projections and vertical cross sections were generated by using the IMARIS software package (Bitplane AG, Zürich, Switzerland) running on an Indigo 2 workstation (Silicon Graphics, Mountain View, Calif.). Images were processed for display by using Photoshop software (Adobe, Mountain View, Calif.).
RESULTS
Cell growth regulation of Gfp expression in P. putida SM1700.
P. putida SM1700 carries a chromosomal insertion of a gfp allele, gfp[AGA], which encodes an unstable Gfp, fused to the ribosomal E. coli promoter rrnBP1 (see above for details concerning this construction). Insertion of this reporter system into the chromosome of P. putida SM1700 did not affect the growth characteristics of the strain. The length of the lag phase and the doubling time in the exponential phase were similar for P. putida SM1700 and its parent strain, JB156, both in LB broth (data not shown) and on minimal medium containing citrate as the only carbon source (Fig. 1A).
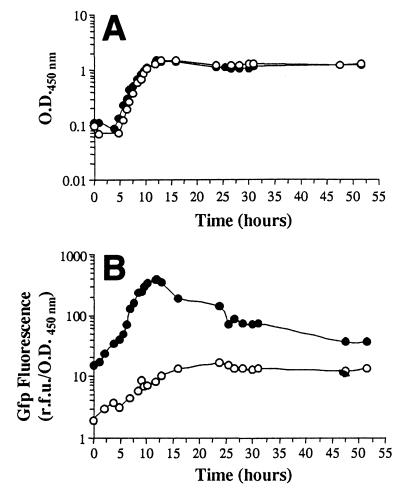
FIG. 1.
Growth phase-dependent expression of the rrnBP1 promoter in P. putida SM1700 growing in batch cultures at 20°C. P. putida JB156 (wild type) (○) and P. putida SM1700 (mini-Tn5-Kmr-rrnBP1::gfp[AGA]) (●) were grown at 20°C on AB minimal media containing 8 mM sodium citrate as the sole carbon source. (A) Growth curves. (B) Intensity of green fluorescence.
In order to determine the relationship between bacterial growth and ribosomal synthesis in P. putida SM1700, we measured the fluorescent signal emitted by cells of this organism growing in batch cultures and in chemostats at 20°C. This temperature was used in the experiments described below for cultivation of barley seedlings colonized by P. putida strains. In batch cultures, Gfp expression depended on the growth phase of the culture; the intensity of the fluorescence emitted by P. putida SM1700 cells increased during the exponential growth phase to about 400 RFU/OD450 unit, but after the cells entered the stationary phase there was a decrease in fluorescence (Fig. 1). After approximately 25 h, the Gfp fluorescence of this strain was less than 100 RFU/OD450 unit (Fig. 1B), and Gfp fluorescence of single cells could not be detected by epifluorescence microscopy. The level of background green autofluorescence of P. putida cells was determined by measuring the fluorescence emitted by P. putida JB156 cells grown under the same culture conditions. The intensity of the fluorescence emitted by this strain increased from 2 to 10 RFU/OD450 unit during the exponential growth phase and then remained relatively stable during the rest of the experiment (Fig. 1). These results are consistent with previously published data for the phenotype of the rrnBP1 promoter in E. coli (4) and in P. putida strains grown at 30°C (52). The fluorescent signal emitted under the same culture conditions by P. putida CRR300, which constitutively expressed a stable Gfp (Gfpmut3*) (16) from the PA1/04/03 promoter (36), was also measured. The level of green fluorescence remained high (between 3,000 and 6,000 RFU/OD450 unit) until the end of the experiment (data not shown).
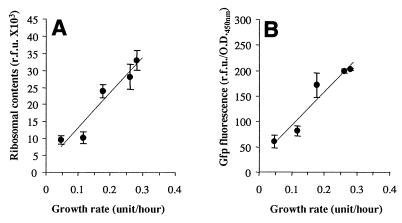
P. putida SM1700 was grown at 20°C in chemostats operated at 5 different dilution rates which resulted in growth rates between 0.05 and 0.28 h−1. After stabilization of a chemostat, samples were taken in order to examine ribosomal synthesis, which was determined by measuring the Gfp fluorescence emitted by the cells, and the ribosomal content, which was determined by quantitative hybridization with fluorescently labeled rRNA probes. We found that the ribosomal content (Fig. 2A) and the Gfp fluorescence intensity (Fig. 2B) depended on the growth rate. At the lowest growth rates (between 0.05 and 0.13 h−1) the level of Gfp fluorescence emission remained below 100 RFU/OD450 unit (Fig. 2B), and Gfp fluorescence of single cells could not be detected by epifluorescence microscopy. However, green fluorescent cells were easily detected by epifluorescence microscopy at growth rates between 0.17 and 0.28 h−1. The background autofluorescence emitted by the cells was estimated by calculating the green fluorescence emitted by cells of the parent strain, P. putida JB156, grown under similar conditions. Autofluorescence remained relatively constant at about 20 RFU/OD450 unit at growth rates between 0.05 and 0.35 h−1 (data not shown). These results are consistent with those obtained by Sternberg et al. (52) at 30°C for similar fusions of the P1 promoter to other gfp alleles that encode Gfp with shorter half-lives.
FIG. 2.
Variations in ribosomal synthesis and ribosomal content as a function of growth rate in P. putida SM1700 growing in chemostats at 20°C. P. putida SM1700 was grown in chemostats at various dilution rates that resulted in growth rates between 0.05 and 0.28 h−1. After stabilization of the chemostats, ribosomal contents (A) and ribosomal synthesis (B) were estimated by quantitative hybridization with fluorescently labeled rRNA probes and by measuring the Gfp fluorescence emitted by the cells, respectively. Each point represents an average based on three independent measurements. The error bars indicate standard deviations from the averages.
Ribosomal contents and volume of P. putida cells in the root systems of barley seedlings grown under sterile conditions.
Stationary-phase cultures of P. putida JB156 (parent strain), CRR300 (mini-Tn5-Kmr-PA1/04/03::gfpmut3*), or SM1700 (mini-Tn5-Kmr-rrnBP1::gfp[AGA]) were used to inoculate sterile barley seeds at a density of approximately 104 CFU/seed. For comparison with starved cells, fractions of the cell cultures were inoculated into M8 buffer and incubated at 20°C. Visualization of the inoculated cells by epifluorescence microscopy showed that all of the cells present in the P. putida CRR300 inoculum emitted green fluorescence; however, Gfp fluorescence of single cells could not be detected in the P. putida JB156 and SM1700 inocula. Coated seeds were germinated at 20°C in the dark in agar plates containing no carbon source or nitrogen source for 3 days. Bacteria were extracted from the root systems of 1- to 3-day-old barley seedlings, and samples were used for plate counting, hybridization, and visualization of Gfp fluorescence emission by epifluorescence microscopy. As a sterility control, uninoculated sterile seeds were germinated under the same conditions, and no bacteria were detected after extraction and dilution plating on LB agar. The lengths of the roots of 1-, 2-, and 3-day-old barley seedlings grown under these conditions were approximately 0.5 to 1.0, 1.5 to 2.5, and 2.0 to 3.0 cm, respectively.
Insertion of the mini-Tn5 cassettes into the chromosomes of P. putida CRR300 and SM1700 did not affect the ability of the strains to colonize the barley rhizosphere compared with parent strain P. putida JB156. During the first day of plant growth, the number of bacteria in the root system reached approximately 107 CFU/root regardless of the strain inoculated. During the following 2 days, the relative increment of bacterial biomass was smaller than the increment observed during the first day of root development, although the numbers of CFU per root increased in similar ways for the three strains to about 108 CFU/root (Table 2).
TABLE 2.
Densities of cells of P. putida strains in the root systems of agar-grown barley seedlings
| Time (days) | Log10 CFU/roota
|
||
|---|---|---|---|
| JB156 | CRR300 | SM1700 | |
| 1 | 7.03 ± 0.19 | 6.80 ± 0.27 | 6.94 ± 0.45 |
| 2 | 7.42 ± 0.32 | 7.51 ± 0.10 | 7.35 ± 0.30 |
| 3 | 7.63 ± 0.76 | 7.80 ± 0.55 | 7.69 ± 0.20 |
The values are averages ± standard deviations based on the values obtained from three different roots.
Fast-growing green fluorescent P. putida SM1700 cells could be visualized after extraction from the root systems of 1-day-old seedlings, although this population represented only a small fraction of the extracted cells. After 2 and 3 days, no green fluorescent cells were extracted from the roots inoculated with this strain, suggesting that the growth rate of the strain in the root systems of the barley seedlings was less than the growth rate that permitted detectable rrnBP1 promoter-directed Gfp expression (about 0.17 h−1 for cells growing exponentially in a chemostat). In addition, green fluorescent cells of this strain were not detected in the M8 buffer suspension, as expected for the phenotype of the rrnBP1 promoter under starvation conditions. In contrast, all of the P. putida CRR300 cells incubated in M8 buffer or extracted from the roots emitted green fluorescence at all sampling times.
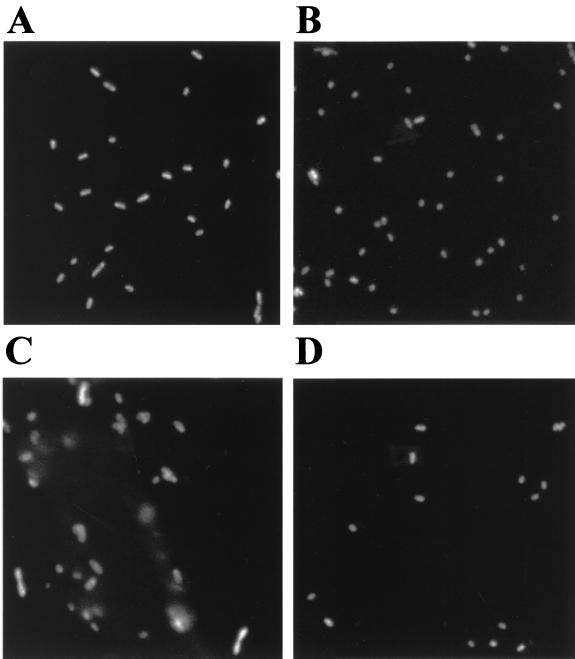
The ribosomal contents of P. putida JB156 and SM170 cells extracted from the root samples during the 3 days of investigation were estimated by analyzing images of hybridized cells. For comparison with cells growing exponentially in a chemostat, the fluorescence intensity was normalized to 1 for the cells growing at the lowest growth rate (0.05 h−1) (Table 3 and Fig. 3A). The calculated relative ribosomal contents of the stationary-phase cells used as the inocula (Fig. 3B) were 0.55 and 0.64 for P. putida JB156 and SM1700, respectively. At every sampling time, the ribosomal contents of the cells incubated in M8 buffer were at these levels for both strains (Table 3). The ribosomal contents of the cells extracted from the root systems corresponded to the relative increment of bacterial biomass. One day after planting, the average ribosomal content increased to about 1.12 for both strains. At this sampling time, at the single cell level the ribosomal contents and volumes of the cells extracted from the rhizosphere were very heterogeneous (Fig. 3C). Approximately 10% of the extracted P. putida SM1700 cells were found to have ribosomal contents that were equal to or greater than the ribosomal contents calculated for the green fluorescent cells growing at a growth rate of 0.17 h−1 (relative ribosomal content, 2.5). During the following 2 days, the average ribosomal content decreased to the level found during starvation on M8 buffer (Table 3 and Fig. 3D), and less than 2% of the extracted cells had relative ribosomal contents of ≥2.5.
TABLE 3.
Ribosomal contents of P. putida cells in the root systems of agar-grown barley seedlings
| Time (days) | Fluorescence intensity (RFU)a
|
|||
|---|---|---|---|---|
| M8 buffer
|
Rhizosphere
|
|||
| JB156 | SM1700 | JB156 | SM1700 | |
| 1 | 0.54 ± 0.05 | 0.61 ± 0.11 | 1.13 ± 0.16 | 1.12 ± 0.16 |
| 2 | 0.46 ± 0.05 | 0.69 ± 0.13 | 0.57 ± 0.04 | 0.76 ± 0.08 |
| 3 | 0.46 ± 0.05 | 0.55 ± 0.04 | 0.49 ± 0.05 | 0.67 ± 0.13 |
Fluorescence intensity was normalized to 1 RFU for the cells growing in a chemostat at a growth rate of 0.05 h−1. The values are averages ± standard deviations based on the values obtained from three different samples.
FIG. 3.
Epifluorescence micrographs of fixed and hybridized P. putida SM1700 cells grown under various conditions. (A) P. putida SM1700 cells growing exponentially in a chemostat at a growth rate of 0.05 h−1. (B) Stationary-phase P. putida SM1700 cells used to inoculate sterile barley seeds at a density of approximately 104 CFU/seed. Coated seeds were placed in agar plates and incubated at 20°C for 3 days. (C and D) Bacteria extracted from the root systems of 1-day-old (C) and 3-day-old (D) barley seedlings. In all cases cells were fixed and hybridized with a CY3-labeled rRNA probe.
Ribosomal contents and volumes of P. putida cells in the root systems of soil-grown barley seedlings.
Stationary-phase cultures of P. putida CRR300 (mini-Tn5-Kmr-PA1/04/03::gfpmut3*), P. putida SM1700 (mini-Tn5-Kmr-rrnBP1::gfp[AGA]), or parent strain P. putida JB156 were introduced into nonsterile soil at a density of approximately 107 CFU/g of soil. After soil inoculation, sterile barley seedlings were planted in some of the soil systems inoculated with each of the strains, and all of the preparations were incubated at room temperature (18 to 20°C). During the 14 days after planting, bacteria were extracted from the bulk soil and rhizosphere soil, and samples were used for plate counting and visualization of green fluorescent cells. The ribosomal contents of P. putida SM1700 cells extracted from soil samples were determined after hybridization.
The survival of the three strains in the bulk soil and rhizosphere soil was influenced by the presence of the plants. In the rhizosphere soil, the number of bacteria remained relatively constant, about 107 CFU/g of soil, during the 14 days of the experiment. In contrast, in the bulk soil the number of culturable bacteria declined slowly and was about 105 CFU/g of soil by the end of the experiment (data not shown).
Green fluorescent P. putida CRR300 cells extracted from the rhizosphere soil and the bulk soil were visualized by epifluorescence microscopy during the 14 days of the experiment. However, the observed difference in survival of the strains in bulk and rhizosphere soils was not matched by a significant difference in ribosomal synthesis. Green fluorescent P. putida SM1700 cells were not detected at any sampling time after extraction from the rhizosphere or bulk soil, indicating that no detectable activation of the P1 ribosomal promoter occurred after the cells were introduced into the soil. In fact, the average ribosomal content and volume of P. putida SM1700 cells extracted both from the rhizosphere soil and the bulk soil declined about 5- and 1.5-fold, respectively, during the first day following introduction into the soil. After this, no significant differences in either ribosomal content or cell volume were observed between the cells extracted from the rhizosphere soil and the cells extracted from the bulk soil (data not shown).
Spatial distribution of active P. putida cells in the roots of barley seedlings.
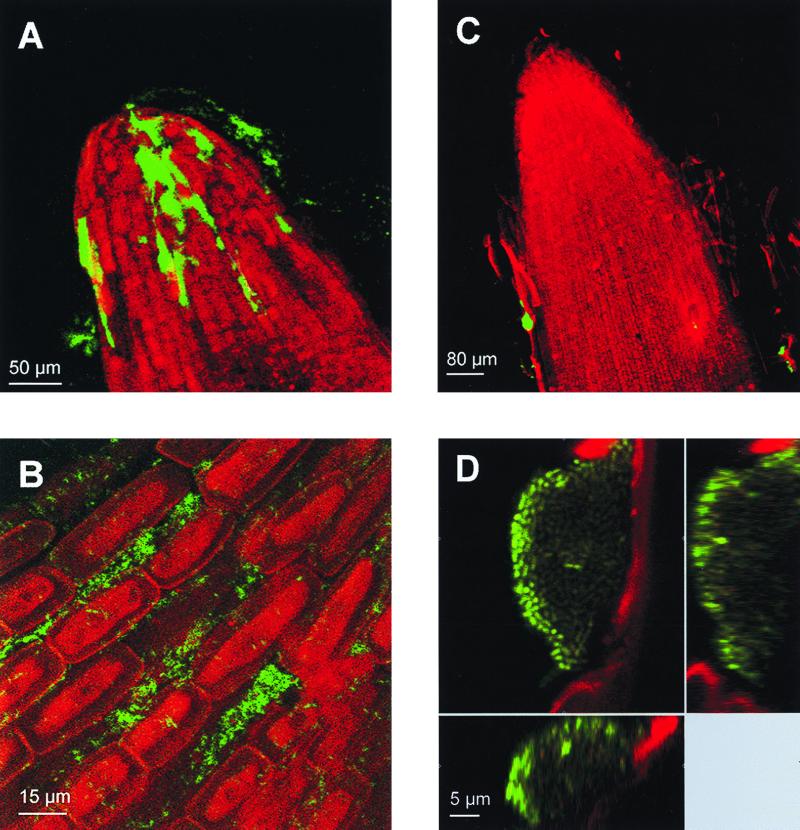
Gfp-tagged P. putida cells colonizing the rhizosphere of young barley seedlings that were 1 to 3 days old were visualized by SCLM in a nondestructive way by using rhizosphere chambers filled with vermiculite (see above for details concerning construction of the rhizosphere chambers). Sterile barley seeds were coated with approximately 104 CFU of P. putida CRR300 or SM1700 per seed, planted in the chambers, and incubated at 20°C. Green fluorescent P. putida CRR300 single cells and microcolonies were observed throughout the entire root systems of 1- to 3-day-old barley seedlings. The bacteria colonized the root hairs (data not shown), as well as the root surfaces and root tips (Fig. 4A), but they were most abundant in the intercellular crevices between neighboring plant root epidermal cells (Fig. 4B).
FIG. 4.
In situ visualization of active P. putida cells on the root surfaces of barley seedlings: visualization of Gfp-tagged P. putida CRR300 (PA1/04/03::gfpmut3*) (A and B) and P. putida SM1700 (rrnBP1::gfp[AGA]) (C and D) cells colonizing the root tips of 2-day-old barley seedlings. The green fluorescence emitted by the cells and the red autofluorescence emitted by the root material were visualized by SCLM. (A and C) xyz scan pictures (magnification, ×200) of root tips colonized by P. putida CRR300 and SM1700, respectively. (B and D) xyz pictures (magnification, ×630) of bacterial cells on the root surfaces. Vertical sections of the colony shown in panel D are shown to the right and below.
Activation of the P1 ribosomal promoter in P. putida SM1700 was visualized in 1- and 2-day-old barley seedlings colonized by this strain. In general, green fluorescence was visualized only on the sloughing root sheath cells (Fig. 4C). Two different patterns of light emission were observed in active microcolonies. In most cases, the intensities of the light emitted by all of the cells forming the microcolonies were similar. However, microcolonies in which there was a reduction in the intensity of the light emitted by the cells located in the center of the microcolony were also observed (Fig. 4D), indicating that the activity of these cells was reduced compared with the activity of the cells located in the periphery of the colony. No green fluorescent cells were observed in the root hair zone or in the crevices between epidermal cells. Active cells were not detected in 3-day-old barley seedlings.
DISCUSSION
Bacterial activity in nature is predominantly associated with surface-bound microbial communities that form complex and heterogeneous assemblages consisting of single- or multiple-species biofilms. It has been a challenge in microbial ecology to develop methods which can be used to determine the physiological activity of single cells colonizing specific surfaces (e.g., intestinal and plant root surfaces). In this study, expression of an unstable reporter Gfp driven by a ribosomal promoter (52) and estimation of ribosomal contents by quantitative hybridization with fluorescently labeled rRNA probes were used to assess the growth of P. putida single cells in the rhizosphere. The roots of young barley seedlings were used as a model system, mainly because colonization of the young barley seedling root system by Pseudomonas cells has been studied in great detail (26, 33, 34). Although our conclusions cannot be directly applied to every other plant-bacterium system, as it is known that rhizosphere colonization is influenced by the nature of the microorganisms, the soil type, and the seeds used (37, 39), they contribute to our understanding of the behavior of P. putida cells when these cells are used as a seed-applied inoculant.
Brennerova and Crowley (8) used a mini-Tn7 cassette containing a transcriptional fusion of the E. coli rrnBP2 promoter to the luxCDABE genes to study the activity of Pseudomonas strains in the rhizosphere. Although this system could be used successfully with P. fluorescens and bioluminescence production did not require the addition of exogenous substrates, integration of the cassette into the chromosome of a P. putida strain resulted in reduced growth of the strain, probably due to the strength of the ribosomal promoter used (8). On the other hand, although detection of bioluminescent bacterial cells in the rhizosphere at the single-cell level has been reported by other authors (45), individual cells that form microcolonies could not be distinguished by this technique.
Insertion of mini-Tn5 cassettes into the chromosome of P. putida CRR300 or SM1700 did not affect the growth characteristics of either strain in batch cultures (Fig. 1A) or the ability of the organisms to colonize the rhizospheres of agar-grown (Table 2) and soil-grown barley plants. It has been shown previously that the Gfp is a useful marker for direct observation of P. fluorescens cells introduced into soil (54, 55) or into the rhizosphere of agar-grown tomato seedlings (6). Using an unstable version of the Gfp that is encoded by the gfp[AGA] allele allowed us to perform a real-time gene expression study of bacterial cells colonizing a plant rhizosphere at the single-cell level. Gfp expression in P. putida SM1700 is strongly dependent on the growth phase and the growth rate of the strain, which is consistent with data obtained for regulation of the rrnBP1 E. coli promoter (4, 24, 52). We found that induction of the rrnBP1 promoter resulting in expression of detectable Gfp at the single-cell level occurred at growth rates between 0.17 and 0.28 h−1. It is important to note, however, that Gfp fluorescence could not be detected in single cells growing in a chemostat at a low growth rate (between 0.05 and 0.12 h−1). Therefore, although this reporter system can be used to detect fast-growing cells, the lack of apparent Gfp expression observed in the rhizosphere of soil-grown barley seedlings should not be taken as an indication of starvation; the rrnBP1 promoter could be active (and the cells could be growing) at levels that do not allow Gfp detection.
Differences in the numbers of bacteria in the rhizosphere and in bulk soil are often explained by the general increase in microbial activity in the rhizosphere that results from growth on carbon substrates provided by root exudates. The activity of P. putida cells colonizing the root systems of barley seedlings grown under sterile conditions corresponded to the relative increment of bacterial biomass (Tables 2 and 3). During the first day of plant growth, bacterial biomass increased rapidly, and the average ribosomal content of the cells colonizing the rhizosphere of barley seedlings was found to be twice as high as the average ribosomal content of starved cells. However, after 2 or 3 days the ribosomal content of the extracted cells was similar to the ribosomal content of starved cells (Table 3 and Fig. 3). This observation strongly suggests that the growth rate of rhizosphere-associated bacteria decreases rapidly once the population reaches a steady-state level. This initial sequence of events was not detected for P. putida cells colonizing the rhizosphere of soil-grown barley seedlings under nonsterile conditions, probably because of immediate competition for nutrients between the introduced P. putida cells and the indigenous soil microorganisms. Nevertheless, our data suggest that even at the low growth rates supported by root exudates, the P. putida strain tested is capable of replicating at rates sufficient to allow colonization of the developing roots.
Constitutive Gfp expression by P. putida CRR300 allowed us to visualize single cells, as well as cell assemblages, on the surfaces of barley seedling roots. In 1- to 3-day-old barley seedlings, most of the bacteria were at the borders of adjacent plant root cells (Fig. 4C), which is consistent with data reported for other Pseudomonas strains (6, 12, 26). All of the cells forming a microcolony emitted green fluorescence at similar intensities (data not shown). Expression of the P1 ribosomal promoter in P. putida SM1700 cells colonizing the barley rhizosphere, which indicated that cells growing at a relatively high growth rate were present, was detected close to the root tips on sloughing root sheath cells and only during the first and second days of plant growth under sterile conditions. Other workers have also shown that the root tips and the sites of lateral root emergence are the only sites in the rhizosphere where relatively high levels of microbial activity occur (8). Active microcolonies composed of cells emitting various amounts of green fluorescent light were observed at the root tips, which probably reflected differences in the activity of the cells during microcolony development (Fig. 4D). The fact that the brightest (most active) cells were located at the edges of the microcolonies suggests that after synthesis, Gfp[GAA] is turned over in the oldest cells localized in the center of a microcolony. This observation is consistent with data reported by Andersen et al. (2) on the development of colonies of P. putida strains expressing other unstable Gfp. Alternatively, the cells located at the periphery of a microcolony could have better access to the nutrients supplied by the mucoid layer covering the microcolonies on plant root surfaces. Electron microscopy studies have shown that there is such a layer over the microcolonies of a P. fluorescens biocontrol strain that colonizes the root surfaces of tomato plants (12).
Our monitoring system for bacterial growth activity thus seems to be useful for in situ monitoring of fast-growing bacterial cells during rhizosphere colonization, which suggests that there are other potential uses of this marker system for studies of specific gene expression in plant-microbe systems.
ACKNOWLEDGMENTS
This work was supported by a grant to S.M. from the Danish Biotechnology Programme. C.R. was supported in part by CEC grant BIO4CT975038.
We thank Jens Bo Andersen for construction of the gfp allele gfp[AGA], Tove Johansen for construction of P. putida SM1700, Tine Rask Licht for help with Cellstat, and Bjarke Christensen for assistance with confocal microscopy. We are grateful to Ole Nybroe and Linda E. Jensen for help with the soil experiments.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen J B, Sternberg C, Poulsen L K, Bjorn S P, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assmus B, Hutzler P, Kirchhof G, Amann R, Lawrence J R, Hartmann A. In situ localization of Azospirillum brasilense in the rhizosphere of wheat with fluorescently labeled, rRNA-targeted oligonucleotide probes and scanning confocal laser microscopy. Appl Environ Microbiol. 1995;61:1013–1019. doi: 10.1128/aem.61.3.1013-1019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett M S, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloem J, Veninga M, Shepherd J. Fully automatic determination of soil bacterium numbers, cell volume, and frequencies of dividing cells by confocal laser scanning microscopy and image analysis. Appl Environ Microbiol. 1995;61:926–936. doi: 10.1128/aem.61.3.926-936.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloemberg G V, O'Toole G A, Lugtenberg B J J, Kolter R. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol. 1997;63:4543–4551. doi: 10.1128/aem.63.11.4543-4551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazil G M, Kenefick L, Callanan M, Haro A, de Lorenzo V, Dowling D N, O'Gara F. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl Environ Microbiol. 1995;61:1946–1952. doi: 10.1128/aem.61.5.1946-1952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennerova M V, Crowley D. Direct detection of rhizosphere-colonizing Pseudomonas sp. using an Escherichia coli rRNA promoter in a Tn7-lux system. FEMS Microbiol Ecol. 1994;14:319–330. [Google Scholar]
- 9.Campbell R, Greaves M P. Anatomy and community structure of the rhizosphere. In: Lynch J M, editor. The rhizosphere. Chichester, United Kingdom: John Wiley & Sons; 1990. pp. 11–34. [Google Scholar]
- 10.Chabot R, Antoun H, Kloepper J W, Beauchamp C J. Root colonization of maize and lettuce by bioluminescent Rhizobium leguminosarum biovar phaseoli. Appl Environ Microbiol. 1996;62:2767–2772. doi: 10.1128/aem.62.8.2767-2772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;264:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 12.Ching-A-Woeng T F C, de Priester W, van der Bij A J, Lugtenberg B J J. Description of the colonization of a gnotobiotic tomato rhizosphere by Pseudomonas fluorescens biocontrol strain WCS365 using scanning electron microscopy. Mol Plant-Microbe Interact. 1997;10:79–86. [Google Scholar]
- 13.Christensen B B, Sternberg C, Andersen J B, Eberl L, Moller S, Givskov M, Molin S. Establishment of new genetic traits in a microbial biofilm community. Appl Environ Microbiol. 1998;64:2247–2255. doi: 10.1128/aem.64.6.2247-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen B B, Sternberg C, Andersen J B, Palmer R J, Jr, Toftgaard Nielsen A, Givskov M, Molin S. Molecular tools for the study of biofilm physiology. Methods Enzymol. 1999;310:20–42. doi: 10.1016/s0076-6879(99)10004-1. [DOI] [PubMed] [Google Scholar]
- 15.Christensen H, Poulsen L K. Detection of Pseudomonas in soil by rRNA targeted in situ hybridization. Soil Biol Biochem. 1994;26:1093–1096. [Google Scholar]
- 16.Cormack B P, Valdivia R H, Falkow S. FACS optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 17.Crowley D E, Brennerova M V, Irwin C, Brenner V, Focht D D. Rhizosphere effects on biodegradation of 2,5-dichlorobenzoate by a bioluminescent strain of root-colonizing Pseudomonas fluorescens. FEMS Microbiol Ecol. 1996;20:79–89. [Google Scholar]
- 18.Crowley D E, Alvey S, Gilbert E S. Rhizosphere ecology of xenobiotic-degrading microorganisms. ACS Symp Ser. 1997;664:19–34. [Google Scholar]
- 19.de Weger L A, Dunbar P, Mahafee W F, Lugtenberg B J J, Sayler G. Use of bioluminescence markers to detect Pseudomonas spp. in the rhizosphere. Appl Environ Microbiol. 1991;57:3641–3644. doi: 10.1128/aem.57.12.3641-3644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Weger L A, Dekkers L C, van der Bij A, Lugtenberg B J J. Use of phosphate-reporter bacteria to study phosphate limitation in the rhizosphere and in bulk soil. Mol Plant-Microbe Interact. 1993;7:32–38. [Google Scholar]
- 21.Dowling D N, O'Gara F. Metabolites of Pseudomonas involved in the biocontrol of plant disease. Trends Biotechnol. 1994;12:133–141. [Google Scholar]
- 22.Foster R C. Microenvironments of soil microorganisms. Biol Fertil Soils. 1988;6:189–203. [Google Scholar]
- 23.Givskov M, Eberl L, Møller S, Poulsen L K, Molin S. Responses to nutrient starvation in Pseudomonas putida: analysis of general cross-protection, cell shape, and molecular content. J Bacteriol. 1994;176:7–14. doi: 10.1128/jb.176.1.7-14.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gourse R L, de Boer H A, Nomura M. DNA determinants of rRNA synthesis in E. coli: growth rate dependent regulation, feedback inhibition, upstream activation, antitermination. Cell. 1986;44:197–205. doi: 10.1016/0092-8674(86)90498-8. [DOI] [PubMed] [Google Scholar]
- 25.Hahn D, Amann R I, Ludwig W, Akkermans A D L, Schleifer K H. Detection of micro-organisms in soil after in situ hybridization with rRNA-targeted, fluorescently labelled oligonucleotides. J Gen Microbiol. 1992;138:879–887. doi: 10.1099/00221287-138-5-879. [DOI] [PubMed] [Google Scholar]
- 26.Hansen M, Kragelund L, Nybroe O, Sørensen J. Early colonization of barley roots by Pseudomonas fluorescens studied by immunofluorescence technique and confocal laser scanning microscopy. FEMS Microbiol Ecol. 1997;23:353–360. [Google Scholar]
- 27.Heijnen C E, Page S, van Elsas J D. Metabolic activity of Flavobacterium strain P25 during starvation and after introduction into bulk soil and the rhizosphere of wheat. FEMS Microbiol Ecol. 1995;18:129–138. [Google Scholar]
- 28.Herrero M V, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holben W E, Jansson J K, Chelm B K, Tiedje J M. Probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler B, de Lorenzo V, Timmis K N. A general system to integrate lacZ fusions into the chromosomes of gram negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet. 1992;233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- 31.Kjeldgaard N O, Kurland C G. The distribution of soluble and ribosomal RNA as a function of growth rate. J Mol Biol. 1963;6:341–348. [Google Scholar]
- 32.Kloepper J W, Liffshitz R, Zablotowicz R M. Free living bacterial inocula for enhancing crop productivity. Trends Biotechnol. 1989;7:39–44. [Google Scholar]
- 33.Kragelund L, Nybroe O. Competition between Pseudomonas fluorescens Ag1 and Alcaligenes eutrophus JMP134 (pJP4) during colonization of barley roots. FEMS Microbiol Ecol. 1996;20:41–51. [Google Scholar]
- 34.Kragelund L, Hosbond C, Nybroe O. Distribution of metabolic activity and phosphate starvation response of lux-tagged Pseudomonas fluorescens reporter bacteria in the barley rhizosphere. Appl Environ Microbiol. 1997;63:4920–4928. doi: 10.1128/aem.63.12.4920-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroer N, Barkay T, Sørensen S, Weber D. Effect of root exudates and bacterial metabolic activity on conjugal gene transfer in the rhizosphere of a marsh plant. FEMS Microbiol Ecol. 1998;25:375–384. [Google Scholar]
- 36.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latour X, Corberand T, Laguerre G, Allard F, Lemanceau P. The composition of fluorescent Pseudomonas populations associated with roots is influenced by plant and soil type. Appl Environ Microbiol. 1996;62:2449–2456. doi: 10.1128/aem.62.7.2449-2456.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lugtenberg B J J, de Weger L A, Bennett J W. Microbial stimulation of plant growth and protection from disease. Curr Opin Biotechnol. 1991;2:457–464. [Google Scholar]
- 39.Maloney P E, van Bruggen A H C, Hu S. Bacterial community structure in relation to the carbon environments in lettuce and tomato rhizospheres and in bulk soil. Microb Ecol. 1997;34:109–117. doi: 10.1007/s002489900040. [DOI] [PubMed] [Google Scholar]
- 40.Møller S, Kristensen C S, Poulsen L K, Carstensen J M, Molin S. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl Environ Microbiol. 1995;61:741–748. doi: 10.1128/aem.61.2.741-748.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Møller S, Sternberg C, Andersen J B, Christensen B B, Ramos J L, Givskov M, Molin S. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol. 1998;64:721–732. doi: 10.1128/aem.64.2.721-732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Sullivan D J, O'Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prosser J I. Molecular marker systems for detection of genetically engineered micro-organisms in the environment. Microbiology. 1994;140:5–17. doi: 10.1099/13500872-140-1-5. [DOI] [PubMed] [Google Scholar]
- 45.Rattray E A S, Prosser J I, Glover L A, Killham K. Characterization of rhizosphere colonization by luminescent Enterobacter cloacae at the population and single-cell levels. Appl Environ Microbiol. 1995;61:2950–2957. doi: 10.1128/aem.61.8.2950-2957.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodríguez G G, Phipps D, Ishiguro K, Ridgway H D. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:3573–3578. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rovira A D, Davey C B. Biology of the rhizosphere. In: Carson E W, editor. The plant root and its environment. Charlottesville: University of Virginia Press; 1974. pp. 153–204. [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Schaechter M, Maaløe O, Kjeldgaard N O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 50.Schloter M, Borlinghaus R, Bode W, Hartmann Q. Direct identification and localization of Azospirillum in the rhizosphere of wheat using fluorescence-labeled monoclonal antibodies and confocal scanning laser microscopy. J Microsc. 1993;171:173–177. [Google Scholar]
- 51.Söderberg K H, Bååth E. Bacterial activity along a young barley root measured by thymidine and leucine incorporation techniques. Soil Biol Biochem. 1998;30:1259–1268. [Google Scholar]
- 52.Sternberg C, Christensen B B, Johansen T, Toftgaard Nielsen A, Andersen J B, Givskov M, Molin S. Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol. 1999;65:4108–4117. doi: 10.1128/aem.65.9.4108-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomashow L S. Biological control of plant root pathogens. Curr Opin Biotechnol. 1996;7:343–347. doi: 10.1016/s0958-1669(96)80042-5. [DOI] [PubMed] [Google Scholar]
- 54.Tombolini R, Jansson J K. Monitoring of GFP-tagged bacterial cells. Methods Mol Biol. 1998;102:285–298. doi: 10.1385/0-89603-520-4:285. [DOI] [PubMed] [Google Scholar]
- 55.Unge A, Tombolini R, Mølbak L, Jansson J. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl Environ Microbiol. 1999;65:813–821. doi: 10.1128/aem.65.2.813-821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winding A, Binnerup S J, Sørensen J. Viability of indigenous soil bacteria assayed by respiratory activity and growth. Appl Environ Microbiol. 1994;60:2869–2875. doi: 10.1128/aem.60.8.2869-2875.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]