Abstract
Ionizing radiation is used in cancer radiation therapy to effectively damage the DNA of tumors. The main damage is due to generation of highly reactive secondary species such as low-energy electrons (LEEs). The accurate quantification of DNA radiation damage of well-defined DNA target sequences in terms of absolute cross sections for LEE-induced DNA strand breaks is possible by the DNA origami technique; however, to date, it is possible only for DNA single strands. In the present work DNA double strand breaks in the DNA sequence 5′-d(CAC)4/5′-d(GTG)4 are compared with DNA single strand breaks in the oligonucleotides 5′-d(CAC)4 and 5′-d(GTG)4 upon irradiation with LEEs in the energy range from 5 to 20 eV. A maximum of strand break cross section was found around 7 and 10 eV independent of the DNA sequence, indicating that dissociative electron attachment is the underlying mechanism of strand breakage and confirming previous studies using plasmid DNA.
Currently, one of the most common ways to treat cancer is radiation therapy using high-energy (MeV) photon, electron, or ion beams.1,2 The primary high-energy radiation leads to water radiolysis with OH radicals and low-energy electrons (LEEs; <20 eV) being the most reactive secondary particles generated along the ionization track of water.3−5 The LEEs recombine or react within femtoseconds with molecules or subunits of biomolecules to produce excited states, radicals, and transient negative ions (TNIs).6 The formation of such TNIs is due to the attachment of an electron to a formerly unoccupied molecular orbital via a resonant Franck–Condon transition localized on the DNA components.7,8 The short-lived TNIs rapidly decay either by autodetachment9 of the extra electron or by dissociation resulting in the formation of a neutral radical and an anion, the latter process being termed dissociative electron attachment (DEA).10 Hence, LEEs are considered efficient radiation damage contributors11 causing single strand breaks (SSBs), double strand breaks (DSBs), base damage, and inter- and intrastrand cross-links. An accurate quantification of DNA radiation damage in the form of absolute cross sections for radiation-induced DNA strand breaks (SBs) is required to (i) provide a fundamental physical basis for the simulation of the dose distributions in patients prior to radiation treatment12 and (ii) to develop new strategies for cancer treatment with radio- and chemotherapy.13,14 Because of the low penetration depth of LEEs, there is a need for highly sensitive physicochemical experiments to quantify LEE-induced strand breaks.15 Previous studies used agarose gel electrophoresis of supercoiled plasmid DNA;16 however, the method does not provide information about the response of specific DNA sequences to LEEs.17 The accurate quantification of LEE-induced strand breaks in well-defined DNA sequences is possible using a recently developed DNA origami technique.18,19
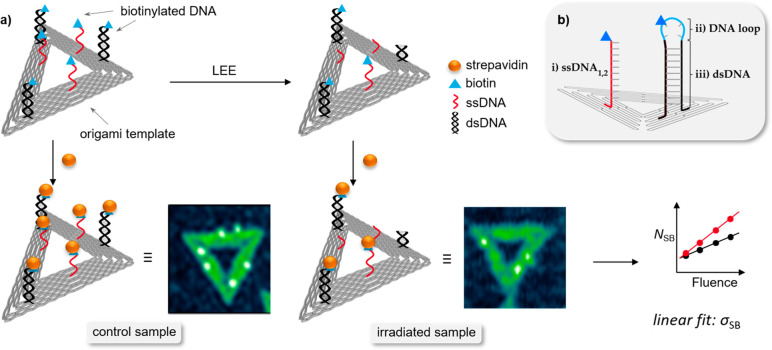
DNA origami triangles serve as a platform for biotinylated oligonucleotide target sequences that are exposed to LEEs of specific energy. Subsequent to the irradiation, the biotin (Bt) label of the remaining intact DNA target sequences can be visualized in atomic force microscopy (AFM) by binding to streptavidin, which appears as a bright spot in AFM images (Figure 1).
Figure 1.
(a) Scheme of the experimental procedure to determine absolute DNA strand break cross sections (σSB). Each DNA origami (gray triangle) carries three biotinylated single (red) and double-stranded (black) DNA target sequences protruding from the template. The intact DNA sequences are visualized with atomic force microscopy (AFM) by treating the irradiated samples with streptavidin (yellow spheres). Bright spots in AFM images indicate streptavidin molecules attached to intact target sequences. Linear fits from the plots of the number of strand breaks (NSB) as a function of the fluence yield the absolute strand break cross sections (σSB). (b) Scheme of protruding target (i) single-stranded DNA, (ii) DNA loop, and (iii) double-stranded DNA stem sequence from the DNA origami triangle.
A missing spot indicates a cleavage of the target sequence due to the interaction with LEEs; that is, an SB occurred. An absolute cross section for DNA SBs can be determined from the slope of the exposure–response curves (Figure 2). Compared to other experimental approaches, the advantage of the DNA origami technique is the relatively simple absolute quantification of strand break yields, the versatility in the choice of target sequences, and the possibility to irradiate two sequences in a single irradiation experiment providing a perfect comparison of the response of two target sequences. In previous work, absolute single strand break cross sections have been determined for homooligomers,20,21 mixed sequences,22 telomeric DNA,23 and ssDNA sensitized with potential radiosensitizers.24−26 In addition to LEE irradiation, also experiments with vacuum-UV radiation21 as well as X-rays and γ rays27 have been performed. However, so far only single strand break cross sections have been determined. This leaves an important gap, because it is mostly the DSBs that are responsible for mutagenic and genotoxic effects in cells.28 Herein, we present a method to quantify LEE-induced DSBs in well-defined DNA sequences based on the DNA origami technology.29 Additionally, we successfully explore the energy dependence of LEE-induced SSBs and DSBs in the range from 5 to 20 eV. This energy range covers the most relevant DEA resonances for DSBs.
Figure 2.
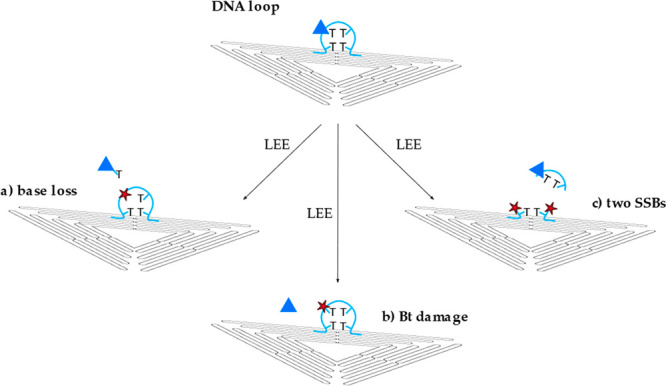
Schematic showing various types of the DNA loop damage (light blue) caused by low-energy electrons. (a) Base loss of the thymine base including the Bt label, (b) damage of the Bt label, and (c) two separated single strand breaks in different positions.
In order to determine absolute cross sections for DSBs using the DNA origami technique, we modified the experimental scheme that was used previously for SSBs.18,19 Triangular DNA origami nanostructures are assembled from a long circular single-stranded scaffold strand and a set of 208 short artificial staple strands. For the irradiation experiments the double-stranded DNA target sequences are extended from the DNA origami platform by introducing a DNA hairpin hpDNA (Figure 1b, parts ii and iii) into one of the staple strands. The stem of the hairpin represents the double-stranded target sequence (dsDNA) while the purpose of the loop (DNA loop) is to ensure that the double-stranded sequence remains closed, i.e., double-stranded. Furthermore, the stem of the DNA hairpin is attached to two staple strands within the DNA origami platform to make sure that the double strand forms correctly. The correct formation of double-stranded DNA extending from the DNA origami platform is confirmed in control experiments reported in the Supporting Information (Figure S1). The sequence of the DNA hairpin hpDNA is (5′-d(CAC)4T(Bt-dT)T2(GTG)4) and includes the DNA loop 5′-d(T(Bt-dT)T2) and the dsDNA stem sequence (5′-d(CAC)4/5′-d(GTG)4).
During the annealing process, hpDNA forms a double strand consisting of 12 DNA base pairs (Figure 1b, part iii; black) and a loop of four nonhybridized thymine bases (Figure 1b, part ii; light blue). The Bt label is covalently bound to one of the thymine bases in the DNA loop. In the case of the respective single strands ssDNA1 (CAC)4 and ssDNA2 (GTG)4 (Figure 1b, part i), the Bt label is attached to the DNA backbone at the 5′ end. Additional control experiments with 5′-d(T(Bt-dT)T2) have been carried out in order to characterize the stability of the single-stranded DNA loop (Figure 2) and the Bt conjugated to the base. These experiments are intended to determine the stability of the thymine-bound Bt upon the irradiation with LEEs between 5 and 20 eV. Possible loss of Bt can occur because of the loss of the thymine base to which the Bt marker is bound (Figure 2a), LEE-induced damage to the Bt label (Figure 2b), or damage of the DNA loop involving two SSBs (Figure 2c) that may be initiated by either 1 or 2 incident electrons. These processes are indistinguishable in the AFM analysis using the DNA origami technique. Nevertheless, if path c is a two-electron process, this will result in a power-law-dependency of NSB with increasing F in the exposure-response curves, which was, however, not observed in the present experiments. Table 1 summarizes the DNA strand break CS of the DNA loop irradiated at electron energies between 5 and 20 eV. The associated exposure–response curves show a slight linear increase in NSB with F, whereby σloop was found in the range from (0.12 ± 0.12) × 10–15 cm2 for 5 eV up to (0.33 ± 0.16) × 10–15 cm2 for 10 eV electron energy.
Table 1. Overview of the Absolute Cross Sections (σloop) for SSBs for the DNA Loop upon Electron Irradiation between 5 and 20 eV.
| DNA SSB Cross Section σloop [10–15 cm2] of the DNA Loop | ||||||
|---|---|---|---|---|---|---|
| energy | 5 eV | 7 eV | 8.4 eV | 10 eV | 11 eV | 12 eV |
| DNA loop | 0.12 ± 0.12 | 0.22 ± 0.15 | 0.25 ± 0.12 | 0.33 ± 0.16 | 0.27 ± 0.16 | 0.19 ± 0.16 |
| energy | 14 eV | 16 eV | 18 eV | 19 eV | 20 eV |
|---|---|---|---|---|---|
| DNA loop | 0.24 ± 0.13 | 0.21 ± 0.23 | 0.19 ± 0.17 | 0.20 ± 0.13 | 0.23 ± 0.16 |
Overall, the Bt label in the loop is subject to LEE-induced DNA damage itself, which has to be considered in the calculation of the final strand break yield. Therefore, the absolute DNA strand break cross section σDSB for hpDNA is corrected to dsDNA for the DNA stem sequence by the DNA loop damage σloop.
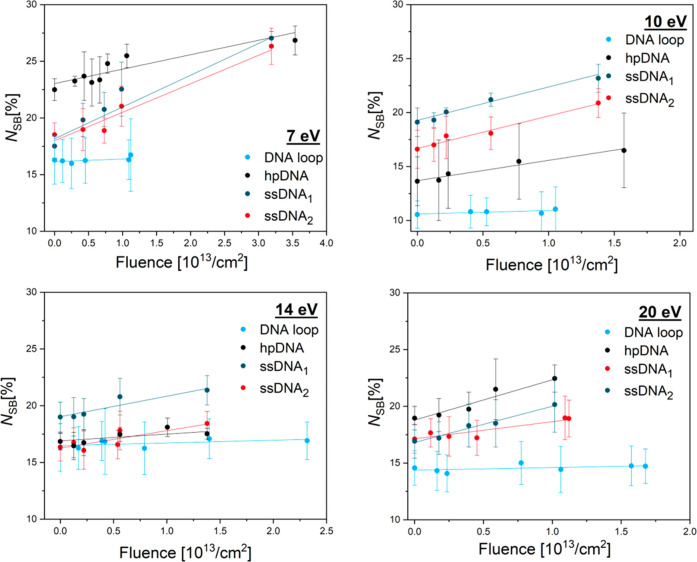
Figure 3 shows examples of exposure-response curves showing the linear dependence of NSB on the fluence at electron energies of 7, 10, 14, and 20 eV for ssDNA1,2, hpDNA and DNA loop. From the slope of the linear fits, we determined the absolute SB cross sections (σSB) for the respective DNA sequence and electron energy. Table 2 summarizes all experimentally determined DNA strand break CSs of the two associated ssDNA1 (CAC)4 and ssDNA2 (GTG)4 and the respective double-stranded hpDNA. Because of the possibility of LEE-induced DNA damage in the DNA loop, σDSB for hpDNA has to be corrected to consider only the damage of the double-stranded stem sequence dsDNA. Therefore, σloop of the loop is subtracted from σDSB of hpDNA for each electron energy and is shown as dsDNA. It needs to be noted that the additivity of cross sections is only a first approximation, because the strand break cross section does not depend linearly on the length of the sequence, and also the environment might have an effect on the strand break cross section. Figure 4 displays all obtained values for σSSB of ssDNA1,2 ((CAC)4, blue; and (GTG)4, red) and σDSB for dsDNA (black-light blue) at different irradiation energies between 5 and 20 eV electrons.
Figure 3.
Number of strand breaks (NSB) as a function of the fluence at 7, 10, 14, and 20 eV electrons for the DNA loop (light blue), double-stranded DNA hairpin hpDNA (black), and the two single-stranded DNA sequences 1 (dark blue) and 2 (red).
Table 2. Summary of σSSB for the ssDNA1 and ssDNA2, as Well as hpDNA, Which Is Subtracted by σloop of DNA Loop Giving Values Named as dsDNA upon Electron Irradiation between 5 and 20 eV.
| DNA SB CS σSSB and σDSB [10–15 cm2] of ssDNA1, ssDNA2, hpDNA, and dsDNA | ||||||
|---|---|---|---|---|---|---|
| energy | 5 eV | 7 eV | 8.4 eV | 10 eV | 11 eV | 12 eV |
| ssDNA1 | 2.58 ± 0.37 | 2.80 ± 0.19 | 2.60 ± 0.17 | 3.10 ± 0.41 | 2.80 ± 0.35 | 2.10 ± 0.31 |
| ssDNA2 | 2.44 ± 0.37 | 2.52 ± 0.36 | 2.43 ± 0.27 | 3.00 ± 0.32 | 2.70 ± 0.27 | 1.90 ± 0.23 |
| hpDNA | 0.98 ± 0.29 | 1.27 ± 0.27 | 1.32 ± 0.30 | 1.88 ± 0.17 | 1.52 ± 0.22 | 0.80 ± 0.19 |
| dsDNA | 0.86 ± 0.29 | 1.05 ± 0.27 | 1.07 ± 0.30 | 1.50 ± 0.17 | 1.25 ± 0.22 | 0.61 ± 0.19 |
| energy | 14 eV | 16 eV | 18 eV | 19 eV | 20 eV |
|---|---|---|---|---|---|
| ssDNA1 | 1.80 ± 0.39 | 2.20 ± 0.27 | 2.55 ± 0.40 | 3.50 ± 0.39 | 3.55 ± 0.37 |
| ssDNA2 | 1.53 ± 0.36 | 2.07 ± 0.37 | 2.44 ± 0.49 | 3.00 ± 0.24 | 3.20 ± 0.32 |
| hpDNA | 0.60 ± 0.29 | 1.42 ± 0.27 | 1.47 ± 0.29 | 1.52 ± 0.30 | 1.51 ± 0.36 |
| dsDNA | 0.36 ± 0.29 | 1.21 ± 0.27 | 1.28 ± 0.29 | 1.32 ± 0.30 | 1.28 ± 0.36 |
Figure 4.
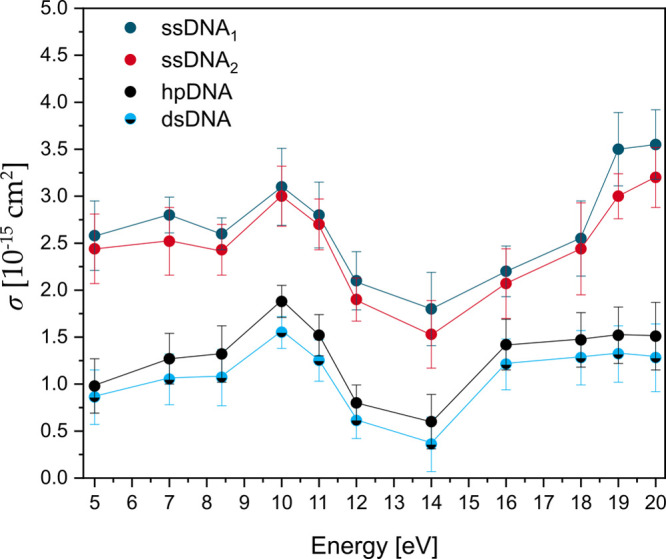
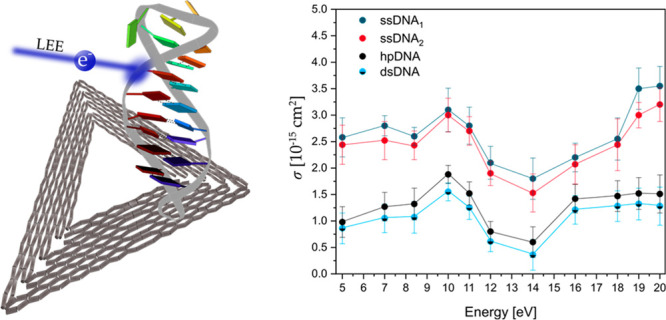
Plot of absolute DNA strand break cross section (σSSB) for the single-stranded DNA sequences ssDNA1 5′-Bt-d(CAC)4 (dark blue) and ssDNA2 5′-Bt-d(GTG)4 (red) and the absolute DNA strand break cross section (σDSB) for the double-stranded hairpin DNA hpDNA (black) and stem sequence dsDNA (light blue-black) corrected by σloop of the DNA loop (Table 2) upon electron irradiation between 5 and 20 eV.
Accordingly, σSSB and σDSB have a very similar shape in terms of energy-dependency. Below about 12 eV, electron attachment occurs at specific electron energies, in which anion resonances appear as large changes in σSB. We find a clear maximum of DSBs and SSBs at 10 eV electron energy using ssDNA1,2 and the respective dsDNA stem sequence. The cross sections for ssDNA1,2 exhibit also another pronounced local maximum around 7 eV. In previous experiments using single-stranded poly adenine and 5′-dTT(ATA)3TT,20,25 only the maximum around 7 eV has been observed, indicating that specific DNA sequences give rise to specific anion resonances resulting in different energy dependence of strand breakage. At energies between 14 and 15 eV the strand break cross sections show clear minima and start to rise only toward 20 eV, where other mechanisms than DEA start to contribute to strand breakage. In previous investigations of LEE-induced DNA damage with plasmid DNA (pGEM 3Zf(−); 3199 base pairs) a similar strong electron energy-dependent signature of SBs was observed with a broad resonance between 7 and 13 eV for DNA single strands, followed by a minimum at energies of 14–15 eV.17 Because of the electron energy distribution in the DNA origami experiments the DSB yield is still nonzero in this energy regime, and the smallest σDSB for dsDNA of (0.60 ± 0.29) × 10–15 cm2 is observed. Figure 3 shows a rise of both the SSB and DSB yields above 14 eV. σDSB reaches a plateau at 16 eV with similar values as at 10 eV, whereas σSSB is rising monotonically and no plateau is observed within the studied energy range for the SSBs. The DNA damage above 14 eV is assigned to the nonresonant mechanisms of DNA strand breakage, i.e., strand breakage due to dipolar dissociation and ionization-induced dissociation.17
Sanche et al. show a ten times higher yield of DNA SSBs compared to DNA DSBs in plasmid DNA.30 A slightly smaller factor is observed in the present DNA origami irradiation experiments, in which σSSB is a factor of 3 higher than σDSB in all investigated electron energies. Overall, the results confirm the experimental findings published by Sanche et al.7
The dsDNA sequence in the hairpin enables approximately one helical turn formed by 12 base pairs. A DSB might be formed by two independent, closely opposed SSBs within one helical turn. But in fact, this is a two-electron process, which would lead to a nonlinearity in the fluence dependence of the number of strand breaks (NSB). However, this behavior could not be observed in the present low-energy electron-induced DNA damage experiments using DNA origami nanostructures. Instead the DSBs must be formed by a single-electron process. There is a proposed pathway yielding a local multiply damage site (LMDS) with two or more damage sites within a few helical turns induced by a single electron.30−32 The LMDS includes oxidized purines and pyrimidines, abasic sites, and strand breaks describing the differences in σSSB and σDSB.33−35 The initial step is the capture of the electron by a positive electron affinity (EA) of an electronically excited state of a base followed by the formation of a core-excited TNI on the base. The TNI can decay by DEA or autoionization. The latter leaves the base in an electronically excited state resulting in a separation of the additional electron and the electronic excitation. Both can cause damage on each of the complementary DNA single strands. The initial base stays in a dissociative state leading to C–O bond scission within the same strand, while the additional electron transfers to the phosphate group on the opposite strand, causing rupture of the C–O bond via DEA. DEA studies support the reaction mechanism proposed by Sanche et al. with a most likely strand breakage in the C–O phosphodiester bond in the DNA backbone by transferring the excess energy from the nucleobase to the DNA backbone.36
The aim of the present study was the first absolute quantification of the energy-dependent strand breakage of a specific sequence of double-stranded DNA in the LEE energy range of 5–20 eV. The strand break cross sections of the complementary single strands ssDNA1 and ssDNA2 (5′-d(CAC)4 and 5′-d(GTG)4) are three times higher than the strand break cross sections of the dsDNA at every energy. The σSB for all sequences investigated in this section exhibits a broad resonant structure peaking at 10 eV. At energies between 14 and 15 eV the strand break cross sections show clear minima and start to rise only toward 20 eV, where other mechanisms like ionization contribute to strand breakage. The resonant structure of the DSB cross section below 14 eV clearly indicates that DEA is the major mechanism responsible for the strand breakage. A similar signature of the SB yield, which is strongly dependent on the electron energy, was shown in several studies using plasmid DNA and could be confirmed with these energy-dependent irradiation experiments also for oligonucleotides. The peak position and its absolute value are influenced by the choice of the nucleotide sequence and its neighboring bases.
Acknowledgments
This research was supported by the European Research Council (ERC; consolidator Grant No. 772752) and by the German Research Foundation (DFG, Project No. 230710387).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpclett.2c00684.
The authors declare no competing financial interest.
Supplementary Material
References
- Gofman J. W.Radiation and Human Health; Sierra Club Books: San Francisco, 1981. [Google Scholar]
- Sevilla M. D.; Becker D.; Kumar A.; Adhikary A. Gamma and Ion-Beam Irradiation of DNA: Free Radical Mechanisms, Electron Effects, and Radiation Chemical Track Structure. Radiat. Phys. Chem. 2016, 128, 60–74. 10.1016/j.radphyschem.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton M. Nature of Radiation Chemistry. J. Phys. Colloid Chem. 1947, 51 (2), 611–625. 10.1021/j150452a029. [DOI] [PubMed] [Google Scholar]
- Kai T.; Yokoya A.; Ukai M.; Fujii K.; Higuchi M.; Watanabe R. Dynamics of Low-Energy Electrons in Liquid Water with Consideration of Coulomb Interaction with Positively Charged Water Molecules Induced by Electron Collision. Radiat. Phys. Chem. 2014, 102, 16–22. 10.1016/j.radphyschem.2014.04.017. [DOI] [Google Scholar]
- Alizadeh E.; Orlando T. M.; Sanche L. Biomolecular Damage Induced by Ionizing Radiation: The Direct and Indirect Effects of Low-Energy Electrons on DNA. Annu. Rev. Phys. Chem. 2015, 66, 379–398. 10.1146/annurev-physchem-040513-103605. [DOI] [PubMed] [Google Scholar]
- Schulz G. J. Resonances in Electron Impact on Diatomic Molecules. Rev. Mod. Phys. 1973, 45 (3), 423–486. 10.1103/RevModPhys.45.423. [DOI] [Google Scholar]
- Boudaïffa B.; Cloutier P.; Hunting D.; Huels M. A.; Sanche L. Resonant Formation of DNA Strand Breaks by Low-Energy (3 to 20 eV) Electrons. Science 2000, 287 (5458), 1658–1660. 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- Kumar A.; Becker D.; Adhikary A.; Sevilla M. D. Reaction of Electrons with DNA: Radiation Damage to Radiosensitization. Int. J. Mol. Sci. 2019, 20 (16), 3998. 10.3390/ijms20163998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran T. F.Electron Transfer Reactions. Electron–Molecule Interactions and their Applications; Elsevier, 1984; pp 1–64. [Google Scholar]
- Ma J.; Kumar A.; Muroya Y.; Yamashita S.; Sakurai T.; Denisov S. A.; Sevilla M. D.; Adhikary A.; Seki S.; Mostafavi M. Observation of Dissociative Quasi-Free Electron Attachment to Nucleoside via Excited Anion Radical in Solution. Nat. Commun. 2019, 10 (1), 102. 10.1038/s41467-018-08005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimblott S. M.; LaVerne J. A. Production of Low-Energy Electrons by Ionizing Radiation. Radiat. Phys. Chem. 2007, 76 (8–9), 1244–1247. 10.1016/j.radphyschem.2007.02.012. [DOI] [Google Scholar]
- Kohanoff J.; McAllister M.; Tribello G. A.; Gu B. Interactions Between Low-Energy Electrons and DNA: A Perspective from First-Principles Simulations. J. Phys.: Condens. Matter 2017, 29 (38), 383001. 10.1088/1361-648X/aa79e3. [DOI] [PubMed] [Google Scholar]
- Rezaee M.; Hill R. P.; Jaffray D. A. The Exploitation of Low-Energy Electrons in Cancer Treatment. Radiat. Res. 2017, 188 (2), 123–143. 10.1667/RR14727.1. [DOI] [PubMed] [Google Scholar]
- Schürmann R.; Vogel S.; Ebel K.; Bald I. The Physico-Chemical Basis of DNA Radiosensitization: Implications for Cancer Radiation Therapy. Chem. Eur. J. 2018, 24 (41), 10271–10279. 10.1002/chem.201800804. [DOI] [PubMed] [Google Scholar]
- Meesungnoen J.; Jay-Gerin J.-P.; Filali-Mouhim A.; Mankhetkorn S. Low-Energy Electron Penetration Range in Liquid Water. Radiat. Res. 2002, 158 (5), 657–660. 10.1667/0033-7587(2002)158[0657:LEEPRI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Boudaïffa B.; Cloutier P.; Hunting D.; Huels M. A.; Sanche L. Cross Sections for Low-Energy (10 – 50 eV) Electron Damage to DNA. Radiat. Res. 2002, 157 (3), 227–234. 10.1667/0033-7587(2002)157[0227:CSFLEE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Huels M. A.; Boudaïffa B.; Cloutier P.; Hunting D.; Sanche L. Single, Double, and Multiple Double Strand Breaks Induced in DNA by 3–100 eV Electrons. J. Am. Chem. Soc. 2003, 125 (15), 4467–4477. 10.1021/ja029527x. [DOI] [PubMed] [Google Scholar]
- Keller A.; Bald I.; Rotaru A.; Cauët E.; Gothelf K. V.; Besenbacher F. Probing Electron-Induced Bond Cleavage at the Single-Molecule Level Using DNA Origami Templates. ACS Nano 2012, 6 (5), 4392–4399. 10.1021/nn3010747. [DOI] [PubMed] [Google Scholar]
- Rackwitz J.; Ranković M. L.; Milosavljević A. R.; Bald I. A Novel Setup for the Determination of Absolute Cross Sections for Low-Energy Electron Induced Strand Breaks in Oligonucleotides – The Effect of the Radiosensitizer 5-Fluorouracil. Eur. Phys. J. D 2017, 71 (2), R287. 10.1140/epjd/e2016-70608-4. [DOI] [Google Scholar]
- Ebel K.; Bald I. Length and Energy Dependence of Low-Energy Electron-Induced Strand Breaks in Poly(A) DNA. Int. J. Mol. Sci. 2020, 21 (1), 111. 10.3390/ijms21010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S.; Ebel K.; Schürmann R. M.; Heck C.; Meiling T.; Milosavljevic A. R.; Giuliani A.; Bald I. Vacuum-UV and Low-Energy Electron-Induced DNA Strand Breaks - Influence of the DNA Sequence and Substrate. ChemPhysChem 2019, 20 (6), 823–830. 10.1002/cphc.201801152. [DOI] [PubMed] [Google Scholar]
- Vogel S.; Rackwitz J.; Schürman R.; Prinz J.; Milosavljević A. R.; Réfrégiers M.; Giuliani A.; Bald I. Using DNA Origami Nanostructures to Determine Absolute Cross Sections for UV Photon-Induced DNA Strand Breakage. J. Phys. Chem. Lett. 2015, 6 (22), 4589–4593. 10.1021/acs.jpclett.5b02238. [DOI] [PubMed] [Google Scholar]
- Rackwitz J.; Bald I. Low-Energy Electron-Induced Strand Breaks in Telomere-Derived DNA Sequences-Influence of DNA Sequence and Topology. Chem. Eur. J. 2018, 24 (18), 4680–4688. 10.1002/chem.201705889. [DOI] [PubMed] [Google Scholar]
- Rackwitz J.; Kopyra J.; Dąbkowska I.; Ebel K.; Ranković M. L.; Milosavljević A. R.; Bald I. Sensitizing DNA Towards Low-Energy Electrons with 2-Fluoroadenine. Angew. Chem. Int. Ed. Engl. 2016, 55 (35), 10248–10252. 10.1002/anie.201603464. [DOI] [PubMed] [Google Scholar]
- Schürmann R.; Tsering T.; Tanzer K.; Denifl S.; Kumar S. V. K.; Bald I. Resonant Formation of Strand Breaks in Sensitized Oligonucleotides Induced by Low-Energy Electrons (0.5–9 eV). Angew. Chem. Int. Ed. Engl. 2017, 56 (36), 10952–10955. 10.1002/anie.201705504. [DOI] [PubMed] [Google Scholar]
- Vogel S.; Ebel K.; Heck C.; Schürmann R. M.; Milosavljević A. R.; Giuliani A.; Bald I. Vacuum-UV Induced DNA Strand Breaks - Influence of the Radiosensitizers 5-Bromouracil and 8-Bromoadenine. Phys. Chem. Chem. Phys. 2019, 21 (4), 1972–1979. 10.1039/C8CP06813E. [DOI] [PubMed] [Google Scholar]
- Sala L.; Zerolová A.; Rodriguez A.; Reimitz D.; Davídková M.; Ebel K.; Bald I.; Kočišek J. Folding DNA Into Origami Nanostructures Enhances Resistance to Ionizing Radiation. Nanoscale 2021, 13 (25), 11197–11203. 10.1039/D1NR02013G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C.; Hoeijmakers J. H.; Kanaar R. Chromosomal Stability and the DNA Double-Stranded Break Connection. Nat. Rev. Genet. 2001, 2 (3), 196–206. 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Ebel K.Quantification of Low-Energy Electron Induced Single and Double Strand Breaks in Well-Defined DNA Sequences Using DNA Origami Nanostructures. Dissertation, University of Potsdam, 2020. [Google Scholar]
- Gao Y.; Zheng Y.; Sanche L. Low-Energy Electron Damage to Condensed-Phase DNA and Its Constituents. Int. J. Mol. Sci. 2021, 22 (15), 7879. 10.3390/ijms22157879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.; Zhou L.; Tian Q.; Zheng Y.; Sanche L. Chemoradiation Cancer Therapy: Molecular Mechanisms of Cisplatin Radiosensitization. J. Phys. Chem. C 2017, 121 (32), 17505–17513. 10.1021/acs.jpcc.7b05271. [DOI] [Google Scholar]
- Shao Y.; Dong Y.; Hunting D.; Zheng Y.; Sanche L. Unified Mechanism for the Generation of Isolated and Clustered DNA Damages by a Single Low Energy (5–10 eV) Electron. J. Phys. Chem. C 2017, 121 (4), 2466–2472. 10.1021/acs.jpcc.6b12110. [DOI] [Google Scholar]
- Sutherland B. M.; Bennett P. V.; Sidorkina O.; Laval J. Clustered Damages and Total Lesions Induced in DNA by Ionizing Radiation: Oxidized Bases and Strand Breaks. Biochemistry 2000, 39 (27), 8026–8031. 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- Sutherland B. M.; Bennett P. V.; Sutherland J. C.; Laval J. Clustered DNA Damages Induced by X Rays in Human Cells. Radiat. Res. 2002, 157 (6), 611–616. 10.1667/0033-7587(2002)157[0611:CDDIBX]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ward J. F. Radiation Mutagenesis: The Initial DNA Lesions Responsible. Radiat. Res. 1995, 142 (3), 362–368. 10.2307/3579145. [DOI] [PubMed] [Google Scholar]
- Simons J. How do Low-Eergy (0.1–2 eV) Electrons Cause DNA-Strand Breaks?. Acc. Chem. Res. 2006, 39 (10), 772–779. 10.1021/ar0680769. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.