Abstract
Sleep plays a significant role in the mental and physical development of children. Emerging evidence in animals and human adults indicates a relationship between sleep and the gut microbiota; however, it is unclear whether the sleep of preschoolers during a key developmental period, associates with features of their gut microbiota. The objective of this study was to assess the relationship between sleep and gut microbiota in preschool-aged children (4.37 ± 0.48 years, n = 143). Sleep measures included total night-time sleep (TST), sleep efficiency (SE), and wake-time after sleep onset (WASO) assessed using actigraphy. Beta-diversity differences between children with low and high TST (p = .048) suggest gut microbiota community differences. Particularly, relative abundance of Bifidobacterium was higher in the high TST group and Bacteroides, was higher in children who had greater SE and less WASO (LDA score >2). In contrast, some Lachnospiraceae members including Blautia and Coprococcus 1 were associated with shorter night-time sleep duration and less efficiency, respectively. We also found a group of fecal metabolites, including specific neuroactive compounds and immunomodulating metabolites were associated with greater sleep efficiency and less time awake at night. Notably, tryptophan and its metabolizing products were higher in children who had higher SE or lower WASO (LDA score >2); concentration of propionate was higher in children with less WASO (p = .036). Overall, our results reveal a novel association between sleep and gut microbiota in preschool-aged children. Longer night-time sleep and greater sleep efficiency were associated with specific commensal bacteria that may regulate sleep through modulating neurotransmitter metabolism and the immune system.
Keywords: actigraphy, Alberta Pregnancy Outcomes and Nutrition (APrON) study, gut microbiota, preschoolers, short-chain fatty acids, sleep, tryptophan
Statement of Significance.
Our study assessed the association between sleep and gut microbiota in preschool-aged children, a developmental period when sleep problems are common. An association between sleep and specific gut microbial taxa including Bifidobacterium and Bacteroides was observed in this preschool cohort; the findings suggest that sleep and gut microbiota crosstalk in preschoolers may be mediated by microbial metabolites such as tryptophan and its derivatives and propionate. Our results warrant future studies to explore the potential of gut microbiota alteration as a strategy to improve sleep in young children.
Introduction
Sleep is essential for child growth and development [1]. Unfortunately, sleep problems, short sleep duration, and night waking are common in children [2–5]. Among children under 5 years of age, sleep problems have been reported by a quarter of parents [1]. Persistent child sleep problems and short sleep duration are associated with important outcomes, including impaired cognitive development, risk of obesity, poor mental health, poor academic performance, and poor social adjustment [6–9].
Recent evidence suggests that the composition and function of the gut microbiota contribute to regulation of sleep [10, 11]. Gut microbiota plays important roles in modulating fundamental processes, such as metabolism and immune system function [12–15], with downstream effects on circadian patterns and sleep. In addition, extensive bi-directional communication between the gut and brain suggests a role for the gut microbiota on sleep regulation [16, 17]. Specifically, the gut bacteria produce a wide range of neurochemicals that can directly and indirectly stimulate the nervous system [18], and potentially affect sleep. For example, serotonin is one of the key neurotransmitters that controls circadian rhythm, sleep, and waking [19, 20], and it is known that the gut microbiota plays an important role in serotonin biosynthesis and regulation [21].
The interrelationship between gut microbiota, microbial metabolites, and sleep has been observed in both animal and human studies. A recent murine study reported that mice whose gut bacteria have been depleted showed significant sleep disturbances (spending less time in non-REM sleep during the light phase and spending more time in non-REM and REM sleep during the dark phase), and they have reduced gut serotonin concentrations [22]. In human adults (males), sleep efficiency and total sleep time were positively correlated with the diversity of the gut microbiota (the complexity or variety of bacterial taxa present in the gut) [23]. The importance of gut-brain connections for sleep is underscored by evidence that perturbing sleep in rodents and human adults also perturbs the composition of the gut microbiota [24, 25].
Despite a small body of evidence supporting a connection between sleep duration and efficiency and gut microbiota in human adults, no study has determined whether sleep is associated with gut microbiota in children at preschool age. The preschool years are a period when both sleep patterns and gut microbiota undergo transition toward an adultlike pattern, yet not completely mature [26, 27]. Characteristics of sleep patterns at preschool age include discontinuation of daytime naps, the consolidation of sleep to the night-time period, and a gradual decrease in sleep duration [28, 29]. Similarly, the gut microbiota reaches a relatively stable stage after three years age, with the presence of adult-like phylum members [27]. However, some characteristics that strongly resemble an earlier age remain, such as enrichment of oligosaccharides degrader Bifidobacterium [30] and lower diversity compared to adults [31]. Understanding the relationship between sleep and gut microbiota during this transitional period may allow development of targeted intervention to promote sleep health through reshaping the gut microbiota. The objective of the study was to assess the association between actigraphy sleep measures and the fecal microbiota composition and metabolites in young children at preschool age.
We hypothesized that sleep duration, sleep efficiency, and wake after sleep onset in preschoolers are associated with gut microbiota profile and function.
Methods
Study design
This is a subanalysis of the Alberta Pregnancy Outcomes and Nutrition (APrON) prospective cohort study [32] that recruited pregnant individuals between 2009 and 2012 in Alberta, Canada. Data in this subanalysis are from a follow-up study conducted at child ages 3–4 years. Ethics approval for this study was obtained from the Health Research Ethics Boards at the University of Calgary (REB14-1702 and REB15-1200). Informed written consent was obtained from the parent or legal guardian and verbal assent was obtained from children prior to data collection.
Participants
Complete details of participants in the APrON study have been reported [32]. Briefly, at the time of recruitment, pregnant women were ≥16 years, <27 weeks of gestation, and planned to deliver their babies within the recruitment region. Women were excluded if they were unable to complete questionnaires in English. The sample is largely White, well educated, with average income for the recruitment region. The sample somewhat underrepresents young, low income, and low education mothers, relative to nationally representative data from pregnant women in Canada [33]. For the current sample, preschool children born to APrON mothers were invited to participate in a laboratory-based neurocognitive assessment (not reported here) and objective measures of child sleep (actigraphy) and provide a fecal sample. Children with antibiotic exposure 2 weeks before fecal sample collection were excluded. The current sample comprises the 143 children who provided both a fecal sample and actigraphy data
Actigraphy measures
Wrist actigraphy data were collected using the Motionlogger Actigraph (Ambulatory Monitoring, Inc.). Children were asked to wear the actigraph on their nondominant wrist for 48 hours, capturing two nights of sleep. Data analysis was completed within the ActionW software using the Cole-Kripke algorithm [34] and a 5-minute threshold for sleep and wake periods [35]. For each participant, we calculated: 1) total night-time sleep (the hours/night spent sleeping while in bed); 2) the total duration of awakenings after sleep onset (WASO); and 3) sleep efficiency (percentage of time in bed/ total time spent sleeping). Parents were asked to keep a diary of night-time sleep. Daytime quiescence was recorded by actigraph; however, a daytime diary was not taken therefore it was not possible to determine if these periods of quiescence were sleep periods.
Microbiota analysis
Fecal samples were collected in the same week as actigraphy measures with assistance of parents at home using a toilet cover along with a sterile 50-ml plastic conical collection tube and plastic applicator. Time of stool sample collection was recorded. Information on usual dietary intake was collected at 3 years of age (approximately 1 year prior to stool sample collection) using a semi-quantitative food frequency questionnaire. Overall diet quality was estimated using a previously published dietary quality score that consists of six diet components (vegetables and fruit, grain products, milk and alternatives, meat and alternatives, candy/snacks, and sugar-sweetened beverages) [36].
Collected fecal samples were temporarily stored in a home freezer for up to 24 h before transport to the study lab in a cooler surrounded by freezer packs. Fecal samples were stored at −80°C until further processing. Fecal DNA was extracted using FastDNA® Spin Kit for Feces (MP Biomedicals, Santa Ana, California, USA) following manufacture’s instruction. 16S rRNA gene amplicon sequencing was performed using the MiSeq platform at the Centre for Health Genomics and Informatics (University of Calgary, Calgary, Canada). The PCR amplification of the V3 and V4 region of the 16S rRNA gene was performed using manufacturer-recommended primers (Forward: 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3’; Reverse: 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGT ATCTAATCC-3’). Paired-end16S rRNA gene amplicon sequence reads were analyzed with QIIME 2 (2018.11.0) [37]. Raw sequence data were demultiplexed and quality filtered using the q2-demux plugin followed by denoising with DADA2 [38] (via q2-dada2). All amplicon sequence variants (ASVs) were aligned with mafft [39] (via q2-alignment) and used to construct a phylogeny with fasttree2 [40] (via q2-phylogeny). Alpha-diversity metrics (a measure of diversity within a gut microbiota community) and beta-diversity metrics (a measure of similarity of two microbiota communities, weighted UniFrac, and unweighted UniFrac) were estimated using q2-diversity. The sampling depth used in diversity analyses was 11 896. Taxonomy was assigned to ASVs using the q2-feature-classifier [41] classify-sklearn naive Bayes taxonomy classifier against the Silva (132) 99% OTUs reference sequences [42].
Fecal metabolomic analysis
Fecal metabolomic analysis was performed at the Calgary Metabolomics Research Facility. Briefly, 100–200 mg of fecal samples were diluted 5 times (w/v) into 50% methanol/water solution. The diluted samples were homogenized using Tissue Lyser II (QIAGEN) and then incubated on ice for 30 minutes for full extraction. After incubation, samples were centrifuged at approximately 13 000 rpm and 500 µl of supernatant were collected for metabolomic analysis. Metabolomics runs were performed on a Q Exactive™ HF Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo-Fisher) coupled to a Vanquish™ UHPLC System (Thermo-Fisher). Chromatographic separation was achieved on a Syncronis HILIC UHPLC column (2.1 mm x 100 mm x 1.7 µm, Thermo-Fisher) using a binary solvent system at a flow rate of 600 µL/min. Metabolite data were analyzed by El-MAVEN software package. Metabolites were identified by matching observed m/z signals (+/–10 ppm) and chromatographic retention times to those observed from commercial metabolite standards (Sigma-Aldrich). Raw untargeted data was generated using the automated feature detection function in EL-MAVEN with a minimum signal intensity threshold of 50 000 signal intensity.
Short-chain fatty acids (SCFAs) were extracted from fecal samples with ice-cold extraction solvent (50% water/acetonitrile, v/v) spiked with stable isotope-labeled internal standards (acetic acid-1,2-13C2, propionic acid-13C3, 1 mM; butyric acid-1,2-13C2, 1 mM) before submitted to LC-MS/MS-based analysis. More details of SCFA sample preparation and analyzing methods were described in our previous study [43].
Statistical analysis
To assess the association between sleep and the gut microbiota, participants were divided into “low” and “high” groups based on the median of TST, SE, and WASO.
Differences in alpha-diversity (Observed OTUs, Shannon, and Faith PD) and SCFAs between low and high sleep groups were assessed using Mann-Whitney test (GraphPad Prism version 5.00). Group differences in beta-diversity of microbiota were assessed using the permutational analysis of variance (PERMANOVA) model with 9999 permutations based on the parameters permutation of residuals under a reduced model and a type III sum of squares (Primer-E v.7; Primer-E Ltd.). Taxonomic differences at genus level and metabolite differences between low and high sleep groups were analyzed using LEfSe (linear discriminant analysis effect size) [44] with LDA score > 2 as significance cut-off.
Results
Demographic characteristics and sleep measures
Participating children have an average age of 4.37 ± 0.48 years old and with approximately equal sex distribution (males, 47.6%). The majority of children were White (88.1%) and from a family with household income > $100 000 CAD per year (63.6%), which was the median household income for the recruitment region.
Children in this study had an average TST of 9.48 hours per night (568.6 ± 51.70 min), SE of 94.43 ± 3.78%, and WASO of 32.97 ± 22.04 min (Table 1). Children were categorized to low and high sleep groups using median split (values above the median were classified as “high”; values equal to and smaller than median were classified as “low”). The majority of children who had longer night-time sleep (high TST) also had high SE (overlap rate 69%) and low WASO (overlap rate 67%), and almost all children who had high SE also had low WASO (overlap rate 99%). The majority of sleep data were collected on consecutive days (57%) during a weekday (75%). Sleep measures collected on weekdays were not different from these collected on weekends and holidays (data not shown). A small proportion of studied children (7.8%) had periods of quiescence during the day (62.4 ± 17.7 min), however, we could not determine if these were naps because we did not have a daytime napping diary.
Table 1.
Participant characteristics and sleep measures (n = 143)
| Age (year), mean (SD, range) | 4.37 (0.48, 3.06–5.00) |
|---|---|
| Sex -male, n (%) | 68 (47.6%) |
| BMI z-score, mean (SD, range) | 0.29 (0.80, –3.00 to 1.98) |
| Ethnicity | |
| Caucasian | 126 (88.1%) |
| Chinese | 6 (4.2%) |
| Latin American | 4 (2.8%) |
| Other | 7 (4.9%) |
| Preterm (n, %) | 4 (2.8%) |
| Delivery mode-Cesarean, n,( %) | 33 (23.1%) |
| Mother's education level, n, ( %) | |
| Completed high school diploma | 6 (4.2%) |
| Completed trade, technical diploma | 22 (15.4%) |
| Completed university degree | 83 (58%) |
| Completed postgraduate degree | 32 (22.4%) |
| Annual household income (CAD), n (%)Ϯ | |
| <$20 000 | 2 (1.4%) |
| $20 000 – $39 999 | 2 (1.4%) |
| $40 000 – $69 9999 | 11 (7.7%) |
| $70 000 – $999 999 | 37 (35.9%) |
| >$100 000 | 91 (63.6%) |
| Actigraphy measures | |
| TST (min), mean (SD, range) | 568.57 (51.70, 369.00–684.50) |
| SE (%), mean (SD, range) | 94.43 (3.78, 79.03–99.92) |
| WASO (min), mean (SD, range) | 32.97 (22.04, 0.50–101.50) |
BMI, body mass index; SE, sleep efficiency; TST, total night sleep time; WASO, wake-time after sleep onset.
Sleep and the fecal microbiota
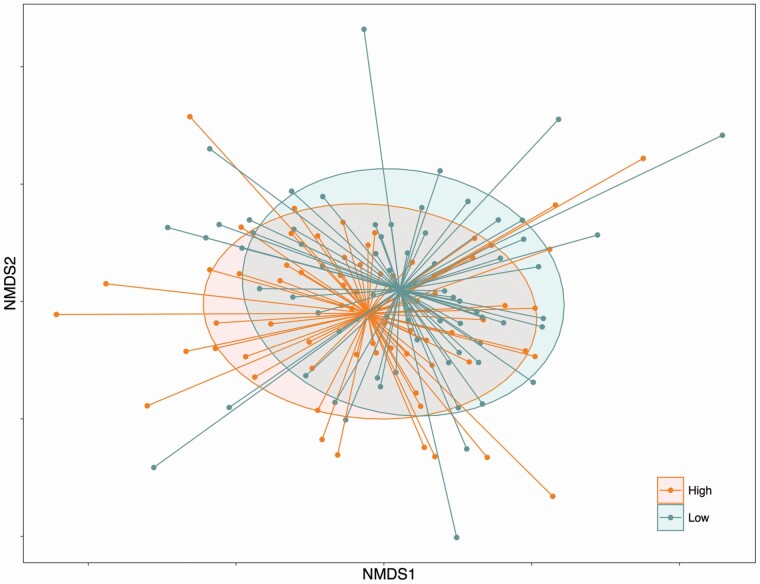
The gut microbial communities (weighted UniFrac distance of beta-diversity, which takes into account relative abundance of taxa) were different between children who had low and high TST (p = .048, Figure 1). Similarly, SE showed a trend toward significant difference with a p value of .07 however beta-diversity was not different for low and high WASO (p = .11). Unweighted UniFrac distance (which takes into account only presence/abundance of taxa) was not different between any sleep measures. No differences in alpha-diversity (Observed OTUs, Shannon and Faith PD) of the fecal microbiota were observed between children who were classified in the low and high groups of TST, SE, and WASO (Supplementary Figure 1).
Figure 1.
Nonmetric multidimensional scaling plot derived from weighted UniFrac distance showing difference in gut microbiota in children with low and high total night-time sleep (PERMANOVA, p = .048).
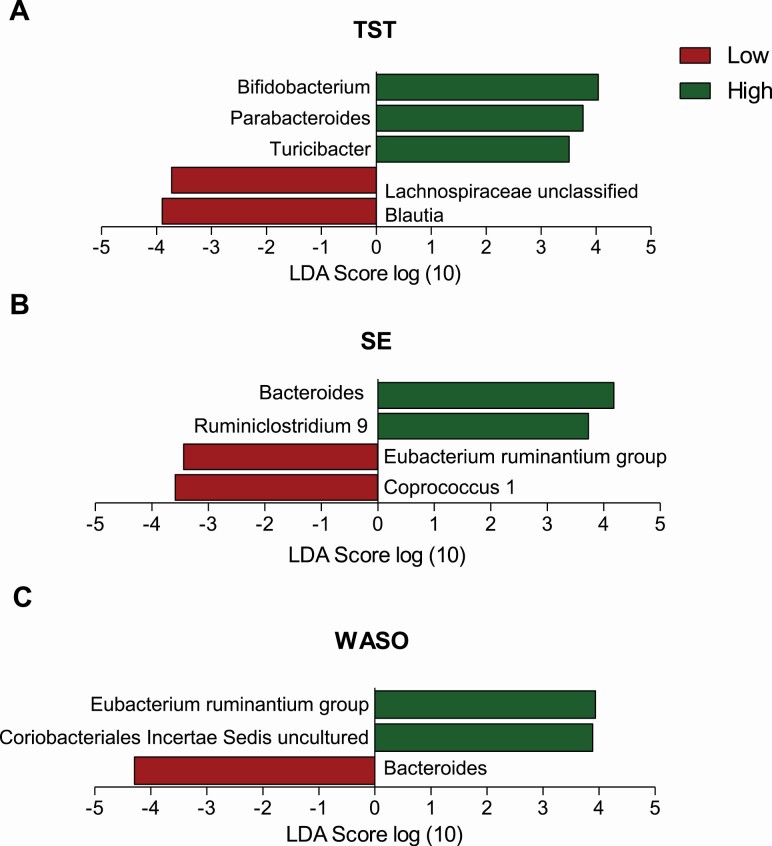
Some taxa differed between the low and high groups for TST, SE, and WASO (Figure 2). Specifically, children who recorded more total night-time sleep (high TST) had higher relative abundance of Bifidobacterium (8.06 ± 5.01% vs. 6.26% ± 4.24%, LDA = 4.04), Parabacteroides (0.91 ± 1.04% vs. 0.68 ± 1.03%, LDA =3.76), and Turicibacter (0.47 ± 0.60% vs. 0.45 ± 0.97%, LDA = 3.51); children who had higher SE showed higher relative abundance of Ruminiclostridium 9 (1.03 ± 0.21% vs. 0.07 ± 1.23%, LDA = 3.73) and Bacteroides (13.25 ± 8.75% vs. 10.08 ± 7.94%, LDA = 4.18). Relative abundance of Bacteroides was also higher in children who had lower WASO scores (10.08 ± 7.94% vs. 9.94 ± 8.03%, for low and high WASO, LDA = 4.29). In contrast, children with low SE or high WASO had lower relative abundance of taxa belonging to Eubacterium ruminantium group compared to their corresponding high or low groups (0.11 ± 0.40% vs. 0.26 ± 0.54%, for low and high SE, LDA = 3.32; 0.074 ± 0.33% vs. 0.28 ± 0.58%, for high and low WASO, LDA = 3.94). Other taxa that were associated with sleep groups included Blautia (9.75 ± 4.32% vs. 10.99 ± 4.43%, for high and low TST, LDA = 5.05) and Lachnospiraceae unclassified (5.21 ± 3.07% vs. 6.09 ± 3.04%, for high and low TST, LDA = 3.73), Coprococcus 1 (0.18 ± 0.22% vs. 0.27 ± 0.29%, for high and low SE, LDA = 3.58), and a taxon belonging to Coriobacteriales (Coriobacteriales Incertae Sedis, 0.12 ± 0.21% vs. 0.074 ± 0.16%, for high and low WASO, LDA = 3.89). Overall diet quality and time of fecal sample collection were not different between low and high sleep groups (Supplementary Table 1).
Figure 2.
Relative abundance of the most discriminant bacterial groups according to LEfSe in children who had low and high total night-time sleep (TST, A), sleep efficiency (SE, B), and wake-time after sleep onset (WASO, C).
Sleep and the fecal metabolites
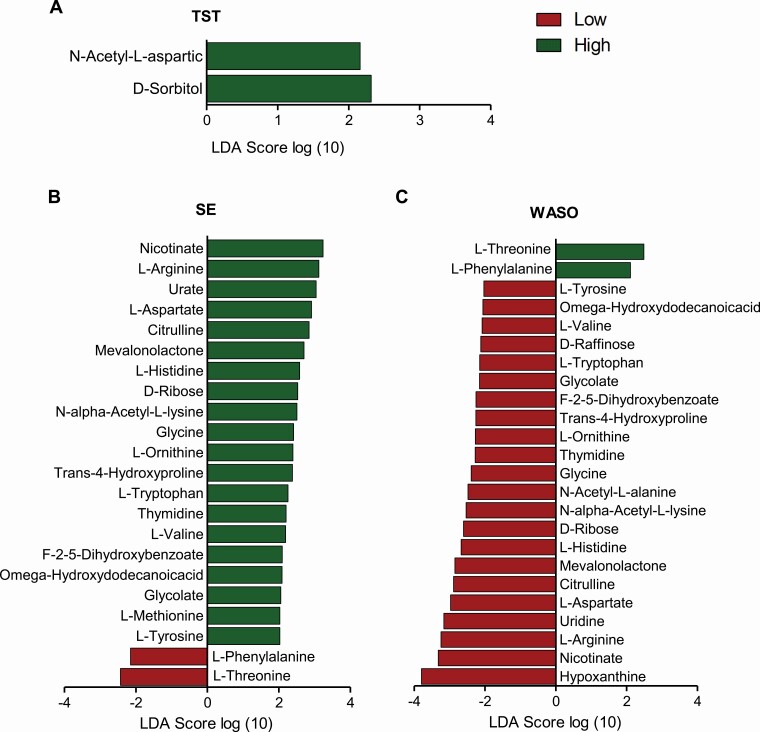
We analyzed the fecal metabolites of the preschoolers to further assess the association between sleep and the function of the gut microbiota. In total, 141 metabolites were detected and 23 metabolites differed as a function of sleep groups, including amino acids, vitamins, monosaccharides, and sugar alcohol (Figure 3). Signal intensity of N-Acetyl-L-aspartic acid (LDA = 2.16) and D-sorbitol (LDA = 2.32) were greater in children with high TST. Moreover, a group of detected metabolites were greater in both high SE and low WASO, including nicotinate (LDA = 3.23 for SE; LDA = 3.32 for WASO), glycine (LDA = 2.41 for SE; LDA = 2.39 for WASO), tryptophan (LDA = 2.26 for SE; LDA = 2.15 for WASO), tyrosine (LDA = 2.41 for SE; LDA = 2.39 for WASO) and L-ornithine (LDA = 2.27 for WASO). In contrast, L- phenylalanine (LDA = 2.15 for SE; LDA = 2.11 for WASO) and L-threonine (LDA = 2.43 for SE; LDA = 2.49 for WASO) were associated with less sleep efficiency and more disruption and they were higher in both low SE and high WASO groups.
Figure 3.
Relative abundance of the most discriminant fecal metabolites according to LEfSe in children who had low and total night-time sleep (TST, A), sleep efficiency (SE, B), and wake-time after sleep onset (WASO, C).
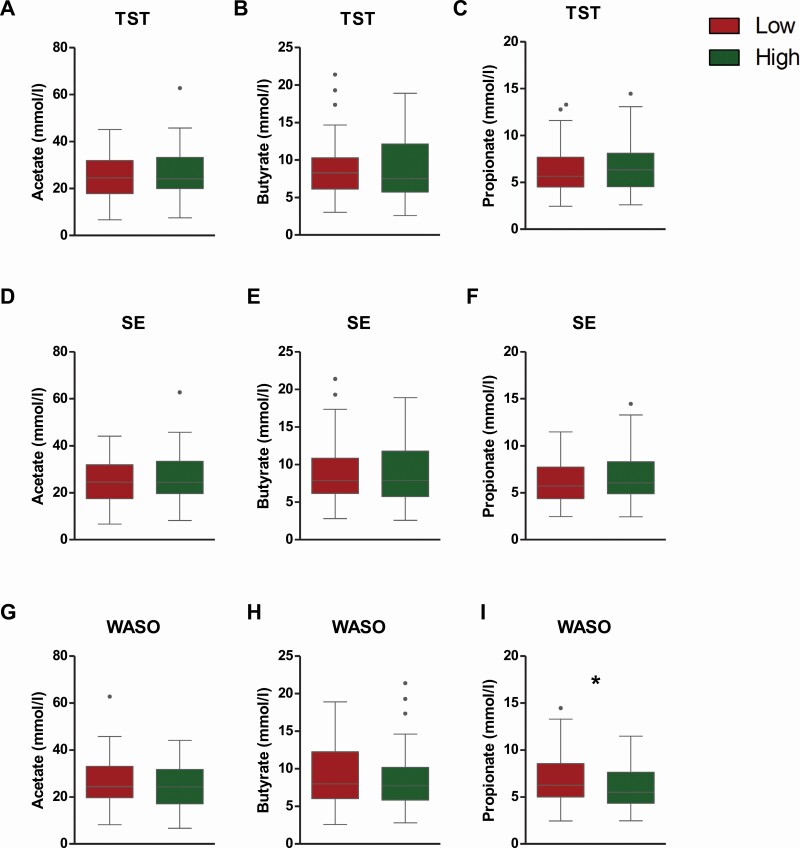
Furthermore, fecal propionate concentrations were higher in children with low WASO compared to the children who had longer wake time (high WASO). Differences in SCFA concentrations were not observed between low and high TST and SE groups (Figure 4).
Figure 4.
Differences of fecal SCFA concentrations in children with low and high total night-time sleep (TST, A-C), sleep efficiency (SE, D-F) and wake-time after sleep onset (WASO, G-I). Mann-Whitney U test; *, p < .05.
Discussion
This is the first study, to our knowledge, investigating the association between objectively assessed sleep and the gut microbiota in children at preschool age. Our results show key differences in gut microbiota composition and metabolites between children with low and high TST, SE, and WASO measures and, therefore, demonstrate the association between sleep and the gut microbiota in preschoolers.
Our results show that children who had a relatively longer night sleep had a significantly different gut microbiota community structure (beta-diversity, weighted UniFrac distance) compared to children who had shorter night sleep. Beta-diversity differences were only significant for TST not for SE or WASO indicating that night sleep duration may have a stronger influence on the overall gut microbiota community than sleep efficiency. The discriminate analysis (LEfSe) further determined which specific microbial members are most likely to explain differences between the low and high sleep groups. We found a group of commensal gut bacteria, including Bifidobacterium and Bacteroides, were separately associated with longer night-time sleep duration, greater sleep efficiency, and less time awake at night in preschoolers. Most of these taxa have been linked with sleep-related neurochemicals in previous studies, such as serotonin [45, 46] and its precursors (e.g. tryptophan and gamma-aminobutyric acid) [47]. This suggests the potential mediating roles of neurotransmitters between gut microbiota and sleep. Importantly, studies in human adults also demonstrated the effect of Bifidobacterium, either administrated as a single probiotic or mixed with other probiotics, in regulating mood [47, 48] and improving sleep quality [49]. In addition to the impact on neurotransmission, the immunoregulating properties of specific taxa may also contribute to regulating sleep. The sleep-immune interaction is a well-established phenomenon [50] and the immunomodulating effect of both Bifidobacterium and Bacteroides, the two relatively abundant taxa in children, are well demonstrated in both in vitro and in vivo studies [51]. Additionally, Bacteroides is one of the key taxa producing the SCFA propionate via the succinate pathway [52]; a high proportion of propionate in fecal samples is linked with longer uninterrupted sleep in infants [53] and propionate was also associated with low WASO in our study. Therefore, the SCFA producing capability may be another factor contributing to the connection of specific taxa and fewer night-time sleep disruptions.
Taken together, the association between the five specific taxa and longer night-time sleep duration, greater sleep efficiency, and fewer night-time sleep disruptions may be due to their functional properties in neurotransmission regulation, immunomodulation, and SCFA production.
On the other hand, we also found five taxa that showed greater abundance in children who had shorter night-time sleep duration, less sleep efficiency, and longer waking time (Figure 2), of which Blautia is the second most abundant genera in the studied children. Taxa associated with shorter night-time sleep duration and sleep efficiency in this study are highly consistent with a recent study in healthy adults, where Smith et al. reported that both Blautia and other Lachnospiraceae members were negatively correlated with SE and TST, whereas Coprococcus was positively correlated with the number of night awakenings [23]. These findings suggest that taxa contributing to shorter sleep duration in adults may also do so in young children. Notably, three of the taxa associated with sleep disruptions in adults, including Blautia, Coprococcus, and the Lachnospiraceae unclassified, belong to the Lachnospiraceae family. Despite the SCFA producing ability of many Lachnospiraceae members [52], the impact of Lachnospiraceae on host physiology is in fact controversial as Lachnospiraceae prevalence is associated with intra- and extraintestinal diseases [54] such as metabolic disorders [55, 56]. Although it is difficult to formulate a plausible mechanism between identified taxa and sleep disruption based on available literature, the consistency of identified taxa related to shorter sleep duration and more night-time sleep disruption in the current and previous studies encourage further investigation on the negative impact of these specific microbial members on sleep.
The differences of fecal metabolites observed between low and high sleep groups provided further insight into the links between sleep and gut microbiota. Several metabolites associated with greater sleep efficiency and less WASO are involved in tryptophan metabolism, including nicotinate, arginine, glycine, tryptophan, and ornithine (KEGG, pathway reference, map00260, map00380, map00400). Tryptophan is an essential aromatic amino acid and a precursor to many microbial and host metabolites including serotonin [57]. Although it is unclear whether gut microbiota-produced tryptophan makes a significant contribution to host physiology [57], tryptophan metabolites generated from direct metabolism by microbes and kynurenine pathway are known to play critical roles in immune responses and neurotransmission [57]. Moreover, effects of some of the metabolites, such as glycine and ornithine, in improving sleep quality have been shown in human trials [58, 59]. On the other hand, the relatively higher level of metabolites associated with low SE and high WASO may be associated with less efficient microbial metabolism. Phenylalanine and threonine are precursors of tyrosine and glycine, respectively. As tyrosine and glycine were higher in the high SE and low WASO groups, it is reasonable to observe that phenylalanine and threonine showed relatively greater signal intensity in low SE and high WASO groups.
The link between sleep and SCFAs is thought to derive mainly from their roles in the neuro-immunoendocrine regulation [60]. Yet, studies investigating the association between sleep and SCFAs are few and the outcomes are inconsistent [61, 62]. The higher level of propionate in low WASO can be explained by the higher relative abundance of Bacteroides in the same group of children because Bacteroides is known to be one of the main propionate producers [52]. The connection of propionate and sleep is supported by previous studies revealing the modulating roles of propionate in gut-brain communication, although the mechanisms of how propionate influences sleep are unknown. Propionate can initiate a gut-brain neural circuit [63] and interact with endothelium of the blood-brain barrier in vitro [64]; effect of propionate reducing anticipatory reward responses was reported in healthy adult men [65]. Our results for propionate are highly consistent with a recent study in infants showing that a higher proportion of propionate in fecal samples was associated with longer uninterrupted sleep [53].
Gut microbiota profile of preschoolers can be affect by diet [66, 67] and even perinatal factors such as gestational age and delivery mode [68]. Although our findings cannot describe the factors that contribute to development of the gut microbiota community, our study showed a clear difference in gut microbiota profile between children who had longer sleep duration and higher efficiency compared to those who shorter sleep duration and lower efficiency. Furthermore, overall dietary quality, gestational age, and delivery mode were not different between low and high sleep groups reported in this study. As such, it seems unlikely that any of these factors influence the observed association between gut microbiota and sleep, and specifically that they are not driving the gut microbiota characteristics in children who showed “high” and “low” sleep measures.
Using a combined technology of 16S rRNA amplicon sequencing and metabolomics, our study not only revealed the relationship between sleep duration and gut microbiota members but also their activities. These findings generate new insights into previously unidentified causes of sleep problems in children at preschool age and may allow the development of a more precise gut microbiota manipulation strategy to improve sleep duration and decrease night-time sleep disruption in children. Yet, this study also has several limitations that should be kept in mind. For instance, only two nights of actigraphy data were collected to assess sleep. This time window reflects a pragmatic choice geared toward encouraging our young participants to complete the study but is shorter than the recommended five or more nights, although pediatric studies with two nights have shown acceptable reliability [69]. Future studies should aim to collect sleep measures over a longer period of time to confirm our findings. Our study is also limited by its focus on night-time sleep. Because participants in the current study were asked to perform many additional data collection steps (not reported here), we decided to not obtain daytime napping diaries from parents (parents did keep a diary of night-time sleep). We were able to detect periods of quiesence consistent with napping in the actigraph record, but we did not include these potential napping periods in the total sleep time beucase we could not be sure that the children were asleep durng this time. Consistent with previously reports [28, 29], the majorty of children in our study did not have any periods of quiesence that could have been a nap (92.2%). Nevertheless, a small number of children may have continued to nap at this age and the data we collected for them reflects only the night-time portion of their sleep and not their total sleep. Additionaly, only one fecal sample was collected in our study. Feacal microbiota compostion and function may have cyclic dynamics and daily variation [70]. However, previous studies have shown that the intraindividual gut microbiome signature is largely stable over different time points [71], indicating that a single gut microbiome time point can still provide valuable insights into the associations between the gut microbiota and sleep assessed in this study. Moreover, the time of fecal sample collection was not different between sleep groups and none of the fecal metabolites associated with sleep differed as a function of stool collection time. Nevertheless, this is the first study investigating the association between sleep and gut microbiota in preschool-aged children and a significant part of the outcomes are consistent with previously reported microbiota-sleep associations in adults.
In conclusion, our results demonstrated that sleep duration, efficiency, and wake after sleep onset in young children are associated with gut microbiota and microbial metabolites such as tryptophan, tryptophan derivatives, and propionate. These microbial metabolites may be key mediators for the sleep and gut microbiota connection. Although the underlying mechanisms of the connection between sleep and gut microbiota remain unclear, our study suggests that further investigation is warranted to elucidate the connection between sleep and specific microbial members and metabolites and to determine if these factors can be harnessed to improve sleep in young children.
Supplementary Material
Acknowledgments
We would like to thank Syed Rizvi who assisted by processing the actigraphy data. Thanks to the participants in the Alberta Pregnancy Outcomes and Nutrition (APrON) study for their time and effort to make this study possible. Metabolomics data were acquired by Ryan Alexander Groves and Marija Drikic at the Calgary Metabolomics Research Facility (CMRF), which is supported by the International Microbiome Centre.
Contributor Information
Yanan Wang, Department of Pediatrics, University of Calgary, Calgary, AB, Canada.
Marcel van de Wouw, Department of Pediatrics, University of Calgary, Calgary, AB, Canada.
Lauren Drogos, Department of Psychology, University of Calgary, Calgary, AB, Canada; Libin Cardiovascular Institute, University of Calgary, Calgary, AB, Canada.
Elnaz Vaghef-Mehrabani, Department of Pediatrics, University of Calgary, Calgary, AB, Canada.
Raylene A Reimer, Alberta Children’s Hospital Research Institute (ACHRI), Calgary, AB, Canada; Faculty of Kinesiology, University of Calgary, Calgary, AB, Canada; Department of Biochemistry and Molecular Biology, Cumming School of Medicine, University of Calgary, Calgary, AB, Canada.
Lianne Tomfohr-Madsen, Department of Pediatrics, University of Calgary, Calgary, AB, Canada; Department of Psychology, University of Calgary, Calgary, AB, Canada; Department of Community Health Sciences, University of Calgary, Calgary, AB, Canada.
Gerald F Giesbrecht, Department of Pediatrics, University of Calgary, Calgary, AB, Canada; Department of Psychology, University of Calgary, Calgary, AB, Canada; Alberta Children’s Hospital Research Institute (ACHRI), Calgary, AB, Canada; Department of Community Health Sciences, University of Calgary, Calgary, AB, Canada.
Funding
This study was funded by the Canadian Institutes of Health Research.
Disclosure Statement
The authors declare no conflicts of interest.
References
- 1. Bathory E, et al. Sleep regulation, physiology and development, sleep duration and patterns, and sleep hygiene in infants, toddlers, and preschool-age children. Curr Probl Pediatr Adolesc Health Care. 2017;47(2):29–42. [DOI] [PubMed] [Google Scholar]
- 2. Byars KC, et al. Prevalence, patterns, and persistence of sleep problems in the first 3 years of life. Pediatrics. 2012;129(2):e276–e284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pollock JI. Night-waking at five years of age: predictors and prognosis. J Child Psychol Psychiatry Allied Discip. 1994;35(4):699–708. [DOI] [PubMed] [Google Scholar]
- 4. Lozoff B, et al. Sleep problems seen in pediatric practice. Pediatrics 1985;75(3):477–483. [PubMed] [Google Scholar]
- 5. Field T. Infant sleep problems and interventions: a review. Infant Behavior & Development 2017;47:40–53. [DOI] [PubMed] [Google Scholar]
- 6. Wu Y, et al. Short sleep duration and obesity among children: A systematic review and meta-analysis of prospective studies. Obesity Res Clinl Pract. 2017;11(2):140–150. [DOI] [PubMed] [Google Scholar]
- 7. Chaput J-P, et al. Systematic review of the relationships between sleep duration and health indicators in school-aged children and youth. Appl Physiol Nutr Metab Physiol Appl Nutr Metabol. 2016;41(6 Suppl 3) : S266–S282. [DOI] [PubMed] [Google Scholar]
- 8. Matricciani L, et al. Children’s sleep and health: a meta-review. Sleep Med Rev. 2019;46:136–150. [DOI] [PubMed] [Google Scholar]
- 9. Touchette E, et al. Associations between sleep duration patterns and behavioral/cognitive functioning at school entry. Sleep 2007;30(9):1213–1219. doi: 10.1093/sleep/30.9.1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krueger JM, et al. Sleep and Microbes. Int Rev Neurobiol. 2016;131:207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farré N, et al. Sleep and circadian alterations and the gut microbiome: associations or causality? Curr Sleep Med Rep. 2018;4(1):50–57. [Google Scholar]
- 12. Fan Y, et al. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. [DOI] [PubMed] [Google Scholar]
- 13. Belkaid Y, et al. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marchesi JR, et al. The gut microbiota and host health: a new clinical frontier. Gut 2016;65(2):330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin AM, et al. The influence of the gut microbiome on host metabolism through the regulation of gut hormone release. Front Physiol. 2019;10:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morais LH, et al. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol. 2021;19(4):241–255. [DOI] [PubMed] [Google Scholar]
- 17. Carabotti M, et al. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol 2015;28(2):203–209. [PMC free article] [PubMed] [Google Scholar]
- 18. Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693:128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ursin R. Serotonin and sleep. Sleep Med Rev. 2002;6(1):55–69. [DOI] [PubMed] [Google Scholar]
- 20. Siegel JM. The neurotransmitters of sleep. J Clin Psychiatry. 2004;65(Suppl 16):4–7. [PMC free article] [PubMed] [Google Scholar]
- 21. Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015;161(2):264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ogawa Y, et al. Gut microbiota depletion by chronic antibiotic treatment alters the sleep/wake architecture and sleep EEG power spectra in mice. Sci Rep. 2020;10(1):19554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Smith RP, et al. Gut microbiome diversity is associated with sleep physiology in humans. PLoS One. 2019;14(10):e0222394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benedict C, et al. Gut microbiota and glucometabolic alterations in response to recurrent partial sleep deprivation in normal-weight young individuals. Molecular Metabolism 2016;5(12):1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El Aidy S, et al. A brief period of sleep deprivation leads to subtle changes in mouse gut microbiota. J Sleep Res. 2020;29(6):e12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thiedke CC. Sleep disorders and sleep problems in childhood. Am Fam Physician. 2001;63(2):277–284. [PubMed] [Google Scholar]
- 27. Derrien M, et al. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019;27(12):997–1010. [DOI] [PubMed] [Google Scholar]
- 28. Jiang F. Sleep and Early Brain Development. Ann Nutr Metab. 2019;75(Suppl 1):44–54. [DOI] [PubMed] [Google Scholar]
- 29. El Shakankiry HM. Sleep physiology and sleep disorders in childhood. Nat Sci Sleep 2011;3:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ringel-Kulka T, et al. Intestinal microbiota in healthy U.S. young children and adults--a high throughput microarray analysis. PloS One. 2013;8(5):e64315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng J, et al. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 2016;10(4):1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaplan BJ, et al. The Alberta Pregnancy Outcomes and Nutrition (APrON) cohort study: rationale and methods. Mat Child Nutr. 2014;10(1):44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leung BM, et al. Comparison of sample characteristics in two pregnancy cohorts: community-based versus population-based recruitment methods. BMC Med Res Methodol. 2013;13:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cole RJ, et al. Automatic sleep/wake identification from wrist activity. Sleep 1992;15(5):461–469. doi: 10.1093/sleep/15.5.461 [DOI] [PubMed] [Google Scholar]
- 35. Meltzer LJ, et al. Use of actigraphy for assessment in pediatric sleep research. Sleep Med Rev. 2012;16(5):463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jarman M, et al. Development of a diet quality index to assess adherence to Canadian dietary recommendations in 3-year-old children. Public Health Nutr. 2020;23(3):385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Katoh K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Price MN, et al. FastTree 2 – Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bokulich NA, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018;6(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van de Wouw M, et al. Associations between the gut microbiota and internalizing behaviors in preschool children. Psychosom Med. 2021. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 44. Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deng Y, et al. Involvement of the microbiota-gut-brain axis in chronic restraint stress: disturbances of the kynurenine metabolic pathway in both the gut and brain. Gut Microbes 2021;13(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fung TC, et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. 2019;4(12):2064–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marotta A, et al. Effects of probiotics on cognitive reactivity, mood, and sleep quality. Front Psychiatry. 2019;10:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pinto-Sanchez MI, et al. Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: a pilot study in patients with irritable bowel syndrome. Gastroenterology 2017;153(2):448–459.e8. [DOI] [PubMed] [Google Scholar]
- 49. Moloney GM, et al. Improvements in sleep indices during exam stress due to consumption of a Bifidobacterium longum. Brain Behav Immun Health 2021;10:100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Besedovsky L, et al. The sleep-immune crosstalk in health and disease. Physiol Rev. 2019;99(3):1325–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng D, et al. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Louis P, et al. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29–41. [DOI] [PubMed] [Google Scholar]
- 53. Heath A-LM, et al. Association between the fecal short-chain fatty acid propionate and infant sleep. Eur J Clin Nutr. 2020;74(9):1362–1365. [DOI] [PubMed] [Google Scholar]
- 54. Vacca M, et al. The controversial role of human gut Lachnospiraceae. Microorganisms 2020;8(4):573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chávez-Carbajal A, et al. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int J Mol Sci . 2019;20(2):438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kostic Aleksandar D, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015;17(2):260–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Agus A, et al. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host & Microbe 2018;23(6):716–724. [DOI] [PubMed] [Google Scholar]
- 58. Kawai N, et al. The sleep-promoting and hypothermic effects of glycine are mediated by NMDA receptors in the suprachiasmatic nucleus. Neuropsychopharmacology. 2015;40(6):1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miyake M, et al. Randomised controlled trial of the effects of L-ornithine on stress markers and sleep quality in healthy workers. Nutr J. 2014;13:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Silva YP, et al. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol. 2020;11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Magzal F, et al. Associations between fecal short-chain fatty acids and sleep continuity in older adults with insomnia symptoms. Sci Rep. 2021;11(1):4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Szentirmai E, et al. Butyrate, a metabolite of intestinal bacteria, enhances sleep. Sci Rep. 2019;9(1):7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. De Vadder F, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014;156(1):84–96. [DOI] [PubMed] [Google Scholar]
- 64. Hoyles L, et al. Microbiome–host systems interactions: protective effects of propionate upon the blood–brain barrier. Microbiome 2018;6(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Byrne CS, et al. Increased colonic propionate reduces anticipatory reward responses in the human striatum to high-energy foods. Am J Clin Nutr. 2016;104(1):5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zmora N, et al. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(1):35–56. [DOI] [PubMed] [Google Scholar]
- 67. Herman DR, et al. Dietary habits of 2- to 9-year-old American children are associated with gut microbiome composition. J Acad Nutr Diet.2020;120(4):517–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fouhy F, et al. Perinatal factors affect the gut microbiota up to four years after birth. Nat Commun. 2019;10(1):1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sadeh A. III. Sleep assessment methods. Monogr Soc Res Child Dev. 2015;80(1):33–48. [DOI] [PubMed] [Google Scholar]
- 70. Thaiss CA, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014;159(3):514–529. [DOI] [PubMed] [Google Scholar]
- 71. Jones RB, et al. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep. 2018;8(1):4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.