Abstract
Repositioning or repurposing drugs account for a substantial part of entering approval pipeline drugs, which indicates that drug repositioning has huge market potential and value. Computational technologies such as machine learning methods have accelerated the process of drug repositioning in the last few decades years. The repositioning potential of type 2 diabetes mellitus (T2DM) drugs for various diseases such as cancer, neurodegenerative diseases, and cardiovascular diseases have been widely studied. Hence, the related summary about repurposing antidiabetic drugs is of great significance. In this review, we focus on the machine learning methods for the development of new T2DM drugs and give an overview of the repurposing potential of the existing antidiabetic agents.
Abbreviations: T2DM, type 2 diabetes mellitus; DM, diabetes mellitus; ASCVD, atherosclerotic cardiovascular disease; GLP-1, glucagon-like peptide 1; TZD, thiazolidinedione; DPP-4, dipeptidyl peptidase 4; SGLT-2, sodium-glucose cotransporter 2; DTI, drug-target interaction; DDI, drug-drug interaction; DDA, drug-disease association; DNNs, deep neural networks; DBNs, deep brief networks; RNNs, recurrent neural networks; AEs, autoencoders; CNNs, convolutional neural networks; GNNs, graph neural networks; AD, Alzheimer’s Disease; PD, Parkinson’s Disease; PI3K/AKT, phosphatidylinositol 3-kinase/AKT; cAMP/PKA, cyclic adenosine monophosphate/protein kinase A; CVD, cardiovascular diseases; CV, cardiovascular; ML, machine learning; DL, deep learning
Keywords: Drug repositioning, Drug repurposing, T2DM, Machine learning, Deep learning, Antidiabetic drugs
1. Introduction
The concept of drug repositioning (also known as drug repurposing) was first proposed in 2004 [1]. The related terminology usage of drug repositioning is vague and unclear due to its imprecise definition, which has been improved in light of drug repositioning development over the years [2], [3]. Drug repositioning, drug repurposing, drug redirecting, or drug reprofiling is the process of finding new uses outside the scope of original drug indications. The active substances used in drug repositioning include marketed drugs with expired patents, withdrawn drugs with safety concerns, and compounds that have failed to achieve the expected effectiveness and safety in clinical trials. Nevertheless, the structural modification of active compounds by de novo drug design methods does not belong to the field of drug repositioning [3]. Although lack of precise definition, it does not deter the pharmaceutical industries and academic research communities from investigating drug repositioning [4]. Compared with conventional drug discovery and development, drug repositioning can save development time and money by skipping several time-consuming and costly stages, thus welcomed by the pharmaceutical industry [1].
Due to the explosive growth of drug research costs, drug repurposing becomes a promising drug discovery way and is commonly used to find treatments for cancer [5], cardiac disease [6], neuropathy [7], infectious diseases [8], psychosis [9], rare disease [10], especially for type 2 diabetes mellitus (T2DM) [11]. T2DM is a non-communicable epidemic disease but rising as a major health issue across the globe. The major classes of antidiabetic medications for T2DM are difficult to reach a reasonable balance in the role of efficacy, side effects, and patient tolerance [12]. Hence, drug repositioning provides a cost-effective and promising approach to finding new effective drugs for T2DM [11]. Besides, metabolic disorders [13] associated with T2DM such as hyperinsulinemia, oxidative stress, inflammation, and excessive glycation can lead to the development of cancer [14], Parkinson's disease [15], Alzheimer's disease [16], and cardiovascular disease [17]. Antidiabetic drugs are proven to have the repositioning potential to treat these diseases [14], [15], [16], [18]. Traditionally, drug candidates which are discovered for new indications by random experiments may cost much research capital. The computational approaches such as machine learning and classical algorithms supply directed and cost-saving approaches for drug repositioning. In this review, we focus on the machine learning approaches used in finding new drugs for T2DM and new indications for antidiabetic medicines.
2. The latest statistics and management of T2DM
Diabetes mellitus (DM) is a non-communicable disease with a rapidly increasing number of patients worldwide. According to the data by the International Diabetes Federation (IDF), about 537 million adults (20–79 years old) are living with diabetes in 2021, and the total number is projected to rise to 643 million by 2030 and 783 million by 2045 [19]. The increasing potential high-risk groups of T2DM (541 million adults) and high mortality (6.7 million deaths in 2021, corresponds to 12.2% of global deaths from all causes in adults), coupled with the excessive global health expenditure (USD 966 billion in 2021 for adults), make diabetes a significant global challenge to the health and well-being of individuals, families, and societies. Especially, T2DM accounts for approximately 90% of all cases of diabetes worldwide [20], [21].
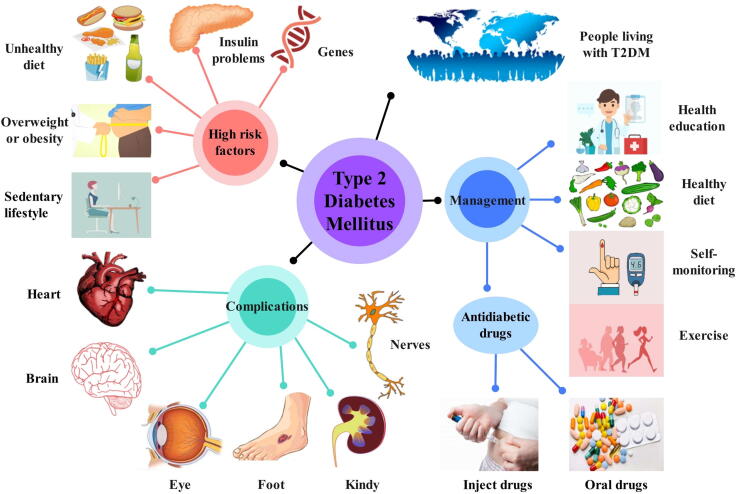
T2DM is a chronic disease caused by the inability of faulty pancreatic β cells to secret a normal amount of insulin to maintain normal body consumption, or peripheral tissue has a decreased susceptibility to insulin. As represented in Fig. 1, T2DM is mostly determined by multiple genes [22] and environmental factors. Too many calories intake and living a sedentary lifestyle may result in overweight and obesity, which are believed to be closely associated with T2DM [23]. T2DM complications [24], [25] are most commonly involved in hypertension, atherosclerotic cardiovascular disease (ASCVD), nephropathy, retinopathy, diabetic neuropathy, and diabetic foot. Diabetes management includes lifestyle intervention, pharmacological therapy, and routine blood glucose monitoring [12]. Antidiabetic medications commonly used in pharmacological therapy contain biguanides, sulfonylureas, glucagon-like peptide 1 (GLP-1) receptor agonists, thiazolidinedione (TZD), dipeptidyl peptidase 4 (DPP-4) inhibitors, sodium-glucose cotransporter 2 (SGLT-2) inhibitors, and α-glucosidase inhibitors. Polypharmacy or monotherapy of antidiabetic drugs is based on the patient’s age, course of the disease, patient tolerance, physical condition, side effects, and drug efficacy. Therefore, the development of new T2DM drugs with better efficacy and fewer side effects is a problem that researchers have been actively addressing. Computational approaches provide a new insight from the perspective of bioinformatics for drug repositioning to find new correspondence identification based on a large number of diseases, genes, targets, drugs, and prior knowledge [26].
Fig. 1.
Schematic diagram of T2DM. It shows the high-risk factors, complications, and management of T2DM.
3. Resources and methods for drug repositioning
3.1. Predicting new interactions for drug repositioning
Generally, there are three steps in drug repositioning strategy before bringing a candidate drug into the market process: discovery of novel drug-target (or drug-drug, drug-disease) pairs, mechanism evaluation of drug action in preclinical animal models, and evaluation of curative effect in clinical trials [26]. Among all these steps, finding a highly credible new relationship between drug molecules, therapeutic targets and diseases is crucial to the subsequent process of drug repositioning. Drugs often respond to disease by acting on key targets that play a specific role in the disease. Drug-target interactions can be measured by some binding or competitive cellular assays, and then the efficacy of the drug on the target can be evaluated from the dose-effect curve and fitted EC50 or IC50 values [27]. However, the cellular assays cannot simulate the complex physiological environment of the body, which may result in the failure of new drug development at later stages [28]. Phenotypic screening in cellular or animal models can identify drugs that alter phenotypes in complex biological systems by measuring model responses, and then determining disease states and mechanisms of action [29]. But phenotypic screening is often limited to one experiment at a time for a specific disease and requires a great deal of effort and time [30]. Machine learning methods are able to extract new potential, possibly unexpected drug-disease (or drug-drug, drug-target) relationships from large amounts of chemical and biomedical complex data [31] such as drugs, protein targets, gene expression data, signaling pathways, electronic health records, clinical medical records, and adverse drug reaction data.
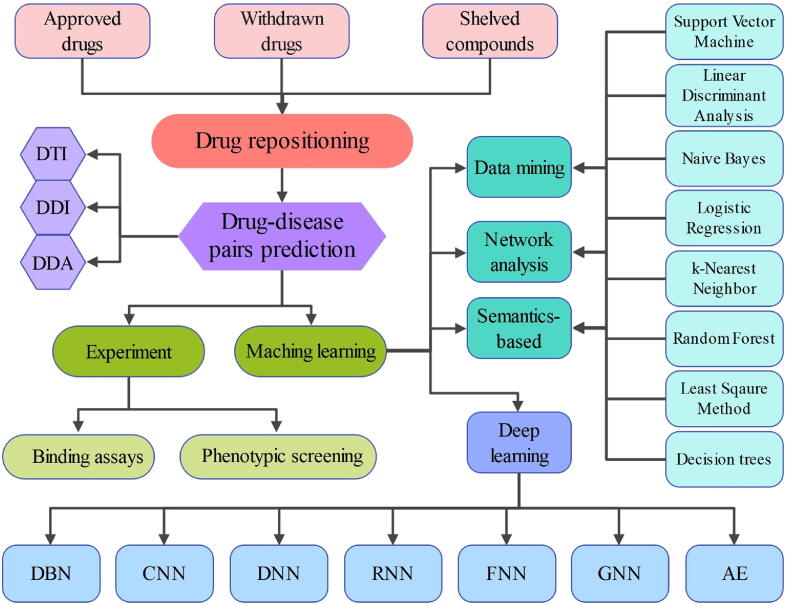
Machine learning (ML) approaches such as support vector machine (SVM), Naive Bayes and random forest (RF) (see Fig. 2) with high prediction accuracy and technological breakthroughs are playing an increasingly important role in drug discovery [32] and repositioning [33], [34]. Data mining, network analysis, and semantic analysis are common methods for ML to predict drug-target interactions (DTI), drug-drug interactions (DDI), and drug-disease associations (DDA). Deep learning (DL), a subfield of machine learning, has gained more attention in drug repositioning in recent years [35], [36]. DL simulates artificial neural networks and establishes high-level learning architectures to learn more useful features and ultimately improve the accuracy of classification or prediction. As compared to traditional machine learning methods, deep learning methods such as convolutional neural networks (CNNs), graph neural networks (GNNs), and autoencoders (AEs) have a powerful ability to automatically extract features as well as an effective feature representation to obtain different levels of information [37], [38], [39].
Fig. 2.
The machine learning methods for drug-disease pairs prediction in drug repositioning. DTI: drug-target interaction, DDI: drug-drug interaction, DDA: drug-disease association, DBN: deep belief network, CNN: convolutional neural network, DNN: deep neural network, RNN: recurrent neural network, FNN: feedforward neural network, GNN: graph neural network, AE: autoencoder.
3.2. Resources for drug repositioning
Data sources for drug repositioning rely on various bioinformatics and pharmacogenomics databases, several excellent reviews have described in detail the data resources, algorithmic resources, and research methods, which are also applicable to T2DM drug repositioning. Tanoli et al. [40] presented an overview of 102 database resources available for drug repositioning in four categories, namely drug-target interaction, disease, chemical, and biomolecular databases, and showed their applications for drug repositioning. Zhang et al. [41] provided a comprehensive collection of databases for drug repositioning and described the types of data available in each database, enumerated data representation methods and software or servers for data transformation; they also concluded with a list of common machine learning methods for drug repositioning. Hodos et al. [30] provided a detailed description of the data types available for drug repositioning, and the representation of different types of data for machine learning input, and also listed three superior algorithmic frameworks for drug repositioning: similarity-based methods, network methods, and matrix factorization. Pan et al. [42] summarized commonly used online open databases and sequence-based and graph-based representation learning methods, as well as a summary of deep learning models for drug repositioning in recent years. Yu et al. [43] summarized in detail the six most commonly used deep learning frameworks: deep brief networks (DBNs), deep neural networks (DNNs), AEs, CNNs, recurrent neural networks (RNNs), and GNNs, in addition, the authors discussed several representative deep learning models based on input data of varying complexity. Several other reviews focused on machine learning methods for processing different types of data for drug repositioning [44], [45], [46].
3.3. Machine learning methodologies in DTI, DDI, DDA prediction
There are three steps involved in predicting a new DTI, DDI or DDA using machine learning methods. First, the input data such as drug side effects, drug chemical structures, and disease genes are pre-processed to obtain training data through feature extraction and feature selection; second, relevant machine learning algorithms are deployed for training; third, predictive models are utilized to obtain drug repositioning results on a test dataset. Data are transformed into a uniform normalized format, such as a computer-readable steering vector or matrix, before being fed into a machine learning model for representation learning. Representation learning [47] (or feature learning) is a collection of techniques for transforming raw data into something that can be effectively exploited by machine learning. The main sequence-based [48] input representations are international chemical identifier (InChI) and Simplified Molecular Input Line Entry System (SMILES), which use specialized syntax to encode three-dimensional chemical structures as strings of text [49], [50]. Graph-based representations [51], [52] can represent the spatial and structural relationships of proteins and biomolecules as well as the interconnected nature between different biological data [53]. Representation learning is mainly divided into supervised and unsupervised learning approaches to extract features of the input data for downstream training tasks.
4. Machine learning models applied in drug repositioning
4.1. Drug-target interactions (DTIs) prediction
New targets may imply new indications. Previous review papers provide various perspectives on machine learning methods for DTIs prediction. Chen et al. [54] summarized machine learning methods into supervised and semi-supervised categories and gave a detailed description of their subclasses. Lavecchia [55] described the theoretical foundations and applications of machine learning algorithms for predicting DTIs in the context of ligand-based virtual screening. Serçinoğlu et al. [56] provided a comprehensive overview of database resources and bioinformatics tools of computational approaches for drug repositioning. Bagherian et al. [57] focused on machine learning methods for DTIs prediction and gave an elaboration on four aspects of similarity/distance-based methods, deep learning methods, matrix factorization methods, and feature-based methods. In addition, some new DTI prediction models with better performance have been published recently, Zeng et al. [58] developed a network-based deep learning model named deepDTnet to predict drug-target interactions (DTI), which used a network embedding method to put vectors representations to low-dimensional vector space and Positive-Unlabeled (PU)-matrix completion algorithm to low-rank matrix completion. And their model showed that topotecan had a good therapeutic effect in a mouse model of multiple sclerosis. Wen et al. [59] applied the deep belief network (DBN) to the construction of the DeepDTIs model, it extracted drugs and targets features from the compound substructure and protein sequences. The DeepDTIs model allows researchers to search for all possible drug-target interactions in the network space, without the limitations of previous target classification. Maryam et al. [60] proposed a model named the Coupled Tensor-Matrix Completion (CTMC), which combined matrix factorization-based method with drug-drug and target-target tensors to repurpose drug molecules, and showed a better output in comparison with other matrix-factorization-based methods in performance evaluations. Bai et al. [61], [62] developed a software named MolAICal that could study the interaction between targets and ligands by deep learning models and classical algorithms [63]. Ceddia et al. [64] proposed a shortest-path enhanced Non-negative Matrix Tri-Factorization method, which used protein-protein interaction networks for shortest-path evaluation of drug-protein pairs, with improved number and accuracy of predictions, also indicated that integrating heterogeneous network data had significant advantages for DTIs prediction.
4.2. Drug-drug interactions (DDIs) prediction
Drugs which have similar characteristics in some respects are likely to share targets and physiological effects to treat the same diseases. The use of two or more drugs in a patient may have a rising risk of adverse patient outcomes due to the DDIs [65]. The discovery of DDIs not only helps to address the drug administration and patient safety caused by adverse drug reactions (ADRs) [66], but also can explore pharmacological functions of drugs and new drug-indication relationships for drug repositioning. In the recently published review [44], the researchers made a comprehensive summary of computational methods for DDIs detection, which included literature-based, machine learning-based, and pharmacovigilance-based data mining methods, as well as a description of data sources for DDIs prediction. Zhang et al. [67] mainly focused on the deep learning-based methods for extracting DDIs and described them in four categories that comprised supervised methods, semi-supervised methods, unsupervised methods, and distant supervision. In general, machine learning is most widely used in DDI prediction. Wang et al. [68] proposed a graph convolutional network with multi-kernel (GCNMK) that applied two known DDIs graph kernels to the graph convolutional layers and used fully connected layers to predict potential DDIs. Zhou et al. [69] utilized the Markov Clustering Algorithm to cluster a set of drugs into several groups according to their drug interaction profiles, new drug-target relationships could be deduced from known targets of a drug with its associated drugs. Mei et al. [70] used drug target genes and signaling pathways without data integration to train a simplified DDI predictive model in a biological background. Yan et al. [71] developed a similarity-based model named DDIGIP which used drug Gaussian interaction profile (GIP) kernel similarity and regularized least squares (RLS) classifier to predict DDIs for both old and novel drugs. Moreover, deep learning methods that combine multiple characteristic information networks of drugs seem to have more accurate prediction behavior for DDIs. Yan et al. [72] proposed a method named NMDADNN that predicted DDIs well in performance testing based on a DNN by integrating serval heterogeneous information sources and extracting the unified drug mapping features.
4.3. Drug-disease associations (DDAs) prediction
The accumulation of vast amounts of biomedicine data and advances in computation approaches have made it easier for researchers to extract unknown DDAs from the complex networks, which can provide new ideas for drug repositioning. Hu et al. [73] used low‑rank tensors and matrix complement technology to construct a prediction model LMFDA which could fuse multiple similar networks and predict potential DDAs. Yang et al. [74] proposed a multi-similarities bilinear matrix factorization (MSBMF) model that combined matrix factorization with the alternating direction method of multipliers algorithm to effectively measured the relationship between drugs and diseases. Wu et al. [75] proposed a model SSGC based on a semi-supervised graph cut algorithm to integrate multiple sources of data thus allowing the comprehensive similarities measurement of drugs and diseases from multi-view and multiple layers.
5. Repositioning potential of antidiabetic drugs
Because T2DM shares the same pathophysiological mechanisms as many other diseases, antidiabetic drug repositioning has been extensively studied for new indications. Moreover, due to the high number of patients with T2DM and the long history of medication use, antidiabetic drugs have relatively complete clinical safety data, making them the drugs of choice for repositioning. The repositioning potential of antidiabetic drugs is mainly involved in cancer, neurodegenerative diseases, and cardiovascular diseases.
5.1. Repositioning antidiabetic drugs for cancer
T2DM and cancer have common potential risk factors [76] including age, sex, obesity, physical activity, diet, alcohol, smoking, etc. In addition, pathological surroundings such as hyperglycemia, hyperinsulinemia [77], inflammation [78], and oxidative stress [79], jointly contribute to the association of T2DM with the development of various cancers [80]. A great deal of evidence proves that antidiabetic drugs are capable of reducing the overall cancer incidence and mortality even though most of the clinical trials are conducted in cancer patients with diabetes. Table 1 summarized the studies on antidiabetic drugs for the treatment of cancer. Metformin is the most widely studied antidiabetic drug repurposed for cancer. Previous and most recent evidence-based meta-analyses of case-control and cohort studies showed that metformin was associated with a decreased risk of overall cancer incidence and mortality [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], the same effect was also produced in site-specific cancer such as breast cancer [92], colorectal cancer [93], liver cancer [94], [95], pancreatic cancer [96], endometrial cancer [97] and lung cancer [98], [99]. Evidence from retrospective analyses has yielded contradictory or insignificant results regarding the association of sulfonylurea drugs with the risk and mortality of cancer [100], [101], [102], [103]. Glyburide is an anti-diabetic sulfonylurea drug extensively used for the treatment of T2DM. The glyburide may be beneficial to the anti-cancer effect by regulating ATP-binding cassette protein super-family and ATP-sensitive potassium channels [104]. A recent non-diabetes mice model study suggested that glyburide might inhibit NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome to attenuate inflammation-related lung tumorigenesis [105]. Another case study showed that gliclazide tended to have a decreased risk of liver cancer [101].
Table 1.
Repositioning studies of antidiabetic drugs in the treatment of cancer.
| Antidiabetic drug classes | Drugs | Cancer | Research methods |
|---|---|---|---|
| Biguanides | Liraglutide | Breast cancer [92] | Meta-analysis |
| Colorectal cancer [93] | |||
| Liver cancer [94], [95] | |||
| Pancreatic cancer [96] | |||
| Endometrial cancer [97] | |||
| Lung cancer [98], [99] | |||
| Sulfonylurea | Glyburide | Lung cancer [105] | Animal study |
| Gliclazide | Liver cancer [101] | Case study | |
| GLP-I receptor agonists | liraglutide | Breast cancer [113] | Meta-analysis |
| Prostate cancer [107] | Cell study | ||
| Endometrial cancer [111] | Cell study | ||
| Exendin-4 | Colorectal cancer [108], [109] | Cell/Animal study | |
| Ovarian cancer [110] | Cell study | ||
| Polycystic ovary syndrome [114] | Meta-analysis | ||
| SGLT-2 inhibitors | Canagliflozin | Breast cancer [116] | Cell study |
| Lung cancer [117] | Cell study | ||
| Dapagliflozin | Breast cancer [116] | Cell study | |
| Sitagliptin | Prostate cancer [118] | Retrospective cohort study | |
| DPP4 inhibitors | Linagliptin | Colorectal cancer [122], [124], [125] | Computational/Meta-analysis/Animal study |
| Gastric cancer [123] | Cell study | ||
| Vildagliptin | Colorectal lung metastases [126] | Animal study |
Various experimental studies on animals, cancer cell lines, and clinical data have shown that GLP-1 receptor agonists of antidiabetic drugs have potential clinical benefits for breast cancer [106], prostate cancer [107], colorectal cancer [108], [109], ovarian cancer [110], endometrial cancer [111], pancreatic cancer [112], [113], and polycystic ovary syndrome [114]. SGLT-2 is one of the transporters for active glucose uptake in cancer cells. SGLT-2 inhibitors have an anti-cancer effect by inhibiting glucose uptake in tumors expressing SGLT-2. An animal study shows canagliflozin inhibits the growth of SGLT-2-expressing liver cancers by reducing intracellular glucose uptake [115]. A cell study in breast cancer proved that SGLT-2 inhibitors could reduce the viability of breast cancer cells by inducing G1 phase arrest and apoptosis based on the adenosine-monophosphate activated protein kinase/mammalian target of rapamycin (AMPK/mTOR) signal pathway [116]. Other evidence indicated that SGLT-2 could also inhibit the proliferation of cancer cells in lung cancer [117] and pancreatic cancer [118].
DPP-4 inhibitors can enhance and prolong the activity of endogenous incretin hormones GLP-1 and glucose-dependent insulinotropic polypeptide (GIP) by inhibiting the activity of DPP-4 enzyme, thereby improving glycaemic control. DPP-4 inhibitors have been shown to be associated with a reduced risk of prostate cancer [119], oral cancer [120], and breast cancer [121]. A repositioning study showed that linagliptin had a therapeutic effect on colorectal cancer based on molecular docking and gene expression profiling methods, and this result was verified by in vivo experiments [122]. Another cell study showed that sitagliptin had a potential inhibitory effect on proliferation and clonality of gastric cancer cells by activating AMPK and inhibiting Yes‐associated protein (YAP) and Melanoma‐associated antigen‐A3 (MAGE‐A3) [123]. Besides, sitagliptin was also associated with a reduced risk of colorectal cancer [124], [125]. Animal studies showed that vildagliptin could suppress the development and growth of colorectal lung metastases [126].
To sum up, although there is no final conclusion that has yet been reached on whether antidiabetic drugs can reduce the cancer risk, the anti-cancer activities have been studied extensively. If the strategy of repositioning antidiabetic drugs for cancer treatment is feasible, it will be cost-effective for both patients and pharmaceutical companies.
5.2. Repositioning antidiabetic drugs for neurodegenerative diseases
In T2DM patients, insulin resistance leads to the problems of impaired insulin metabolism, impaired glucose metabolism, neuroinflammation, and oxidative stress. The pathological manifestations of neurodegenerative diseases are mainly involved in the deposition of neuritic plaques, formation of intracellular neurofibrillary tangles (NFTs), vasculopathy and inflammation-related damage, and mitochondrial structural and functional abnormalities. Alzheimer’s Disease (AD) [127] and Parkinson’s Disease (PD) [128] are the two major diseases of neurodegenerative disorders. Many studies demonstrate that the repositioning of antidiabetic drugs is feasible for neurodegenerative disorders [129], [130]. Recent researches provide a detailed review of the epidemiological evidence in regard to the associations between T2DM and neurodegenerative diseases [127], [128], [131], as well as the potential mechanism of antidiabetic drugs for the treatment of neurodegenerative disorders [131], [132].
GLP-1 receptor agonists have a significant neuroprotective and neurotrophic role by intervening in signaling pathways of phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) and cyclic adenosine monophosphate/protein kinase A (cAMP/PKA), as well as regulating intracellular calcium homeostasis and inflammatory processes [133]. A meta-analysis [134] and several clinical trials [135], [136] have demonstrated the therapeutic potential of exenatide in improving motor symptoms in PD patients. A larger phase 3, placebo-controlled and 96-week clinical trial is ongoing to investigate the time-accumulating effect of exenatide on reduced motor deterioration in patients with PD [137]. NCT03456687 (https://clinicaltrials.gov) is a phase I clinical trial designed to study the effects of exenatide on motor function and brain in PD patients. NCT04269642 is a placebo-controlled, parallel comparison, phase IIa clinical study to evaluate the efficacy and safety of sustained-release (SR) exenatide (PT320) in the treatment of patients with early PD. NCT04305002 is a randomized, parallel controlled, phase 2 clinical trial designed to reveal the effect of exenatide on disease progression in early PD patients with T2DM. An animal study shows liraglutide can suppress neuroinflammation by increasing p-AMPK expression and reducing NF-κB protein levels in the PD mice model [138]. A randomized and placebo-controlled phase II clinical trial (NCT02953665) indicated that liraglutide had significant improvement in motor and cognitive symptoms of PD patients.
In summary, PD and AD as new fields of drug repositioning of antidiabetic drugs have gained extensive research that indicates the close links between the pathological mechanisms of diabetes and neurodegenerative diseases. GLP-1 receptor agonists such as exenatide and liraglutide have entered late-stage clinical trials. The development of innovative treatment modalities to treat neurodegenerative diseases with substantial unmet medical needs is of great interest. But the longer follow-up duration, better-designed, and much larger enrolled population clinical trials are further needed for the safety and efficacy verification of the repositioned drugs in order that those drugs can be effectively used to treat new indications.
5.3. Repositioning antidiabetic drugs for cardiovascular diseases
People with diabetes are more likely to have cardiovascular disease (CVD) than people without diabetes (https://www.idf.org/our-activities/care-prevention/cardiovascular-disease.html). As a major feature of T2DM, insulin resistance has been considered an independent risk factor for cardiovascular events [139]. In addition, lipid metabolism obstruction associated with T2DM provides a pro-inflammatory and hyperlipidemia environment that is another important factor in cardiovascular disease [140]. Moreover, chronic and persistent hyperglycemia caused by T2DM has a link to the accumulation of advanced glycation end-products (AGEs) that further damage macrovascular and microvascular [141], thus leading to CVD-like atherosclerosis. Based on the common molecular mechanism between T2DM and CVD, studies have gained popularity on the relationship between the use of antidiabetic drugs and the risk of CVD [142].
SGLT-2 inhibitors can decrease the rates of cardiovascular mortality and major adverse cardiovascular events (MACE) [18], [143]. A meta-analysis showed that both SGLT-2 inhibitors and GLP-1 receptor agonists reduced the risk of myocardial infarction and cardiovascular death in T2DM patients with known atherosclerotic cardiovascular disease (ASCVD). Meanwhile, SGLT-2 inhibitors have a prominent effect on preventing hospitalization for heart failure and progression of kidney disease whereas GLP-1 receptor agonists reduce the risk of stroke [144]. Other lines of evidence also revealed that SGLT-2 inhibitors and GLP-1 receptor agonists reduced cardiovascular events and had a beneficial effect on kidney outcomes [145], [146]. DPP-4 inhibitors showed insignificant results about MACE in T2DM patients in clinical trials which were designed for testing CV safety/efficacy and cardiovascular benefits [147]. But another clinical trial had different results that patients with T2DM treatment with DPP-4 inhibitors were associated with an increased risk of hospitalization for heart failure [148].
The international guidelines recommend that SGLT-2 inhibitors or GLP-1 receptor agonists as part of glycemic management in T2DM patients who have been diagnosed with ASCVD. SGLT-2 inhibitors are particularly recommended for T2DM patients with chronic kidney disease (CKD) and ASCVD patients with heart failure concerns [149]. Antidiabetic agents that have beneficial effects on the cardiovascular system can reduce the rate of cardiovascular-related disability and premature mortality in T2DM patients, so the studies of antidiabetic drug repositioning have positive significance to improve the quality of life and management of T2DM patients.
6. Conclusion
In this article, we described the current situation of T2DM and some machine learning approaches in the candidate drug discovery stage of drug repositioning. In addition, we summarized some reliable evidence of antidiabetic drugs in repositioning treatment for cancer, neurodegenerative disorders, and cardiovascular diseases. Drug repositioning can find new uses for old drugs that failed to show the efficacy of predetermined indications. Generic drugs with good performance can be repurposed for new indications and ultimately maximize the therapeutic and economic benefits. Repositioning antidiabetic drugs with good efficacy and safety in the treatment of cancer, neurodegenerative diseases, and cardiovascular diseases is a promising way for drug discovery. Machine learning, especially deep learning approaches with powerful data processing and intelligent prediction ability, can efficiently and accurately find meaningful drug-disease relationships in drug repositioning, which is the primary computational technology of future drug discovery and research.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to “Tencent AI Lab Rhino-Bird Focused Research Program (Tencent AI Lab RBFR2022006)” who provides the grant for this manuscript. We also thank the Supercomputing Center of Lanzhou University and Gansu Computation Center for supporting this paper.
References
- 1.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 2.Langedijk J., et al. Drug repositioning and repurposing: terminology and definitions in literature. Drug Discov Today. 2015;20(8):1027–1034. doi: 10.1016/j.drudis.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Jourdan J.P., et al. Drug repositioning: a brief overview. J Pharm Pharmacol. 2020;72(9):1145–1151. doi: 10.1111/jphp.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oprea T.I., et al. Drug Repurposing from an Academic Perspective. Drug Discov Today Ther Strateg. 2011;8(3–4):61–69. doi: 10.1016/j.ddstr.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolini F., Sukhatme V.P., Bouche G. Drug repurposing in oncology–patient and health systems opportunities. Nat Rev Clin Oncol. 2015;12(12):732–742. doi: 10.1038/nrclinonc.2015.169. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari R., Lüscher T.F. Reincarnated medicines: using out-dated drugs for novel indications. Eur Heart J. 2016;37(33):2571–2576. doi: 10.1093/eurheartj/ehw051. [DOI] [PubMed] [Google Scholar]
- 7.Fava M. The promise and challenges of drug repurposing in psychiatry. World Psychiatry. 2018;17(1):28–29. doi: 10.1002/wps.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farha M.A., Brown E.D. Drug repurposing for antimicrobial discovery. Nat Microbiol. 2019;4(4):565–577. doi: 10.1038/s41564-019-0357-1. [DOI] [PubMed] [Google Scholar]
- 9.Parsons C.G. CNS repurposing – Potential new uses for old drugs: Examples of screens for Alzheimer's disease. Parkinson's disease and spasticity Neuropharmacology. 2019;147:4–10. doi: 10.1016/j.neuropharm.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 10.van den Berg S., et al. Drug Repurposing for Rare Diseases: A Role for Academia. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.746987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turner N., et al. Repurposing Drugs to Target the Diabetes Epidemic. Trends Pharmacol Sci. 2016;37(5):379–389. doi: 10.1016/j.tips.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhury A., et al. Clinical Review of Antidiabetic Drugs: Implications for Type 2 Diabetes Mellitus Management. Front Endocrinol (Lausanne) 2017;8:6. doi: 10.3389/fendo.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeFronzo R.A., et al. Type 2 diabetes mellitus. Nat Rev Dis Primers. 2015;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 14.Shlomai G., et al. Type 2 Diabetes Mellitus and Cancer: The Role of Pharmacotherapy. J Clin Oncol. 2016;34(35):4261–4269. doi: 10.1200/JCO.2016.67.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foltynie T., Athauda D. Repurposing anti-diabetic drugs for the treatment of Parkinson's disease: Rationale and clinical experience. Prog Brain Res. 2020;252:493–523. doi: 10.1016/bs.pbr.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Yarchoan M., Arnold S.E. Repurposing Diabetes Drugs for Brain Insulin Resistance in Alzheimer Disease. Diabetes. 2014;63(7):2253–2261. doi: 10.2337/db14-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giacco F., Brownlee M. Oxidative stress and diabetic complications. Circ Res. 2010;107(9):1058–1070. doi: 10.1161/CIRCRESAHA.110.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zinman B., et al. Empagliflozin, Cardiovascular Outcomes, and Mortality in Type 2 Diabetes. N Engl J Med. 2015;373(22):2117–2128. doi: 10.1056/NEJMoa1504720. [DOI] [PubMed] [Google Scholar]
- 19.Sun H., et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract. 2022;183 doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holman N., Young B., Gadsby R. Current prevalence of Type 1 and Type 2 diabetes in adults and children in the UK. Diabet Med. 2015;32(9):1119–1120. doi: 10.1111/dme.12791. [DOI] [PubMed] [Google Scholar]
- 21.Bruno G., et al. Incidence of type 1 and type 2 diabetes in adults aged 30–49 years: the population-based registry in the province of Turin. Italy. 2005;28(11):2613. doi: 10.2337/diacare.28.11.2613. [DOI] [PubMed] [Google Scholar]
- 22.Kwak S.H., Park K.S. Recent progress in genetic and epigenetic research on type 2 diabetes. Exp Mol Med. 2016;48 doi: 10.1038/emm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stinkens R., et al. Targeting fatty acid metabolism to improve glucose metabolism. Obes Rev. 2015;16(9):715–757. doi: 10.1111/obr.12298. [DOI] [PubMed] [Google Scholar]
- 24.Faselis C., et al. Microvascular Complications of Type 2 Diabetes Mellitus. Curr Vasc Pharmacol. 2020;18(2):117–124. doi: 10.2174/1570161117666190502103733. [DOI] [PubMed] [Google Scholar]
- 25.Viigimaa M., et al. Macrovascular Complications of Type 2 Diabetes Mellitus. Curr Vasc Pharmacol. 2020;18(2):110–116. doi: 10.2174/1570161117666190405165151. [DOI] [PubMed] [Google Scholar]
- 26.Pushpakom S., et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 27.Keiser M.J., et al. Relating protein pharmacology by ligand chemistry. Nat Biotechnol. 2007;25(2):197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 28.Langhans S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front Pharmacol. 2018;9:6. doi: 10.3389/fphar.2018.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng W., Thorne N., McKew J.C. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today. 2013;18(21–22):1067–1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodos R.A., et al. In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med. 2016;8(3):186–210. doi: 10.1002/wsbm.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellazzi R., et al. Data analysis and data mining: current issues in biomedical informatics. Methods Inf Med. 2011;50(6):536–544. doi: 10.3414/ME11-06-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vamathevan J., et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ekins S. The Next Era: Deep Learning in Pharmaceutical Research. Pharm Res. 2016;33(11):2594–2603. doi: 10.1007/s11095-016-2029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gawehn E., Hiss J.A., Schneider G. Deep Learning in Drug Discovery. Mol Inform. 2016;35(1):3–14. doi: 10.1002/minf.201501008. [DOI] [PubMed] [Google Scholar]
- 35.Pinoli, P., et al. Predicting Drug Synergism by Means of Non-Negative Matrix Tri-Factorization. IEEE/ACM Trans Comput Biol Bioinform, 2021. PP. [DOI] [PubMed]
- 36.Karimi M., et al. DeepAffinity: interpretable deep learning of compound-protein affinity through unified recurrent and convolutional neural networks. Bioinformatics. 2019;35(18):3329–3338. doi: 10.1093/bioinformatics/btz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Korotcov A., et al. Comparison of Deep Learning With Multiple Machine Learning Methods and Metrics Using Diverse Drug Discovery Data Sets. Mol Pharm. 2017;14(12):4462–4475. doi: 10.1021/acs.molpharmaceut.7b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aliper A., et al. Deep Learning Applications for Predicting Pharmacological Properties of Drugs and Drug Repurposing Using Transcriptomic Data. Mol Pharm. 2016;13(7):2524–2530. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanoli Z., et al. Exploration of databases and methods supporting drug repurposing: a comprehensive survey. Brief Bioinform. 2021;22(2):1656–1678. doi: 10.1093/bib/bbaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W., et al. Recent Advances in the Machine Learning-Based Drug-Target Interaction Prediction. Curr Drug Metab. 2019;20(3):194–202. doi: 10.2174/1389200219666180821094047. [DOI] [PubMed] [Google Scholar]
- 42.Pan X., et al. Deep learning for drug repurposing: Methods, databases, and applications. WIREs Computational Molecular. Science. 2022 [Google Scholar]
- 43.Yu J.L., Dai Q.Q., Li G.B. Deep learning in target prediction and drug repositioning: Recent advances and challenges. Drug Discov Today. 2021 doi: 10.1016/j.drudis.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 44.Qiu, Y., et al. A Comprehensive Review of Computational Methods for Drug-drug Interaction Detection. IEEE/ACM Trans Comput Biol Bioinform, 2021. PP. [DOI] [PubMed]
- 45.Ding H., et al. Similarity-based machine learning methods for predicting drug-target interactions: a brief review. Brief Bioinform. 2014;15(5):734–747. doi: 10.1093/bib/bbt056. [DOI] [PubMed] [Google Scholar]
- 46.Kim J., et al. Comprehensive Survey of Recent Drug Discovery Using Deep Learning. Int J Mol Sci. 2021;22(18) doi: 10.3390/ijms22189983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bengio Y., Courville A., Vincent P. Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell. 2013;35(8):1798–1828. doi: 10.1109/TPAMI.2013.50. [DOI] [PubMed] [Google Scholar]
- 48.D'Souza S., Prema K.V., Balaji S. Machine learning models for drug-target interactions: current knowledge and future directions. Drug Discov Today. 2020;25(4):748–756. doi: 10.1016/j.drudis.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Weininger, D., et al. SMILES. 2. Algorithm for generation of unique SMILES notation. 1989. 29(2). 97-101.
- 50.Weininger, D.J.J.o.c.i. and c. sciences, SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. 1988. 28(1): p. 31-36.
- 51.Chuang K.V., Gunsalus L.M., Keiser M.J. Learning Molecular Representations for Medicinal Chemistry. J Med Chem. 2020;63(16):8705–8722. doi: 10.1021/acs.jmedchem.0c00385. [DOI] [PubMed] [Google Scholar]
- 52.Fasoulis R., Paliouras G., Kavraki L.E. Graph representation learning for structural proteomics. Emerg Top Life Sci. 2021;5(6):789–802. doi: 10.1042/ETLS20210225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamora-Resendiz, R. and S.J.b. Crivelli. Structural learning of proteins using graph convolutional neural networks. 2019. 610444.
- 54.Chen R., et al. Machine Learning for Drug-Target Interaction Prediction. Molecules. 2018;23(9) doi: 10.3390/molecules23092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lavecchia A. Machine-learning approaches in drug discovery: methods and applications. Drug Discov Today. 2015;20(3):318–331. doi: 10.1016/j.drudis.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 56.Serçinoğlu, O. and P.O. Sarica, In Silico Databases and Tools for Drug Repurposing, in In Silico Drug Design. 2019. p. 703-742.
- 57.Bagherian M., et al. Machine learning approaches and databases for prediction of drug-target interaction: a survey paper. Brief Bioinform. 2021;22(1):247–269. doi: 10.1093/bib/bbz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng X., et al. Target identification among known drugs by deep learning from heterogeneous networks. Chem Sci. 2020;11(7):1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen M., et al. Deep-Learning-Based Drug-Target Interaction Prediction. J Proteome Res. 2017;16(4):1401–1409. doi: 10.1021/acs.jproteome.6b00618. [DOI] [PubMed] [Google Scholar]
- 60.Bagherian M., et al. Coupled matrix-matrix and coupled tensor-matrix completion methods for predicting drug-target interactions. Brief Bioinform. 2021;22(2):2161–2171. doi: 10.1093/bib/bbaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai, Q., Research and development of MolAICal for drug design via deep learning and classical programming. arXiv 2020. doi: https://arxiv.org/abs/2006.09747
- 62.Bai Q., et al. MolAICal: a soft tool for 3D drug design of protein targets by artificial intelligence and classical algorithm. Brief Bioinform. 2021;22(3):bbaa161. doi: 10.1093/bib/bbaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai Q., et al. Application advances of deep learning methods for de novo drug design and molecular dynamics simulation. WIREs Comput Mol Sci. 2022;12:e1581. doi: 10.1002/wcms.1581. [DOI] [Google Scholar]
- 64.Ceddia G., et al. Matrix Factorization-based Technique for Drug Repurposing Predictions. IEEE J Biomed Health Inform. 2020;24(11):3162–3172. doi: 10.1109/JBHI.2020.2991763. [DOI] [PubMed] [Google Scholar]
- 65.Becker M.L., et al. Hospitalisations and emergency department visits due to drug-drug interactions: a literature review. Pharmacoepidemiol Drug Saf. 2007;16(6):641–651. doi: 10.1002/pds.1351. [DOI] [PubMed] [Google Scholar]
- 66.Chee B.W., Berlin R., Schatz B. Predicting adverse drug events from personal health messages. AMIA Annu Symp Proc. 2011;2011:217–226. [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang T., Leng J., Liu Y. Deep learning for drug-drug interaction extraction from the literature: a review. Brief Bioinform. 2020;21(5):1609–1627. doi: 10.1093/bib/bbz087. [DOI] [PubMed] [Google Scholar]
- 68.Wang F., et al. Predicting drug-drug interactions by graph convolutional network with multi-kernel. Brief Bioinform. 2021 doi: 10.1093/bib/bbab511. [DOI] [PubMed] [Google Scholar]
- 69.Zhou B., et al. Drug repurposing based on drug-drug interaction. Chem Biol Drug Des. 2015;85(2):137–144. doi: 10.1111/cbdd.12378. [DOI] [PubMed] [Google Scholar]
- 70.Mei S., Zhang K. A machine learning framework for predicting drug-drug interactions. Sci Rep. 2021;11(1):17619. doi: 10.1038/s41598-021-97193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan C., et al. DDIGIP: predicting drug-drug interactions based on Gaussian interaction profile kernels. BMC Bioinf. 2019;20(Suppl 15):538. doi: 10.1186/s12859-019-3093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yan X.Y., et al. Prediction of the Drug-Drug Interaction Types with the Unified Embedding Features from Drug Similarity Networks. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.794205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu P., et al. Learning from low-rank multimodal representations for predicting disease-drug associations. BMC Med Inform Decis Mak. 2021;21(Suppl 1):308. doi: 10.1186/s12911-021-01648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang M., et al. Computational drug repositioning based on multi-similarities bilinear matrix factorization. Brief Bioinform. 2021;22(4) doi: 10.1093/bib/bbaa267. [DOI] [PubMed] [Google Scholar]
- 75.Wu G., Liu J., Wang C. Predicting drug-disease interactions by semi-supervised graph cut algorithm and three-layer data integration. BMC Med Genomics. 2017;10(Suppl 5):79. doi: 10.1186/s12920-017-0311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giovannucci E., et al. Diabetes and cancer: a consensus report. CA Cancer J Clin. 2010;60(4):207–221. doi: 10.3322/caac.20078. [DOI] [PubMed] [Google Scholar]
- 77.Ferguson R.D., et al. Hyperinsulinemia enhances c-Myc-mediated mammary tumor development and advances metastatic progression to the lung in a mouse model of type 2 diabetes. Breast Cancer Res. 2012;14(1):R8. doi: 10.1186/bcr3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo Y., et al. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38(7):904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Reuter S., et al. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abudawood M. Diabetes and cancer: A comprehensive review. J Res Med Sci. 2019;24:94. doi: 10.4103/jrms.JRMS_242_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Decensi A., et al. Metformin and cancer risk in diabetic patients: a systematic review and meta-analysis. Cancer Prev Res (Phila) 2010;3(11):1451–1461. doi: 10.1158/1940-6207.CAPR-10-0157. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Z.J., Li S. The prognostic value of metformin for cancer patients with concurrent diabetes: a systematic review and meta-analysis. Diabetes Obes Metab. 2014;16(8):707–710. doi: 10.1111/dom.12267. [DOI] [PubMed] [Google Scholar]
- 83.Lega I.C., et al. The effect of metformin on mortality following cancer among patients with diabetes. Cancer Epidemiol Biomarkers Prev. 2014;23(10):1974–1984. doi: 10.1158/1055-9965.EPI-14-0327. [DOI] [PubMed] [Google Scholar]
- 84.Wu L., et al. Pharmacologic Therapy of Diabetes and Overall Cancer Risk and Mortality: A Meta-Analysis of 265 Studies. Sci Rep. 2015;5:10147. doi: 10.1038/srep10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu F., et al. Metformin therapy and risk of colorectal adenomas and colorectal cancer in type 2 diabetes mellitus patients: A systematic review and meta-analysis. Oncotarget. 2017;8(9):16017–16026. doi: 10.18632/oncotarget.13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thakkar B., et al. Metformin and sulfonylureas in relation to cancer risk in type II diabetes patients: a meta-analysis using primary data of published studies. Metabolism. 2013;62(7):922–934. doi: 10.1016/j.metabol.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 87.Gandini S., et al. Metformin and cancer risk and mortality: a systematic review and meta-analysis taking into account biases and confounders. Cancer Prev Res (Phila) 2014;7(9):867–885. doi: 10.1158/1940-6207.CAPR-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Noto H., et al. Cancer risk in diabetic patients treated with metformin: a systematic review and meta-analysis. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0033411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soranna D., et al. Cancer risk associated with use of metformin and sulfonylurea in type 2 diabetes: a meta-analysis. Oncologist. 2012;17(6):813–822. doi: 10.1634/theoncologist.2011-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stevens R.J., et al. Cancer outcomes and all-cause mortality in adults allocated to metformin: systematic review and collaborative meta-analysis of randomised clinical trials. Diabetologia. 2012;55(10):2593–2603. doi: 10.1007/s00125-012-2653-7. [DOI] [PubMed] [Google Scholar]
- 91.Zhang P., et al. Association of metformin use with cancer incidence and mortality: a meta-analysis. Cancer Epidemiol. 2013;37(3):207–218. doi: 10.1016/j.canep.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 92.Rahmani J., et al. The effect of metformin on biomarkers associated with breast cancer outcomes: a systematic review, meta-analysis, and dose-response of randomized clinical trials. Clin Transl Oncol. 2020;22(1):37–49. doi: 10.1007/s12094-019-02108-9. [DOI] [PubMed] [Google Scholar]
- 93.Ng C.W., et al. Metformin and colorectal cancer: a systematic review, meta-analysis and meta-regression. Int J Colorectal Dis. 2020;35(8):1501–1512. doi: 10.1007/s00384-020-03676-x. [DOI] [PubMed] [Google Scholar]
- 94.Zhou J., et al. Meta-analysis: The efficacy of metformin and other anti-hyperglycemic agents in prolonging the survival of hepatocellular carcinoma patients with type 2 diabetes. Ann Hepatol. 2020;19(3):320–328. doi: 10.1016/j.aohep.2019.11.008. [DOI] [PubMed] [Google Scholar]
- 95.Hu H., et al. The Effect of Metformin on Aminotransferase Levels, Metabolic Parameters and Body Mass Index in Nonalcoholic Fatty Liver Disease Patients: A Metaanalysis. Curr Pharm Des. 2021;27(29):3235–3243. doi: 10.2174/1381612827666210315144821. [DOI] [PubMed] [Google Scholar]
- 96.Shi Y.Q., et al. Relationships are between metformin use and survival in pancreatic cancer patients concurrent with diabetes: A systematic review and meta-analysis. Medicine (Baltimore) 2020;99(37) doi: 10.1097/MD.0000000000021687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gong H., Chen Y., Zhou D. Prognostic significance of metformin treatment in endometrial cancer: a meta-analysis. Pharmazie. 2020;75(8):401–406. doi: 10.1691/ph.2020.0346. [DOI] [PubMed] [Google Scholar]
- 98.Xiao K., et al. The effect of metformin on lung cancer risk and survival in patients with type 2 diabetes mellitus: A meta-analysis. J Clin Pharm Ther. 2020;45(4):783–792. doi: 10.1111/jcpt.13167. [DOI] [PubMed] [Google Scholar]
- 99.Brancher S., et al. The role of metformin on lung cancer survival: the first systematic review and meta-analysis of observational studies and randomized clinical trials. J Cancer Res Clin Oncol. 2021;147(10):2819–2836. doi: 10.1007/s00432-021-03728-x. [DOI] [PubMed] [Google Scholar]
- 100.Gao R., Yang T., Xu W. Enemies or weapons in hands: investigational anti-diabetic drug glibenclamide and cancer risk. Expert Opin Investig Drugs. 2017;26(7):853–864. doi: 10.1080/13543784.2017.1333104. [DOI] [PubMed] [Google Scholar]
- 101.Lee J.Y., et al. Incident Hepatocellular Carcinoma Risk in Patients Treated with a Sulfonylurea: A Nationwide, Nested, Case-Control Study. Sci Rep. 2019;9(1):8532. doi: 10.1038/s41598-019-44447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang X., et al. Use of sulphonylurea and cancer in type 2 diabetes-The Hong Kong Diabetes Registry. Diabetes Res Clin Pract. 2010;90(3):343–351. doi: 10.1016/j.diabres.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 103.Monami M., et al. Sulphonylureas and cancer: a case-control study. Acta Diabetol. 2009;46(4):279–284. doi: 10.1007/s00592-008-0083-2. [DOI] [PubMed] [Google Scholar]
- 104.Qian X., et al. Glibenclamide exerts an antitumor activity through reactive oxygen species-c-jun NH2-terminal kinase pathway in human gastric cancer cell line MGC-803. Biochem Pharmacol. 2008;76(12):1705–1715. doi: 10.1016/j.bcp.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 105.Li M., et al. Glyburide attenuates B(a)p and LPS-induced inflammation-related lung tumorigenesis in mice. Environ Toxicol. 2021;36(8):1713–1722. doi: 10.1002/tox.23293. [DOI] [PubMed] [Google Scholar]
- 106.Zhao W., et al. Liraglutide inhibits the proliferation and promotes the apoptosis of MCF-7 human breast cancer cells through downregulation of microRNA-27a expression. Mol Med Rep. 2018;17(4):5202–5212. doi: 10.3892/mmr.2018.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eftekhari S., Montazeri H., Tarighi P. Synergistic anti-tumor effects of Liraglutide, a glucagon-like peptide-1 receptor agonist, along with Docetaxel on LNCaP prostate cancer cell line. Eur J Pharmacol. 2020;878 doi: 10.1016/j.ejphar.2020.173102. [DOI] [PubMed] [Google Scholar]
- 108.Wenjing H., et al. Exendin-4 does not modify growth or apoptosis of human colon cancer cells. Endocr Res. 2017;42(3):209–218. doi: 10.1080/07435800.2017.1292525. [DOI] [PubMed] [Google Scholar]
- 109.Koehler J.A., Kain T., Drucker D.J. Glucagon-like peptide-1 receptor activation inhibits growth and augments apoptosis in murine CT26 colon cancer cells. Endocrinology. 2011;152(9):3362–3372. doi: 10.1210/en.2011-1201. [DOI] [PubMed] [Google Scholar]
- 110.Kosowska A., et al. Exenatide modulates tumor-endothelial cell interactions in human ovarian cancer cells. Endocr Connect. 2017;6(8):856–865. doi: 10.1530/EC-17-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kanda R., et al. Expression of the glucagon-like peptide-1 receptor and its role in regulating autophagy in endometrial cancer. BMC Cancer. 2018;18(1):657. doi: 10.1186/s12885-018-4570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Monami M., et al. Safety issues with glucagon-like peptide-1 receptor agonists (pancreatitis, pancreatic cancer and cholelithiasis): Data from randomized controlled trials. Diabetes Obes Metab. 2017;19(9):1233–1241. doi: 10.1111/dom.12926. [DOI] [PubMed] [Google Scholar]
- 113.Cao C., Yang S., Zhou Z. GLP-1 receptor agonists and pancreatic safety concerns in type 2 diabetic patients: data from cardiovascular outcome trials. Endocrine. 2020;68(3):518–525. doi: 10.1007/s12020-020-02223-6. [DOI] [PubMed] [Google Scholar]
- 114.Han Y., Li Y., He B. GLP-1 receptor agonists versus metformin in PCOS: a systematic review and meta-analysis. Reprod Biomed Online. 2019;39(2):332–342. doi: 10.1016/j.rbmo.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 115.Kaji K., et al. Sodium glucose cotransporter 2 inhibitor canagliflozin attenuates liver cancer cell growth and angiogenic activity by inhibiting glucose uptake. Int J Cancer. 2018;142(8):1712–1722. doi: 10.1002/ijc.31193. [DOI] [PubMed] [Google Scholar]
- 116.Zhou J., et al. Sodium-glucose co-transporter-2 (SGLT-2) inhibition reduces glucose uptake to induce breast cancer cell growth arrest through AMPK/mTOR pathway. Biomed Pharmacother. 2020;132 doi: 10.1016/j.biopha.2020.110821. [DOI] [PubMed] [Google Scholar]
- 117.Yamamoto L., et al. Sodium-glucose cotransporter 2 inhibitor canagliflozin attenuates lung cancer cell proliferation in vitro. Diabetol Int. 2021;12(4):389–398. doi: 10.1007/s13340-021-00494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu D., et al. Inhibitory effects of canagliflozin on pancreatic cancer are mediated via the downregulation of glucose transporter-1 and lactate dehydrogenase A. Int J Oncol. 2020;57(5):1223–1233. doi: 10.3892/ijo.2020.5120. [DOI] [PubMed] [Google Scholar]
- 119.Tseng C.H. Sitagliptin may reduce prostate cancer risk in male patients with type 2 diabetes. Oncotarget. 2017;8(12):19057–19064. doi: 10.18632/oncotarget.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tseng C.H. Sitagliptin and oral cancer risk in type 2 diabetes patients. Oncotarget. 2017;8(57):96753–96760. doi: 10.18632/oncotarget.18239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tseng C.H. Sitagliptin May Reduce Breast Cancer Risk in Women With Type 2 Diabetes. Clin Breast Cancer. 2017;17(3):211–218. doi: 10.1016/j.clbc.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 122.Li Y., et al. Repositioning of Hypoglycemic Drug Linagliptin for Cancer Treatment. Front Pharmacol. 2020;11:187. doi: 10.3389/fphar.2020.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang Q., et al. Sitagliptin affects gastric cancer cells proliferation by suppressing Melanoma-associated antigen-A3 expression through Yes-associated protein inactivation. Cancer Med. 2020;9(11):3816–3828. doi: 10.1002/cam4.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dicembrini I., et al. Risk of cancer in patients treated with dipeptidyl peptidase-4 inhibitors: an extensive meta-analysis of randomized controlled trials. Acta Diabetol. 2020;57(6):689–696. doi: 10.1007/s00592-020-01479-8. [DOI] [PubMed] [Google Scholar]
- 125.Femia A.P., et al. Long-term treatment with Sitagliptin, a dipeptidyl peptidase-4 inhibitor, reduces colon carcinogenesis and reactive oxygen species in 1,2-dimethylhydrazine-induced rats. Int J Cancer. 2013;133(10):2498–2503. doi: 10.1002/ijc.28260. [DOI] [PubMed] [Google Scholar]
- 126.Jang J.H., et al. Suppression of lung metastases by the CD26/DPP4 inhibitor Vildagliptin in mice. Clin Exp Metastasis. 2015;32(7):677–687. doi: 10.1007/s10585-015-9736-z. [DOI] [PubMed] [Google Scholar]
- 127.Tumminia A., et al. Type 2 Diabetes Mellitus and Alzheimer’s Disease: Role of Insulin Signalling and Therapeutic Implications. Int J Mol Sci. 2018;19(11) doi: 10.3390/ijms19113306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cheong J.L.Y., et al. The Association Between Type 2 Diabetes Mellitus and Parkinson’s Disease. Journal of Parkinson's Disease. 2020;10(3):775–789. doi: 10.3233/JPD-191900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Athauda D., Foltynie T. Insulin resistance and Parkinson's disease: A new target for disease modification? Prog Neurobiol. 2016;145–146:98–120. doi: 10.1016/j.pneurobio.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 130.Zhang L., et al. The Novel Dual GLP-1/GIP Receptor Agonist DA-CH5 Is Superior to Single GLP-1 Receptor Agonists in the MPTP Model of. Parkinson's Disease. 2020;10(2) doi: 10.3233/JPD-191768. [DOI] [PubMed] [Google Scholar]
- 131.Boccardi V., Murasecco I., Mecocci P. Diabetes drugs in the fight against Alzheimer's disease. Ageing Res Rev. 2019;54 doi: 10.1016/j.arr.2019.100936. [DOI] [PubMed] [Google Scholar]
- 132.Kandimalla R., Thirumala V., Reddy P.H. Is Alzheimer's disease a Type 3 Diabetes? A critical appraisal. Biochim Biophys Acta Mol Basis Dis. 2017;1863(5):1078–1089. doi: 10.1016/j.bbadis.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu X.Y., et al. Potential new therapeutic target for Alzheimer's disease: Glucagon-like peptide-1. Eur J Neurosci. 2021;54(10):7749–7769. doi: 10.1111/ejn.15502. [DOI] [PubMed] [Google Scholar]
- 134.Mulvaney, C.A., et al., GLP-1 receptor agonists for Parkinson's disease. Cochrane Database Syst Rev, 2020. 7(7): p. Cd012990. [DOI] [PMC free article] [PubMed]
- 135.Athauda D., et al. Exenatide once weekly versus placebo in Parkinson's disease: a randomised, double-blind, placebo-controlled trial. The Lancet. 2017;390(10103):1664–1675. doi: 10.1016/S0140-6736(17)31585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aviles-Olmos I., et al. Exenatide and the treatment of patients with Parkinson's disease. J Clin Invest. 2013;123(6):2730–2736. doi: 10.1172/JCI68295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vijiaratnam N., et al. Exenatide once weekly over 2 years as a potential disease-modifying treatment for Parkinson's disease: protocol for a multicentre, randomised, double blind, parallel group, placebo controlled, phase 3 trial: The 'Exenatide-PD3' study. BMJ Open. 2021;11(5) doi: 10.1136/bmjopen-2020-047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cao B., et al. Neuroprotective effects of liraglutide against inflammation through the AMPK/NF-κB pathway in a mouse model of Parkinson's disease. Metab Brain Dis. 2021 doi: 10.1007/s11011-021-00879-1. [DOI] [PubMed] [Google Scholar]
- 139.Adeva-Andany M.M., et al. Insulin resistance is a cardiovascular risk factor in humans. Diabetes Metab Syndr. 2019;13(2):1449–1455. doi: 10.1016/j.dsx.2019.02.023. [DOI] [PubMed] [Google Scholar]
- 140.Athyros V.G., et al. Diabetes and lipid metabolism. Hormones (Athens) 2018;17(1):61–67. doi: 10.1007/s42000-018-0014-8. [DOI] [PubMed] [Google Scholar]
- 141.Stirban A., Gawlowski T., Roden M. Vascular effects of advanced glycation endproducts: Clinical effects and molecular mechanisms. Mol Metab. 2014;3(2):94–108. doi: 10.1016/j.molmet.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schubert M., et al. Repurposing Antidiabetic Drugs for Cardiovascular Disease. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.568632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Neal B., et al. Canagliflozin and Cardiovascular and Renal Events in Type 2 Diabetes. N Engl J Med. 2017;377(7):644–657. doi: 10.1056/NEJMoa1611925. [DOI] [PubMed] [Google Scholar]
- 144.Zelniker T.A., et al. Comparison of the Effects of Glucagon-Like Peptide Receptor Agonists and Sodium-Glucose Cotransporter 2 Inhibitors for Prevention of Major Adverse Cardiovascular and Renal Outcomes in Type 2 Diabetes Mellitus. Circulation. 2019;139(17):2022–2031. doi: 10.1161/CIRCULATIONAHA.118.038868. [DOI] [PubMed] [Google Scholar]
- 145.Palmer S.C., et al. Sodium-glucose cotransporter protein-2 (SGLT-2) inhibitors and glucagon-like peptide-1 (GLP-1) receptor agonists for type 2 diabetes: systematic review and network meta-analysis of randomised controlled trials. BMJ. 2021;372 doi: 10.1136/bmj.m4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kristensen S.L., et al. Cardiovascular, mortality, and kidney outcomes with GLP-1 receptor agonists in patients with type 2 diabetes: a systematic review and meta-analysis of cardiovascular outcome trials. The Lancet Diabetes & Endocrinology. 2019;7(10):776–785. doi: 10.1016/S2213-8587(19)30249-9. [DOI] [PubMed] [Google Scholar]
- 147.Scheen A.J. Cardiovascular Effects of New Oral Glucose-Lowering Agents: DPP-4 and SGLT-2 Inhibitors. Circ Res. 2018;122(10):1439–1459. doi: 10.1161/CIRCRESAHA.117.311588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Scirica B.M., et al. Heart failure, saxagliptin, and diabetes mellitus: observations from the SAVOR-TIMI 53 randomized trial. Circulation. 2014;130(18):1579–1588. doi: 10.1161/CIRCULATIONAHA.114.010389. [DOI] [PubMed] [Google Scholar]
- 149.Davies, M.J., et al., Management of Hyperglycemia in Type 2 Diabetes, 2018. A Consensus Report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetes Care, 2018. 41(12): p. 2669-2701. [DOI] [PMC free article] [PubMed]