Abstract
Trichoderma harzianum was cotransformed with genes encoding green fluorescent protein (GFP), β-glucuronidase (GUS), and hygromycin B (hygB) resistance, using polyethylene glycol-mediated transformation. One cotransformant (ThzID1-M3) was mitotically stable for 6 months despite successive subculturing without selection pressure. ThzID1-M3 morphology was similar to that of the wild type; however, the mycelial growth rate on agar was reduced. ThzID1-M3 was formed into calcium alginate pellets and placed onto buried glass slides in a nonsterile soil, and its ability to grow, sporulate, and colonize sclerotia of Sclerotinia sclerotiorum was compared with that of the wild-type strain. Wild-type and transformant strains both colonized sclerotia at levels above those of indigenous Trichoderma spp. in untreated controls. There were no significant differences in colonization levels between wild-type and cotransformant strains; however, the presence of the GFP and GUS marker genes permitted differentiation of introduced Trichoderma from indigenous strains. GFP activity was a useful tool for nondestructive monitoring of the hyphal growth of the transformant in a natural soil. The green color of cotransformant hyphae was clearly visible with a UV epifluorescence microscope, while indigenous fungi in the same samples were barely visible. Green-fluorescing conidiophores and conidia were observed within the first 3 days of incubation in soil, and this was followed by the formation of terminal and intercalary chlamydospores and subsequent disintegration of older hyphal segments. Addition of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) substrate to recovered glass slides confirmed the activity of GUS as well as GFP in soil. Our results suggest that cotransformation with GFP and GUS can provide a valuable tool for the detection and monitoring of specific strains of T. harzianum released into the soil.
Trichoderma spp. have received considerable attention as potential biological control agents against a wide range of soil-borne plant-pathogenic fungi (7, 23) in the greenhouse (8, 16, 20, 24) and field (12, 13, 15, 19). However, the efficacy of Trichoderma spp. as biocontrol agents in natural soils may be limited by soil fungistasis (23), competition by other soil microorganisms (17), poor plant root colonization (2), or unfavorable environmental conditions. Identification and quantification of ecological factors affecting the establishment and the population dynamics of introduced Trichoderma strains in natural habitats may provide more predictable and effective biocontrol of plant diseases.
Although several methods have been used to study the occurrence and distribution of Trichoderma in soils (1, 25), few methods have allowed quantitative evaluation of population dynamics and survival. For example, Knudsen and coworkers (11, 18) quantified the influences of temperature, soil matric potential, nutrient source, and antagonistic bacteria on the hyphal growth and biocontrol efficacy of pelletized T. harzianum in soils. However, it was not possible to differentiate the hyphal growth of this fungal agent from that of indigenous Trichoderma strains (3, 18). Knudsen et al. (18) pointed out that the use of dilution plating for numerical estimation of fungal populations does not differentiate among the different propagules (hyphal fragments, conidia, and chlamydospores) that may generate colonies when plated on agar, and thus it is not a true estimate of fungal biomass. The use of mutant strains resistant to specific fungicides may partially overcome problems related to nonspecific recovery (1, 2), but this method does not allow for in situ monitoring of growth dynamics and survival structures of introduced Trichoderma strains or in situ differentiation of introduced Trichoderma strains from indigenous strains.
Recently, genetic engineering of biocontrol agents with reporter or marker genes has provided useful tools for detection and monitoring of introduced biocontrol agents in natural environments (14, 21). For example, the selectable hygromycin B phosphotransferase (hygB) gene, coding for resistance to this antibiotic, has been used to detect fungal biocontrol agents in the rhizosphere and phyllosphere (21, 22). The β-glucuronidase (GUS) marker gene also is a promising tool for ecological studies of biocontrol agents, because of the low background activity of GUS in fungi and plants, the relative ease and sensitivity of detection (29), and the apparent lack of influence of GUS expression on biocontrol efficacy (36). However, some background GUS activity may be present in unsterile systems or natural soils. Aspergillus niger has some indigenous acidic GUS activity, and T. harzianum strain T3 showed indigenous acidic β-galactosidase activity (36). Therefore, for study of growth patterns and sporulation of an introduced fungus in natural ecosystems, the GUS system may have limited usefulness.
The green fluorescent protein (GFP) of the jellyfish Aequorea victoria also has been developed as a reporter for gene expression (5). It has been successfully cloned and expressed in several organisms (6, 9, 31). GFP was shown to be a useful tool for studying plant-fungus interactions in vivo (34) and has been used to assess the colonization and dispersal of Aureobasidium pullulans in the phyllosphere (37). GFP requires only UV or blue light and oxygen to induce green fluorescence. An exogenous substrate, such as GUS requires, is not needed for the detection of GFP, thus avoiding problems related to cell permeability and substrate uptake (10, 31).
The objectives of this study were (i) to produce a mitotically stable cotransformant of T. harzianum isolate ThzID1 expressing the hygB gene as well as both marker genes, GUS and GFP, and (ii) to determine whether the cotransformant provides a useful tool for in situ monitoring of T. harzianum hyphal growth and activity in soil, including the ability to colonize the resting form (sclerotia) of the plant-pathogenic fungus Sclerotinia sclerotiorum.
MATERIALS AND METHODS
Transformation vectors and preparation of plasmid DNA.
Plasmids pAN7-2, containing the Escherichia coli hygB gene (27), and pNOM102, containing the E. coli GUS gene (29), were obtained from H. Leung (Department of Plant Pathology, Washington State University). Both plasmids contained the constitutive Aspergillus nidulans glyceraldehyde-3-phosphate dehydrogenase (gpd) promoter. Plasmid pTEFEGFP containing the E. coli engineered GFP (EGFP) gene (37) was provided by D. Cullen (Forest Products Laboratory, U.S. Department of Agriculture, University of Wisconsin—Madison). It contains the Aureobasidium pullulans translation elongation factor promoter and Aspergillus awamori glucoamylase terminator. Each plasmid was propagated in E. coli strain HB101, and purification of plasmid DNA was performed by the method of Sambrook et al. (30).
Protoplast preparation.
Protoplasts were generated by combining the protocols described by Thrane et al. (36) and Sivan et al. (33) as detailed below, except that potato dextrose broth (PDB) was replaced by glucose yeast extract broth (GYEB; 15 g of glucose, 3 g of yeast extract, 1 liter of water). Conidia (2 × 106 to 5 × 106) of T. harzianum isolate ThzID1 were added to 100 ml of GYEB for 23 h at 26 C with shaking (100 rpm). The resultant mycelium was collected on four layers of cheesecloth and washed with 1.2 M MgSO4. A 1-g portion of fresh mycelium was transferred to a 50-ml flask containing 20 ml of sterile-filtered (0.45-μm-pore-size filter) lysing enzyme (Sigma L-2265) solution (7.5 mg per ml in 1.2 M MgSO4–10 mM sodium phosphate buffer [pH 5.8]). The flask was incubated for 3 to 4 h at 26°C with shaking at 100 rpm. Protoplasts were separated from mycelial debris by filtering through six layers of cheesecloth and washed twice in 1.2 M MgSO4 by centrifugation at 500 × g for 5 min at 4°C. The protoplasts then were rinsed, suspended in STC medium (0.6 M sorbitol, 10 mM Tris-HCl [pH 7.5], 10 mM CaCl2), and kept on ice until used.
Transformation of protoplasts.
The transformation procedures were based on the methods of Penttilä et al. (26) and Shi et al. (32). Circular plasmid DNA (1 μg each of pAN7-2, pNOM102, and pTEFEGFP) in 25 μl of Tris-EDTA buffer (30) was mixed into 100 μl of protoplast suspension (1.2 × 108/ml) in a 50-ml centrifuge tube. The mixture was incubated for 20 min on ice. In a dropwise manner, 2 ml of polyethlene glycol (PEG) solution (60% [wt/vol] PEG 3350 in 25 mM CaCl2–25 mM Tris-HCl [pH 7.5]) was added after incubation. The suspension was gently combined with 30 ml of ice-cold STC medium. The protoplasts were concentrated by centrifugation at 2,500 rpm for 10 min, and the pelleted protoplasts were resuspended in 4 ml of PDB containing 1 M sorbitol. After incubation at 26 C for 9 h, 0.5 ml of the suspension was plated with 10 ml of molten potato dextrose agar (PDA). The solidified plate was overlaid with 10 ml of molten PDA (0.7%) containing 300 μg of hygromycin B per ml (150-μg/ml final concentration).
Colonies were chosen after incubation at 25°C for 5 to 7 days and transferred to PDA containing 150 μg of hygromycin B per ml. Colonies were tested for GUS activity as follows. A mycelial segment from each colony was placed in a microtiter plate containing 200 μl of 10mM sodium phosphate buffer (pH 7.5) supplemented with 4 μl of substrate (4 mg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid [X-Gluc; Sigma Chemical Co. or Clonetech] in 1 ml of 50 mM sodium phosphate buffer [pH 7.5]) and incubated in the dark for 48 h (36).
Colonies that developed blue pigment were examined for GFP activity. Cotransformants that showed both GUS and GFP activities were selected for further tests.
Selection of stable cotransformants.
To eliminate isolates containing heterokaryons, each transformant was incubated on a PDA slant containing 150 μg of hygromycin B per ml, under a fluorescent light, for 7 days at 25°C. Conidia of each transformant then were collected through four layers of cheesecloth and plated on PDA containing 150 μg of hygromycin B per ml. After incubation for 2 to 3 days, several distinct colonies (i.e., single-spore isolates) were transferred to new PDA slants containing 150 μg of hygromycin B per ml. After testing for GUS and GFP activities, one isolate was chosen to conduct the selection process again; this was repeated five times. To test the stability of cotransformants selected in this manner, a mycelial plug of each cotransformant was transferred to a PDA plate without antibiotic and the plate was incubated at 25°C. After 4 days of incubation, cotransformants were subcultured on new PDA plates without antibiotic for an additional 40 days. A final subculture of each cotransformant was tested to confirm HygBr and GUS and GFP activity.
Southern blot analysis.
Wild-type and cotransformant isolates were grown from conidia in PDB without antibiotic. Genomic DNA was isolated from 72-h-old thalli that were lyophilized and ground in liquid nitrogen (28). Genomic and plasmid DNAs were digested with restriction enzymes as follows: pAN7-2 with HindIII-EcoRI, pNOM102 with NcoI, and pTEFEGFP with HindIII-BamHI. DNA digests were electrophoresed in 1% agarose and blotted on nylon membranes (Boehringer Mannheim, Indianapolis, Ind.) using standard techniques (30). DNA fragments of each plasmid were separately labeled with digoxigenin-dUTP (Boehringer Mannheim) and used to probe genomic blots of wild-type and cotransformant M3 as specified by the manufacturer. Hybridization signals were detected colorimetrically with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as substrates for alkaline phosphatase (Boehringer Mannheim).
Growth on PDA and in soil.
Mycelial disks (5 mm in diameter) of wild-type and cotransformant strains were transferred to PDA plates with or without hygromycin B and incubated at 25°C for 3 days in the dark. Radial growth was measured under a light microscope and recorded daily, beginning 24 h after incubation. There were five replicates for each strain. Hyphal extension of cotransformant ThzID1-M3 also was measured in soil. Palouse silt loam soil was obtained from the University of Idaho Parker Farm near Moscow. Soil analysis (University of Idaho Analytical Services Laboratory) indicated that the soil contained 20% sand, 20% clay, and 60% silt by weight, with 82.2 μg of plant-available iron per g. Soil pH in soil-water (2:1) solution was approximately 5.9. The soil was sieved through a 2-mm mesh and air dried prior to use. The soil was adjusted to a soil moisture content of −100 kPa.
Alginate pellets of the transformant strain were made using previously described methods (18). ThzID1-M3 was grown on PDA for 7 days, and three 1-cm2 pieces from the culture margins were placed in 500 ml of PDB in a 1-liter flask. The flasks were incubated at approximately 22°C on a rotary shaker (120 rpm), with 12 h of light per day, for 1 week. The mycelial biomass was then strained, rinsed with sterile distilled water, and added to 100 ml of 1% aqueous sodium alginate solution. The mixture of fungus-alginate solution was added as drops to 0.25 M aqueous CaCl2. Pellets formed in the CaCl2 solution were removed, rinsed with sterile distilled water, and allowed to air dry on waxed paper. The pellets were stored in glass beakers at 4°C.
A glass petri dish (15 cm in diameter) was about half filled with soil, and a glass slide precoated with 1.8% water agar was placed on the soil surface. A single pellet was glued in the middle of the glass slide with cyanoacrylate glue, which previously was determined not to influence the growth of Trichoderma (G. R. Knudsen, unpublished results). The glass slide and pellet were covered with soil to fill the plate, and then the plates were placed in a covered plastic container lined with moist paper towels and incubated at 25°C for 3, 5, and 10 days. At each sampling time, glass slides were removed and examined using epifluorescence microscopy. Radial growth was quantified by measuring colony diameters from captured video images at magnifications of ×250 or ×400. The experiment was repeated once, with five replicates each time.
Colonization of sclerotia in soil.
To produce sclerotia of S. sclerotiorum, mycelial disks grown on PDA for 7 days were transferred to sterilized sliced carrots in 2-liter Erlenmeyer flasks. After 6 to 8 weeks of incubation at 16°C, sclerotia were harvested, rinsed with sterile distilled water, and air dried for 2 to 3 days. The sclerotia were attached to plastic toothpicks with cyanoacrylate glue and allowed to dry overnight. All sclerotia then were surface disinfected with a sterile solution (10% ethanol–10% bleach in water) for 1 min and stored at 4°C before use.
A quantity of a Palouse silt loam soil was prepared as described above. Pellets of ThzID1 and ThzID1-M3 were produced as described above. A plastic container (25 by 25 by 10 cm) was filled with 1 kg of soil to which five pellets of each isolate had been added. Twenty sclerotia were randomly placed in the soil at the depth of 2 cm. The containers were covered and sealed with plastic film to maintain a relatively constant moisture content. Treatments were as follows: untreated (no-Trichoderma) control, ThzID1 alginate pellets, and ThzID1-M3 alginate pellets. There were four replicates. After 7 and 14 days of incubation at 25°C, half of the sclerotia from each container were recovered, surface sterilized, and placed on PDA containing 50 μg of streptomycin per ml. The plates were incubated at 25°C for 7 days, and sclerotia were observed to determine whether S. sclerotiorum, Trichoderma spp., or other fungi were growing from them. For treatment with cotransformant ThzID1-M3, mycelia grown from sclerotia were examined for GUS and GFP activities. Proportions of sclerotia colonized by Trichoderma spp. or ThzID1-M3 were recorded.
GFP and GUS expression of the cotransformant after inoculation into soil.
A pellet (as described above) of cotransformant ThzID1-M3 and a pellet of the wild-type ThzID1 were glued onto a glass microscope slide, 0.5 cm apart. The glass slide was incubated in a glass petri dish (15 cm in diameter) containing soil as described above. After 2, 3, and 5 days of incubation at 25°C, the glass slides were carefully removed and observed microscopically. GFP activity of cotransformant ThzID1-M3 was observed as described above, using epifluorescence. To detect GUS activity, 400 μl of 10mM sodium phosphate buffer (pH 7.5) supplemented with 16 μl of X-Gluc substrate was carefully placed on the glass slide with a glass coverslip and then incubated at 37°C for 4 h in the dark prior to observation.
Statistical analysis.
All experiments were conducted as completely randomized designs. Analysis of variance was performed for each experiment, and the least-significant-difference (LSD) test was used for means separation, where appropriate, using PROC GLM (SAS Institute Inc., Cary, N.C.).
RESULTS AND DISCUSSION
Fungal cotransformation and stability.
Before transformation, the effect of the dose of hygromycin B on the regeneration frequency of protoplasts was determined. Regeneration of untransformed protoplasts was completely inhibited at 150 μg of hygromycin B per ml (data not shown). Therefore, growth at this concentration was used as the selection criterion for hygromycin B-tolerant transformants. The average frequency of transformation to hygromycin tolerance was 15 transformants per μg of DNA. All transformants were examined for GUS and GFP activities. A total of five expressed both GUS and GFP activities. In PEG-mediated transformations, protoplasts generated from young mycelia of Trichoderma typically contain 2 to 12 nuclei per protoplast (35). During transformation, protoplasts fuse to form aggregates, which develop into thalli containing more than 30 nuclei per cell (33). Sivan et al. (33) reported that stabilization of transformants by single-conidium selection was essential to obtain stable transformants, due to the heterokaryotic nature of putative transformants. To select a monokaryon from each cotransformant, several colonies developed from a single conidium of each cotransformant were chosen and tested for GUS and GFP activities. Four cotransformants showed the ability to express both GUS and GFP activities. To test the stability of expression in cotransformants, these cotransformants were subcultured successively on PDA without selection pressure, up to 10 times. Cotransformant ThzID1-M3 was the only isolate that exhibited stable expression of all three foreign genes.
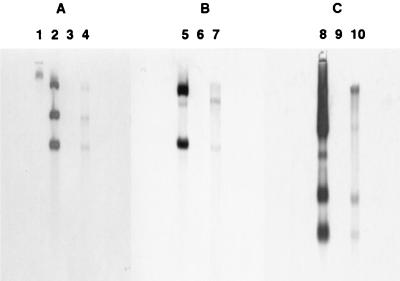
Southern hybridizations were performed to confirm integration of the foreign plasmid DNA into the genome of cotransformant ThzID1-M3. Total genomic DNAs from cotransformant ThzID1-M3 and wild-type ThzID1 were isolated, digested, and probed with the HindIII-EcoRI pAN7-2 fragment, the NcoI-NcoI pNOM102 fragment, and the HindIII-BamHI pTEFEGFP fragment. There were no background signals of the wild-type genomic DNA (Fig. 1). This isolate showed no reversion to the wild-type phenotype over a 6-month period.
FIG. 1.
Southern blot analysis of genomic DNAs from T. harzianum wild-type ThzID1 and cotransformant ThzID1-M3. (A) Probed with HindIII-EcoRI fragments from pAN7-2 including the hygromycin B resistance gene. (B) Probed with NcoI-NcoI fragments from pNOM102 including the GUS gene. (C) Probed with HindIII-BamHI fragments from pTEFEGFP including the GFP gene. Lanes: 1, λ-HindIII; 2, 3, and 4: pAN7-2, wild type, and cotransformant, respectively, digested with HindIII-EcoRI; 5, 6, and 7, pNOM102, wild type, and cotransformant, respectively, digested with NcoI-NcoI; 8, 9, and 10, pTEFEGFP, wild type, and cotransformant, respectively, digested with HindIII-BamHI.
Hyphal growth and colonization on sclerotia.
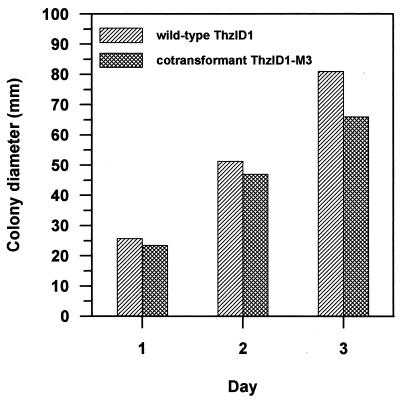
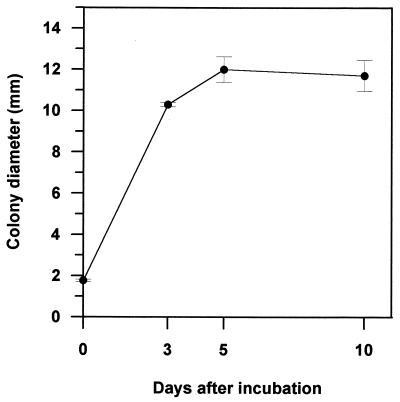
The morphology of cotransformant ThzID1-M3 was similar to that of the wild type (data not shown). However, the mycelial growth of the cotransformant on PDA was slower than that of the wild type (Fig. 2). The wild-type ThzID1 did not grow on 100 μg of hygromycin B per ml, whereas the cotransformant grew at 300 μg of hygromycin B per ml (data not shown). GFP was a useful tool for monitoring the growth of introduced T. harzianum in soil. It provided a nondestructive sampling for visualization of hyphae in situ. The mean colony diameter increased rapidly up to 5 days in soil but subsequently decreased by day 10 (Fig. 3). Previous work in our laboratory demonstrated several environmental factors that influence radial growth and density of hyphae originating from alginate pellets of T. harzianum (11, 18, 19). However, previous experiments necessarily were conducted using steamed soil, because it was impossible to differentiate hyphae from different sources in raw soil. When compared with those results, our present results indicate a lower growth rate in nonsterile soil, possibly resulting from reduced growth ability of the cotransformant and/or from the influence of the soil microbiota in an untreated soil.
FIG. 2.
Growth of T. harzianum wild-type ThzID1 and cotransformant ThzID1-M3 on PDA after 1, 2, and 3 days of incubation at 25°C in the dark. All values for days and isolates were significantly different (P < 0.05) according to an LSD analysis.
FIG. 3.
Colony diameter of T. harzianum ThzID1-M3 originating from alginate pellets in soil at 3, 5, and 10 days. Colony diameters were measured at a magnification of ×250 or ×400 under an epifluorescence microscope. Vertical bars represent ±1 standard error of the mean.
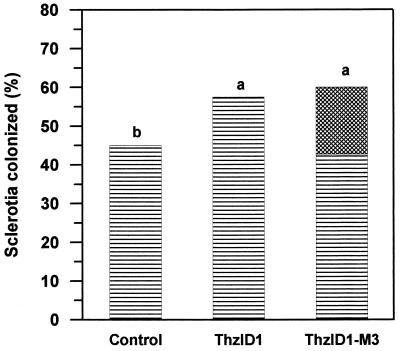
Using alginate pellets, the ability of cotransformant ThzID1-M3 to colonize sclerotia of S. sclerotiorum, compared to the wild-type ThzID1, in soil was not significantly different after 7 or 14 days (P > 0.05). Therefore, only results after 14 days of incubation are presented (Fig. 4). Treatments with alginate pellets of either the wild-type ThzID1 or cotransformant ThzID1-M3 resulted in higher percentages of sclerotia colonized by Trichoderma spp. compared to nontreated controls (P < 0.05). There were no significant differences between treatments with the introduced wild type compared to the cotransformant (mean, 58 and 60%, respectively; P > 0.05). However, while approximately 60% of sclerotia were colonized by Trichoderma spp. in cotransformant treatments, the recombinant phenotype (GFP + GUS) accounted only for about 18% of sclerotial colonization, with the remaining 42% being attributable to (presumably indigenous) Trichoderma spp. without the recombinant phenotype. Bin et al. (3) reported that incorporation of pellets of T. harzianum into field soil significantly increased the colonization of sclerotia by Trichoderma spp. Knudsen et al. (19) concluded that, based on sampled populations of Trichoderma propagules in field trials, the majority of sclerotial colonization resulted from hyphal growth of introduced T. harzianum. However, in the present study, we were able for the first time to quantify the relative contribution of an introduced Trichoderma strain to total sclerotial colonization, because of its unique phenotype. Our results indicate a relatively smaller contribution of the introduced strain to total colonization than was previously estimated, but it should be noted that soil environmental conditions in previous studies (3, 19) probably were less conducive to activation of dormant propagules of indigenous Trichoderma, since they were hotter and drier.
FIG. 4.
Percentage of sclerotia colonized by Trichoderma spp. in soil at 14 days. Five alginate pellets of T. harzianum ThzID1 or ThzID1-M3 were applied in a 25-cm2 pot containing 1 kg of natural soil. The dashed portion of ThzID1-M3 treatment represents the proportion of sclerotia colonized by T. harzianum ThzID1-M3. Means followed by the same letter are not significantly different (P > 0.05) according to an LSD analysis.
GFP and GUS expression of the cotransformant after inoculation into soil.
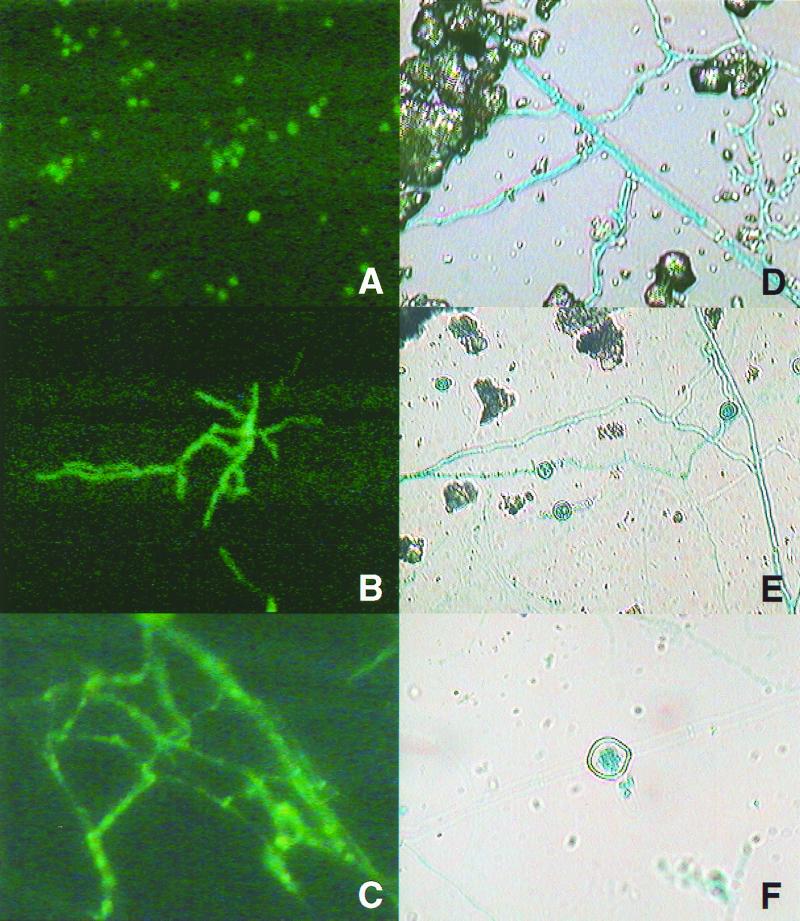
GFP expression in soil was observed using epifluorescence with a BPF510 illuminator slide set including 2X KP 490 and B 229 for the excitation filter and a G 247 barrier filter. Thus viewed, hyphae of the wild-type T. harzianum were barely visible. In contrast, the cotransformant exhibited distinct green fluorescence (Figs. 5A to C). Hyphal growth from pellets was first observed after 2 days of incubation and continued for approximately 5 days. GFP activity of hyphae revealed that green fluorescence was more intense near septa in older hyphae (Fig. 5C). After 5 days of incubation, most structures apart from pellets of ThzID1-M3 were terminal or intercalary chlamydospores. Older thick-walled chlamydospores were hardly seen due to low intensity of GFP activity. Conidia or conidiophores of ThzID1-M3 were visible mostly around pellets only after 3 days of incubation.
FIG. 5.
Photomicrographs of GFP (A to C) and GUS (D to F) activities of T. harzianum ThzID1-M3 in natural soil. (A) Conidia; (B) conidial germination; (C and D) hyphal growth; (E and F) chlamydospore formation. Magnifications, ×400 (A to D and F) and ×250 (E).
To view GUS activity, glass slides first were removed and treated with the X-Gluc substrate. After a 4-h incubation at 37°C in the dark, the glass slides were examined microscopically. GUS was a less useful tool for in situ monitoring of growth or structures of the introduced fungus under these experimental conditions, since addition of the X-Gluc substrate to glass slides resulted in some sample disruption. Low activity of GUS was also observed in most resting conidia and several hyphal segments. In hyphae of cotransformant ThzID1-M3 (Fig. 5D), chlamydospores were terminal or intercalary. Older hyphae appeared to be degraded partially or completely after forming a chlamydospore under the experimental conditions (Fig. 5E and F). These chlamydospores may play an important role as survival structures of introduced Trichoderma spp. in natural ecosystems (23), but little is known about their survival and ecological importance.
In conclusion, cotransformation with GFP and GUS provides a potentially useful tool for monitoring hyphal growth patterns and population dynamics of Trichoderma isolates introduced into natural systems and will help provide insight into the important abiotic and biotic factors affecting biocontrol efficacy. Significant biotic factors may include soil bacteria (3, 11, 15), indigenous soil fungi, and soil microfauna (4) that can influence the growth and efficacy of the introduced fungus. These are areas for future investigation.
ACKNOWLEDGMENTS
We thank D. Cullen and H. Leung for providing vectors. Matt Morra, Maury Wiese, and three anonymous reviewers provided helpful comments and criticisms.
Footnotes
Published as Idaho Agricultural Experiment Station paper 99712.
REFERENCES
- 1.Abd-El Moity T H, Papavizas G C, Shatla M N. Induction of new isolates of Trichoderma harzianum tolerant to fungicides and their experimental use for control of white rot of onion. Phytopathology. 1982;72:396–400. [Google Scholar]
- 2.Ahmad J S, Baker R. Rhizosphere competence of Trichoderma harzianum. Phytopathology. 1987;77:182–189. [Google Scholar]
- 3.Bin L, Knudsen G R, Eschen D J. Influence of an antagonistic strain of Pseudomonas fluorescens on growth and ability of Trichoderma harzianum to colonize sclerotia of Sclerotinia sclerotiorum in soil. Phytopathology. 1991;81:994–1000. [Google Scholar]
- 4.Bollen G J, Middelkoop J, Hofman T W. Effects of soil fauna on infection of potato sprouts by Rhizoctonia solani. In: Beemster A B R, Bollen G J, Gerlagh M, Ruissen M A, Schippers B, Tempel A, editors. Biotic interactions and soil-borne diseases. Proceedings of the First Conference of the European Foundation for Plant Pathology. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1991. pp. 27–34. [Google Scholar]
- 5.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 6.Cheng L, Fu J, Tsukamoto A, Hawley R G. Use of green fluorescent protein variants to monitor gene transfer and expression in mammalian cells. Nat Biotechnol. 1996;14:606–609. doi: 10.1038/nbt0596-606. [DOI] [PubMed] [Google Scholar]
- 7.Chet I. Trichoderma—application, mode of action, and potential as biocontrol agent of soilborne plant pathogenic fungi. In: Chet I, editor. Innovative approaches to plant disease control. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 137–160. [Google Scholar]
- 8.Chet I, Baker R. Isolation and biocontrol potential of Trichoderma hamatum from soil naturally suppressive to Rhizoctonia solani. Phytopathology. 1981;71:286–290. [Google Scholar]
- 9.Cormack B P, Bertram G, Egerton M, Gow N A R, Falkow S, Brown A J P. Yeast-enhanced green fluorescent protein (yEGFP): a reporter of gene expression in Candida albicans. J Gen Microbiol. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 10.Cubitt A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 11.Dandurand L M, Knudsen G R. Influence of Pseudomonas fluorescens on hyphal growth and biocontrol activity of Trichoderma harzianum in the spermosphere and rhizosphere of pea. Phytopathology. 1993;83:265–270. [Google Scholar]
- 12.Elad Y, Chet I, Henis Y. Biological control of Rhizoctonia solani in strawberry fields by Trichoderma harzianum. Plant Soil. 1981;60:245–254. [Google Scholar]
- 13.Elad Y, Hadar Y, Hadar E, Chet I, Henis Y. Biological control of Rhizoctonia solani by Trichoderma harzianum in carnation. Plant Dis. 1981;65:675–677. [Google Scholar]
- 14.Green H, Jensen D F. A tool for monitoring Trichoderma harzianum. II. The use of a GUS transformant for ecological studies in the rhizosphere. Phytopathology. 1995;85:1436–1440. [Google Scholar]
- 15.Hadar Y, Harman G E, Taylor A G. Evaluation of Trichoderma koningii and T. harzianum from New York soils for biological control of seed rot caused by Pythium spp. Phytopathology. 1984;74:106–110. [Google Scholar]
- 16.Harman G E, Chet I, Baker R. Factors affecting Trichoderma hamatum applied to seeds as a biocontrol agent. Phytopathology. 1981;71:569–572. [Google Scholar]
- 17.Hubbard J P, Harman G E, Hadar Y. Effect of soilborne Pseudomonas spp. on the biological control agent, Trichoderma hamatum, on pea seeds. Phytopathology. 1983;73:655–659. [Google Scholar]
- 18.Knudsen G R, Bin L. Effects of temperature, soil moisture, and wheat bran on growth of Trichoderma harzianum from alginate pellets. Phytopathology. 1990;80:724–727. [Google Scholar]
- 19.Knudsen G R, Eschen D J, Dandurand L M, Bin L. Potential for biocontrol of Sclerotinia sclerotiorum through colonization of sclerotia by Trichoderma harzianum. Plant Dis. 1991;75:466–470. [Google Scholar]
- 20.Kuter G A, Nelson E B, Hoitink H A J, Madden L V. Fungal population in container media amended with composted hardwood bark suppressive and conductive to Rhizoctonia damping-off. Phytopathology. 1983;73:1450–1456. [Google Scholar]
- 21.Lo C-T, Nelson E B, Hayes C K, Harman G E. Ecological studies of transformed Trichoderma harzianum strain 1295-22 in the rhizosphere and on the phylloplane of creeping bentgrass. Phytopathology. 1998;88:129–136. doi: 10.1094/PHYTO.1998.88.2.129. [DOI] [PubMed] [Google Scholar]
- 22.Migheli Q, Herrera-Estrella A, Avataneo M, Gullino M L. Fate of transformed Trichoderma harzianum in the phylloplane of tomato plants. Mol Ecol. 1994;3:153–159. [Google Scholar]
- 23.Papavizas G C. Trichoderma and Gliocladium: biology, ecology, and potential for biocontrol. Annu Rev Phytopathol. 1985;23:23–54. [Google Scholar]
- 24.Papavizas G C, Lewis J A, Abd-El Moity T H. Evaluation of new biotypes of Trichoderma harzianum for tolerance to benomyl and enhanced biocontrol capabilities. Phytopathology. 1982;72:126–132. [Google Scholar]
- 25.Papavizas G C, Lumsden R D. Improved medium for isolation of Trichoderma spp. from soil. Plant Dis. 1982;66:1019–1020. [Google Scholar]
- 26.Penttilä M, Nevalainen H, Rättö M, Salminen E, Knowles J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 1987;61:155–164. doi: 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- 27.Punt P J, Oliver R P, Dingemense M A, Pouwels P H, van den Hondel C A M J J. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene. 1987;56:117–124. doi: 10.1016/0378-1119(87)90164-8. [DOI] [PubMed] [Google Scholar]
- 28.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 29.Roberts I N, Oliver R P, Punt P J, van den Hondel C A M J J. Expression of Escherichia coli β-glucuronidase gene in industrial and phytopathogenic filamentous fungi. Curr Genet. 1989;15:177–180. doi: 10.1007/BF00435503. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Sheen J, Hwang S, Niwa Y, Kobayaski H, Galbraith D W. Green-fluorescent protein as a new vital marker in plant cells. Plant J. 1995;8:777–784. doi: 10.1046/j.1365-313x.1995.08050777.x. [DOI] [PubMed] [Google Scholar]
- 32.Shi Z, Christian D, Leung H. Enhanced transformation in Magnaporthe grisea by restriction enzyme mediated integration of plasmid DNA. Phytopathology. 1995;85:329–333. [Google Scholar]
- 33.Sivan A, Stasz T E, Hemmat M, Hayes C K, Harman G E. Transformation of Trichoderma spp. with plasmids conferring hygromycin B resistance. Mycologia. 1992;84:687–694. [Google Scholar]
- 34.Spellig T, Bottin A, Kahmann R. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol Gen Genet. 1996;252:503–509. doi: 10.1007/BF02172396. [DOI] [PubMed] [Google Scholar]
- 35.Stasz T E, Harman G E, Weeden N F. Protoplast preparation and fusion in two biocontrol stains of Trichoderma harzianum. Mycologia. 1988;80:141–150. [Google Scholar]
- 36.Thrane C, Lübeck M, Green H, Degefu Y, Allerup S, Thrane U, Jensen D F. A tool for monitoring Trichoderma harzianum. I. Transformation with the GUS gene by protoplast technology. Phytopathology. 1995;85:1428–1435. [Google Scholar]
- 37.Vanden Wymelenberg A J, Cullen D, Spear R N, Schoenike B, Andrews J H. Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. BioTechniques. 1997;23:686–690. doi: 10.2144/97234st01. [DOI] [PubMed] [Google Scholar]