Abstract
The human rhomboid-5 homolog-1 (RHBDF1) is a multi-transmembrane protein present mainly on the endoplasmic reticulum. RHBDF1 has been implicated in the activation of epidermal growth factor receptor (EGFR)–derived cell growth signals and other activities critical to cellular responses to stressful conditions, but details of this activation mechanism are unclear. Here, we report a RHBDF1 mRNA transcript alternative splicing variant X6 (RHBDF1 X6 or RHX6) that antagonizes RHBDF1 activities. We found that while the RHBDF1 gene is marginally expressed in breast tumor-adjacent normal tissues, it is markedly elevated in the tumor tissues. In sharp contrast, the RHX6 mRNA represents the primary RHBDF1 variant in normal breast epithelial cells and tumor-adjacent normal tissues but is diminished in breast cancer cells and tumors. We demonstrate that, functionally, RHX6 acts as an inhibitor of RHBDF1 activities. We show that artificially overexpressing RHX6 in breast cancer cells leads to retarded proliferation, migration, and decreased production of epithelial–mesenchymal transition-related adhesion molecules. Mechanically, RHX6 is able to inhibit the maturation of TACE, a protease that processes pro-TGFα, a pro-ligand of EGFR, and to prevent intracellular transportation of pro-TGFα to the cell surface. Additionally, we show that the production of RHX6 is under the control of the alternative splicing regulator RNA binding motif protein-4 (RBM4). Our findings suggest that differential splicing of the RHBDF1 gene transcript may have a regulatory role in the development of epithelial cell cancers.
Keywords: RHBDF1, RHX6, RBM4, EGFR, alternative splicing, breast cancer
Abbreviations: RHBDF1, rhomboid 5 homolog 1; RHX6, rhomboid 5 homolog 1, transcript variant X6; RBM4, RNA binding motif protein 4; EGFR, epidermal growth factor receptor; TACE, tumor necrosis factor alpha converting enzyme; pro-TGFα, pro-transforming growth factor α; HSC70, heat shock cognate protein 70; co-IP, co-immunoprecipitation; BafA, bafilomycin A; EMT, epithelial-mesenchymal transition
The human rhomboid-5 homolog-1 (RHBDF1) gene is a member of the rhomboid gene family (1, 2, 3). Members of this gene family can be divided into two categories based on whether or not they are proteolytically active. Proteolytically active rhomboid proteins are represented by Drosophila rhomboid-1, which is required for the processing of pro–epidermal growth factor (pro-EGF) to yield the mature ligand of epidermal growth factor receptor (EGFR), the latter being a known oncogene in epithelial cell cancers (4). Proteolytically inactive rhomboid proteins, referred to as inactive rhomboids (iRhoms), include RHBDF1 (5, 6, 7). RHBDF1 has been reported to take part critically in biological activities involved in cell survival (8), such as the mediation of G-protein–coupled receptor (GPCR) ligand-induced transactivation of EGFR-derived growth signals by facilitating the transportation of pro-TGFα, a pro-ligand of EGFR, from the endoplasmic reticulum to the cell surface (9, 10). RHBDF1 was also shown to be an essential component of a protein machinery pivotal to the maintenance of hypoxia-inducible factor-1α (HIF1α) in breast cancer cells under hypoxic conditions (11). Other activities of RHBDF1 include promoting the shedding of the TNF receptor by the protease ADAM17 (12), disrupting epithelial cell apicobasal polarity (13), as well as a regulatory role on proteasome activities under endoplasmic reticulum stress (14). RHBDF1 is also shown to promote fibrotic stroma growth in tumors by facilitating endothelial–mesenchymal transition (EndMT) (15). It thus seems plausible that RHBDF1 is a multifaceted protein that functions in assisting the activation of cell growth signaling pathways.
Alternative splicing is an essential mechanism of gene expression and function diversification (16, 17). In humans a great number of multiexon genes are known to produce differential splicing variants, often with different functions (18, 19). Alternative splicing events that occur in cancers may affect the functions of genes important to cancer disease development (20, 21). Denotations of the RHBDF1 gene transcript indicate different variants (22), including changes to the 5′ UTR, the amino terminal region, a segment in the middle of the protein, the carboxyl terminus, and the 3′ UTR of the transcript. It is unclear, however, whether these variants actually exist.
In this study, we demonstrate the existence of a RHBDF1 gene splicing variant, namely RHBDF1 transcript variant X6 (RHBDF1 X6 or RHX6). We report that the RHX6 transcript exists in breast cancer cells and tumors and that it is capable of inhibiting the functions of RHBDF1 in relation to EGFR activation. We show that the production of RHX6 is under the control of the splicing regulator RNA binding motif protein-4 (RBM4). These findings are consistent with the view that the functions of the RHBDF1 gene are subject to gene splicing regulations.
Results
Detection of RHBDF1 gene-splicing variant X6 in human breast cancer cells and clinical specimens
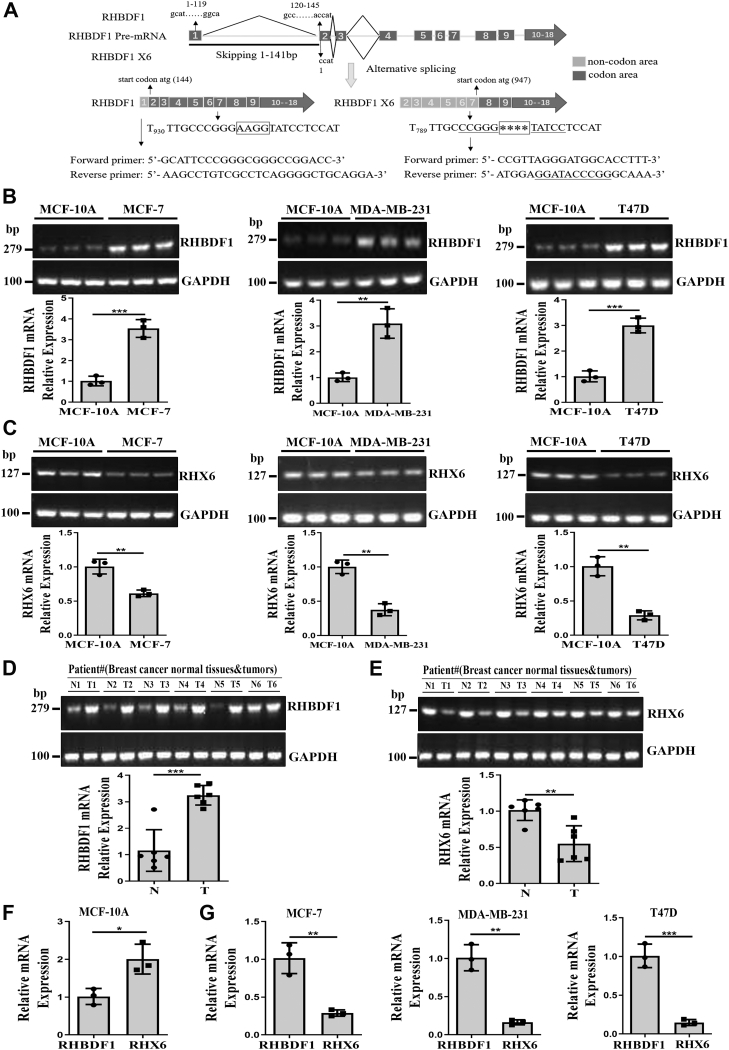
Analysis of database entries indicates that an alternative splicing variant of the RHBDF1 gene transcript exists (22). The variant, herein referred to as RHBDF1 X6 (RHX6), is missing exon 1 and part of exon 2 of the 18 exons in the full-length transcript. Translation of the RHX6 mRNA begins at exon 7, resulting in a protein that is 316 amino acid residues shorter at the N terminus compared to the 855 amino acid residues of RHBDF1. Additionally, compared with the sequence of the RHBDF1 mRNA (Fig. S1), four bases (AAGG) are missing in the RHX6 mRNA (Fig. 1A), providing an opportunity for specific identification. Note that this sequence is consistent with that documented previously (22); however, it is different from that of RHBDF1 (iRhom-1) variant X6 currently recorded in the NCBI database.
Figure 1.
Detection of variant RHX6 in human breast cancer cells and clinical specimens.A, RHBDF1 pre-mRNA undergoes alternative splicing to yield RHBDF1 mRNA and RHX6 mRNA. RHBDF1 mRNA contains all of the 1 to 18 exons, whereas RHX6 mRNA is missing exon 1 and part of exon 2. Translation RHX6 mRNA begins at exon 7. Four bases (AAGG) in the RHBDF1 mRNA are missing in the RHX6 mRNA (marked with asterisks ∗∗∗∗). The unique sequence at this site is utilized for PCR primers to detect RHX6 mRNA. B and C, RHBDF1 and RHX6 mRNA levels in MCF-7, MDA-MB-231, and T47D cells compared with MCF-10A cells (triplicated wells; experiment repeated three times). D and E, RHBDF1 and RHX6 mRNA levels in tumor tissues compared with adjacent normal tissues (number of clinical specimens n = 6; experiment repeated three times). F and G, RHX6 mRNA levels in MCF-10A, MCF-7, MDA-MB-231, and T47D cells compared with RHBDF1 mRNA levels (triplicated wells; experiment repeated three times). Data are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student's t test).
We determined the existence of RHBDF1 and RHX6 in three different breast cancer cell lines MDA-MB-231, MCF-7, and T47D in comparison with immortalized but nontumorigenic human breast epithelial cell line MCF-10A. RHBDF1 mRNA levels in MCF-7, MDA-MB-231, and T47D cells were significantly higher, about 3.5-, 3.1-, and 3-fold, respectively, than that in MCF-10A cells (Fig. 1B). In sharp contrast, RHX6 mRNA levels were markedly lowered in the cancer cells, about 61.1%, 37.5%, and 28.8%, respectively, of that in MCF-10A cells (Fig. 1C). We then determined RHBDF1 and RHX6 mRNA levels in six pairs of frozen specimens of breast cancer tissues and the corresponding adjacent normal tissues. We found that RHBDF1 mRNA levels on average were higher in the tumor tissues, about 2.8-fold of that in the adjacent normal tissues (Fig. 1D), whereas RHX6 mRNA levels were again lower in the tumor tissues, about 54.2% of that in the adjacent normal tissues (Fig. 1E). We then directly compared RHBDF1 and RHX6 expression in the same cells and found that RHX6 mRNA level was 2-fold of RHBDF1 mRNA levels in MCF-10A cells (Fig. 1F), whereas it was 28.4%, 16.1%, and 14.6% of RHBDF1 mRNA levels in MCF-7, MDA-MB-231, and T47D cells, respectively (Fig. 1G). These findings suggest that the process of RHBDF1 gene alternative splicing in normal breast epithelial cells and breast cancer cells give rise to opposite outcomes: RHX6 is more abundant in normal breast epithelial cells than in breast cancer cells, whereas RHBDF1 is more abundant in breast cancer cells than in normal breast epithelial cells.
Inhibitory effect of RHX6 overexpression on breast cancer cell migration, colony formation, proliferation, and adhesion molecule expressions
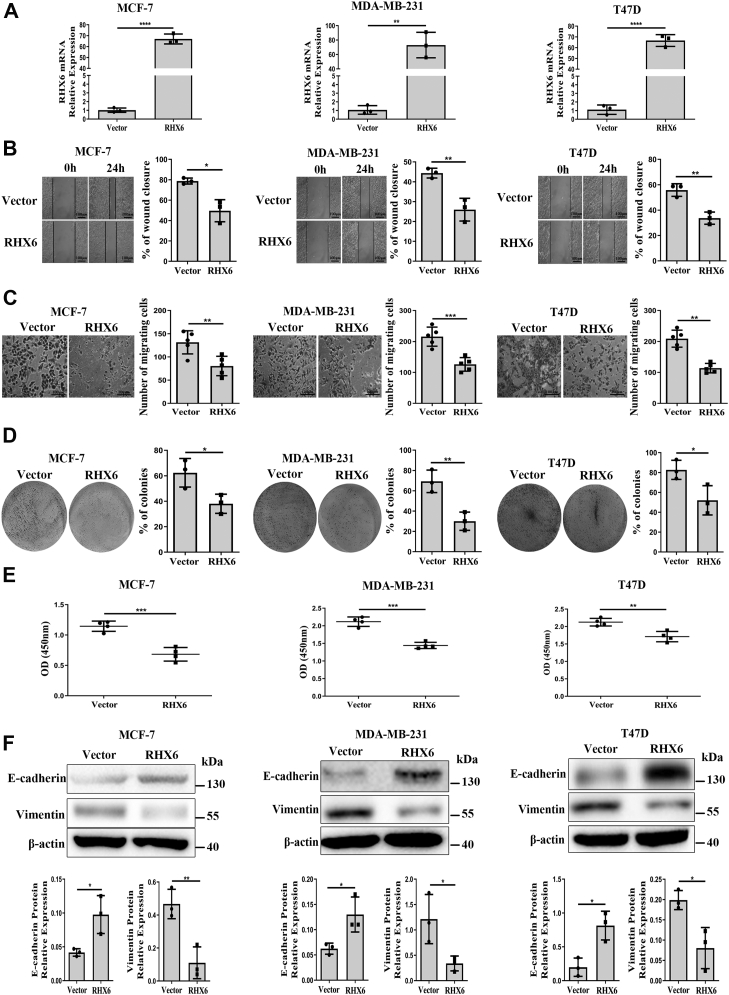
We linked the RHX6 variant to an enhanced GFP tag (Fig. S2, A and B) and transiently transfected breast cancer cells with the construct, which led to an increase of RHX6 mRNA levels to greater than 60 times compared to that in the empty vector–transfected cells (Fig. 2A). Results from an in vitro wound healing assay indicated that RHX6 overexpression led to an about 60% reduction of the wound closure rate (Fig. 2B). Additionally, by using an assay that measured the ability of the cells to infiltrate through polycarbonate-coated filters, we found that RHX6-overexpression in the cancer cells resulted in an about 60% reduction in the infiltration rates compared to that of the control cells (Fig. 2C). Moreover, we carried out a colony formation assay to measure the impact of RHX6 on the ability of the cells to undergo anchorage-independent growth and found that RHX6 overexpression in MCF-7, MDA-MB-231, and T47D cells resulted in substantially decreased extents of colony formation to 61.0%, 43.3%, and 62.9%, respectively, of that of the control cells (Fig. 2D). Furthermore, we determined the proliferation rates of RHX6-overexpressing MCF-7, MDA-MB-231, and T47D cells and found them to have decreased to 59.6%, 68.1%, and 80.5%, respectively, of that of the control cells (Fig. 2E). These findings indicate that raising RHX6 levels in breast cancer cells exerts an inhibitory effect on cell migration, colony formation, and proliferation.
Figure 2.
RHX6inhibitsthe migration, proliferation, and EMT in breast cancer cells.A, the mRNA levels of RHX6 in MCF-7, MDA-MB-231, and T47D cells transfected with RHX6 (triplicated wells; experiment repeated three times). B, the wound closure rate of MCF-7, MDA-MB-231, and T47D cells transfected with either vector or RHX6; the scale bar represents 100 μm; (experiment repeated three times). C, the number of migrating cells in MCF-7, MDA-MB-231, and T47D cells transfected with either vector or RHX6; the scale bar represents 100 μm; (experiment repeated three times). D, the percentage of colony formation of MCF-7, MDA-MB-231, and T47D cells transfected with either vector or RHX6 (Images of the whole plate were shown; experiment repeated three times). E, the absorbance value of MCF-7, MDA-MB-231, and T47D cells transfected with either vector or RHX6 (experiment repeated three times). F, the protein levels of E-cadherin and vimentin in MCF-7, MDA-MB-231, and T47D cells transfected with either vector or RHX6 (experiment repeated three times). Data are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student's t test). EMT, epithelial–mesenchymal transition.
Knowing that RHBDF1 promotes epithelial–mesenchymal transition (EMT) in colorectal cancer cells (23) and EndMT in breast cancer cells (15), we determined if raising RHX6 levels in breast cancer cells has any effect on EMT by measuring changes in cell adhesion molecules such as E-cadherin and vimentin. We found that E-cadherin protein levels increased by 2.3-, 2.1-, and 4.1-fold while vimentin protein levels decreased by 23.6%, 28.0%, and 40.4%, respectively, in RHX6-overexpressing MCF-7, MDA-MB-231, and T47D cells in comparison to that in the control cells (Fig. 2F). These findings suggest that RHX6 overexpression inhibits the ability of the cancer cells to undergo EMT.
Splicing regulator RBM4 is involved in the generation of RHBDF1 variants
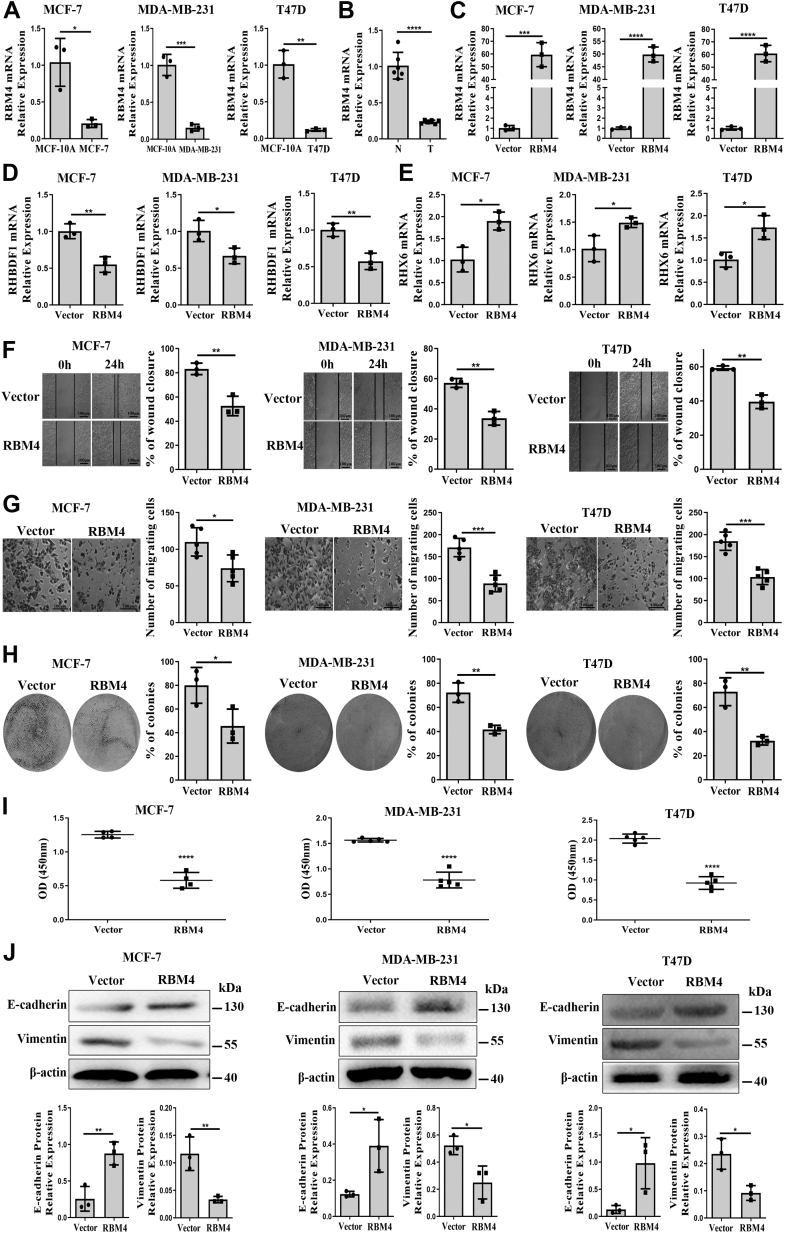
Since the splicing regulator RBM4 is known to have a major role in the regulation of alternative splicing (24), we determined the expression levels of RBM4 in breast cancer cells by using quatitative PCR (qPCR) in comparison with that in MCF-10A cells. We found that RBM4 mRNA levels in MCF-10A cells were considerably higher by 5-, 6.4-, and 8.7-fold, respectively, over that in MCF-7, MDA-MB-231, and T47D cells (Fig 3A). When repeating the experiments with breast cancer specimens, we found RBM4 levels in adjacent normal tissues to be 4.3-fold of that in tumor tissues (Fig. 3B). Additionally, we found that RBM4 overexpression in MCF-7, MDA-MB-231, and T47D cells (Fig. 3C) resulted in a substantial decrease of the RHBDF1 mRNA to 54.9%, 66.3%, and 66.7%, respectively, of that in the control cells (Fig. 3D). On the other hand, RBM4 overexpression led to a marked increase of the RHX6 mRNA to 1.9-, 1.5-, and 1.7-fold, respectively, in these cells over that in the control cells (Fig. 3E). Consistently, raising RBM4 levels in MCF-7, MDA-MB-231, and T47D cells led to a reduction of the closure rates in the wound healing assay to 63.2%, 59%, and 66.8%, respectively, of that the control cells (Fig. 3F). In good agreement, the infiltration assay showed that RBM4 overexpression caused a decrease of the migration rate of MCF-7, MDA-MB-231, and T47D cells to 67.3%, 52%, and 56.1%, respectively, over the control cells (Fig. 3G). Moreover, the abilities of colony formation of RBM4-overexpressing MCF-7, MDA-MB-231, and T47D cells decreased to 57.1%, 57.6%, and 44.3%, respectively, of that of the control cells (Fig. 3H). When assessed with proliferation assays, RBM4 overexpression in MCF-7, MDA-MB-231, and T47D cells led to a reduction of the net increase of cell numbers to 49%, 50%, and 45.4%, respectively, compared to that of the vector-control cells (Fig. 3I). Regarding EMT-related adhesion molecules, we found that RBM4 overexpression resulted in marked increased E-cadherin expression but significantly decreased vimentin expression (Fig. 3J). These data indicate that raising RBM4 levels is accompanied with the promotion of RHX6 production and the suppression of RHBDF1 production simultaneously.
Figure 3.
RBM4 increasesRHX6 expression and reduced breast cancer cells migration, proliferation and EMT.A, the mRNA levels of RBM4 in MCF-7, MDA-MB-231, and T47D compared with MCF-10A (triplicated wells; experiment repeated three times). B, the mRNA levels of RBM4 in tumor tissues compared with adjacent normal tissues. (number of clinical specimens n = 6; experiment repeated three times). C–E, RBM4, RHBDF1, and RHX6 mRNA levels in MCF-7, MDA-MB-231, and T47D cells transfected with vector or RBM4 (triplicated wells; experiment repeated three times). F, analyzed the wound closure rate of MCF-7, MDA-MB-231, and T47D cells, which overexpressed vector or RBM4; the scale bar represents 100 μm; (experiment repeated three times). G, analyzed the number of migrating cells in MCF-7, MDA-MB-231, and T47D cells, which overexpressed vector or RBM4; the scale bar represents 100 μm; (experiment repeated three times). H, analyzed the percentage of colony formation in MCF-7, MDA-MB-231, and T47D cells, which overexpressed vector or RBM4 (Images of the whole plate were shown; experiment repeated three times). I, analyzed the absorbance value of MCF-7, MDA-MB-231, and T47D cells, which overexpressed vector or RBM4 (experiment repeated three times). J, the protein levels of E-cadherin and vimentin in MCF-7, MDA-MB-231, and T47D cells after treatment with vector or RBM4 (experiment repeated tree times). Data are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student's t test). EMT, epithelial–mesenchymal transition.
RBM4 gene silencing results in suppression of RHX6 function
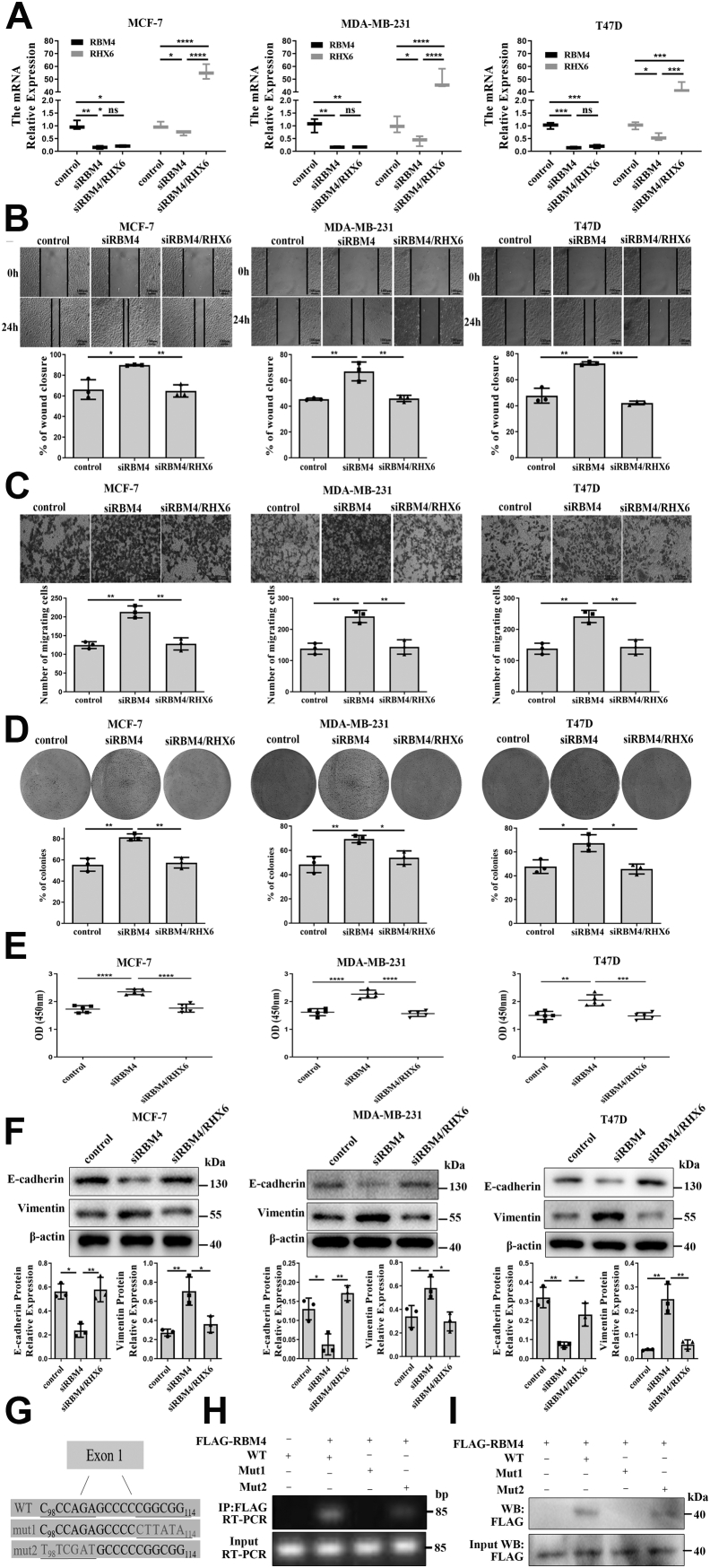
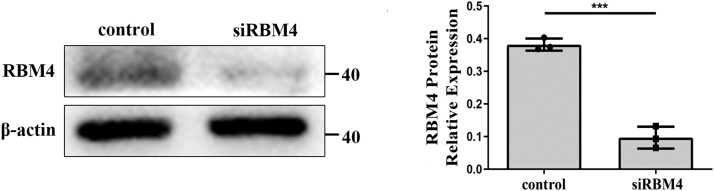
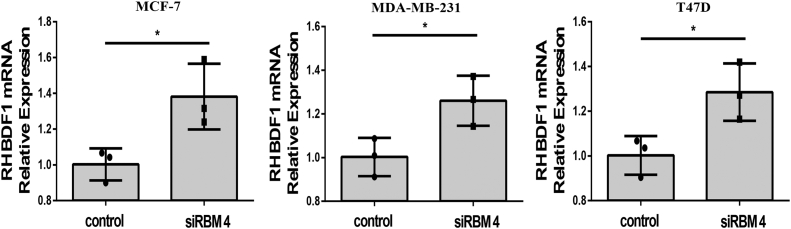
We treated the cancer cells with siRBM4 alone or together with a RHX6-overexpressing plasmid. The siRBM4 treatment led to an about 90% reduction of RBM4 mRNA compared to the results from scrambled siRNA treatment and was followed by a decline of 40 to 60% of RHX6 mRNA (Fig. 4A). Simultaneously, overexpressing RHX6, which led to an about 50-fold increase of RHX6, had little impact on RBM4 levels, however (Fig. 4A). Interestingly, RBM4 gene silencing gave rise to increased RHBDF1 levels by 130 to 140% compared to that in the control cells (Fig. S4). The cancer cells treated with siRBM4 displayed 1.5-fold increase of cell migration rates compared to control siRNA-treated cells. On the other hand, the migration rates of the cells simultaneously treated with siRBM4 and RHX6 were the same as that of control siRNA-treated cells (Fig. 4B). Similar effects were observed regarding the rates of cell infiltration and proliferation (Fig. 4, C–E), as well as the production of E-cadherin and vimentin (Fig. 4F). These findings indicate that inhibition of RMB4 gene expression results in a shift in RHBDF1 gene splicing toward declined RHX6 production but enhanced RHBDF1 production.
Figure 4.
RBM4 attenuatesmigration, proliferation, and EMT of breast cancer cells through RHX6.A, relative mRNA levels of RBM4 and RHX6 in MCF-7, MDA-MB-231, and T47D cells transfected with control siRNA, siRBM4, or both siRBM4 and a RHX6-expressing, as determined by qPCR (triplicated wells; experiment repeated three times). B, the wound closure rate of MCF-7, MDA-MB-231, and T47D cells treated with control siRNA, siRBM4, or both siRBM4 and a RHX6-expressing; the scale bar represents 100 μm; (experiment repeated three times). C, the number of migrating cells in MCF-7, MDA-MB-231, and T47D cells treated with control siRNA, siRBM4, or both siRBM4 and a RHX6-expressing; the scale bar represents 100 μm; (experiment repeated three times). D, the percentage of colony formation of MCF-7, MDA-MB-231, and T47D cells treated with control siRNA, siRBM4, or both siRBM4 and a RHX6-expressing (Images of the whole plate were shown; experiment repeated three times). E, the absorbance value of MCF-7, MDA-MB-231, and T47D cells treated with control siRNA, siRBM4, or both siRBM4 and a RHX6-expressing (experiment repeated three times). F, the protein levels of E-cadherin and vimentin in MCF-7, MDA-MB-231, and T47D cells treated with control siRNA, siRBM4, or both siRBM4 and a RHX6-expressing (experiment repeated three times). G, schematic representations of biotin-labeled single-stranded RNA molecules denoting the WT and the two mutants concerning RBM4-binding site CGGCGG. H, RIP experiments using 293T cells cotransfected with FLAG-RBM4 and one of the biotin-labeled single-stranded RNA molecules as indicated. IP: anti-FLAG antibody; RT-PCR: primers against one of the coprecipitated single-stranded RNA molecules (experiment repeated three times). I, reverse verification experiments using 293T cells transfected with FLAG-RBM4 and magnetic beads coated with biotin-labeled single-stranded RNA as indicated; FLAG-RBM4 detected by Western blotting analysis with an anti-FLAG antibody (experiment repeated three times). Data are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student's t test). EMT, epithelial–mesenchymal transition; RIP, RNA immunoprecipitation.
Interaction of RBM4 with the RHBDF1 gene transcript
It has been shown that a nucleotide motif CGGCGG is the potential binding site for RBM4 on gene transcripts (24, 25). This motif exists on exon-1 of the RHBDF1 gene transcript. We thus designed three biotin-labeled single-stranded RNA molecules resembling RHBDF1 pre-mRNA for RNA immunoprecipitation (RIP) experiments. The single-stranded RNA molecules contained, respectively, the wild-type CGGCGG sequence (WT), a replacement of the CGGCGG motif with a random sequence of CTTATA (mut1), and a random CCCAGA-to-TTCGAT mutation at a position five nucleotides ahead of the CGGCGG motif (mut2) (Fig. 4G). We then transfected 293T cells with FLAG-labeled RBM4 and one of the three RNA molecules. RIP experiments using cell lysates demonstrated that the RBM4 protein was able to co-immunoprecipitate with either WT or mut2 RNA but not mut1 RNA (Fig. 4H). The specificity of RBM4 binding to the CGGCGG motif was confirmed in a reverse verification experiment by subjecting the cell lysates of FLAG-RBM4-transfected 293T cells to immunoprecipitation with magnetic beads coated with one of the three biotin-labeled single-stranded RNA (Fig. 4I). These data demonstrate that the splicing regulator RBM4 is able to interact with the CGGCGG motif on the RHBDF1 gene transcript.
RHX6 inhibits RHBDF1-mediated activation of the EGFR signaling pathway
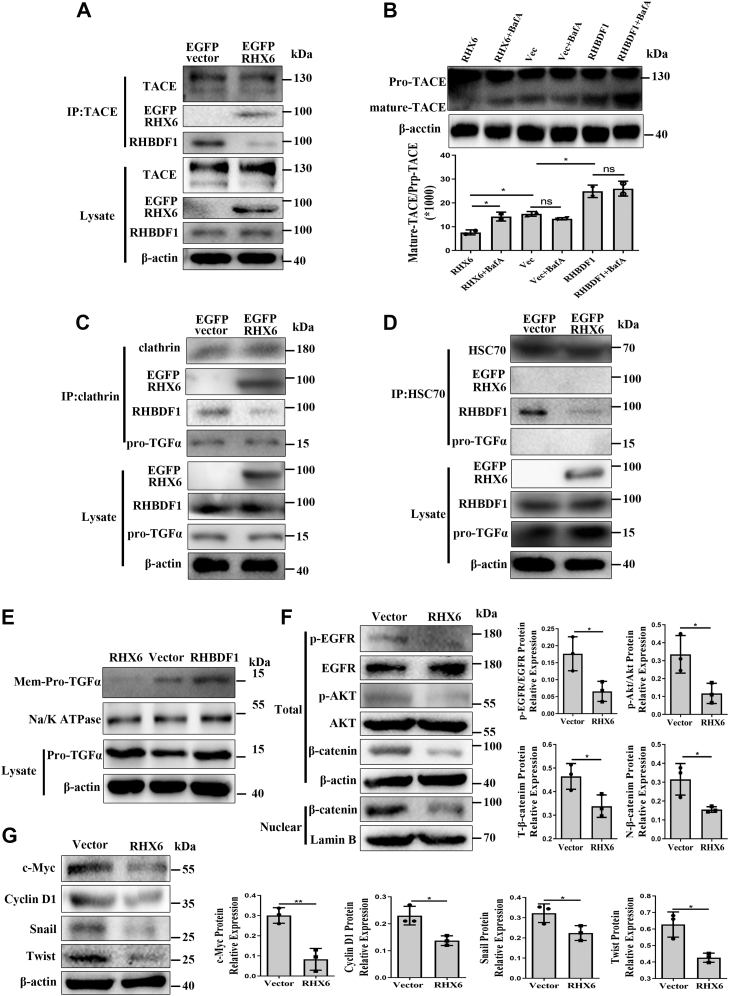
We determined whether RHX6 would compete with RHBDF1 to inhibit TNFα-converting enzyme (TACE) maturation and pro-TGFα membrane transport. Coimmunoprecipitation experiments showed that competition of RHX6 with RHBDF1 for binding to TACE led to declined production of mature TACE (Fig. 5A). We then cultured RHX6- or RHBDF1-transfected MCF-7 cells in the presence or absence of lysosomal inhibitor bafilomycin A (BafA). We found that RHBDF1 overexpression was associated with upregulated levels of mature TACE, whereas RHX6 overexpression resulted in lowered levels of mature TACE in comparison with that in the control cells (Fig. 5B). Additionally, BafA treatment gave rise to a significant enhancement of mature TACE protein production in RHX6-transfected cells but not in either RHBDF1- or mock-transfected cells (Fig. 5B). These findings suggest that RHX6 may have promoted lysosome degradation of TACE.
Figure 5.
RHX6 inhibits TACE maturation and pro-TGFα membrane transport.A, co-IP analyses the interaction of RHBDF1 and RHX6 with TACE in empty vector–transfected or RHX6-transfected MCF-7 cells. (experiment repeated three times). B, mature TACE protein levels in MCF-7 cells transfected with either empty vector, RHX6, or RHBDF1, and cultured for 4 h in the presence or absence of lysosomal inhibitor in triplicated wells (experiment repeated three times). C, co-IP analyses the interaction of RHBDF1 and RHX6 with clathrin in empty vector–transfected or RHX6-transfected MCF-7 cells. (experiment repeated three times). D, co-IP analyses the interaction of RHBDF1 and RHX6 with HSC70 in empty vector–transfected or RHX6-transfected MCF-7 cells. (experiment repeated three times). E, Western blotting analysis of membrane-associated pro-TGFα in empty vector–transfected, RHX6-transfected, or RHBDF1-transfected MCF-7 cells; Na/K ATPase marks the membrane preparation of the cells (experiment repeated three times). F and G, the protein levels of p-EGFR, EGFR, p-AKT, AKT, total β-catenin, nuclear β-catenin, c-myc, Cyclin D1, twist, and snail protein levels in MCF-7 cells transfected with either empty vector or RHX6 (experiment repeated three times). Data are mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (Student's t test). co-IP, co-immunoprecipitation; TACE, TNFα converting enzyme.
Furthering the study, we determined whether RHX6 would compete with RHBDF1 for binding to clathrin and HSC70, two proteins that are known to be critical in RHBDF1-facilitated TACE maturation and pro-TGFα transport (9). We found that clathrin was able to bind to RHX6 and pro-TGFα, and that overexpressing RHX6 led to diminished clathrin binding to RHBDF1 (Fig. 5C). Interestingly, although RHX6 did not seem to interact directly with HSC70 under the experimental conditions, we found that the extent of RHBDF1 binding to HSC70 significantly declined in RHX6-overexpressing cells (Fig. 5D). Moreover, the amount of pro-TGFα associated with Na/K-ATPase–marked cell membranes also decreased markedly in RHX6-overexpressing cells (Fig. 5E). Consistently, Western blotting analyses showed that the activation of the EGFR signaling pathway represented by EGFR and AKT phosphorylation was effectively inhibited in RHX6-overexpressing cells. Furthermore, the amount of total and nuclear β-catenin protein, which would undergo degradation when EGFR and AKT was not activated (26, 27, 28), also declined in RHX6-overexpressing cells (Fig. 5F). Consistently, the expression levels of the c-myc, Cyclin D1, Twist, and Snail genes, signals downstream of EGFR activation that promote cell migration and proliferation (29, 30), declined in RHX6-overexpressing cells (Fig. 5G). These findings suggest that RHX6 interfere with RHBDF1-facilitated TACE and pro-TGFα maturation and in turn inhibit RHBDF1-mediated activation of EGFR signals.
Discussion
Our findings indicate that, despite of a deletion of nearly one-third of the full length of the primary structure at the N terminus compared to that of the RHBDF1 protein, the RHX6 protein can interfere with the functions of RHBDF1, inhibiting RHBDF1 activities we measured that are associated with EGFR signal activation. It is expected that the expression patterns of RHBDF1 and RHX6 be correlated to each other as the results of alternative gene splicing; however, the specific patterns of the abundances of the two gene products reveal an underlying mechanism that may have a role in cancer development: RHX6 mRNA levels in breast cancer patients are higher in tumor-adjacent normal tissues than in tumor tissues, whereas RHBDF1 mRNA is nearly absent in normal tissues but is prominent in tumor tissues (8). Our study further reveals that the splicing regulator RBM4 takes part in the modulation of RHBDF1 gene splicing, with an outcome leaning toward enhanced RHX6 expression levels. This finding is of importance because it indicates a possibility that the RBM4-RHX6 axis be targeted in the development of new approaches aiming at managing EGFR activities in epithelial cancers.
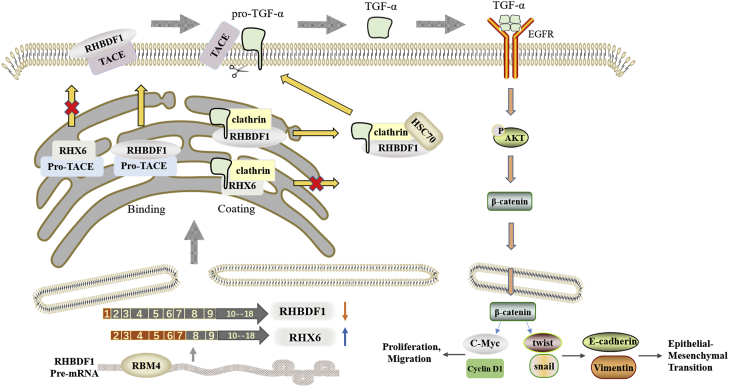
We focused on the activation of EGFR for an initial assessment of RHX6 function (Fig. 6). The maturation of TACE and the intracellular transportation of pro-TGFα are critical to the activation of the EGFR signaling pathway. We reported previously (9) that RHBDF1 is required in clathrin-coated vesicle (CCV)–dependent pro-TGFα membrane trafficking in breast cancer cells upon stimulation by GPCR agonists. Mechanistically, RHBDF1 interaction with Auxilin-2, a CCV protein, is required for the recruitment of HSC70 to CCV in order for clathrin uncoating to take place. Raising RHX6 protein levels in breast cancer cells prevents RHBDF1-assisted maturation of TACE. High RHX6 levels also lead to an inhibition of RHBDF1-facilitated CCV-dependent transport of pro-TGFα. In addition to EGFR activation, we and others have shown previously that RHBDF1 is involved in the maintenance of the stability of HIF1α in a hypoxic microenvironment (11), disruption of apicobasal polarity of breast epithelial cells (13), and promotion of EndMT (15). It would be of interest to find out if RHX6 has a potential role in competing with RHBDF1 in modulating these activities.
Figure 6.
Schematic representation of RHX6-mediated inhibition of the activation of EGFR signaling pathway. RHX6 interference with RHBDF1 binding to TACE and clathrin leads to diminished mature TACE. Clathrin bound to RHX6 is unable to recruit HSC70, thus unable to carry out pro-TGFα transportation to the cell surface, failing to activate EGFR signaling pathway. TACE, TNFα converting enzyme.
Our finding that the splicing factor RBM4 takes part in the regulation of RHBDF1 gene splicing is also of importance in advancing the understanding of breast cancer development. RBM4 is reported to have the role of a tumor suppressor (31, 32, 33), whose expression in tumors is subdued (34, 35). In good agreement with these findings, we demonstrate that silencing of the RBM4 gene results in downregulation of RHX6, along with enhancement in cancer cell proliferation, migration, and possibly EMT. On the other hand, artificially overexpressing RHX6 leads to marked inhibition of these activities. It is thus plausible that one mechanism by which RMB4 asserts its tumor-suppressing role is through differential splicing regulation of the RHBDF1 gene.
In summary, our findings are consistent with the view that RHBDF1 gene differential splicing, controlled by RBM4, results in a gene variant RHX6, which can inhibit GPCR ligand-initiated, RHBDF1-mediated EGFR activation. Since a large N-terminal section, nearly one-third of the full length of RHBDF1, is missing in RHX6, it is apparent that the structural moieties permitting RHX6 interference with RHBDF1 are composed by the rest two-third of the RHBDF1 protein. Further investigations on the relationship between RHX6 and RHBDF1 may give rise to in-depth insights into the development of breast cancer and other epithelial cancers.
Experimental procedures
Reagents and antibodies
Anti-phospho-EGFR (Tyr1068) (D7A5), EGFR (D38B1), TACE antibody, β-catenin (D10A8) XP rabbit mAb, Snail (C15D3) rabbit mAb, phospho-Akt (Thr450) (D5G4) rabbit mAb, Na/K-ATPase antibody #3010, E-cadherin (24E10) rabbit mAb #3195, and Akt antibody are purchased from Cell Signaling Technology. Anti-Twist antibody (Twist2C1a), anti-clathrin heavy chain (ab21679), and anti-IRHOM1 antibody are purchased from Abcam. HSC70 polyclonal antibody, c-myc mAb, cyclin D1 mAb, RBM4 polyclonal antibody, GFP tag polyclonal antibody, FLAG tag polyclonal antibody, beta actin mAb, lamin B1 polyclonal antibody, horseradish peroxidase–conjugated Affinipure goat antimouse IgG(H + L), horseradish peroxidase–conjugated Affinipure goat anti-rabbit IgG(H + L) are purchased from Proteintech. pCMV3-FLAG-RBM4 and pEGFP-C2 plasmids are purchased from Sino Biological and Clontech, respectively. Human-RBM4-siRNA is purchased from genepharma. Sulfo-NHS-LC-Biotin and Lipofectamine 2000 are purchased from Thermo Fisher Scientific. Magnetic beads are purchased from Bimake. PrimeSTAR Max DNA Polymerase and pMD 19-T Vector Cloning Kit are purchased from TaKaRa. FastPure Gel DNA Extraction Mini Kit is purchased from Vazyme. EndoFree Plasmid Mini Kit is purchased from CWBIO. XhoI, ACC65I, and T4 DNA ligase are purchased from NEB. Radioimmunoprecipitation assay (RIPA) buffer (high), PMSF, and Cocktail are purchased from Solarbio. Agarose, cell-counting kit-8, crystal violet staining solution, GelRed, BCA protein concentration determination kit, Nuclear and Cytoplasmic Protein Extraction Kit are purchased from Beyotime. EasyScript First-Strand complementary DNA (cDNA) Synthesis SuperMix and PerfectStart Green qPCR SuperMix are purchased from Transgen. Dulbecco's modified Eagle's medium (DMEM)/F-12, RPMI1640 medium, DMEM (high glucose), fetal bovine serum (FBS), horse serum, and trypsin are purchased from Gibco. Penicillin–streptomycin solution is purchased from Hyclone. Transwell plates of 8 μm pore size are purchased from Corning. BafA1 (#S1413) is purchased from SelleckChem. The three biotin-labeled single-stranded RNAs are synthesized by GENEWIZ.
Human breast tissue samples
We select six cases of primary breast cancer in the full information database of patients in the Department of Breast Cancer Pathology and Research Laboratory, Tianjin Medical University Cancer Hospital, Tianjin, China. All the patients do not undergo neoadjuvant chemotherapy and radiotherapy before surgical operation. The studies in this work are abide by the Declaration of Helsinki principles. The studies are approved by Tianjin Medical University Cancer Hospital of the review board and all breast cancer patients are fully informed and signed informed consent. All sections are diagnosed with invasive breast cancer by two senior pathologists. Pathologic diagnosis of five patients is invasive ductal carcinoma-no special type (IDC-NOS), and one patient is invasive ductal carcinoma-no special type (IDC-NOS) mixed with invasive micropapillary carcinoma. The detail clinicopathological information of six breast cancer patients are collected in the Table S1.
Molecular cloning
RHX6 transcript is obtained from MCF-10A cells by using PCR. The PCR reaction (94 °C for 2 min, 94 °C for 20 s, primer melting temperature (60–65 °C) for 15 s, 72 °C for 90 s, 35 cycles, 72 °C for 10 min, and 4 °C forever) is carried out with a S1000 Thermal Cycler from MCF-10A cDNA (300 ng) using cloning primers (10 μM). The PCR products are run on an agarose gel and visualized. The amplified gene is ligated into the T vector and sequenced. The primer sequences are as follows: RHX6 forward primer: TTCTTTGCCCGGGTATCC TCCA; reverse primer: TCAGTGGAGCTGAGCGTCCA.
The RHX6 CDS area is also obtained from MCF-10A cells. The PCR reaction (94 °C for 2 min, 94 °C for 20 s, primer melting temperature (60–65 °C) for 15 s, 72 °C for 90 s, 35 cycles, 72 °C for 10 min, and 4 °C forever) is carried out with a S1000 Thermal Cycler using cloning primers with unique restriction enzyme sites. The PCR products are run on an agarose gel and visualized. PCR products are then digested with enzymes corresponding to the unique restriction sites XhoI and ACC65I and ligated into the pEGFP-C2 vectors that are previously linearized using the same restriction enzymes. The same plasmid vector, pEGFP-C2 with a GFP tag, is used for all RHX6 overexpression studies. The primer sequences are as follows: forward primer: CCGCTCGAGCATGCTGCCCTTGGAGCGAGG; reverse primer: CGCGGTACCT CAGTGGAGCTGAGCGTCCAGTT.
Cell culture and transfection
Human MCF-10A, MCF-7, MDA-MB-231, T47D, and 293T cell lines are purchased from American Type Culture Collection (ATCC). MCF-10A is cultured in DMEM (F12) medium containing 20% (v/v) horse serum and penicillin (100 U/ml)–streptomycin (0.1 mg/ml) with 5% CO2 at 37 °C; MCF-7, T47D, HEK-293T, and MDA-MB-231 are cultured in DMEM and RPMI1640, respectively, containing 10%(v/v) FBS and penicillin (100 U/ml)–streptomycin (0.1 mg/ml) with 5% CO2 at 37 °C. The RHX6 and RBM4 overexpression and RBM4 knockdown cell lines are established by transiently transfecting the constructed RHX6 plasmids, the purchased RBM4 plasmids, and siRBM4 into the cells by Lipofectamine 2000. The medium is changed after 6 h, and the follow-up detection is performed after 24 to 48 h of transfection. The randomized siRNA sequence is GCGUACGCCUUACACCAUGAGUUAU. The siRBM4 sequence is described previously (36). Treatment of MCF-7 cells with siRBM4 resulted in an about 74% decrease of the RBM4 protein (Fig. S3).
Quantitative real-time PCR
Trizol is used to extract RNA from cultured cells or clinical tissues, then EasyScript First-Strand cDNA Synthesis SuperMix kit is used to reverse it into cDNA, and finally PerfectStart Green qPCR SuperMix for gene amplification is used. The primer sequences are as follows: RHX6 forward primer: CCGTTGGGATGGCACCTTT; reverse primer: ATGGAGGATACCCGGGCAAA. RHBDF1 forward primer: GCATTCCCGGGCGGGCCGGACC; reverse primer: AAGCCTGTCGCCTCAGGG GCTGCAGGA. RBM4 forward primer: GAGCGCTCGATGCCTACTAC; reverse primer: GGTCGGCAACAGGTGTCTAT. GAPDH forward primer: GCGATGCTGG CGCTGAGTAC; reverse primer: ATGATGACCCTTTTGGCTCCCC.
RT-PCR
The PCR reaction (94 °C for 2 min, 94 °C for 20 s, primer melting temperature (60–65 °C) for 15 s, 72 °C for 90 s, 35 cycles, 72 °C for 10 min, and 4 °C forever) is carried out with a S1000 Thermal Cycler from MCF-10A, MCF-7, MDA-MB-231, and T47D cDNA (300 ng) using qPCR primers (10 μM). The PCR products are run on an agarose gel and visualized.
Western blot analysis
Protein lysates are mixed with the loading buffer and heated at 100 °C for 5 min. The protein is separated by 10% SDS-PAGE electrophoresis and then transferred to the polyvinylidene difluoride membrane, blocked with 5% skimmed milk powder for 2 h, and incubated with the primary antibody overnight. The polyvinylidene difluoride membrane is incubated with the secondary antibody for 2 h at room temperature (RT), and the protein expression is detected by enhanced chemiluminescence solution. The expression is analyzed by Chemiluminescence imaging system (Tanon) and Gel-Pro Analyzer software V.4.0 (Media Cybernetics).
Wound-healing experiment assay
After 48 h of cell transfection, the degree of fusion basically is reached more than 90%. A 200 μl pipette tip is used to gently draw a horizontal line and a vertical line at the bottom. The cells are washed twice with PBS and then observed under a microscope. The pictures of scratch are taken at the junction of the horizontal and vertical lines. After 24 h, the scratch is observed and pictured at the same position. Analyzing is done with ImageJ (National Institutes of Health).
Cell migration assay
Cells are transfected as required and then digested 48 h later. The transwell chamber is placed in a 24-well plate, 200 μl of serum-free cell suspension is added to the upper chamber, and 700 μl of culture medium containing 10% FBS is added to the lower chamber. After 24 h, the culture medium is discarded and then the cells are fixed with 4% paraformaldehyde for 20 min, washed twice with PBS, and stained with 1% crystal violet for 30 min. After washing with PBS, the cells are wiped off in the upper chamber with a cotton swab and counted and observed under a microscope (Axiovert200M, Zeiss).
Colony formation assay
The cells are taken during the logarithmic growth phase and transfected as required every 72 h. The culture medium is discarded after 14 days, the cells are washed twice with PBS, fixed with 4% paraformaldehyde for 30 min, washed, and stained with 1% crystal violet staining solution for 30 min. Finally, the dye solution is washed, and pictures are taken and counted.
Cell proliferation assay
The cells are plated in a 96-well plate and transfected with the required plasmid and siRNA. After culturing for 48 h, 20 μl of cell-counting kit-8 is added to each well, incubated for 4 h in a cell incubator, and the absorbance value is measured at 450 nm wavelength with a Bio-Rad microplate reader.
RNA immunoprecipitation (RIP)
Thirty-seven percent formaldehyde is added to the cells transfected with WT RNA, RBM4 and WT RNA, RBM4 and mutant RNA1, and RBM4 and mutant RNA2 for crosslinking. After 10 min of slow rotation culture, glycine solution is added to 0.25 M to terminate the crosslinking. The cells are washed twice with precooled PBS and collected in a centrifuge tube. Centrifuge at 3000 rpm for 4 min at 4 °C. The cells are resuspended in RIPA containing protease inhibitors and placed on ice for later use. The protein A/G immunoprecipitation magnetic beads and FLAG antibody are rotated and incubated for 2 h at RT and then the beads are washed twice with RIPA containing protease inhibitors. Then, the antibody-coated beads are mixed with the protein lysis solution and incubated for 2 h with rotation at RT. The beads are washed three times with RIPA containing protease inhibitors and then incubated at 70 °C for 45 min to reverse crosslinking, and Trizol is added to extract RNA for subsequent verification.
RNA pull down
After washing the cells transfected with RBM4 three times with PBS, the cells are collected in a centrifuge tube. Centrifuge at 3000 rpm for 4 min at 4 °C. The cells are resuspended in RIPA containing protease inhibitors and placed on ice for later use. The Streptavidin Magnetic Beads are incubated with biotin-labeled WT RNA, mutant RNA1, and mutant RNA2 for 2 h at RT and then the beads are washed twice times. The RNA-binding beads are then mixed with the protein lysis solution and rotated at RT for 2 h. The beads are washed three times with RIPA containing protease inhibitors, then the protein loading buffer is added and heated at 100 °C for 10 min. After centrifugation, the supernatant is taken for protein detection.
Co-immunoprecipitation
The cells transfected with vector or RHX6 are lysed on ice with RIPA containing a cocktail of phosphatase inhibitors and PMSF. Magnetic beads are incubated with TACE, clathrin, and HSC70 antibodies for 2 h at RT with rotation. The magnetic beads are incubated and lysate with rotation at RT for 2 h; protein loading buffer is added and heated at 100 °C for 10 min and then SDS-PAGE and Western blot analysis are performed.
Biotinylation of cell plasma membrane proteins
The cells are washed with ice-cold PBS and then incubated with sulpho-NHS-SS-biotin–based Cell Surface Protein Isolation Kit for 15 min at 4 °C. The reaction is terminated with 100 mM glycine in PBS. The cells are rinsed with PBS and lysed in RIPA lysis buffer with added protease inhibitor cocktail. The lysates are centrifuged for 10 min at 13,000 rpm. Biotinylated cell membrane proteins are precipitated with Streptavidin Sepharose solutions. The pellets are gently rinsed with PBS, dissolved in SDS sample buffer, incubated at 65 °C for 10 min, and analyzed by Western blot.
Statistical analysis
The data are analyzed by analysis of a two-tailed Student's t test in GraphPad Prism 5.0 (GraphPad Software). Differences with p value < 0.05 are considered statistically significant.
Data availability
The data that support the findings of this study are available in the methods and/or supplementary material of this article.
Supporting information
This article contains supporting information (22).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
R. J. and L. L. conceptualization; R. J. and L. L. methodology; R. J., Y. C., and C. Z. validation; R. J. and H. Z. formal analysis; R. J., Q. S., J. Z., and L. F. resources; R. J., Y. S., and L. L. writing–original draft; L. L. funding acquisition.
Funding and addition information
This study was funded in part by grants from the National Natural Science Foundation of China, China (Grants 81874167 and 82073064 to L. Y. L.) and the Haihe Laboratory of Cell Ecosystem Innovation Fund.
Edited by Qi-Qun Tang
Supporting information
Supplemental Figure S1.
Supplemental Figure S2.
Supplemental Figure S3.
Supplemental Figure S4.
References
- 1.Freeman M. Rhomboids: 7 years of a new protease family. Semin. Cell Dev. Biol. 2009;20:231–239. doi: 10.1016/j.semcdb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Lemberg M.K., Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthew, Freeman Rhomboid proteases and their biological functions. Annu. Rev. Genet. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- 4.Sigismund S., Avanzato D., Lanzetti L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018;12:3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman M. The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu. Rev. Cell Dev. Biol. 2014;30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- 6.Freeman M. Rhomboids, signalling and cell biology. Biochem. Soc. Trans. 2016;44:945–950. doi: 10.1042/BST20160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dulloo I., Muliyil S., Freeman M. The molecular, cellular and pathophysiological roles of iRhom pseudoproteases. Open Biol. 2019;9:190003. doi: 10.1098/rsob.190003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Z., Zou H., Tian F., Grandis J.R., Mixson A.J., Lu P.Y., et al. Human rhomboid family-1 gene silencing causes apoptosis or autophagy to epithelial cancer cells and inhibits xenograft tumor growth. Mol. Cancer Ther. 2008;7:1355–1364. doi: 10.1158/1535-7163.MCT-08-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Bai T.R., Gao S., Zhou Z., Peng X.M., Zhang L.S., et al. Human rhomboid family-1 modulates clathrin coated vesicle-dependent pro-transforming growth factor α membrane trafficking to promote breast cancer progression. EBioMed. 2018;36:229–240. doi: 10.1016/j.ebiom.2018.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou H., Thomas S.M., Yan Z.-W., Grandis J.R., Vogt A., Li L.-Y. Human rhomboid family-1 gene RHBDF1 participates in GPCR-mediated transactivation of EGFR growth signals in head and neck squamous cancer cells. FASEB J. 2009;23:425–432. doi: 10.1096/fj.08-112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Z., Liu F., Zhang Z.-S., Shu F., Zheng Y., Fu L., et al. Human rhomboid family-1 suppresses oxygen-independent degradation of hypoxia-inducible factor-1 in breast cancer. Cancer Res. 2014;74:2719–2730. doi: 10.1158/0008-5472.CAN-13-1027. [DOI] [PubMed] [Google Scholar]
- 12.Maney S.K., McIlwain D.R., Polz R., Pandyra A.A., Sundaram B., Wolff D., et al. Deletions in the cytoplasmic domain of iRhom1 and iRhom2 promote shedding of the TNF receptor by the protease ADAM17. Sci. Signal. 2015;8:ra109. doi: 10.1126/scisignal.aac5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng X.M., Shan G., Deng H.T., Cai H.X., Zhou Z., Rong X., et al. Perturbation of epithelial apicobasal polarity by rhomboid family-1 gene overexpression. FASEB J. 2018;32:5577–5586. doi: 10.1096/fj.201800016R. [DOI] [PubMed] [Google Scholar]
- 14.Lee W.J., Kim Y.D., Park J., Shim S.M., Lee J., Hong S.H., et al. iRhom1 regulates proteasome activity via PAC1/2 under ER stress. Sci. Rep. 2015;5:11559. doi: 10.1038/srep11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao S., Zhang L.S., Wang L., Xiao N.N., Zhang Z.S. RHBDF1 promotes AP-1-activated endothelial-mesenchymal transition in tumor fibrotic stroma formation. Signal. Transduct. Target. Ther. 2021;6:273. doi: 10.1038/s41392-021-00597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelemen O., Convertini P., Zhang Z., Wen Y., Shen M. Function of alternative splicing. Gene Amsterdam. 2013;514:1–30. doi: 10.1016/j.gene.2012.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H.K., Pham M.H.C., Ko K.S., Rhee B.D., Han J. Alternative splicing isoforms in health and disease. Pfluegers Archiv: Eur. J. Physiol. 2018;470:995–1016. doi: 10.1007/s00424-018-2136-x. [DOI] [PubMed] [Google Scholar]
- 18.Yang X., Coulombe-Huntington J., Kang S., Sheynkman G.M., Vidal M. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016;164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y., Rio D.C. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oltean S., Bates D.O. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33:5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 21.Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int. J. Cell Biol. 2013;2013:463786. doi: 10.1155/2013/463786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshua P., Kenton K. Alternative splice variants of rhomboid proteins: comparative analysis of database entries for select model organisms and validation of functional potential. F1000Res. 2018;7:139. doi: 10.12688/f1000research.13383.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan H., Wei R., Xiao Y., Song Y., Wang J., Yu H., et al. RHBDF1 regulates APC-mediated stimulation of the epithelial-to-mesenchymal transition and proliferation of colorectal cancer cells in part via the Wnt/β-catenin signalling pathway. Exp. Cell Res. 2018;368:24–36. doi: 10.1016/j.yexcr.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Chen D., Qian H., Tsai Y.S., Shao S., Liu Q., et al. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer cell. 2014;26:374–389. doi: 10.1016/j.ccr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Y., Yu J., Han W., Fan X., Qian H., Wei H., et al. A splicing isoform of TEAD4 attenuates the Hippo–YAP signalling to inhibit tumour proliferation. Nat. Commun. 2016;7 doi: 10.1038/ncomms11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anju A., Kingshuk D., Natalia L., Swati S., Muzaffer C., Graham C., et al. The AKT/I kappa B kinase pathway promotes angiogenic/metastatic gene expression in colorectal cancer by activating nuclear factor-kappa B and beta-catenin. Oncogene. 2005;24:1021–1031. doi: 10.1038/sj.onc.1208296. [DOI] [PubMed] [Google Scholar]
- 27.Sharma M., Chuang W.W., Sun Z. Phosphatidylinositol 3-kinase/akt stimulates androgen pathway through GSK3β inhibition and nuclear β-catenin accumulation. J. Biol. Chem. 2002;277:30935–30941. doi: 10.1074/jbc.M201919200. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki M., Shigematsu H., Nakajima T., Kubo R., Motohashi S., Sekine Y., et al. Synchronous alterations of Wnt and epidermal growth factor receptor signaling pathways through aberrant methylation and mutation in non small cell lung cancer. Clin. Cancer Res. 2007;13:6087–6092. doi: 10.1158/1078-0432.CCR-07-0591. [DOI] [PubMed] [Google Scholar]
- 29.Shang S., Hua F., Hu Z.W. The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8:33972–33989. doi: 10.18632/oncotarget.15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez D.M., Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.None RBM4-regulated alternative splicing suppresses tumorigenesis. Cancer Discov. 2014;4:1253. doi: 10.1158/2159-8290.CD-RW2014-198. [DOI] [PubMed] [Google Scholar]
- 32.Yong H., Zhao W., Zhou X., Liu Z., Tang Q., Shi H., et al. RNA-binding motif 4 (RBM4) suppresses tumor growth and metastasis in human gastric cancer. Med. Sci. Monit. 2019;25:4025–4034. doi: 10.12659/MSM.914513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W.Y., Quan W., Yang F., Wei Y.X., Chen J.J., Yu H., et al. RBM4 modulates the proliferation and expression of inflammatory factors via the alternative splicing of regulatory factors in HeLa cells. Mol. Genet. Genomics. 2020;295:95–106. doi: 10.1007/s00438-019-01606-3. [DOI] [PubMed] [Google Scholar]
- 34.Yong H., Zhu H., Zhang S., Zhao W., Wang W., Chen C., et al. Prognostic value of decreased expression of RBM4 in human gastric cancer. Sci. Rep. 2016;6:28222. doi: 10.1038/srep28222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Yong H., Fu J., Gao G., Fu M. miR-504 promoted gastric cancer cell proliferation and inhibited cell apoptosis by targeting RBM4. J. Immunol. Res. 2021;2021:5555950. doi: 10.1155/2021/5555950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su C.H., Hung K.Y., Hung S.C., Tarn W.Y. RBM4 regulates neuronal differentiation of mesenchymal stem cells by modulating alternative splicing of pyruvate kinase M. Mol. Cell Biol. 2017;37 doi: 10.1128/MCB.00466-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in the methods and/or supplementary material of this article.