Abstract
On the basis of the knowledge that the proline-rich hot spot PPPRPP region of P(151)PSNPPPRPP(160), an oligopeptide derived from the cytosolic portion of p22phox (p22), binds to the single functional bis-SH3 domain of the regulatory protein p47phox (p47), we designed a mimetic of the tripeptide PPP based on NMR and X-ray crystallographic data for the p22(151−161) peptide PPSNPPPRPPA with a peptide construct. Incorporation of the synthetic pseudo-triproline mimetic Pro-Pro-Cyp in a molecule derived from molecular modeling studies led to only a 7-fold diminution in activity in a surface plasmon resonance assay relative to the same molecule containing the natural Pro-Pro-Pro tripeptide. The alternative sequence corresponding to a Pro-Cyp-Pro insertion was inactive. This is a first example of the use of a triproline mimetic to interfere with the formation of the p47–p22 complex, which is critical for the activation of NOX, leading to the production of reactive oxygen species as superoxide anions.
Keywords: Triproline mimetic, protein−protein interaction, cytosolic proteins, hot spots
The NADPH oxidases (reduced forms of nicotinamide adenine dinucleotide phosphate oxidases) are a family of membrane-associated multicomponent enzymes present in phagocytes and macrophages.1−5 Also known as NOX, they are widely distributed in eukaryotic organisms and play a crucial role in maintaining the delicate balance of physiological redox processes responsible for the maintenance of host defense mechanisms against invading pathogens as well as in recruiting inflammatory cells to the site of infection.6−8 This is accomplished through reduction of molecular oxygen across membranes and release of reactive oxygen species (ROS) in the form of superoxide anions (O2•–) that destroy the biochemical machinery of invading microorganisms.9 Among the seven isoforms of these enzymes, NOX2 (NADPH 2, or gp91phox), a phagocystic oxidase, has garnered much attention because of its ubiquitous presence in tissues and organs, where ROS production can trigger physiological responses that ultimately lead to innate and adaptive immunity.10
NADPH oxidase comprises six subunits, including the trans membrane p22 and NOX2, which in combination with flavocytochrome becomes the catalytic site of the oxidase complex. Regulatory cytosolic proteins such as p47 undergo a series of phosphorylations, eventually forming reversible contacts with other multidomain proteins.11
NOX2 becomes catalytically active in vivo when transmembrane protein p22 binds to the single functional bis-SH3 domain of the regulatory protein p47, leading to an enzyme complex that enhances the production of ROS.12−14 Under normal physiological conditions, ROS production mediated by NOX2 is highly regulated to defend against invading pathogens without causing damage to host tissue. Upon the action of external stimuli, the dormant or resting-state form of NADPH oxidase is activated through a series of reversible protein–protein interactions (PPIs) among its regulatory proteins to produce ROS in such a manner that host tissue is not damaged.15 However, the beneficial gatekeeper role of NOX2 toward external harmful stimuli can be jeopardized by the overproduction of ROS under certain pathological conditions, causing oxidative stress and a cascade of physiological events resulting in serious diseases such as cancer, cystic fibrosis, schizophrenia, Alzheimer’s disease, and a host of cardiac- and lung-related diseases.1,16−19 Since the activation of the catalytic site of the NOX2 enzyme complex involves the interaction of the essential regulatory p47 and p22 proteins to produce ROS in an inducible manner, it has been suggested that interfering with this specific PPI can be exploited to render NOX2 as a druggable target.1,20,21
Developing small-molecule inhibitors of p47 presents obvious challenges because of the relatively large surfaces of the interacting proteins at the site of contact.22 Interestingly, a number of natural products such as apocynin, celastrol, and glyotoxin have been reported as NOX2 inhibitors.1,21 Among the synthetic compounds, ebselen, a selenoindazole discovered by high-throughput screening, continues to attract attention.20 PR-39, a peptide of bacterial origin, has been reported to inhibit NADPH oxidase activity by binding to the Src homology domains of p47.23 A series of pyrazolopyridinediones showed selective NOX4 activity but were inert toward NOX2.24 Recently, dimeric 2-aminoquinolines bridged by a ethylenedioxa ether linker exhibiting in vitro Ki activity in the 20 μM range were obtained using a fragment-based approach.25
Interfering with PPIs through the use of small peptides or peptidomimetics has been a successful strategy in drug discovery.26 In such cases, identifying so-called “hot spots” in protein partners engaged in PPIs has been the norm. Given the available structural information related to NOX, we considered a peptidomimetic approach to interfere with the PPI of p47 with p22 as a proof of concept.
The structure of p47 has been investigated by elegant X-ray crystallographic studies27 and in solution by NMR spectroscopy.22 Valuable insights into the conformation of p47 have identified critical regions for recognition, and relevant amino acid sequences, including proline-rich domains (PRDs), have been proposed. The structure of a p47–p22 complex comprising the conserved tandem SH3 domain of p47 bound to an oligopeptide P(151)PSNPPPRPP(160) derived from the cytosolic portion of p22 shows that binding involves the proline-rich sequence PPPRPP of p22.28
The prospects of developing a small-molecule inhibitor of the PPIs between p47 and the hotspots in p22 to control overproduction of ROS represents a major challenge because of the enormous conformational change of the cytosolic p47 component. In fact, p47 needs to adopt an activated conformation by phosphorylation of serine residues in order to bind to p22 prior to translocation of the complex from the cytosol to the membrane.28 As a rule, developing peptidomimetics of peptides with fewer than six residues is a good starting point, whereas peptide sequences consisting of 6–15 residues are considered challenging.29,30
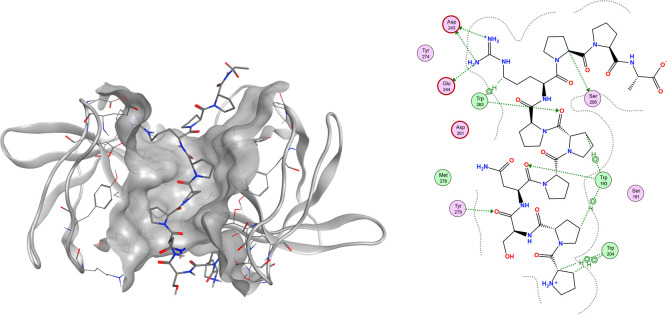
To optimize the bioactive peptide sequence, the hot spots in the 16 amino acid sequence in p22(151–166) that would potentially interact with regions of p47 were identified using X-ray crystallography, surface plasmon resonance (SPR), and an Ala scan. A cocrystal structure of the extended conformation of p47 with the truncated undecapeptide PPSNPPPRPPA (1) is shown in Figure 1 (PDB: 7YXW).
Figure 1.
Structure of p22(151–161) peptide PPSNPPPRPPA (1) cocrystallized with a tandem protein construct.
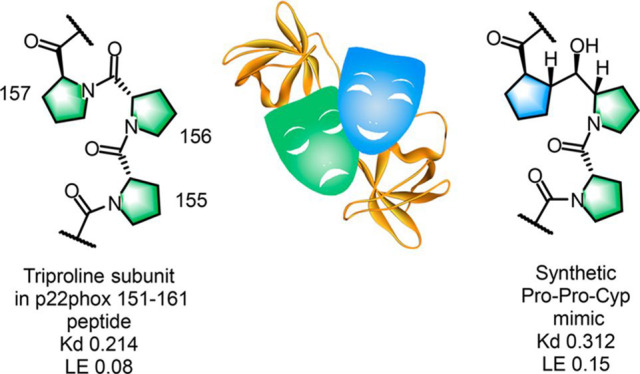
To improve the crystallizability of the protein and to avoid the domain swap observed by Rittinger,28 a single A200G mutation was introduced. The N-terminal Pro-Pro (151–152) is involved in a hydrophobic interaction with Trp(204) and Trp(193), while the Ser-Asn (153–154) residue acts more like a linker to the central triproline unit. Of particular interest is the spatial alignment of the Pro-Pro-Pro (155–157) triad, which is engaged in hydrophobic and hydrogen-bonding interactions with Trp(193) and Trp(263), respectively.
To determine the individual roles of the nine central amino acids in the 16 amino acid sequence PPSNPPPRPPAEARKK, we conducted an Ala scan from Ser(153) to Glu(162) in conjunction with binding affinity and free enthalpy changes. We were pleased to find that the PPPR(155–158) sequence was the hot spot of p22, and hence, it was considered as a suitable candidate for peptidomimetic design. Furthermore, the spatial disposition of the central Pro(156) within the triproline triad and the ionic interaction of Arg(158) with Asp(243) and Glu(244) were critical in the conceptual design of the intended triproline mimetic. The Ala scan revealed that replacement of Pro(159) in the C-terminal part of PPSNPPPRPPAEARKK has no impact on binding, while Pro(160) has an important role in occupying a hydrophobic region. Taken together, these studies indicated that the pentapeptide EARKK could be removed, leaving the proline-rich undecapeptide PPSNPPPRPPA (1) representing p22(151–161) as a truncated construct. Having garnered sufficient structural information from the X-ray and Ala scan studies, we considered undecapeptide 1 to be a starting point in search of truncated analogues and potential small-molecule peptidomimetics. The main challenge was to replace the natural amino acid sequence of the PPPR(155–158) hot spot involved in most of the essential interactions with p47 with a mimetic molecule that would adopt a well-defined geometry in the binding groove comprising two SH3 domains.
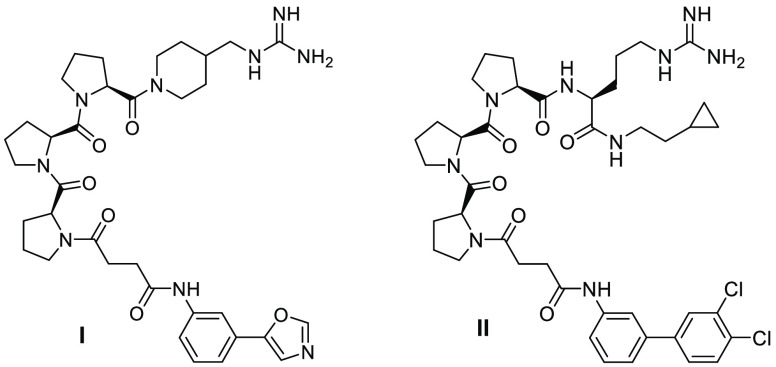
On the basis of these results and our understanding of the role of each amino acid, the following structural optimizations were performed to design a truncated peptidomimetic (data not shown): Pro(151) and Pro(152) were replaced by lipophilic groups; Ser(153) and Asp(154) were replaced by a suitable linker; Arg(158) was truncated or left unmodified; and Pro(159) and Pro(160) were replaced by lipophilic side chains. Two of the optimized triproline peptidomimetics are presented as I and II in Figure 2. The substantial reduction of molecular weight and the maintained affinity of II resulted in a significantly better ligand efficiency compared with the p22(151–166) peptide (vide infra). Compound II was chosen as a surrogate for further modifications.
Figure 2.
Structures of triproline mimetics I and II.
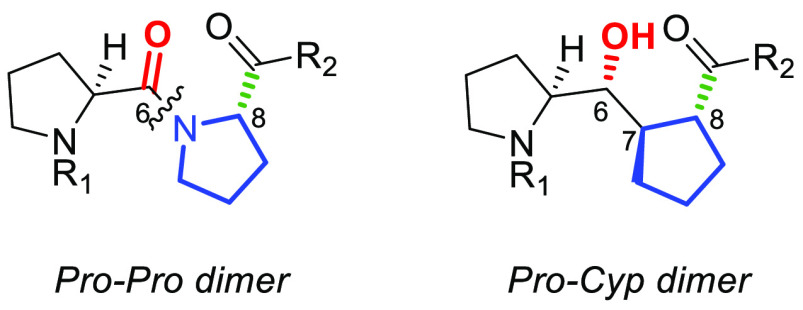
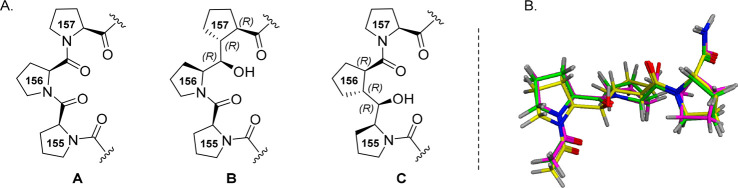
As part of our ongoing interest in the design and synthesis of proline-derived motifs and peptidomimetics,31−35 we recently reported the stereocontrolled synthesis of a series of diastereomeric diproline (Pro-Pro) mimetics represented by a generic pseudo-diproline dimeric Pro-Cyp structure (Figure 3).36 The conceptual design element considered replacing the second Pro in the Pro-Pro dimer with a cyclopentanecarboxylic acid (Cyp) in which the stereochemical combinations of three contiguous stereogenic centers were unambiguously established. The use of Cyp as a proline surrogate has been previously reported sporadically as an effort to simulate a diproline motif, but seldom in the context of a medicinally relevant target.37−44 For the purposes of a triproline mimetic for the Pro-Pro-Pro (155–157) sequence in undecapeptide 1 representing p22 (Figure 1), we envisaged two variants, one in which the Cyp unit would replace the C-terminal Pro(157) (B) and another in which the Cyp unit would replace the central Pro(156) (C) (Figure 4A). Superposition of the Pro-Pro-Pro subunit (A) representing the natural PPP segment with the two pseudo-triproline motifs B and C showed excellent overlap (Figure 4B).
Figure 3.
Pro-Pro and pseudo-diproline Pro-Cyp motifs.
Figure 4.
(A) Structures of Pro-Pro-Pro trimer A, Pro-Pro-Cyp trimer B, and Pro-Cyp-Pro trimer C. (B) Gas-phase optimized geometries of A (green), B (magenta), and C (yellow). Oxygen atoms ares shown in red and nitrogen atoms in blue.
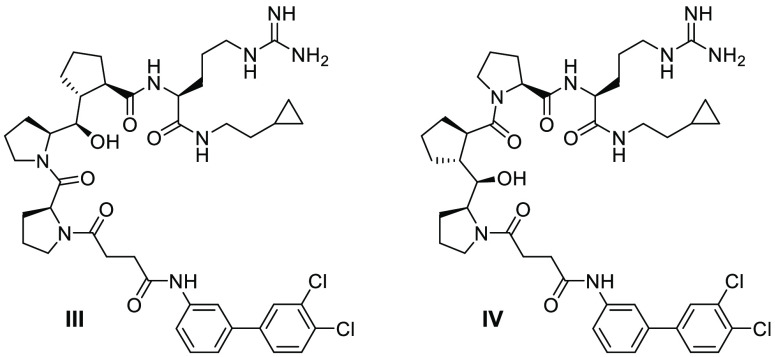
The appropriate stereoisomer of Cyp was selected on the basis of docking the candidate compounds into p47 protein. The Cyp diproline unit was placed in the two possible positions corresponding to the triproline core of compound II. All possible diastereomers were modeled, and thus, a total of 16 isomers were docked. Compound II maintains all of the specific and necessary hydrogens bonds and salt bridges as p22(151–161). The cyclopropylethyl chain in II replacing the last proline in undecapeptide 1 sits on a hydrophobic surface. The succinamide moiety takes the place of the first four amino acids of the undecapeptide, while the appended 3,4-dichlorobiphenyl ring is well-positioned relative to the hydrophobic surface formed by Trp(204) and Trp(193). The Pro-Pro-Cyp 2S,6R,7R,8R isomer III, replacing both Pro-Pro(155–156) and Pro-Pro(156–157), maintained the helical structure and most of the specific interactions with the protein. It was therefore chosen for synthesis and evaluation. When Pro-Pro(155–156) was replaced, rotation of the amide bond between Cyp and the linker was observed in some of the docking poses as a result of the subtle conformation change of Cyp compared with diproline, causing strain on the linker. In either case, the Cyp-containing pseudo-triproline unit would expose a hydrophobic surface similarly to proline and provide possible polar interactions with the stereodefined OH group replacing an amide carbonyl. Preliminary data on the two pseudo-triproline analogues I and II as internal reference compounds revealed that the latter would be a better analogue to use as a control. Pro-Pro-Cyp B and Pro-Cyp-Pro C motifs were elaborated to include an N-terminal 3,4-dichlorobiphenyl succinamide and a C-terminal hydrophobic cyclopropylethylarginine amide, leading to the fully decorated analogues III and IV, respectively (Figure 5), each having a 6R,7R,8R stereochemistry comprising three contiguous stereogenic centers.
Figure 5.
Structures of triproline mimetics III and IV.
We were pleased to find that analogue III, in which Pro(157) was replaced by a cyclopentanecarboxamide, was only 7 times less active in the SPR assay relative to triproline analogue II. The SPR ligand efficiencies (SPR LEs) of control analogue II and triproline mimetic analogue III were comparable to that of p22(151–166). In contrast, analogue IV was totally inactive. Thus, replacement of the pivotal Pro(157) in undecapeptide model 1, a surrogate for p22, with a cyclopentanecarboxamide in the form of synthetic mimetic III does not negatively affect the PPI with p47. Moreover, it appears that the presence of the middle Pro(156) in the Pro-Pro-Cyp triad in III is essential for activity (Figures 5, 6 and Table 1). The replacement of the Pro(156) amide carbonyl in the PPPR segment of undecapeptide 1 with an R-configured secondary alcohol facing the solvent in the active analogue III is beneficial, since it does not disrupt the specific hydrogen-bonding interactions in the complex.
Figure 6.
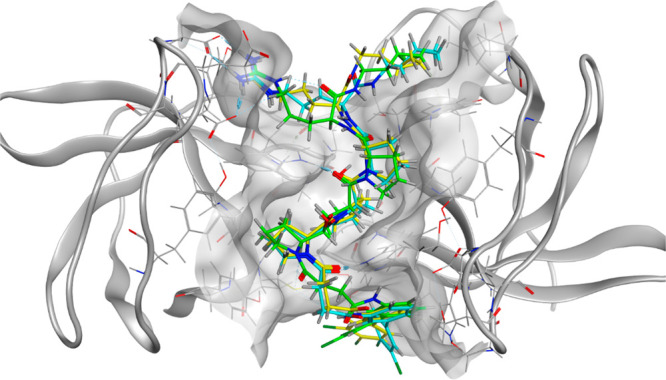
Superimposition of pseudopeptides II–IV in the p47 SH3 domain.
Table 1. Experimental Affinities of Triproline Mimetic Analogues.
| compound | SPR Kd (μM) | SPR LE |
|---|---|---|
| p22(151–166) | 0.214 | 0.08 |
| I | 5.71 | 0.15 |
| II | 0.043 | 0.18 |
| III | 0.312 | 0.15 |
| IV | >30 | NA |
The side chains of peptidomimetic III could potentially be further optimized to improve the affinity and drug-likeness. The arginine amino acid or the guanidine derivatives seem to be a bottleneck for cellular penetration. Because of their strong basic character, they are invariably in a protonated state under physiological conditions, which may disfavor membrane penetration. The lipophilic side chain and linker connected to the N-terminal unit in III could be optimized to reduce the flexibility.45
Although the proline side chains in undecapeptide 1 are only capable of hydrophobic interactions and the backbone forms two hydrogen bonds with p47, the essential helical structure is difficult to mimic perfectly. Further studies are required to develop better di- or triproline mimetics in proline-rich polypeptides to achieve higher affinity while improving drug-likeness.
In conclusion, inasmuch as the design and synthesis of pseudo-triproline mimetic III was part of a proof-of-concept study, the result represents a first example of a rationally designed and novel synthetic triproline mimetic to exhibit submicromolar in vitro activity against the binding of p47 with p22, two critical proteins involved in the activation of NOX1 and NOX2 in relation to regulation of ROS overproduction. Large numbers of biologically relevant PPIs in eukaryotic cells28 involve linear motifs46 in which proline-rich domains encompassed within structurally distinct polyproline type II helices play critical roles in recognition and specificity with other proteins acting as partners.47,48 Incorporation of our Pro-Cyp dipeptide mimetic as a pseudo-diproline or extended versions of it with Pro-Cyp combinations with optimized N- and C-terminal residues to simulate the action of natural oligoproline sequences in designated proteins would augur well for the development of new drug prototypes as modulators of PPIs in medicinally relevant projects.49−51
Acknowledgments
Generous financial assistance from the NSERC/Servier Senior Industrial Research Chair Program is gratefully acknowledged. We thank Dr. Sofiane Hocine for assistance with the artwork for the supplementary cover and the graphical abstract.
Glossary
Abbreviations
- Cyp
cyclopentane carboxylic acid
- NADPH
nicotinamide adenine dinucleotide phosphate
- NOX
NADPH oxidase
- ROS
reactive oxygen species
- PPI
protein–protein interaction
- PRD
proline-rich sequence recognition domain
- SH3
SRC homology 3
- SPR
surface plasmon resonance
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.2c00094.
Synthetic schemes; general information; experimental procedures; 1H NMR and HRMS characterization for all compounds; additional 13C NMR description, NMR spectra, and HPLC for I–IV; general procedures for crystallography, modeling, and surface plasmon resonance; ala-scan raw data (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Diebold B. A.; Smith S. M. E.; Li Y.; Lambeth J. D. NOX2 as a Target for Drug Development: Indications, Possible Complications, and Progress. Antioxid. Redox Signaling 2015, 23, 375–405. 10.1089/ars.2014.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babior B. M.; Lambeth J. D.; Nauseef W. The Neutrophil NADPH Oxidase. Arch. Biochem. Biophys. 2002, 397, 342–344. 10.1006/abbi.2001.2642. [DOI] [PubMed] [Google Scholar]
- Vignais P. V. The Superoxide-Generating NADPH Oxidase: Structural Aspects and Activation Mechanism. Cell. Mol. Life Sci. 2002, 59, 1428–1459. 10.1007/s00018-002-8520-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babior B. M. NADPH Oxidase: An Update. Blood 1999, 93, 1464–1476. 10.1182/blood.V93.5.1464. [DOI] [PubMed] [Google Scholar]
- Groemping Y.; Rittinger K. Activation and Assembly of the NADPH Oxidase: A Structural Perspective. Biochem. J. 2005, 386, 401–416. 10.1042/BJ20041835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard K.; Krause K. H. The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiol. Rev. 2007, 87, 245–313. 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- Lambeth J. D. NOX Enzymes and the Biology of Reactive Oxygen. Nat. Rev. Immunol. 2004, 4, 181–189. 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- Sumimoto H. Structure, Regulation and Evolution of Nox-Family NADPH Oxidases That Produce Reactive Oxygen Species. FEBS J. 2008, 275, 3249–3277. 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- Rada B.; Leto T. L. Oxidative Innate Immune Defenses by Nox/Duox Family NADPH Oxidases. Contrib Microbiol. 2008, 15, 164–187. 10.1159/000136357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsburger F.; Filippova A.; Rasti D.; Seredenina T.; Lam M.; Maghzal G.; Mahiout Z.; Jansen-Dürr P.; Knaus U. G.; Doroshow J.; Stocker R.; Krause K. H.; Jaquet V. Pharmacological Characterization of the Seven Human NOX Isoforms and Their Inhibitors. Redox Biol. 2019, 26 (May), 101272. 10.1016/j.redox.2019.101272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R.; Geng X.; Li F.; Ding Y. NOX Activation by Subunit Interaction and Underlying Mechanisms in Disease. Front. Cell. Neurosci. 2017, 10, 1–13. 10.3389/fncel.2016.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto H.; Kage Y.; Nunoi H.; Sasaki H.; Nose T.; Fukumaki Y.; Ohno M.; Minakami S.; Takeshige K. Role of Src Homology 3 Domains in Assembly and Activation of the Phagocyte NADPH Oxidase. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 5345–5349. 10.1073/pnas.91.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leto T. L.; Adams A. G.; De Mendez I. Assembly of the Phagocyte NADPH Oxidase: Binding of Src Homology 3 Domains to Proline-Rich Targets. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 10650–10654. 10.1073/pnas.91.22.10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumimoto H.; Hata K.; Mizuki K.; Ito T.; Kage Y.; Sakaki Y.; Fukumaki Y.; Nakamura M.; Takeshige K. Assembly and Activation of the Phagocyte NADPH Oxidase. J. Biol. Chem. 1996, 271, 22152–22158. 10.1074/jbc.271.36.22152. [DOI] [PubMed] [Google Scholar]
- Dröge W. Free Radicals in the Physiological Control of Cell Function. Physiol. Rev. 2002, 82, 47–95. 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- Quinn M. T.; Ammons M. C. B.; DeLeo F. R. The Expanding Role of NADPH Oxidases in Health and Disease: No Longer Just Agents of Death and Destruction. Clin. Sci. 2006, 111, 1–20. 10.1042/CS20060059. [DOI] [PubMed] [Google Scholar]
- Cave A. C.; Brewer A. C.; Narayanapanicker A.; Ray R.; Grieve D. J.; Walker S.; Shah A. M. NADPH Oxidases in Cardiovascular Health and Disease. Antioxid. Redox Signaling 2006, 8, 691–728. 10.1089/ars.2006.8.691. [DOI] [PubMed] [Google Scholar]
- Hernandes M. S.; D’Avila J. C.; Trevelin S. C.; Reis P. A.; Kinjo E. R.; Lopes L. R.; Castro-Faria-Neto H. C.; Cunha F. Q.; Britto L. R. G.; Bozza F. A. The Role of Nox2-Derived ROS in the Development of Cognitive Impairment after Sepsis. J. Neuroinflammation 2014, 11, 36. 10.1186/1742-2094-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A.; Scholer-Dahirel A.; Mechta-Grigoriou F. The Role of Reactive Oxygen Species and Metabolism on Cancer Cells and Their Microenvironment. Semin. Cancer Biol. 2014, 25, 23–32. 10.1016/j.semcancer.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Smith S. M. E.; Min J.; Ganesh T.; Diebold B.; Kawahara T.; Zhu Y.; McCoy J.; Sun A.; Snyder J. P.; Fu H.; Du Y.; Lewis I.; Lambeth J. D. Ebselen and Congeners Inhibit NADPH Oxidase 2-Dependent Superoxide Generation by Interrupting the Binding of Regulatory Subunits. Chem. Biol. 2012, 19, 752–763. 10.1016/j.chembiol.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. A.; Neupane G. P.; Lee E. S.; Jeong B. S.; Park B. C.; Thapa P. NADPH Oxidase Inhibitors: A Patent Review. Expert Opin. Ther. Pat. 2011, 21, 1147–1158. 10.1517/13543776.2011.584870. [DOI] [PubMed] [Google Scholar]
- Ogura K.; Nobuhisa I.; Yuzawa S.; Takeya R.; Torikai S.; Saikawa K.; Sumimoto H.; Inagaki F. NMR Solution Structure of the Tandem Src Homology 3 Domains of P47 Phox Complexed with a P22phox-Derived Proline-Rich Peptide. J. Biol. Chem. 2006, 281 (6), 3660–3668. 10.1074/jbc.M505193200. [DOI] [PubMed] [Google Scholar]
- Shi J.; Ross C. R.; Leto T. L.; Blecha F. PR-39, a Proline-Rich Antibacterial Peptide That Inhibits Phagocyte NADPH Oxidase Activity by Binding to Src Homology 3 Domains of P47phox. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 6014–6018. 10.1073/pnas.93.12.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laleu B.; Gaggini F.; Orchard M.; Fioraso-Cartier L.; Cagnon L.; Houngninou-Molango S.; Gradia A.; Duboux G.; Merlot C.; Heitz F.; Szyndralewiez C.; Page P. First in Class, Potent, and Orally Bioavailable NADPH Oxidase Isoform 4 (Nox4) Inhibitors for the Treatment of Idiopathic Pulmonary Fibrosis. J. Med. Chem. 2010, 53, 7715–7730. 10.1021/jm100773e. [DOI] [PubMed] [Google Scholar]
- Solbak S. M. Ø.; Zang J.; Narayanan D.; Høj L. J.; Bucciarelli S.; Softley C.; Meier S.; Langkilde A. E.; Gotfredsen C. H.; Sattler M.; Bach A. Developing Inhibitors of the P47phox-P22phox Protein-Protein Interaction by Fragment-Based Drug Discovery. J. Med. Chem. 2020, 63 (3), 1156–1177. 10.1021/acs.jmedchem.9b01492. [DOI] [PubMed] [Google Scholar]
- Sillerud L. O.; Larson R. S. Design and Structure of Peptide and Peptidomimetic Antagonists of Protein-Protein Interaction. Curr. Protein Pept. Sci. 2005, 6 (2), 151–169. 10.2174/1389203053545462. [DOI] [PubMed] [Google Scholar]
- Yuzawa S.; Suzuki N. N.; Fujioka Y.; Ogura K.; Sumimoto H.; Inagaki F. Crystallization and Preliminary Crystallographic Analysis of the Autoinhibited Form of the Tandem SH3 Domain of P47phox. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003, 59 (8), 1479–1480. 10.1107/S0907444903011636. [DOI] [PubMed] [Google Scholar]
- Groemping Y.; Lapouge K.; Smerdon S. J.; Rittinger K. Molecular Basis of Phosphorylation-Induced Activation of the NADPH Oxidase. Cell 2003, 113, 343–355. 10.1016/S0092-8674(03)00314-3. [DOI] [PubMed] [Google Scholar]
- Lau J. L.; Dunn M. K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- Hummel G.; Reineke U.; Reimer U. Translating Peptides into Small Molecules. Mol. BioSyst. 2006, 2, 499–508. 10.1039/b611791k. [DOI] [PubMed] [Google Scholar]
- Hocine S.; Berger G.; Hanessian S. Design and Synthesis of Backbone-Fused, Conformationally Constrained Morpholine-Proline Chimeras. J. Org. Chem. 2020, 85, 4237–4247. 10.1021/acs.joc.9b03413. [DOI] [PubMed] [Google Scholar]
- Vilchis-Reyes M. A.; Hanessian S. Proline Methanologues: Design, Synthesis, Structural Properties, and Applications in Medicinal Chemistry. Top Heterocycl Chem. 2015, 48, 51–96. 10.1007/7081_2015_191. [DOI] [Google Scholar]
- Berger G.; Vilchis-Reyes M.; Hanessian S. Structural Properties and Stereochemically Distinct Folding Preferences of 4,5-cis and trans-Methano-l-proline Oligomers: The Shortest Crystalline PPII-Type Helical Proline-Derived Tetramer. Angew. Chem., Int. Ed. 2015, 54, 13268–13272. 10.1002/anie.201506208. [DOI] [PubMed] [Google Scholar]
- Hanessian S.; Auzzas L. The Practice of Ring Constraint in Peptidomimetics Using Bicyclic and Polycyclic Amino Acids. Acc. Chem. Res. 2008, 41, 1241–1251. 10.1021/ar8000052. [DOI] [PubMed] [Google Scholar]
- Hanessian S. Structure-Based Organic Synthesis of Drug Prototypes: A Personal Odyssey. ChemMedChem 2006, 1, 1300–1330. 10.1002/cmdc.200600203. [DOI] [PubMed] [Google Scholar]
- Garsi J.-B.; Aguiar P. M.; Hanessian S. Design of Pseudodiproline Dimers as Mimetics of Pro-Pro Units: Stereocontrolled Synthesis, Configurational Relevance, and Structural Properties. J. Org. Chem. 2021, 86, 16834–16847. 10.1021/acs.joc.1c02061. [DOI] [PubMed] [Google Scholar]
- Hanko R.; Rabe K.; Dally R.; Hoppe D. Stereoselektive Synthese von Cyclischen Hydroxyalkyl-Dipeptidisosteren über Metallierte N,N-Dialkylcarbamidsäure-2-alkenylester. Angew. Chem. 1991, 103, 1725–1727. 10.1002/ange.19911031242. [DOI] [Google Scholar]
- Jarho E. M.; Venäläinen J. I.; Huuskonen J.; Christiaans J. A. M.; Garcia-Horsman J. A.; Forsberg M. M.; Järvinen T.; Gynther J.; Männistö P. T.; Wallén E. A. A. A Cyclopent-2-Enecarbonyl Group Mimics Proline at the P2 Position of Prolyl Oligopeptidase Inhibitors. J. Med. Chem. 2004, 47, 5605–5607. 10.1021/jm049503w. [DOI] [PubMed] [Google Scholar]
- Dai N.; Wang X. J.; Etzkorn F. A. The Effect of a Trans-Locked Gly-Pro Alkene Isostere on Collagen Triple Helix Stability. J. Am. Chem. Soc. 2008, 130, 5396–5397. 10.1021/ja711021m. [DOI] [PubMed] [Google Scholar]
- Wang X. J.; Xu B.; Mullins A. B.; Neiler F. K.; Etzkorn F. A. Conformationally Locked Isostere of PhosphoSer-Cis-Pro Inhibits Pin1 23-Fold Better than PhosphoSer-Trans-Pro Isostere. J. Am. Chem. Soc. 2004, 126, 15533–15542. 10.1021/ja046396m. [DOI] [PubMed] [Google Scholar]
- Wang X. J.; Hart S. A.; Xu B.; Mason M. D.; Goodell J. R.; Etzkorn F. A. Serine-Cis-Proline and Serine-Trans-Proline Isosteres: Stereoselective Synthesis of (Z)- and (E)-Alkene Mimics by Still-Wittig and Ireland-Claisen Rearrangements. J. Org. Chem. 2003, 68, 2343–2349. 10.1021/jo026663b. [DOI] [PubMed] [Google Scholar]
- Maassen A.; Gebauer J. M.; Theres Abraham E.; Grimm I.; Neudörfl J.-M.; Kühne R.; Neundorf I.; Baumann U.; Schmalz H.-G. Triple-Helix-Stabilizing Effects in Collagen Model Peptides Containing PPII-Helix-Preorganized Diproline Modules. Angew. Chem., Int. Ed. 2020, 59, 5747–5755. 10.1002/anie.201914101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasta J. D.; Choudhary A.; Jensen K. H.; McGrath N. A.; Raines R. T. Prolyl 4-Hydroxylase: Substrate Isosteres in Which an (E)- or (Z)-Alkene Replaces the Prolyl Peptide Bond. Biochemistry 2017, 56, 219–227. 10.1021/acs.biochem.6b00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandur N. G.; Harms K.; Koert U. First Stereoselective Synthesis of a Pro-Pro E-Alkene Dipeptide Isostere. Synlett 2005, 5, 773–776. 10.1055/s-2005-863731. [DOI] [Google Scholar]
- Flaten G. E.; Kottra G.; Stensen W.; Isaksen G.; Karstad R.; Svendsen J. S.; Daniel H.; Svenson J. In Vitro Characterization of Human Peptide Transporter HPEPT1 Interactions and Passive Permeation Studies of Short Cationic Antimicrobial Peptides. J. Med. Chem. 2011, 54, 2422–2432. 10.1021/jm1015704. [DOI] [PubMed] [Google Scholar]
- Davey N. E.; Van Roey K.; Weatheritt R. J.; Toedt G.; Uyar B.; Altenberg B.; Budd A.; Diella F.; Dinkel H.; Gibson T. J. Attributes of Short Linear Motifs. Mol. BioSyst. 2012, 8, 268–281. 10.1039/C1MB05231D. [DOI] [PubMed] [Google Scholar]
- Freund C.; Schmalz H. G.; Sticht J.; Kuhne R. Proline Rich Sequence Recognition Domain (PRD): Ligands, Function and Inhibition. Handb. Exp. Pharmacol. 2008, 186, 407–429. 10.1007/978-3-540-72843-6_17. [DOI] [PubMed] [Google Scholar]
- Zarrinpar A.; Bhattacharyya R. P.; Lim W. A. The Structure and Function of Proline Recognition Domains. Sci. STKE 2003, 2003, re8. 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- Kay B. K.; Williamson M. P.; Sudol M. The Importance of Being Proline: The Interaction of Proline-Rich Motifs in Signaling Proteins with Their Cognate Domains. FASEB J. 2000, 14, 231–241. 10.1096/fasebj.14.2.231. [DOI] [PubMed] [Google Scholar]
- Opitz R.; Müller M.; Reuter C.; Barone M.; Soicke A.; Roske Y.; Piotukh K.; Huy P.; Beerbaum M.; Wiesner B.; Beyermann M.; Schmieder P.; Freund C.; Volkmer R.; Oschkinat H.; Schmalz H.-G.; Kühne R. A Modular Toolkit to Inhibit Proline-Rich Motif–Mediated Protein–Protein Interactions. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 5011–5016. 10.1073/pnas.1422054112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.; Zhou Q.; He J.; Jiang Z.; Peng C.; Tong R.; Shi J. Recent Advances in the Development of Protein–Protein Interactions Modulators: Mechanisms and Clinical Trials. Signal Transduction Targeted Ther. 2020, 5, 1–23. 10.1038/s41392-020-00315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.