Abstract
Background
Fusarium oxysporum f. sp. cucumerinum (FOC) is the causal agent of cucumber Fusarium wilt, which can cause extensive damages and productivity losses. Cucurbita ficifolia Bouché (Cucurbitaceae) is usually used as rootstock for cucumber because of its excellent resistance to Fusarium wilt. Our previous study found that C.ficifolia has high FOC resistance, the underlying mechanism of which is unclear.
Results
Transcriptome and proteome profiling was performed on the basis of RNA-Seq and isobaric tag for relative and absolute quantitation technology to explore the molecular mechanisms of the response of Cucurbita ficifolia Bouché to Fusarium oxysporum f. sp. cucumerium infection. Comparative analyses revealed that 1850 genes and 356 protein species were differentially regulated at 2d and 4d after FOC inoculation. However, correlation analysis revealed that only 11 and 39 genes were differentially regulated at both the transcriptome and proteome levels after FOC inoculation at 2d and 4d, respectively. After FOC inoculation, plant hormones signal transduction, transcription factors were stimulated, whereas wax biosynthesis and photosynthesis were suppressed. Increased synthesis of oxidative-redox proteins is involved in resistance to FOC.
Conclusions
This study is the first to reveal the response of C. ficifolia leaf to FOC infection at the transcriptome and proteome levels, and to show that FOC infection activates plant hormone signaling and transcription factors while suppressing wax biosynthesis and photosynthesis. The accumulation of oxidative-redox proteins also plays an important role in the resistance of C. ficifolia to FOC. Results provide new information regarding the processes of C. ficifolia leaf resistance to FOC and will contribute to the breeding of cucumber rootstock with FOC resistance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08674-7.
Keywords: Cucurbita ficifolia Bouché, FOC, iTRAQ, RNA-sequencing, Defense response
Background
Fusarium oxysporum f. sp. cucumerinum (FOC) is the causal agent of cucumber Fusarium wilt which can cause serious economic losses, limit production, and decrease fruit quality. FOC infects cucumber through the roots and rapidly invades the aboveground parts via vascular tissues, thus resulting in plant water and nutrient transport blockage and plant wilt [1, 2]. Fusarium wilt is difficult to control, because it is aggravated by intensive farming practices and FOC can survive in the soil for several years [3, 4]. Effective management approaches for controlling cucumber wilt include the use of resistant cultivars, germicides, and grafting to resistant rootstock. However, FOC-resistant varieties are difficult to cultivate, grafting to resistant rootstock is widely used for cucumber to limit the effects of soil-borne pathogens in winter greenhouses and under protected structures [5, 6].
Cucurbita ficifolia Bouché (Cucurbitaceae) is a species of Cucurbita. It originated from Central America and South America, and is thus far mainly cultivated in the low latitude plateau areas, such as Yunnan in China. The color of its mature seeds is black. Great variability is observed in the seed colors of other Cucurbita speies. C. ficifolia which is also called as ‘Black Seeded’ figleaf gourd is usually used as the rootstock for cucumber because of its excellent resistance to Fusarium wilt and salt stress [7, 8].
Pathogen infection triggers complex signaling networks in plant cells. Salicylic acid (SA), jasmonic acid (JA)and ethylene (ET) are the main phytohormones related to host–pathogen interactions; and they modulate each other through a complex network of regulatory interactions [9–11]. Abscisic acid (ABA) and auxin are also key components of the immune response of plants [12]. The ABA content in wilted cucumber plants is higher than that healthy ones after infection with FOC, and ABA may play a crucial regulatory role in modifying stomatal behavior, that results in cucumber wilting due to water loss [13, 14]. F. oxysporum f. sp. melonis race 1.2(FOM1.2) is the most virulent and yield-limiting pathogen of melon. The melon genotype NAD is highly resistant to FOM1.2. Transcriptome analysis revealed that the resistance of genotype NAD is mainly signaled by JA and ET pathways mediated by ABA and auxin after FOM1.2 infection [15]. After phytohormones signaling is activated, downstream transcription factors (TFs) are also triggered, causing changes in the expression of related genes [16, 17]. TFs orchestrate the dynamic interplay between defense genes and the biosynthesis of chemical metabolites during host–pathogen interaction [16].
Next-generation sequencing technology is widely used to systematically reveal plant responses to biotic stresses. It has enriched the knowledge on mRNAs in multiple adverse environments [18]. Proteome profiling can reveal the dynamics of proteins, post-translational modifications, and biological pathways in plants in response to biological stress [19]. The complementary transcriptome and proteome analysis is widely used to resolve plant responses to various biotic stresses. Cylindrocladium leaf blight caused by Calonectria pseudoreteaudii is one of the most severe diseases in eucalyptus plantations and nurseries. The combined transcriptome and proteome analysis of the leaves of resistant eucalyptus cultivars revealed that the JA and sugar signaling pathways were activated after C. pseudoreteaudii infection, whereas photosynthesis and protein metabolism were suppressed [20]. Ciboria carunculoides is one of the disease pathogens that are most relevant to the economic and field losses of mulberry fruit in China. The combined transcriptome and proteome analyses of mulberry fruit at the early and middle stages of C. carunculoides infection revealed that plant hormone signaling pathways, TFs, and secondary metabolites were stimulated, whereas photosynthesis and cellular growth-related metabolism were inhibited [21]. However, studies on the response to C. ficifolia to F. oxysporum have not been reported.
The transcriptome and proteome of C. ficifolia infected with FOC were comparatively analysed by using RNA-seq and isobaric tag for relative and absolute quantitation technology. Comprehensive genome-wide analyses uncovered several interesting insights into C. ficifolia and FOC interactions. This work could further provide a reference for the rootstock breeding of cucumber with fungal pathogen resistance.
Results
Overview of transcriptome and proteome analyses
Given the unavailability of the genomic data of C. ficifolia and the limited read length of the Illumina-seq platform, we first used PacBio sequencing to splice the reference genome. A total of 62 169 unigenes with an average length of 1160 nt were obtained. Sequence alignment by using BLAST showed that 40 031 (64.39%) transcripts were exhibited gene annotation. The unigenes were aligned to the COG database to predict their possible functions. Gene ontology (GO) functional annotations were obtained accordance with Nr annotation information.
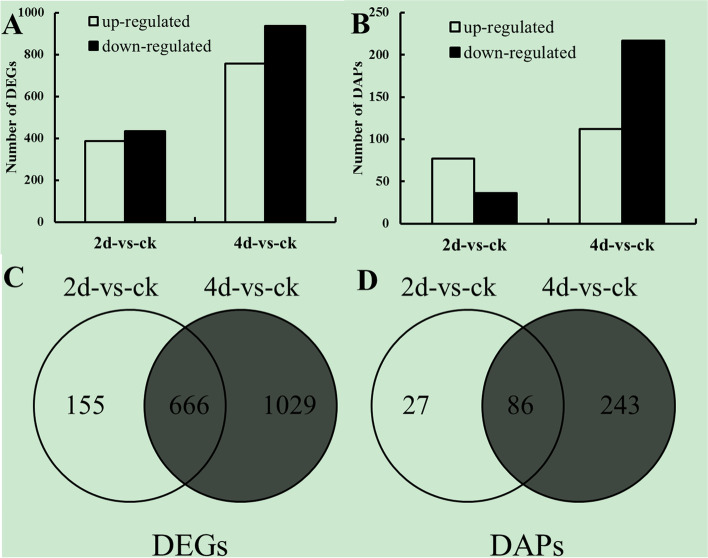
Approximately 48 million clean reads were obtained from each Illumina-seq sample. Genes with expression changes of no less than 2-folds (log2 ratio ≥ 1) and false discovery rate (FDR) < 0.05 were identified as differentially expressed genes (DEGs). In total, 1850 genes in C. ficifolia leaves were found to be differentially expressed after FOC infection. A total of 821 (387 up-regulated and 434 down-regulated) and 1695 (758 up-regulated and 937down-regulated) DEGs were identified to be responsive to FOC infection in 2d-vs-ck and 4d-vs-ck, respectively (Fig. 1; Additional files 1 and 2). Of the 1850 DEGs, 666 were affected by FOC infection in both 2d-vs-ck and 4d-vs-ck with consistent trends (both up- regulated or down—regulated), (Fig. 1).
Fig. 1.
DEGs and DAPsin C. ficifolia leaf after infection by FOC. Number of DEGs (A) and DAPs (B) at 2d and 4d compared with control. Venn diagrams showing DEGs (C) and DAPs (D) classified by their expression pattern
Comparative proteomic analyses were performed on C. ficifolia leaf samples at 2 and 4 d post infection (dpi) and the control treatment. A total of 231 157 spectra were obtained, in which 62 490 distinct peptides, and 2491 protein species were identified. Protein species with a log2 ratio ≥ 1 and FDR < 0.05 were identified as differentially accumulated proteins (DAPs). In C. ficifolia leaves, 356 protein species showed differential expression after FOC infection. A total of 113 (77 up-regulated and 36 down-regulated) and 329 (112 up-regulated and 217 down-regulated) DAPs were identified to be responsive to FOC infection in 2d-vs-ck and 4d-vs-ck respectively (Fig. 1; Additional files 3 and 4). Of the 356 DAPs, only 86 were regulated by FOC in both 2d-vs-ck and 4d-vs-ck (Fig. 1; Additional files 3 and 4).
Phytohormone signal transduction pathways and TFs involved in defense of C. ficifolia at the transcriptional level
Of the 666 DEGs affected by FOC infection in both 2d-vs-ck and 4d-vs-ck, more than twenty genes were related to plant hormone signaling pathways. Nine genes associated with ET signaling were differentially regulated by FOC infection. Among these genes, four were ethylene-responsive TFs, of which three were up-regulated; one, namely 1-aminocyclopropane-1-carboxylate oxidase, was up-regulated. Seven genes involved in auxin signaling, except a gene encoding auxin-responsive protein, were both up-regulated by FOC infection in 2d-vs-ck and 4d-vs-ck. The ABA receptor PYL4 was up-regulated at the mRNA level in 2d-vs-ck and 4d-vs-ck, and Protein MOTHER of FT and TFL1 which respond to ABA were also up-regulated in 2d-vs-ck and 4d-vs-ck. Two genes respond to JA or cytokinin, respectively, also showed consistent trend in 2d-vs-ck and 4d-vs-ck (Table 1). Twelve DEGs (six up- regulated and six down-regulated) in both 2d-vs-ck and 4d-vs-ck were annotated as transcription factors, with most belong to MYB, bHLH, LUX and GLK TF families (Table 1). Four resistance-related genes, including universal stress protein A-like protein, isochorismate synthase, protein ENHANCED DISEASE RESISTANCE 4 and wound-induced protein were both up-regulated in 2d-vs-ck and 4d-vs-ck (Table 1).
Table 1.
DEGs in C. ficifolia leaf after infection by FOC at 2 dpi and 4 dpi
| Gene ID | 2d-VS-CK log2FC | 4d-VS-CK log2FC | Gene annotation |
|---|---|---|---|
| Ethylene | |||
| CL780Contig2 | -4.07 | -1.78 | Protein REVEILLE 7, response to ethylene |
| comp69483_c1_seq15_1 | -3.57 | -3.06 | Protein REVEILLE 7, response to ethylene |
| CL4715Contig1 | -1.89 | -2.01 | Protein REVEILLE 6, response to ethylene |
| comp43670_c0_seq2_3 | 6.05 | 6.41 | Protein REVEILLE 6, response to ethylene |
| CL28656Contig1 | 2.42 | 2.76 | Ethylene-responsive transcription factor ERF106 |
| CL42540Contig1 | 2.72 | 2.40 | Ethylene-responsive transcription factor ERF106 |
| CL27086Contig1 | 3.08 | 3.28 | Ethylene-responsive transcription factor ERF053 |
| CL21678Contig1 | -2.90 | -2.50 | Ethylene-responsive transcription factor 4 |
| CL3070Contig1 | 2.13 | 2.95 | 1-aminocyclopropane-1-carboxylate oxidase homolog 3 |
| Auxin | |||
| CL30348Contig1 | -2.21 | -3.69 | Auxin-responsive protein SAUR50 |
| CL1073Contig1 | 2.17 | 3.11 | Auxin-repressed 12.5 kDa protein |
| CL2472Contig1 | 2.28 | 3.19 | Auxin-repressed 12.5 kDa protein |
| CL24742Contig1 | 1.92 | 1.82 | Auxin response factor 6 |
| comp51974_c0_seq1_1 | 2.08 | 2.02 | Auxin response factor 6 |
| CL7285Contig1 | 2.02 | 2.64 | IAA-amino acid hydrolase ILR1,auxin metabolic process |
| CL24970Contig1 | 3.42 | 4.92 | Glutathione S-transferase,auxin-activated signaling pathway |
| Abscisic acid | |||
| CL10966Contig1 | 3.91 | 3.15 | Protein MOTHER of FT and TFL1,response to abscisic acid |
| CL24926Contig1 | 2.06 | 1.76 | Abscisic acid receptor PYL4 |
| Other hormones | |||
| comp63305_c0_seq6_2 | 2.51 | 2.35 | Protein NRT1/PTR FAMILY 6.2,response to jasmonic acid |
| CL621Contig1 | -2.39 | -3.04 | Cytokinin dehydrogenase 1 |
| Transcription factor | |||
| CL4801Contig1 | 2.46 | 2.57 | Transcription factor SRM1 |
| comp63308_c1_seq4_1 | 1.88 | 2.22 | Transcription factor SRM1 |
| CL2579Contig1 | 7.78 | 7.76 | Transcription factor PCL1 |
| CL24093Contig1 | -2.25 | -3.34 | Transcription factor MYB1R1 |
| CL26072Contig1 | 4.77 | 5.46 | Transcription factor LUX |
| CL37562Contig1 | 6.00 | 6.98 | Transcription factor LUX |
| comp62782_c2_seq8_2 | -1.83 | -1.57 | Transcription factor ILR3 |
| CL1Contig37 | -5.03 | -5.08 | Transcription factor HY5 |
| comp75510_c0_seq1_2 | 2.14 | 2.00 | Transcription factor BOA |
| CL51358Contig1 | -2.70 | -2.82 | Transcription factor bHLH66 |
| CL19223Contig1 | -2.42 | -1.96 | Transcription activator GLK1 |
| CL1942Contig2 | -2.21 | -1.89 | Transcription activator GLK1 |
| Resistance-related genes | |||
| CL25265Contig1 | 3.62 | 4.36 | Universal stress protein A-like protein |
| CL478Contig1 | 3.32 | 4.82 | Isochorismate synthase, chloroplastic |
| comp67592_c0_seq158_1 | 2.47 | 1.95 | Isochorismate synthase, chloroplastic |
| CL29528Contig1 | 3.87 | 4.57 | Protein ENHANCED DISEASE RESISTANCE 4 |
| CL601Contig2 | 1.95 | 2.15 | Wound-induced protein 1 |
| Photosynthesis | |||
| CL5872Contig1 | -2.77 | -2.97 | Photosystem II 22 kDa protein, chloroplastic |
| CL8698Contig1 | -2.29 | -2.76 | Photosystem II 22 kDa protein, chloroplastic |
| CL50394Contig1 | -1.81 | -1.77 | Magnesium-protoporphyrin IX monomethyl ester cyclase |
| CL4989Contig1 | -2.15 | -3.37 | Magnesium-chelatase subunit ChlH, chloroplastic |
| CL25245Contig1 | -1.73 | -2.31 | Chlorophyll a-b binding protein 13, chloroplastic |
A total of 1029 DEGs were found in 4d-vs-ck. However, 666 DEGs were affected by FOC infection in both 2d-vs-ck and 4d-vs-ck and were enriched in plant hormone signal transduction pathways, TFs and resistance-related. Most of the transcript species related to the plant hormone signal transduction pathway were involved in ET, auxin, ABA, and JA signaling. A gene responsive to brassinosteroids was also down-regulated in 4d-vs-ck (Table 2). Fifteen DEGs (nine up-regulated and six down-regulated) were annotated as TFs in 4d-vs-ck (Table 2), with most belonging to the MYB, WARK, TCP and NAC TF families. Seventeen resistance-related genes were found in 4d-vs-ck. They included disease resistance, pathogenesis-related and TMV resistance proteins. Two DEGs annotated as pathogenesis-related proteins showed increased up-regulation with the log2FC of 8.3 and 9.0.
Table 2.
DEGs in C. ficifolia leaf after infection by FOC only at 4 dpi
| Gene ID | 4d-VS-CK log2FC | Gene annotation |
|---|---|---|
| Ethylene | ||
| CL12896Contig1 | 3.92 | Ethylene-responsive proteinase inhibitor 1 |
| comp51941_c0_seq1_3 | -1.50 | Ethylene receptor 2 |
| CL23139Contig1 | 1.94 | 1-aminocyclopropane-1-carboxylate oxidase 5 |
| Jasmonic acid | ||
| CL19963Contig1 | 2.59 | 3-ketoacyl-CoA thiolase 2, peroxisomal |
| CL21247Contig1 | 2.56 | 3-ketoacyl-CoA thiolase 2, peroxisomal |
| ABA | ||
| comp51694_c0_seq3_2 | 2.01 | ABSCISIC ACID-INSENSITIVE 5-like protein 6 |
| CL23454Contig1 | 2.10 | Abscisic acid receptor PYL8 |
| CL23796Contig1 | -1.81 | Abscisic acid receptor PYL4 |
| Auxin | ||
| CL46582Contig1 | -1.51 | Auxin-induced protein 22D |
| CL38635Contig1 | 2.05 | Auxin transport protein BIG |
| CL28225Contig1 | -4.01 | Auxin efflux carrier component 5 |
| Other hormones | ||
| CL25910Contig1 | 1.92 | Gibberellin 20 oxidase 1 |
| CL14729Contig1 | -1.40 | Protein EXORDIUM, response to brassinosteroid |
| CL30190Contig1 | -1.41 | Salicylic acid-binding protein 2 |
| Resistance-related | ||
| CL5641Contig1 | 3.37 | Adrenodoxin-like protein 2, mitochondrial |
| CL28332Contig1 | -2.41 | Peroxidase 47 |
| CL304Contig1 | -1.89 | Peroxidase 39 |
| CL52011Contig1 | 3.12 | Disease resistance protein RPS6 |
| CL33669Contig1 | 1.36 | MLO-like protein 6 |
| CL4874Contig1 | 1.25 | MLO-like protein 12 |
| CL18414Contig1 | -3.36 | Pathogenesis-related protein PR-1 |
| CL26231Contig1 | 8.30 | Pathogenesis-related protein P2 |
| CL10878Contig1 | 9.06 | Pathogenesis-related protein 1 |
| CL1932Contig1 | 3.14 | Pathogenesis-related genes transcriptional activator PTI6 |
| CL51123Contig1 | -1.27 | Protein ENHANCED DISEASE RESISTANCE 2-like |
| CL8780Contig1 | -2.77 | Putative disease resistance RPP13-like protein 1 |
| CL512Contig2 | -1.12 | S-norcoclaurine synthase 2 |
| comp53156_c0_seq3_1 | 1.57 | Wound-induced protein 1 |
| CL48028Contig1 | 1.94 | Universal stress protein A-like protein |
| CL19588Contig1 | 1.52 | TMV resistance protein N |
| CL21402Contig1 | 2.24 | TMV resistance protein N |
| Transcription factor | ||
| CL30475Contig1 | 1.40 | MYB family transcription factor EFM |
| CL50886Contig1 | 1.85 | MYB family transcription factor EFM |
| CL32159Contig1 | -1.18 | Transcription factor MYB59 |
| comp75186_c0_seq1_2 | -2.36 | Transcription factor MYB44 |
| CL54017Contig1 | 4.45 | NAC domain-containing protein 79 |
| CL26897Contig1 | 3.15 | Probable WRKY transcription factor 70 |
| CL2070Contig1 | 3.32 | Probable WRKY transcription factor 69 |
| comp64969_c0_seq1_1 | 1.17 | Transcription factor TCP4 |
| CL45926Contig1 | 2.78 | Transcription factor TCP20 |
| CL28172Contig1 | -1.71 | Transcription factor SRM1 |
| CL25039Contig1 | 1.86 | Transcription factor GTE12 |
| CL38022Contig1 | -1.73 | Transcription factor EMB1444 |
| comp44643_c0_seq12_3 | -1.34 | Transcription factor DIVARICATA |
| CL10732Contig1 | -4.66 | Transcription factor bHLH92 |
| Wax biosynthetic process | ||
| CL13276Contig1 | -2.12 | 3-ketoacyl-CoA synthase 6 |
| CL6623Contig1 | -1.52 | 3-ketoacyl-CoA synthase 6 |
| CL26021Contig1 | -1.97 | 3-ketoacyl-CoA synthase 4 |
| comp31671_c0_seq2_2 | -1.77 | 3-ketoacyl-CoA synthase 2 |
| comp37543_c1_seq2_3 | -1.94 | 3-ketoacyl-CoA synthase 12 |
| CL141Contig1 | -1.38 | 3-ketoacyl-CoA synthase 11 |
| CL51488Contig1 | -1.53 | 3-ketoacyl-CoA synthase 10 |
| CL26445Contig1 | -1.62 | Acyl-[acyl-carrier-protein] desaturase, chloroplastic |
| CL23269Contig1 | 1.87 | Long chain acyl-CoA synthetase 4 |
| CL26279Contig1 | -1.47 | Omega-hydroxypalmitate O-feruloyl transferase |
| CL778Contig1 | -4.27 | Omega-hydroxypalmitate O-feruloyl transferase |
| CL26276Contig1 | -2.45 | Protein ECERIFERUM 1 |
| CL32066Contig1 | -2.54 | Protein ECERIFERUM 1 |
| CL1392Contig2 | -1.83 | Protein HOTHEAD |
| comp50398_c2_seq5_3 | -3.10 | Protein HOTHEAD |
| CL50895Contig1 | -1.71 | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 2 |
| Photosynthesis | ||
| CL42418Contig1 | -1.46 | Photosystem II reaction center W protein, chloroplastic |
| CL7255Contig1 | -1.06 | Photosystem II protein psbY-2, chloroplastic |
| CL30417Contig1 | -1.64 | Photosystem II 5 kDa protein, chloroplastic |
| CL46717Contig1 | -12.75 | Photosystem I reaction center subunit psaK, chloroplastic |
| CL21783Contig1 | -2.04 | Photosystem I reaction center subunit N, chloroplastic |
| CL22180Contig1 | -1.19 | Photosystem I reaction center subunit III, chloroplastic |
| CL782Contig1 | -1.60 | Photosystem I reaction center subunit III, chloroplastic |
| comp67722_c4_seq2_2 | -1.27 | Chlorophyll a-b binding protein of LHCII type I |
| CL31939Contig1 | -1.44 | Chlorophyll a-b binding protein of LHCII type 1 |
| CL13308Contig1 | -2.15 | Chlorophyll a-b binding protein CP29.1, chloroplastic |
| CL20903Contig1 | -2.34 | Chlorophyll a-b binding protein CP29.1, chloroplastic |
| comp69242_c0_seq1_2 | -13.19 | Chlorophyll a-b binding protein CP29.1, chloroplastic |
| CL30759Contig1 | -2.02 | Chlorophyll a-b binding protein CP26, chloroplastic |
| CL55345Contig1 | -1.76 | Chlorophyll a-b binding protein CP24 10A, chloroplastic |
| CL25433Contig1 | -1.05 | Chlorophyll a-b binding protein 8, chloroplastic |
| CL11837Contig1 | -1.38 | Chlorophyll a-b binding protein 7, chloroplastic |
| CL54964Contig1 | -1.28 | Chlorophyll a-b binding protein 7, chloroplastic |
| CL55558Contig1 | -13.46 | Chlorophyll a-b binding protein 4, chloroplastic |
| CL23730Contig1 | -2.31 | Chlorophyll a-b binding protein 3, chloroplastic |
| CL38334Contig1 | -2.17 | Chlorophyll a-b binding protein 3, chloroplastic |
| CL2559Contig1 | -1.59 | Chlorophyll a-b binding protein 151, chloroplastic |
| CL2559Contig2 | -1.86 | Chlorophyll a-b binding protein 151, chloroplastic |
Wax biosynthetic process and photosynthesis are partially repressed by FOC at 4dpi
Among the 1029 DEGs that were up- or down-regulated only in 4d-vs-ck, many related to wax biosynthetic process, carbon fixation and photosynthesis were down-regulated. DEGs annotated as 3-ketoacyl-CoA synthase, long chain acyl-CoA synthase and protein ECERIFERUM, which are related to wax biosynthetic process, were both down-regulated. Several genes encoding key enzymes associated with carbon fixation were down-regulated after FOC infection. These genes included sedoheptulose-1,7-bisphosphatase, glyceraldehyde-3-phosphate dehydrogenase and fructose-1,6-bisphosphatase. A total of 22 down-regulated DEGs were involved in photosynthesis and were related to photosystems I and II and chlorophyll binding proteins. Two DEGs that were annotated as photosystem I reaction center subunit and chlorophyll a-b binding protein were highly down-regulated, with the log2FC of -12.75 and -13.19, respectively (Table 2).
Oxidative-redox proteins involved in defense of C. ficifolia at the translational level
There were 86 DAPs which regulated by FOC in both 2d-vs-ck and 4d-vs-ck, six proteins were annotated as oxidative-redox proteins (Table 3). Six oxidative-redox proteins were one catalase isozyme, two ferredoxins, and three peroxidases. All of the oxidative-redox proteins showed increased accumulation at 2dpi and 4dpi. Three photosystem proteins, one photosystem I reaction center subunit N and two oxygen-evolving enhancer proteins also were up-regulated after FOC infected. Sixteen ribosomal proteins showed decreased accumulation at 2dpi and 4dpi.
Table 3.
DAPs in C. ficifolia leaf after infection by FOC at 2 dpi and 4 dpi
| ID | 2d-VS-CK log2FC | 4d-VS-CK log2FC | Annotation |
|---|---|---|---|
| Oxidative-redox proteins | |||
| CL21412Contig1 | 1.59 | 1.92 | Ferredoxin-1, chloroplastic |
| CL29295Contig1 | 2.09 | 2.24 | Ferredoxin-2, chloroplastic |
| CL34362Contig1 | 2.03 | 2.98 | Catalase isozyme 3 |
| CL22454Contig1 | 3.47 | 4.07 | Peroxidase 21 |
| CL7963Contig1 | 1.61 | 2.01 | Peroxidase 15 |
| comp72441_c0_seq1_2 | 2.06 | 2.75 | Peroxidase 2 |
| Photosystem proteins | |||
| CL21783Contig1 | 2.05 | 2.44 | Photosystem I reaction center subunit N |
| CL1Contig19 | 1.38 | 1.83 | Oxygen-evolving enhancer protein 3–2 |
| comp66617_c2_seq1_2 | 1.65 | 2.36 | Oxygen-evolving enhancer protein 1 |
| Ribosomal protein | |||
| CL10872Contig1 | -1.18 | -2.34 | 60S ribosomal protein L27-3 |
| CL12198Contig1 | -1.06 | -1.36 | 60S ribosomal protein L19-2 |
| CL22024Contig1 | -1.12 | -2.17 | 60S ribosomal protein L24 |
| CL23985Contig1 | -1.28 | -1.50 | 40S ribosomal protein S15a-1 |
| CL25275Contig1 | -1.03 | -1.80 | 40S ribosomal protein S10-3 |
| CL26010Contig1 | -1.10 | -1.91 | 60S ribosomal protein L9 |
| CL27757Contig1 | -1.22 | -1.75 | 30S ribosomal protein S3 |
| CL42728Contig1 | -1.14 | -2.35 | 60S acidic ribosomal protein P2 |
| CL43344Contig1 | -1.13 | -1.93 | 60S ribosomal protein L35 |
| CL43617Contig1 | -1.22 | -1.84 | 40S ribosomal protein S13 |
| CL43782Contig1 | -1.15 | -2.09 | 40S ribosomal protein S6-2 |
| CL47329Contig1 | -1.03 | -1.64 | 60S ribosomal protein L22-2 |
| CL47423Contig1 | -1.31 | -2.06 | 40S ribosomal protein S16 |
| CL52891Contig1 | -1.89 | -3.29 | 40S ribosomal protein S2-4 |
| CL53777Contig1 | -1.15 | -2.39 | 60S ribosomal protein L7a-2 |
| comp72390_c0_seq1_2 | -1.11 | -1.75 | 50S ribosomal protein L35, chloroplastic |
Correlation analysis of transcriptome and proteome data
Transcriptome and proteome data were compared on the basis of the log2-transformed protein species accumulation and log2-gene expression ratios. The DAPs were associated with the corresponding DEGs in accordance with annotations or id correspondences. If they could not be directly related, they were associated in accordance with gene names (protein names) or the BLAST sequences of homologous pairs. A low correlation was observed between transcriptome data and proteome data at 2di and 4di (Fig. 2, Additional files 5 and 6). Compared with control, only 11 genes showed correlated regulation at both the transcription and translation levels at 2dpi. Among these genes, only nine had consistent trends and two had opposite trends at the transcription and translation levels (Additional file 5). At 4 d after FOC infection, 39 genes showed a correlated regulation between transcriptome and proteome data. Of these genes, 19 had the consistent trends and 20 had the opposite trends at the transcription and translation levels (Additional file 6). Two genes that were annotated as pathogenesis-related protein and one gene that was annotated as peroxidase were up-regulated at the transcription and translation levels at 4dpi.
Fig. 2.
Cluster analysis of the DEGs and DAPs with correlated regulation in the transcriptome and proteome datasets
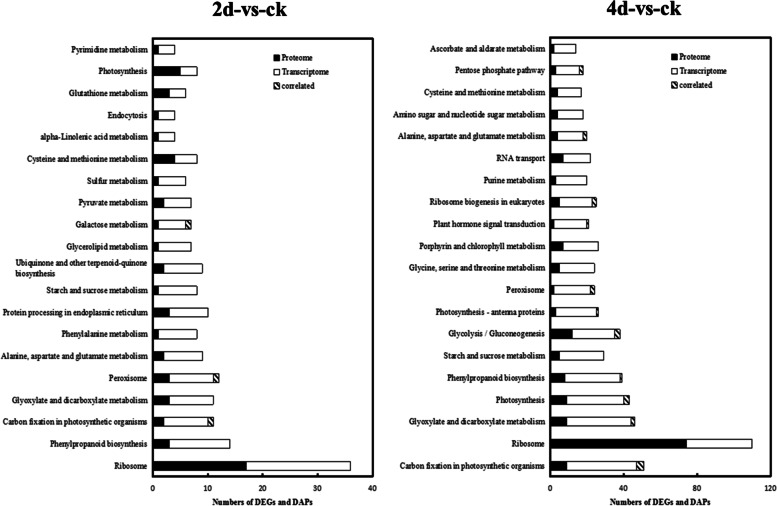
KEGG pathway enrichment analysis revealed that DEGs and DAPs were mainly related to the ribosome, phenylpropanoid biosynthesis, carbon fixation in photosynthetic organisms, peroxisome, glyoxylate and dicarboxylate metabolism, and glycolysis/gluconeogenesis pathways (Fig. 3).
Fig. 3.
KEGG pathway enrichment analysis of DEGs and DAPs in 2d and 4d infected C. ficifolia leaf compared with control
Real-time polymerase chain reaction validation
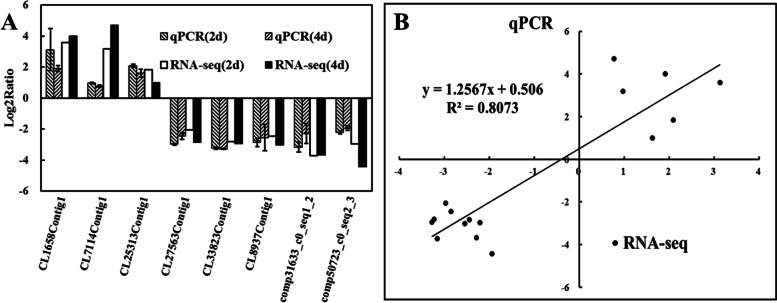
Eight DEGs were selected for real-time polymerase chain reaction analysis to validate the RNA-seq results. These genes showed different expression patterns in C. ficifolia leaves at 2 dpi and 4 dpi. The expression patterns of these genes obtained through qRT-PCR confirm to a large extent the transcriptome data (R2 = 0.8073; Fig. 4).
Fig. 4.
Validation of the transcriptome data. A Results from RNA-seq analysis were compared with those from qPCR. Bars represent mean ± SD (n = 3). B Comparison of log2 expression of 8 selected differentially regulated genes at 2d and 4d infected C. ficifolia leaf as measured by RNA-seq and qPCR. Positive and negative log2 expression ratios represent up- and down-regulation, respectively, compared with control
Discussion
Biotic stress, especially pathogen infection, is generally believed to trigger complex molecular changes in plants. The complete image of molecular dynamics can be revealed via transcriptome and proteome profiling [22]. Leaf samples were obtained at different infection stages to investigate the mechanism of C. ficifolia response to FOC infection.
Leaf response is critical for resolving FOC resistance in C. ficifolia at early stage
FOC infects plants through the roots and rapidly invades the aboveground parts via vascular tissues, thus resulting in plant water and nutrient transport blockage and wilting. In cucumber, which is sensitive to FOC, leaves exhibited stomata closure, decreased transpiration rate, and water loss that then resulted in cell death, during the early stages of FOC infection [13]. However, at the early stage of FOC infection, C. ficifolia showed significantly increased stomatal conductance, transpiration rate and net photosynthetic rate. Our previous study also found that the expression level of HQRGA2, a homologous sequence of NBS disease-resistance gene (Genbank ID: MG946756) significantly increased and declined at 2 and 4dpi [23]. These results indicated that in C. ficifolia, the response of the aboveground parts at the early infection stage is the key mechanism of resistance to FOC. We selected leaf samples of C. ficifolia with short-term infection (2 and 4 days) for Pacbio and Illumina sequencing to obtain sufficient information regarding the response of C. ficifolia to FOC.
Phytohormone and TFs involved in defense of C. ficifolia to FOC
Phytohormones play important role in defense and immune responses. Although SA, JA, and ET are the main phytohormones associated with immune responses against pathogens, ABA, gibberellic acid, Cytokinin, and auxin, are also key parts of the defense response of plants [10]. A number of DEGs related to hormone signaling were found in C. ficifolia after FOC infection (Tables 1 and 2). ET is the principal modulator of the defense response to pathogens. The synthesis of ET begins with the amino acid methionine, which is first converted into S-adenosylmethionine and then into 1-aminocyclopropane-1-carboxylic acid (ACC). ACC is then converted into ET by ACC oxidase (ACO) [24]. Genes encoding ACO can be transcriptionally up-regulated, resulting in ET biosynthesis activation in plants challenged by pathogens. Genes encoding ACO are up-regulated in Arabidopsis infected with Botrytis cinerea [10]. In tomato, ET is also required for the xylem occlusion response to counter the further spread of F. oxysporum f.sp. lycopersici [11]. A DEGs annotated as ACO was both up-regulated by FOC infection in 2d-vs-ck and 4d-vs-ck (Table 1). Another ACO was up-regulated only in 4d-vs-ck. Auxin and JA synergize to promote resistance to necrotrophic pathogens [22]. Seven DEGs with annotations related to auxin were both up-regulated by FOC infection in 2d-vs-ck and 4d-vs-ck (Table 1). ABA is closely linked to resistance to a variety of abiotic stress, especially drought and salinity. The exogenous application of ABA can reduce the resistance of potato to Phytophthora infestans and the resistance of tobacco to Peronospora tabacina [25, 26]. These finding indicated that ABA can depress plant resistance to pathogens, especially fungi. ABA signaling in plants involves perception by a receptor complex that is formed by PYRABACTIN RESISTANCE 1 (PYR) and PYR1-LIKE (PYL) proteins [27]. The ABA receptor PYL4 was both up-regulated by FOC infection in 2d-vs-ck and 4d-vs-ck (Table 1). The ABA receptor PYL8 was up-regulated by FOC infection only in 4d-vs-ck. These results indicated that the ET, auxin, and ABA signal transduction pathways may play a pivotal role in the response to FOC infection.
Biotic stresses trigger a wide range of plant responses, TFs function in the promoter region of stress-related genes; the induction or reduction the expression of these genes may change plant tolerance to biotic stress [28]. Nearly 30 DEGs were TFs and were up-regulated by FOC infection in 2d-vs-ck and 4d-vs-ck. Most of them belonged to the MYB, bHLH, WRKY, NAC, LUX and GLK TF families and presented high increment or decrement after FOC infection (Tables 1 and 2). MYB TFs play an essential role in defense responses in plants. SpMYB (Solanum pimpinellifolium L3708) expression is significantly induced after infection by F. oxysporum. Overexpression SpMYB in tobacco increased resistance to F. oxysporum and the transgenic plants had lower malonaldehyde content but increased peroxidase, superoxide dismutase, and phenylalanine ammonia-lyase activities [29]. WRKY TFs are the global regulators of plant defense signaling. A previous study on two chickpea (Cicer arietinum L.) genotypes with contrasing resistance against F. oxysporum f. sp. ciceri Race1 (Foc1) demonstrated that CaWRKY40 triggered defense to Foc1. In chickpea, overexpressed CaWRKY40 induced resistance to Foc1 by binding to promoters and positively regulated the transcription of CaDefensin and CaWRKY33 [30].
Many reports have indicated that NAC TFs are the principal modulators of plant defense, and systemic acquired resistance [31]. Numerous examples have shown that the expression of the NAC gene change after pathogen infection. The StNAC (Solanum tuberosum) gene was induced after Phytophthora infestans infection [32]. In rice seedlings, 19 and 13 NAC genes were up-regulated after RSV and RTSV infection, respectively [33]. The NAC domain-containing protein 79 was up-regulated by FOC infection only in 4d-vs-ck (Table 2). A number of NAC proteins activate PR genes to regulate plant defense responses [31]. In Arabidopsis, the overexpression of ATAF2, the NAC TF, increased susceptibility to F. oxysporum by inhibiting PR genes expression [34]. In Arabidopsis, cold stimulated the activation of the NAC TF NTL6 which induced PR genes and enhanced disease resistance [35]. Three PR genes were found in C. ficifolia only in 4d-vs-ck. One was down-regulated and two were up-regulated. However, only two up-regulated PR genes were up-regulated at the transcription and translation levels in 4d-vs-ck (Additional file 6). These results indicated that NAC TFs may regulate defense responses to FOC by activating PR proteins in C. ficifolia.
Wax biosynthetic process and photosynthesis are repressed by FOC
After FOC infection, the numbers of genes related to wax biosynthesis and photosynthesis decreased. The wax biosynthesis was significantly reduced by FOC in 4d-vs-ck. (Table 2). Two genes that were annotated as omega-hydroxypalmitate O-feruloyl transferase were down-regulated (Table 2). Omega-hydroxypalmitate O-feruloyl transferase is a pivotal enzyme in the biosynthesis of unsaturated fatty acids, which are the precursors of wax [36]. Seven genes that were annotated as 3-ketoacyl-CoA synthase (KCS) were also down-regulated (Table 2). In potato, stably silencing the KCS gene StKCS6, through RNA interference, decreased the accumulation of peridermal wax [37]. Although FOC infects the plant from root, FOC infection down-regulated 22 DEGs that were related to photosynthesis in 4d-vs-ck. Similar situation was encountered in chickpea infected by F. oxysporum f. sp. ciceri race 1 (FOC1). The photosynthetic stability of susceptible plants was hampered by the down regulation key photosynthetic genes and the photosynthetic stability of resistant chickpea also decreased at later time points [38].
Three photosynthesis-related DAPs show increased accumulation at 2dpi and 4dpi. This is contrary to the transcriptome results, and one explanation is that photosynthesis can help plants defect pathogens through providing of carbon skeleton and energy [39]. The same pattern was observed in the proteomic profile of Pinus monticola infected by Cronartium ribicola in compatible and incompatible interaction [40]. In contrast, photosynthesis-related proteins showed increased accumulation in the early stages (72 h) and decreased accumulation in the later stages(45 d) of cocao infected by the pathogen Moniliophthora perniciosa [41].
Oxidative-redox proteins are induced to resist FOC
The rapid accumulation of reactive oxygen species (ROS) is the earliest typical event in a plant–pathogen interaction [42, 43]. ROS are toxic for both host and pathogens, therefore, the balance between production and removal of ROS are important during stress response [44]. Plants use anti-oxidative enzymes to eliminate ROS. Six oxidative-redox proteins including ferredoxin, catalase isozyme and peroxidase were both up-regulated after FOC infection at 2dpi and 4dpi. In many plant species, up-regulated peroxidases are in line with resistance [45]. Moniliophthora perniciosa is the causal agent of cacao (Theobroma cacao L.) witches’ broom disease (WBD). The cacao genotypes with WBD resistance showed up regulation of oxidative stress proteins twice as large as sensitive genotypes, particularly in proteins related to ROS detoxification [41]. It was also demonstrated that the upregulation of detoxification proteins promoted resistance of Citrus genotypes to Huanglong disease [46]. These results reveal that accumulation of oxidative-redox proteins plays an important role in the resistance of C. ficifolia to FOC.
Conclusions
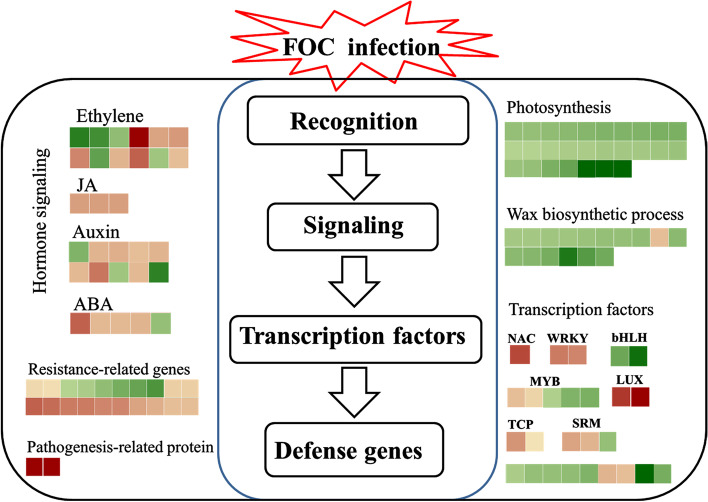
This study is the first to determine the response of C. ficifolia leaves to FOC infection at the transcription and translation levels. It revealed that FOC infection activated phytohormone signaling and TFs but inhibited wax biosynthesis and photosynthesis (Fig. 5). The accumulation of oxidative-redox proteins also plays an important role in the resistance of C. ficifolia to FOC. The results provide new information regarding the processes of C. ficifolia leaf resistance to FOC and will contribute the rootstock breeding of cucumber with resistance to fungal pathogen.
Fig. 5.
Biotic stress–related genes altered in response to FOC infection. Genes that were differentially expressed between control and infected leaf are indicated by colored squares, based on their pattern of expression at the gene level at 2d and 4d. Negative values represent repressed genes (green) and positive values represent up-regulated genes (red)
Methods
Cultivation of plants and pathogenic fungi
This experiment was conducted in a greenhouse at Yunnan Agricultural University, Kunming, and southern China. C. ficifolia Bouché was used as the material in this study. The seeds were collected from local growers in Kunming, Yunnan Province. The seeds identified by Prof. Zhengan Yang, and then kept in the laboratory for long-term use. All C.ficifolia seeds were initially sterilized in 10% hydrogen peroxide for 1 h, rinsed five times with sterile deionized water, and then subjected to accelerated germination in a constant temperature incubator at 28 °C in the dark. After germination, the seeds were transplanted into pots containing peat soil and perlite (V:V = 1:1) and kept in a plant growth chamber under a 12 -h -light/12 –h -dark photoperiod and at the ambient temperature of 25 °C ± 2 °C.
FOC was provided by the Department of Plant Nutrition, Nanjing Agricultural University. The strains were well maintained and were transferred into potato dextrose agar medium for 7 days before inoculation. Agar disks cut from the 7-day-old cultures were filtered through two layers of sterile gauze to remove mycelial fragments and then diluted to the concentration of 1 × 108 conidia/mL with sterile distilled water.
When the third true leaf had emerged, C.ficifolia seedlings were inoculated with 10 mL of endoconidia suspension (fungal infection) or with 10 mL of sterile water (control) through root dipping. Endoconidia suspension (10 mL) was added into the pots of the seedlings with fungal infection to ensure successful FOC infection. Leaves were harvested from the control and infected plants at 2 and 4 dpi for RNA extraction. The leaves from three seedlings were harvested together as one sample, and three biological replicates were used for RNA-seq, iTRAQ and RT-PCR analyses. They were immediately frozen in liquid nitrogen and stored at − 80 °C.
RNA sequencing and DEGs identification
Total RNA was isolated by using TRIzol reagent (Invitrogen, USA) in accordance with the manufacturer’s instructions. A minimum of 50 mg of the total RNA of each sample were transported to oeBiotech (Shanghai, China) for sequencing.
Equal amounts of the total RNA of the control and infected C. ficifolia plants at 2 and 4 dpi were pooled in a combined sample for PacBio library construction and sequencing. Three biological replicates were used for PacBio sequencing. Twelve cycles of PCR amplification were performed by using PrimeSTAR GXL DNA Polymerase (Clontech, USA). After purification with AMPure PB Beads, the cDNA products were then used for the construction of SMRTbell template libraries. One SMRT cell was sequenced on a PacBio Sequel instrument by using a Sequencing kit 2.1 with 10 h movie recordings. Sequencing reads were subjected to circular consensus sequences by using SMRT Analysis Software (https://www.pacb.com/products-and-ser vices/analytical-software/devnet/). PacBio reads were classified into full-length and nonfull-length sequences, and then were corrected with Illumina data generated from the same C. ficifolia RNA samples by using LoRDEC [47]. The isoform was clustered to obtain unigenes (identity = 98%) by using CD-HIT [48].
Nine cDNA libraries (the control and infected plants at 2 and 4 dpi with three biological replicates were named ck1, ck2, ck3, 2d-1, 2d-2, 2d-3, 4d-1, 4d-2, and 4d-3)were constructed for RNA-seq on the Illumina HiSeq™ 2500 platform. Reads appearing in three biological replicates of Illumina were mapped to PacBio sequence for further analysis and normalized to obtain the normalized gene expression level on the basis of Fragments Per kb per Million reads(FPKM) by using bowtie2 [49], and the formula is shown as follows:
FRKM(A) = 109C/NL.
Where FPKM(A) is the expression of gene A, C is the number of reads that are uniquely aligned to gene A, N is the total number of reads that are uniquely aligned to all genes, and L is the number of bases in gene A.
DEGs between the infection and control were screened on the basis of the general method with expression changes no less than two folds (log2 ratio ≥ 1) and false discovery rates (FDRs) < 0.05 [50]. Gene set enrichment analysis with GO data was performed on these DEGs by using Goseq [51]. These DEGs were mapped to KEGG pathway to identify key genes involved in resistance to FOC [52]. The raw Illumina sequencing data had deposit in SAR with the number SRX9738784 to SRX9738792, the raw PacBio sequencing data had deposit in SAR with the number SRX9778938 and can be obtained directly through the link:https://www.ncbi.nlm.nih.gov/sra/?term=cucurbita+ficifoliaand. The commands and parameters used for running bioinformatics programs/pipelines in this manuscript are shown in Additional file 7.
Protein extraction and DAPs identification
The TCA/acetone method was used to extract the total protein of each sample. The samples were ground into powder in liquid nitrogen. The powder was added to phenol extraction buffer, incubated for 10 min, shacked for 40 min, and centrifuged at 15,000 × g for 1 min. The precipitate was dried and resuspended in phenol extraction buffer. Then, the protein was extracted in accordance with the phenol extraction method [21].
iTRAQ analysis was completed at oeBiotech (Shanghai, China). An iTRAQ 8-plex kit was used to label digested protein samples according to the manufacturer's protocol. All of the samples were pooled and separated via liquid chromatography after labeling. Mass spectrometer data were acquired with a Triple TOF 5600 System (AB SCIEX, USA).
Protein species were identified by using Mascot software (version 2.3.02, Matrix Science Inc., 231 Boston, MA, USA) against PacBio sequences. Peptides with significant scores (≥ 20) at the 99% confidence interval were considered as identified. The identified peptide sequences were then assembled into a set of accurately identified proteins. The quantitative protein species ratios were weighted and normalized by using the default parameters of the Mascot software package. A twofold cutoff value was used to identify DAPs with P value < 0.05 [53].
The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifer PXD028075.
Quantitative RT-PCR analysis
Total RNA was isolated from the infected and control of C. ficifolia as mentioned above. Reverse transcription process and qRT-PCR was carried out following previously described methods [54]. The specific primers used for real-time PCR are shown in Additional file 8.
Supplementary Information
Additional file 1. DEGs in C. ficifolia leaf after infection by FOC at 2 dpi compared with control.
Additional file 2. DEGs in C. ficifolia leaf after infection by FOC at 4 dpi compared with control.
Additional file 3. DAPs in C. ficifolia leaf after infection by FOC at 2 dpi compared with control.
Additional file 4. DAPs in C. ficifolia leaf after infection by FOC at 4 dpi compared with control.
Additional file 5. DEGs and DAPs in C. ficifolia leaf after infection by FOC at 2 dpi.
Additional file 6. DEGs and DAPs in C. ficifolia leaf after infection by FOC at 4dpi.
Additional file 7. Commands and parameters used for running bioinformatics programs/pipelines.
Additional file 8. Primer sequences used for qRT-PCR.
Acknowledgements
We thank National Natural Science Foundation of China (No.32060083, No.31460516) and Zhang Xiaolan Expert Workstation in Yunnan Province (No.202205AF150021) for financial support.
About this supplement
This article has been published as part of BMC Genomics Volume 23 Supplement 1, 2022: The 20th International Conference on Bioinformatics (InCoB 2021): genomics. The full contents of the supplement are available online at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-23-supplement-1.Provided
Abbreviations
- ABA
Abscisic acid
- ACC
1-Aminocyclopropane-1-carboxylic acid
- ACO
1-Aminocyclopropane-1-carboxylic acid oxidase
- DAPs
Differentially accumulated proteins
- DEGs
Differentially expressed genes
- dpi
Day post infection
- ET
Ethylene
- FDR
False discovery rate
- FOC
Fusarium oxysporum f. sp. Cucumerinum
- iTRAQ
Isobaric tag for relative and absolute quantitation technology
- JA
Jasmonic acid
- ROS
Reactive oxygen species
- SA
Salicylic acid
- TFs
Transcription factors
Authors’ contributions
Z.Y. and J.X. designed the experiments. J.X. and Y.D. performed the experiments. J.X., T.G., S.H. and X.Y. analyzed the data. J.X., Y.D., K.Z. and J.Z. wrote the article. All authors read and approved the final manuscript.
Funding
This work was supported by the National Nature Science Foundation of China (No.32060083, No. 31460516) and Zhang Xiaolan Expert Workstation in Yunnan Province (No. 202205AF150021). Publication costs are funded by the National Nature Science Foundation of China (No. 32060083) and Zhang Xiaolan Expert Workstation in Yunnan Province (No. 202205AF150021). The funding bodies played no role in the design of the study, the collection, analysis, and interpretation of data, or in the writing of the manuscript.
Availability of data and materials
The raw PacBio and Illumina sequencing data had deposit in SRA with the number SRX9778938 and SRX9738784 to SRX9738792. The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifer PXD028075.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interest
The authors declare that they have no conflict interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Junjun Xie and Yumei Ding contributed equally to this work with the first author.
Contributor Information
Junjun Xie, Email: mylan1@126.com.
Yumei Ding, Email: dym@yaas.org.cn.
Ting Gao, Email: 1071121957@qq.com.
Shuilian He, Email: heshuilian2006@163.com.
Kai Zhao, Email: 34381456@qq.com.
Xuehu Yang, Email: 1348950849@qq.com.
Jie Zhang, Email: zhangjie@ynau.edu.cn.
Zhengan Yang, Email: 454483788@qq.com, Email: yangzhengan@ynau.edu.cn.
References
- 1.Lan C, Ruan H, Yang X, Yao J, Jiang J. Development of a loop-mediated isothermal amplification assay for sensitive and specific detection of Fusarium oxysporum f. sp. cucumerinum Owen. Phytoparasitica. 2018;46:283–93.
- 2.Zhang D, Meng KX, Hao YH, Fan HY, Cui N, Wang SS, Song TF. Comparative proteomic analysis of cucumber roots infected by Fusarium oxysporum f. sp. cucumerium Owen. Physiol Mol Plant Pathol. 2016;96:77–84. doi: 10.1016/j.pmpp.2016.09.002. [DOI] [Google Scholar]
- 3.Shi L, Du N, Yuan Y, Shu S, Sun J, Guo S. Vinegar residue compost as a growth substrate enhances cucumber resistance against the Fusarium wilt pathogen Fusarium oxysporum by regulating physiological and biochemical responses. Environ Sci Pollut Res. 2016;23(18):18277–18287. doi: 10.1007/s11356-016-6798-7. [DOI] [PubMed] [Google Scholar]
- 4.Edel-Hermann V, Lecomte C. Current status of fusarium oxysporum formae speciales and races. Phytopathology. 2019;109(4):512–530. doi: 10.1094/PHYTO-08-18-0320-RVW. [DOI] [PubMed] [Google Scholar]
- 5.Lee JM, Kubota C, Tsao SJ, Bie ZL, Echevarria PH, Morra L, Oda M. Current status of vegetable grafting: diffusion, grafting techniques, automation. Scientia Horticulturae. 2010;127:0–105.
- 6.Anthony PK, William PW, William BR, Paula AA. Cucurbit rootstocks resistant to fusarium oxysporum f. sp. niveum remain resistant when coinfected by meloidogyne incognita in the field. Plant Dis. 2019;103(6):1383–1390. doi: 10.1094/PDIS-10-18-1869-RE. [DOI] [PubMed] [Google Scholar]
- 7.Moya-Hernández A, Bosquez-Molina E, Serrato-Díaz A, Blancas-Flores G, Alarcón-Aguilar FJ. Analysis of genetic diversity of Cucurbita ficifolia Bouché from different regions of Mexico, using AFLP markers and study of its hypoglycemic effect in mice. S Afr J Bot. 2018;116:110–115. doi: 10.1016/j.sajb.2018.02.409. [DOI] [Google Scholar]
- 8.Huang Y, Tang R, Cao Q, Bie ZL. Improving the fruit yield and quality of cucumber by grafting onto the salt tolerant rootstock under NaCl stress. Sci Hortic. 2009;122:26–31. doi: 10.1016/j.scienta.2009.04.004. [DOI] [Google Scholar]
- 9.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 10.Shigenaga AM, Argueso CT. No hormone to rule them all : Interactions of plant hormones during the responses of plants to pathogens. Seminars in Cell Developmental Biology. 2016;56:174–189. doi: 10.1016/j.semcdb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Broekaert WF, Delaure SL, De Bolle MFC, Cammue BPA. The role of ethylene in host-pathogen interactions. Annu Rev Phytopathol. 2006;44:393–416. doi: 10.1146/annurev.phyto.44.070505.143440. [DOI] [PubMed] [Google Scholar]
- 12.Swarupa V, Ravishankar KV, Rekha A. Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta. 2014;239:735–751. doi: 10.1007/s00425-013-2024-8. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, Ling N, Dong X, Zhu Y, Shen Q, Guo S. Thermographic visualization of leaf response in cucumber plants infected with the soil-borne pathogen Fusarium oxysporum f. sp. cucumerinum. Plant Physiol Biochem. 2012;61:153–161. doi: 10.1016/j.plaphy.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Wu F. Differentially expressed transcripts from cucumber (Cucumis sativus L) root upon inoculation with Fusarium oxysporum f. sp. cucumerinum Owen. Physiol Mol Plant Pathol. 2010;74(2):142–150. doi: 10.1016/j.pmpp.2009.10.005. [DOI] [Google Scholar]
- 15.Silvia SM, Bagnaresi P, Sestili S, Biselli C, Zechini A, Orrù L, Cattivelli L, Ficcadenti N. Transcriptome analysis of the melon-fusarium oxysporum f. sp. melonis race 12 pathosystem in susceptible and resistant plants. Front Plant Sci. 2017;8:362. doi: 10.3389/fpls.2017.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe GA, Major IT, Koo AJ. Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol. 2018;69:387–415. doi: 10.1146/annurev-arplant-042817-040047. [DOI] [PubMed] [Google Scholar]
- 17.Ye J, Zhang L, Zhang X, Wu X, Fang R. Plant defense networks against insect-borne pathogens. Trends Plant Sci. 2021;26(3):272–287. doi: 10.1016/j.tplants.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elmore JM, Griffin BD, Walley JW. Advances in functional proteomics to study plant-pathogen interactions. Curr Opin Plant Biol. 2021;63:102061. doi: 10.1016/j.pbi.2021.102061. [DOI] [PubMed] [Google Scholar]
- 20.Chen QZ, Guo WS, Feng LZ, Ye XZ, Xie WF, Huang XP, Liu JY. Transcriptome and proteome analysis of Eucalyptus infected with Calonectria pseudoreteaudii. J Proteomics. 2015;115:117–131. doi: 10.1016/j.jprot.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Dai F, Wang Z, Li Z, Luo G, Wang Y, Tang C. Transcriptomic and proteomic analyses of mulberry (Morus atropurpurea) fruit response to Ciboria carunculoides. J Proteomics. 2019;193:142–153. doi: 10.1016/j.jprot.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Lubberstedt T, Xu M. The Genetic and Molecular Basis of Plant Resistance to Pathogens. J Genet Genomics. 2013;40:23–35. doi: 10.1016/j.jgg.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Ding Yumei, Zhang Jie, Xie Junjun, Yao Chunxin, Zhou Xiaogang, Zhou Liying, Yang Zhengan, Zhang Xingguo. Expression analysis of HQRGA2 and differences of Anti-oxidant enzymes in three varieties of Cucurbita ficifolia under stress of Fusarium oxysporum f. sp. Cucumerrinum. Plant Physiol J. 2019;55(3):359–366. [Google Scholar]
- 24.Argueso CT, Hansen M, Kieber JJ. Regulation of ethylene biosynthesis. J Plant Growth Regul. 2007;26:92–105. doi: 10.1007/s00344-007-0013-5. [DOI] [Google Scholar]
- 25.Henfling J, Bostock R, Kuc J. Effect of abscisic-acid on rishitin and lubiminaccumulation and resistance to Phytophthora infestans and Cladosporium cucumerinum in potato tuber tissue slices. Phytopathology. 1980;70:1074–1078. doi: 10.1094/Phyto-70-1074. [DOI] [Google Scholar]
- 26.Salt SD, Tuzun S, Kuc J. Effects of beta-ionone and abscisic-acid on the growth of tobacco and resistance to blue mold—mimicry of effects of stem infection by Peronospora tabacina Adam. Physiol Mol Plant Pathol. 1986;28:287–297. doi: 10.1016/S0048-4059(86)80071-6. [DOI] [Google Scholar]
- 27.de Zelicourt A, Colcombet J, Hirt H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016;21(8):677–685. doi: 10.1016/j.tplants.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins : regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Luan Y, Li J, Yin Y. Expression of a tomato MYB gene in transgenic tobacco increases resistance to Fusarium oxysporum and Botrytis cinerea. Eur J Plant Pathol. 2015;144(3):607–617. doi: 10.1007/s10658-015-0799-0. [DOI] [Google Scholar]
- 30.Chakraborty J, Ghosh P, Sen S, Nandi AK, Das S. CaMPK9 increases the stability of CaWRKY40 transcription factor which triggers defense response in chickpea upon Fusarium oxysporum f. sp. ciceri Race1 infection. Plant Mol Biol. 2019;100:411–31. [DOI] [PubMed]
- 31.Nuruzzaman M, Sharoni AM, Kikuchi S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol. 2013;4:1–16. doi: 10.3389/fmicb.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collinge M, Boller T. Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol. 2001;46:521–529. doi: 10.1023/A:1010639225091. [DOI] [PubMed] [Google Scholar]
- 33.Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Delessert C, Kazan K, Wilson IW, Van Der Straeten D, Manners J, Dennis ES, Dolferus R. The transcriptionfactor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005;43:745–757. doi: 10.1111/j.1365-313X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- 35.Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim JM, Park CM. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61:661–671. doi: 10.1111/j.1365-313X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- 36.Lotfy S, Javelle F, Negrel J. Distribution of hydroxycinnamoyl-CoA: ω-hydroxypalmitic acid O -hydroxycinnamoyltransferase in higher plants. Phytochemistry. 1995;40:389–391. doi: 10.1016/0031-9422(95)00284-E. [DOI] [Google Scholar]
- 37.Serra O, Marçal S, Hohn C, Franke R, Schreiber L, Prat S, Molinas M, Figueras M. Silencing of StKCS6 in potato periderm leads to reduced chain lengths of suberin and wax compounds and increased peridermal transpiration. J Exp Bot. 2009;60:697–707. doi: 10.1093/jxb/ern314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhar A, Gupta S, Chatterjee M, Sen S, Das S. Differential expressions of photosynthetic genes provide clues to the resistance mechanism during Fusarium oxysporum, fsp ciceri, race 1 (Foc1) infection in chickpea ( Cicer arietinum, L) Eur J Plant Pathol. 2017;148:533–549. doi: 10.1007/s10658-016-1109-1. [DOI] [Google Scholar]
- 39.Major IT, Nicole MC, Duplessis S, Séguin A. Photosynthetic and respiratory changes in leaves of poplar elicited by rust infection. Photosynth Res. 2010;104:41–48. doi: 10.1007/s11120-009-9507-2. [DOI] [PubMed] [Google Scholar]
- 40.Zamany A, Liu JJ, Ekramoddoullah AK. Comparative proteomic profiles of Pinus monticola needles during early compatible and incompatible interactions with Cronartium ribicola. Planta. 2012;236:725–1746. doi: 10.1007/s00425-012-1715-x. [DOI] [PubMed] [Google Scholar]
- 41.Santos EC, Pirovani CP, Correa S, Micheli F, Gramacho KP. The pathogen Moniliophthora perniciosa promotes differential proteomic modulation of cacao genotypes with contrasting resistance to witches broom disease. BMC Plant Biol. 2020;20:1. doi: 10.1186/s12870-019-2170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Averyanov A. Oxidative burst and plant disease resistance. Front Biosci. 2009;1:142–152. doi: 10.2741/s14. [DOI] [PubMed] [Google Scholar]
- 43.Torres MA. ROS in biotic interactions. Physiol Plant. 2010;138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 44.Bailey-Serres JRM. The roles of reactive oxygen species in plant cells. Plant Physiol. 2006;141:311. doi: 10.1104/pp.104.900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Passardi F, Cosio C, Penel C, Dunand C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005;24:255–265. doi: 10.1007/s00299-005-0972-6. [DOI] [PubMed] [Google Scholar]
- 46.Federico M, Reagan RL, Dolan D, Veronica F, Abhaya MD. Proteomic analysis highlights the role of detoxification pathways in increased tolerance to Huanglongbing disease[J] BMC Plant Biol. 2016;16(1):167. doi: 10.1186/s12870-016-0858-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30(24):3506–3514. doi: 10.1093/bioinformatics/btu538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–329. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gan CS, Chong PK, Pham TK, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) J Proteome Res. 2007;6:821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 54.Xie JJ, Lei B, Niu ML, Huang Y, Q. Kong Q, Bie ZL. High throughput sequencing of small RNAs in the two cucurbita germplasm with different sodium accumulation patterns identifies novel microRNAs involved in salt stress response. PLoS One. 2015;10(5):e0127412. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. DEGs in C. ficifolia leaf after infection by FOC at 2 dpi compared with control.
Additional file 2. DEGs in C. ficifolia leaf after infection by FOC at 4 dpi compared with control.
Additional file 3. DAPs in C. ficifolia leaf after infection by FOC at 2 dpi compared with control.
Additional file 4. DAPs in C. ficifolia leaf after infection by FOC at 4 dpi compared with control.
Additional file 5. DEGs and DAPs in C. ficifolia leaf after infection by FOC at 2 dpi.
Additional file 6. DEGs and DAPs in C. ficifolia leaf after infection by FOC at 4dpi.
Additional file 7. Commands and parameters used for running bioinformatics programs/pipelines.
Additional file 8. Primer sequences used for qRT-PCR.
Data Availability Statement
The raw PacBio and Illumina sequencing data had deposit in SRA with the number SRX9778938 and SRX9738784 to SRX9738792. The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifer PXD028075.