Abstract
Standard doses of antibiotics do not efficiently treat chronic infections of the soft tissue and bone. In this Personal View, we advocate for improving treatment of these infections by taking the infectious microenvironment into account. The infectious microenvironment can cause sensitive bacteria to lose their susceptibility to antibiotics that are effective in standard laboratory susceptibility testing. We propose that bacteria behave substantially different in standard laboratory conditions than they do in actual infections. The infectious microenvironment could impose changes in growth and metabolic activity that result in increased protection against antibiotics. Therefore, we advocate that improved antibiotic treatment of chronic infection is achievable when antibiotics are recommended on the basis of susceptibility testing in relevant in vitro conditions that resemble actual infectious microenvironments. We recommend establishing knowledge of the relevant conditions of the chemical and physical composition of the infectious microenvironment. Recent advances in RNA sequencing, metabolomics, and microscopy have made it possible for the characterisation of the microenvironment of infections and to validate the clinical relevance of in vitro conditions to actual infections.
Introduction
We have long believed that laboratory models have provided insights into bacterial behaviour in the human body. Since the times of Robert Koch and Louis Pasteur, two pioneers in microbiology, bacteria isolated from people with an infection were cultured in liquid media or on agar plates with great success. Many of these methods are still used, including in the pharmaceutical industry in which bacteria grown in laboratory media are used to screen for and identify promising antimicrobials, and in clinical microbiology to evaluate the susceptibility of bacteria to antibiotics.1–3 Yet, foundational work in behavioural microbiology has shown that bacteria display intricate phenotypes dictated by a complex and variable surrounding microenvironment,4–7 leading to the question: can the course and therapeutic outcome of bacterial infections in humans be predicted by studying bacteria grown in test tubes? Nevertheless, this form of reductionism has been the foundation of microbiology research during the past 150 years.8 In this Personal View, we propose that if we can understand and exploit the environmental conditions within an infection, we might know how and why to treat with specific drugs, rather than just when.
Infectious microenvironment and why it matters
Although the local microenvironment of an infected body site changes from the healthy situation,9,10 the chemical composition and physical properties associated with these changes are still far from fully characterised. This has implications for understanding the behaviour of bacteria and other microorganisms within healthy and diseased sites, the status of the immune response, and the efficacy of administered antibiotics. For example, recent studies have provided compelling evidence that the structured microbial communities within some human infections behave substantially different than those in the laboratory.6,11–13
When microbial pathogens and the host immune cells that are released in response become locally concentrated, the concerted metabolism alters the chemical microenvironment (figure). These local changes have consequences for bacterial persistence14–16 by reducing antibiotic susceptibility, diversifying the physiological states occupied by the microorganisms, and compromising the efficacy of immune cell function. At the site of infection (figure), bacteria might be planktonic or in multicellular aggregates, possibly attached to an implanted device. The milieu comprises a dense accumulation of host cells (some of which might be dead or inactive), microorganisms and their extracellular polymeric substances, and host polymers such as a fibrotic capsule or extracellular DNA. This structure is permeated by concentration gradients in metabolic substrates, such as oxygen or glucose (decreasing from the exterior towards the implant or infection centre), and metabolic products, such as lactate, virulence factors, and cytokines (increasing from the exterior towards the implant or infection centre). The varied chemical and biochemical microenvironments encompass conditions in which microbial cells might be protected from being killed by antibiotics or antimicrobial peptides (eg, due to diminished metabolic activity or growth) and where immune cells might be less effective (eg, due to local hypoxia and bacterial toxins). The microenvironment could also be mechanically altered: the deposition and alteration of bacterial and host polymers can be expected to affect transport properties and physically restrict motility and function of immune cells (figure).
Figure: The infectious microenvironment.
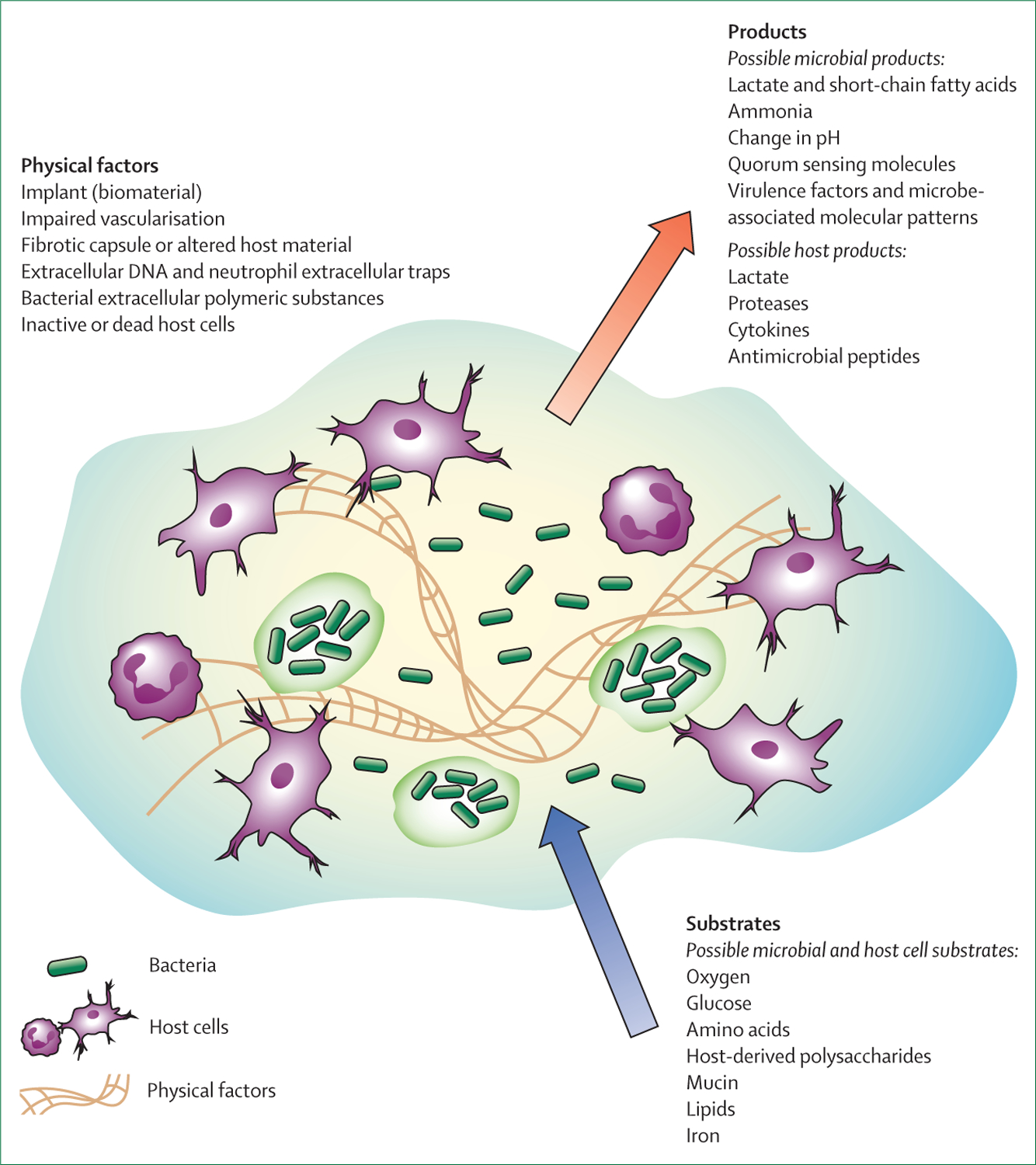
The biochemical and physical microenvironment can be profoundly altered at the site of localised infection.
Antibiotic treatment is more than just minimal inhibitory concentration breakpoints
Microbial antibiotic susceptibility depends strongly on the metabolic and physiological state of the cell,17 which in turn is governed by the chemical microenvironment. Local starvation of a nutrient or electron acceptor required for growth can cause bacteria to enter a non-growing state in which their relative inactivity renders them invulnerable to many antimicrobial agents.18–20 Even actively growing microorganisms could become less susceptible when their metabolism switches—eg, from aerobic respiration to denitrification or fermentation. For instance, rapidly growing Escherichia coli cells grown for 6 h,21 were decimated by kanamycin when challenged on lysogeny broth medium (8·4 log reduction) but scarcely affected when the same medium was supplemented with glucose (1·2 log reduction). Thus, although standard antibiotic regimens devised from laboratory studies of bacterial antibiotic susceptibility under a single optimised growth condition are sufficient to resolve most acute and short-term infections of well vascularised body sites, these doses of antibiotics do not efficiently treat chronic soft tissue and bone infections, with or without implants.
This scarcity of pathogen eradication by antimicrobial chemotherapy has been ascribed to the development of tolerant aggregated bacterial consortia termed biofilms. Biofilms are defined as a coherent cluster of bacterial cells imbedded in a matrix, which are more tolerant to most antimicrobials and the host defence than planktonic bacterial cells.22 Although it was originally proposed that biofilm tolerance arose as a direct result of bacterial aggregation, recent investigations suggest that the microenvironment shapes bacterial behaviour, thus resulting in antibiotics that do not work.23,24 These studies suggest that the altered microenvironment might be as or more important in determining the chronicity of an infection than biofilm formation itself.
The cellular innate immune response to infection, including neutrophils and macrophages, is also strongly influenced by the local microenvironment. For example, molecular oxygen, which is essential for the generation of reactive oxygen species, is one of the crucial weapons used by phagocytes to destroy bacteria. In hypoxic or anoxic environments, this killing mechanism is scarce or disabled.25
The infectious microenvironment is both complex and dynamic in nature. A strong reciprocal coupling can be anticipated: the microenvironment determines pathogen metabolism and growth, which then reshapes the microenvironment that constrains the host response that further modifies the microenvironment. Recent studies have revealed metabolic interactions between host and pathogen,24,26 and between different species of microorganisms.27 As the microenvironment courses along a trajectory, shifts in the microbial ecology of the site and evolution of populations by selection of mutants will naturally follow.28
In addition, distinguishing between colonisation and infection is crucial, since only infection is recognised to provoke an inflammatory host response, whereas colonisation, including by our own microbiota, might induce beneficial interactions. Thus, an introduced pathogen could create an infection or a perturbed host environment (eg, because of inflammation around an implant) resulting in an environment susceptible to infection. Once the interaction is initiated, the environment is reciprocally and continually reshaped by both host and pathogen, and is termed the infectious microenvironment (figure).29 We have expanded this term to describe the change from a balanced microenvironment within the healthy host, to an environment that is at risk of being colonised and infected by microorganisms. Thus, we define the infectious microenvironment as an environment that either promotes colonisation by pathogens or alteration of the microbiota to a pathogenic state, and that once colonised, provides protection from antibiotics and immune function.
We know from several studies that insertion of an implant into a body, surgical interventions, and impaired vascularisation due to pathological changes, favour infection and impair the delivery and function of antibiotics.5,30–32 Thus, the infectious microenvironment is initially created when the normal balance is disturbed, such as when an incision is made by a surgeon or an implant is inserted. We hypothesise that the infectious microenvironment determines susceptibility to antimicrobial chemotherapy and host immune response efficacy. A corollary would be that the outcome of antimicrobial chemotherapy and clearance by the immune defences cannot be accurately modelled in the laboratory without capturing key features of the infectious microenvironment. The implication is that we cannot simply grow bacteria in common laboratory conditions to understand the effectiveness of an antimicrobial treatment strategy, and we might be missing out on new antibiotics that could be effective in vivo but not in vitro.
Antimicrobial susceptibility testing of cultured pathogens has traditionally been on the basis of disk diffusion or minimal inhibitory concentrations breakpoints related to the pharmacokinetic and pharmacodynamic properties of most antimicrobials, including a focus on specialised compartments, such as the spinal fluid. These protocols have been used to predict which antimicrobials to use with variable success. Standardisation of antimicrobial breakpoints such as EUCAST or CLSI have proven reproducible and are an effective and thorough method for optimising treatment of acute infections. However, the use of the same defined conditions—usually rich growth media and organisms in exponential growth—conceals the huge dependence of antimicrobial efficacy on growth conditions. Substrates and conditions, such as oxygen, carbon sources, redox potential, pH, virulence factors, viscosity, material properties, and the growth status of the microorganisms, can have profound effects on antibiotic efficacy.17 Therefore, conventional antimicrobial tests are most likely only informative in instances where bacteria are growing rapidly, as has been proposed in some acute infections. This drawback is not overcome with the use of clinical bacterial isolates since these are still highly responsive to the growth conditions of the assay, but do not have the characteristics of the infectious microenvironment.11 Lack of consideration for the infectious microenvironment is also problematic for the recent focus on whole-genome sequencing of clinical isolates to predict antibiotic susceptibility. The genotype of clinical isolates only reflects the functional capacity of the bacterium and is limited in its ability to accurately predict complex ecological responses, including antibiotic tolerance.11 Thus, although all methods have their strengths, we must also recognise their weaknesses.
For more on EUCAST see https://www.eucast.org/
For more on CLSI see https://clsi.org/
Awareness of the infectious microenvironment and parallels to the tumour microenvironment
Awareness of the infectious microenvironment and its role in treatment and disease is growing. Although many articles now acknowledge the existence of an altered microenvironment, most do not take an interdisciplinary approach focused on integrating the contributions of microorganisms, immune status, and the chemical and physical properties of the environment. Only a few articles encompass the complexity and highlight the necessity for increased awareness of the infectious microenvironment for its role in pathogenesis and treatment failure.33–35
In the field of cancer, there is a deep appreciation for the role of the tumour microenvironment in determining pathogenesis and efficacy of chemotherapy. A closer resemblance to the tumour microenvironment has been achieved in cultures of cells grown as three dimensional spheroids and has advanced drug testing in cancer therapy.36 Strong negative effects of the tumour microenvironment on the outcome of chemotherapy have been well accepted for decades in treatment of tumours.37 Some stressors, mainly hypoxia, exist in the tumour microenvironment. Intratumoural hypoxia results from the changed metabolism and extensive growth of tumour cells, and from delayed angiogenesis and oxygen supply.38 The metabolic consequences of hypoxia include specific impairments of protein and lipid synthesis that are counterproductive to cell growth and proliferation.39 Hypoxia promotes chemoresistance in cancer40 and represents an independent prognostic factor for several types of cancers.41 Diminished availability of oxygen could cause reduced growth, which is connected to increased chemoresistance42 resembling the low susceptibility to antibiotics in bacteria with slow growth. Additional strategies similar to mechanisms that promote protection of bacteria against antibiotics are also induced by hypoxia in tumours. By inducing activation of hypoxia-inducible factors, hypoxia might stimulate efflux pumps, DNA damage inhibition, and antioxidative defence leading to chemoresistance in tumour cells.37 This insight into the significance of the microenvironment for the responsiveness of cancer cells to chemotherapy has been realised with three dimensional cultures of patient-derived cancer organoids (PDTO) for drug testing. By simulating the tumour microenvironment with PDTO, the outcome of in vitro drug exposure tests was correlated with the individual therapy response,43,44 which qualified PDTO models as a central strategy in personalised medicine programmes.45 Parallels to the progress in tumour research could serve as inspiration to incorporate in vivo microenvironment in future optimisations of antibiotic therapy of infectious bacteria. This strategy to identify optimal treatment of bacteria isolated from chronic infections could help close the gap between the outcome of conventional susceptibility testing and the clinical outcome.46,47
Recommendations for models, methods, and interdisciplinary approaches
As a scientific community, we propose that it is essential to take a step back and recognise the shortcomings of our current infection models, both in vitro and in vivo. We also need to develop versatile and realistic in vitro and in vivo models, and understand that in vitro models can be more powerful than animal models depending on the research question.48 Our models must capture the infectious microenvironment by using in vivo microscopy images, chemical measurements, human infection transcriptomes, and other data that describe the infectious microenvironment. In short, we need to understand the infection ecology.
In addition, it is important to validate targets, whether they are diagnostic, therapeutic, or preventative, with the use of the most current and direct methods available, including RNA sequencing, metabolomics, immunohistochemistry, and advanced microscopy on patient samples. When developing therapeutics, it is essential to determine whether the target is expressed and essential within the infection or only in the laboratory model in which it was studied. It is also crucial to develop an in-depth understanding and description of the infectious microenvironment in different types of infections and anatomical sites, sampling directly from these infections rather than inferring this environment from serum or in vitro measurements. This in vivo behaviour has recently been done by assessing the transcriptome of bacteria during human infection, determining which genes are differentially expressed in humans compared with in vitro models, and updating the in vitro models accordingly by adjusting variables, such as oxygen and nutrients, so that the bacterial transcriptomes more closely resemble in vivo conditions.6
In conclusion, if 150 years of targeted immunological and microbiological research has left us with little understanding of the infectious microenvironment, how do we rectify these shortcomings? We propose that this requires an interdisciplinary, holistic approach focused on cataloguing individual components of the infectious microenvironment and rethinking models to incorporate these components. A paradigm shift is needed to solve the complex problems of infection and antibiotic tolerance. Researchers, clinicians, universities, private foundations, drug companies, politicians, and the general public must embrace and invest in a holistic view of health science. We all have a role to play because these problems affect us all.
Acknowledgments
We would like to acknowledge Jill Story for preparing the figure. KPR received grants supporting the basis of this work from the US National Institutes of Health (NIH; R21 AI137462-01A1) and the Ted Nash Long Life Foundation; PSS received grants from NIH (1R01NR16986) and the Lundbeck Foundation; MW received grants from NIH (1R01GM116547, R01DE023193, 5R01DE020100); and TB received grants from the Lundbeck Foundation.
Footnotes
Declaration of interests
We declare no competing interests.
References
- 1.Leclercq R, Canton R, Brown DF, et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect 2013; 19: 141–60. [DOI] [PubMed] [Google Scholar]
- 2.Parker AE, Hamilton MA, Goeres DM. Reproducibility of antimicrobial test methods. Sci Rep 2018; 8: 12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Høiby N, Bjarnsholt T, Moser C, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 2015; 21 (suppl 1): S1–25. [DOI] [PubMed] [Google Scholar]
- 4.Kragh KN, Alhede M, Jensen PO, et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun 2014; 82: 4477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabbe A, Jensen PO, Bjarnsholt T, Coenye T. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol 2019; 27: 850–63. [DOI] [PubMed] [Google Scholar]
- 6.Cornforth DM, Dees JL, Ibberson CB, et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci USA 2018; 115: E5125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci USA 2014; 111: 7819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blevins SM, Bronze MS. Robert Koch and the ‘golden age’ of bacteriology. Int J Infect Dis 2010; 14: e744–51. [DOI] [PubMed] [Google Scholar]
- 9.Worlitzsch D, Tarran R, Ulrich M, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 2002; 109: 317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Maltesen RG, Larsen LH, et al. In vivo gene expression in a Staphylococcus aureus prosthetic joint infection characterized by RNA sequencing and metabolomics: a pilot study. BMC Microbiol 2016; 16: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi E, Falcone M, Molin S, Johansen HK. High-resolution in situ transcriptomics of Pseudomonas aeruginosa unveils genotype independent patho-phenotypes in cystic fibrosis lungs. Nat Commun 2018; 9: 3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolpen M, Kragh KN, Bjarnsholt T, et al. Denitrification by cystic fibrosis pathogens—Stenotrophomonas maltophilia is dormant in sputum. Int J Med Microbiol 2015; 305: 1–10. [DOI] [PubMed] [Google Scholar]
- 13.DePas WH, Starwalt-Lee R, Van Sambeek L, Ravindra Kumar S, Gradinaru V, Newman DK. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 2016; 7: e00796–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown SA, Palmer KL, Whiteley M. Revisiting the host as a growth medium. Nat Rev Microbiol 2008; 6: 657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith H. What happens to bacterial pathogens in vivo? Trends Microbiol 1998; 6: 239–43. [DOI] [PubMed] [Google Scholar]
- 16.Stewart PS. Biophysics of biofilm infection. Pathog Dis 2014; 70: 212–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart PS. Antimicrobial tolerance in biofilms. Microbiol Spectr 2015; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol 2017; 15: 453–64. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen D, Joshi-Datar A, Lepine F, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 2011; 334: 982–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernier SP, Lebeaux D, DeFrancesco AS, et al. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 2013;9: e1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuroff TR, Bernstein H, Lloyd-Randolfi J, Jimenez-Taracido L, Stewart PS, Carlson RP. Robustness analysis of culturing perturbations on Escherichia coli colony biofilm beta-lactam and aminoglycoside antibiotic tolerance. BMC Microbiol 2010; 10: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burmolle M, Ren D, Bjarnsholt T, Sorensen SJ. Interactions in multispecies biofilms: do they actually matter? Trends Microbiol 2014; 22: 84–91. [DOI] [PubMed] [Google Scholar]
- 23.Stewart PS, White B, Boegli L, et al. Conceptual model of biofilm antibiotic tolerance that integrates phenomena of diffusion, metabolism, gene expression, and physiology. J Bacteriol 2019; 201: e00307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heim CE, Bosch ME, Yamada KJ, et al. Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat Microbiol 2020; 5: 1271–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hajdamowicz NH, Hull RC, Foster SJ, Condliffe AM. The impact of hypoxia on the host-pathogen interaction between neutrophils and Staphylococcus aureus. Int J Mol Sci 2019; 20: 5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traven A, Naderer T. Central metabolic interactions of immune cells and microbes: prospects for defeating infections. EMBO Rep 2019; 20: e47995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Røder HL, Olsen NMC, Whiteley M, Burmolle M. Unravelling interspecies interactions across heterogeneities in complex biofilm communities. Environ Microbiol 2020; 22: 5–16. [DOI] [PubMed] [Google Scholar]
- 28.Rossi E, La Rosa R, Bartell JA, et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol 2021; 19: 331–42. [DOI] [PubMed] [Google Scholar]
- 29.Yang JH, Bhargava P, McCloskey D, Mao N, Palsson BO, Collins JJ. Antibiotic-induced changes to the host metabolic environment inhibit drug efficacy and alter immune function. Cell Host Microbe 2017; 22: 757–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen LK, Bjarnsholt T, Kragh KN, et al. In vivo gentamicin susceptibility test for prevention of bacterial biofilms in bone tissue and on implants. Antimicrob Agents Chemother 2019; 63: e01889–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bue M, Hanberg P, Koch J, et al. Single-dose bone pharmacokinetics of vancomycin in a porcine implant-associated osteomyelitis model. J Orthop Res 2018; 36: 1093–98. [DOI] [PubMed] [Google Scholar]
- 32.Jensen PO, Kolpen M, Kragh KN, Kuhl M. Microenvironmental characteristics and physiology of biofilms in chronic infections of CF patients are strongly affected by the host immune response. APMIS 2017; 125: 276–88. [DOI] [PubMed] [Google Scholar]
- 33.Lewis ME, Belland RJ, AbdelRahman YM, et al. Morphologic and molecular evaluation of Chlamydia trachomatis growth in human endocervix reveals distinct growth patterns. Front Cell Infect Microbiol 2014; 4: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lobritz MA, Belenky P, Porter CB, et al. Antibiotic efficacy is linked to bacterial cellular respiration. Proc Natl Acad Sci USA 2015;112: 8173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sønderholm M, Kragh KN, Koren K, et al. Pseudomonas aeruginosa aggregate formation in an alginate bead model system exhibits in vivo-like characteristics. Appl Environ Microbiol 2017; 83: e00113–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edmondson R, Broglie JJ, Adcock AF, Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev Technol 2014; 12: 207–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohwer N, Cramer T. Hypoxia-mediated drug resistance: novel insights on the functional interaction of HIFs and cell death pathways. Drug Resist Updat 2011; 14: 191–201. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Jin M-Z, Yang Z-Y, Jin W-L. Microglia in neurodegenerative diseases. Neural Regen Res 2021; 16: 270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeBerardinis RJ. Is cancer a disease of abnormal cellular metabolism? New angles on an old idea. Genet Med 2008; 10: 767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizin-Towle L, Hall EJ. Studies with bleomycin and misonidazole on aerated and hypoxic cells. Br J Cancer 1978; 37: 254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer 2002; 2: 38–47. [DOI] [PubMed] [Google Scholar]
- 42.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst 2007;99: 1441–54. [DOI] [PubMed] [Google Scholar]
- 43.Steele NG, Chakrabarti J, Wang J, et al. An organoid-based preclinical model of human gastric cancer. Cell Mol Gastroenterol Hepatol 2019; 7: 161–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vlachogiannis G, Hedayat S, Vatsiou A, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018; 359: 920–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambardella V, Tarazona N, Cejalvo JM, et al. Personalized medicine: recent progress in cancer therapy. Cancers (Basel) 2020; 12: 1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen PØ, Møller SA, Lerche CJ, et al. Improving antibiotic treatment of bacterial biofilm by hyperbaric oxygen therapy: not just hot air. Biofilm 2019; 1: 100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coenye T, Goeres D, Van Bambeke F, Bjarnsholt T. Should standardized susceptibility testing for microbial biofilms be introduced in clinical practice? Clin Microbiol Infect 2018; 24: 570–72. [DOI] [PubMed] [Google Scholar]
- 48.Cornforth DM, Diggle FL, Melvin JA, Bomberger JM, Whiteley M. Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio 2020; 11: e03042–19. [DOI] [PMC free article] [PubMed] [Google Scholar]


