Abstract
Comparative sequence analysis of a 16S rRNA gene clone library from the chemocline of the meromictic Lake Cadagno (Switzerland) retrieved two clusters of sequences resembling sulfate-reducing bacteria within the family Desulfovibrionaceae. In situ hybridization showed that, similar to sulfate-reducing bacteria of the family Desulfobacteriaceae, bacteria of one cluster with similarity values to the closest cultured relatives of between 92.6 and 93.1% resembled free cells or cells loosely attached to other cells or debris. Bacteria of the second cluster closely related to Desulfocapsa thiozymogenes DSM7269 with similarity values between 97.9 and 98.4% were generally associated with aggregates of different small-celled phototrophic sulfur bacteria, suggesting a potential interaction between the two groups of bacteria.
Lake Cadagno is a meromictic lake in the Piora valley in the south of Switzerland characterized by a high salinity of the monimolimnion and a permanent chemocline at a depth between 9 and 14 m separating the aerobic epilimnion from the anaerobic, sulfidogenic hypolimnion (21, 29). Due to the infiltration of water through dolomite rich in gypsum, the water chemistry of Lake Cadagno is dominated by inorganic sulfur compounds with high concentrations of sulfate and steep gradients of sulfide in the chemocline (11, 14). A turbidity maximum in the chemocline is correlated with elevated numbers of bacteria (up to 107 cells ml−1), indicating that a bacterial community making use of these gradients is present (26, 27).
In a previous study using in situ hybridization with 16S and 23S rRNA targeted oligonucleotide probes, we demonstrated that the bacterial community in the chemocline of Lake Cadagno mainly consisted of Proteobacteria (26, 27). Averaged over the whole chemocline, cells hybridizing with probes ALF1b, BET42a, GAM42a, and SRB385, targeting respective members of the α, β, γ, and δ subdivisions of Proteobacteria, respectively accounted for 23, 17, 45, and 15% of the DAPI-stained bacteria (26, 27). Phototrophic sulfur bacteria belonging to the γ subdivision of Proteobacteria were most prominent, averaging 33% of the bacteria (7, 21). In situ hybridization identified all large-celled phototrophic sulfur bacteria as Chromatium okenii, while small-celled phototrophic sulfur bacteria consisted of four major populations forming a tight cluster with Amoebobacter purpureus and Lamprocystis roseopersicina (27). Small-celled phototrophic sulfur bacteria were usually found in aggregates, together with cells that hybridized with probe SRB385 targeting sulfate-reducing bacteria of the family Desulfovibrionaceae (26, 27). Since the populations of small-celled phototrophic sulfur bacteria displayed different distribution profiles in the chemocline, indicating different ecophysiological adaptations (27), we were interested to see whether the associated sulfate-reducing bacteria also resembled different populations and whether these were associated with specific populations of phototrophic sulfur bacteria.
For this purpose, representative clones of 82 phylotypes of a 16S rRNA gene clone library from the chemocline of Lake Cadagno that was generated in Escherichia coli and screened for phylotype distribution by restriction analysis in a previous study (3) were analyzed by whole-cell hybridization with Cy3-labeled probes SRB385 or SRB385Db (22) to retrieve clones representing sulfate-reducing bacteria of the families Desulfovibrionaceae and Desulfobacteriaceae, respectively. Five-microliter samples of paraformaldehyde-fixed E. coli cultures spotted onto gelatin-coated slides were hybridized in the presence of nonlabeled competitor probe with 20% formamide in hybridization buffer at 53°C for 2 h as described by Zarda et al. (30). Following hybridization, the slides were washed in buffer without formamide (5 mM EDTA, 20 mM Tris [pH 7.0], 215 mM NaCl, and 0.001% sodium dodecyl sulfate) at 55°C for 20 min. and were examined by epifluorescence microscopy (27). Thirteen clones hybridized with probe SRB385, and 10 hybridized with SRB385Db. Since we were interested in sulfate-reducing bacteria associated with phototrophic sulfur bacteria, further analyses focused on clones hybridizing to probe SRB385. Eight clones with rDNA fragments showing distinct restriction patterns were selected, and the fragments were reamplified and sequenced as described elsewhere (27). The sequences were aligned initially with a subset of bacterial 16S rDNA sequences obtained from the Ribosomal Database Project (16) by using the CLUSTAL W service at EBI (12). Phylogenetic relationships were estimated by using the Phylogeny Inference Package (PHYLIP, version 3.573c). Kimura two-parameter evolutionary distances were calculated by using the DNADIST program, and a phylogenetic tree was derived by using the FITCH program with random order input of sequences and the global rearrangement option (5). The absence of chimeras was verified by submitting our sequences to the RDP program CHECK_CHIMERA (16).
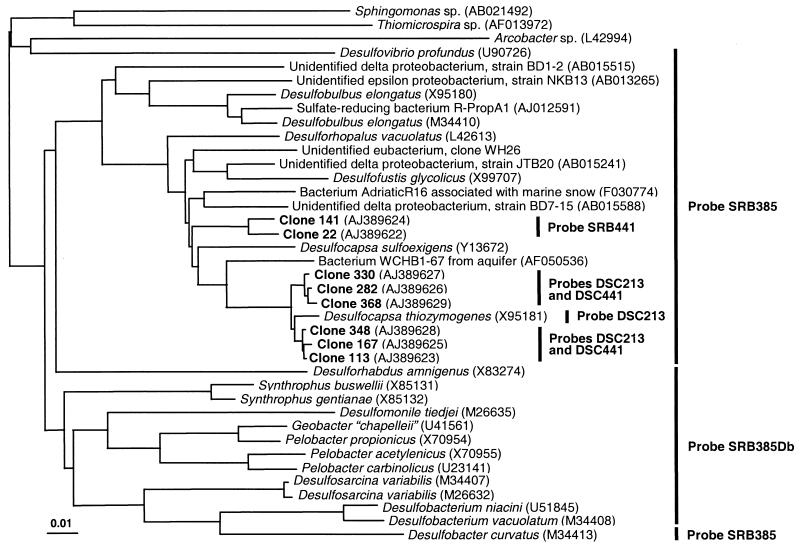
Comparative sequence analysis revealed the presence of two distinct clusters within the δ subdivision of Proteobacteria (Fig. 1). Sequences of one cluster consisting of six clones were closely related to that of Desulfocapsa thiozymogenes DSM7269, with similarity values between 97.9 and 98.4%. The closest cultured relatives to the second cluster of two clones were Desulfofustis glycolicus DSM9705, D. thiozymogenes DSM7269, Desulfocapsa sulfoexigens DSM10523, and Desulforhopalus vacuolatus DSM9700, with similarity values between 92.6 and 93.1%. Similarity values of all clones to other sulfate-reducing bacteria that should be detectable by hybridization with probe SRB385, e.g., Desulfobulbus elongatus, Desulfobacter curvatus, Desulfuromonas pigra, and Desulfovibrio sp., as well as to other phylogenetic groups, were generally below 90%.
FIG. 1.
Neighbor-joining tree based on the aligned sequences of selected clones from the 16S rRNA gene library of the chemocline of Lake Cadagno and of bacteria selected from the EMBL and GenBank databases. The distance scale indicates the expected number of changes per sequence position. Bars and probe designations indicate target groups of sulfate-reducing bacteria for specific oligonucleotide probes.
These results indicate a limited complexity of populations of sulfate-reducing bacteria detectable with probe SRB385. However, since PCR-based approaches for the analysis of microbial diversity in heterogeneous environments might be influenced by several constraints (reviewed in reference 28), our results do not necessarily reflect the abundance of the target sequences in the original sample (24). We therefore tried to confirm the relevance of the sequence data in the original sample by in situ hybridization. Based on comparative analysis of sequences retrieved and those of reference organisms, two oligonucleotide probes, DSC213 (5′ CCT CCC TGT ACG ATA GCT, positions 213 to 230 according to E. coli numbering [2]) and DSC441 (5′ ATT ACA CTT CTT CCC ATC C, positions 441 to 459), were designed to target the cluster of six clones. Probe DSC213 also targeted the closely related D. thiozymogenes DSM7269 (Fig. 1). A third probe, SRB441 (5′ CAT GCA CTT CTT TCC ACT T, positions 441 to 459), was designed to specifically bind to sequences of the two other clones.
Probe specificity with reference to available 16S rRNA sequences was checked with the ARB program (23) and in the EMBL and GenBank databases by using FASTA (19). Pure cultures of sulfate-reducing bacteria such as Desulfotomaculum orientis DSM765 and Desulfovibrio desulfuricans DSM642, of bacteria from other phyla like C. okenii DSM169, Chromatium vinosum DSM180, L. roseopersicina DSM229, A. purpureus DSM4197, Amoebobacter roseus DSM235, Burkholderia cepacia DSM50181, Brevundimonas diminuta DSM1635, and Campylobacter jejuni DSM4688, and of water samples from Lake Cadagno were used to test probe specificity and to establish appropriate in situ hybridization conditions for the specific detection. The specificity of the hybridization was then adjusted by the addition of 30% (probes DSC213 and DSC441) and 5% (probe SRB441) formamide to the hybridization buffer and by a reduction of NaCl in the washing buffer to 124 and 762 mM, respectively (30).
Aliquots (3 μl) of paraformaldehyde-fixed water samples (n = 3) spotted onto gelatin-coated slides (8) were hybridized and concomitantly stained with DAPI according to the method of Zarda et al. (30). The analysis was performed in a top-to-bottom approach, initially detecting members of the domain Bacteria (probe EUB338) (1), then sulfate-reducing bacteria of the families Desulfovibrionaceae (probe SRB385) and Desulfobacteriaceae (probe SRB385Db) (22), followed by different populations of sulfate-reducing bacteria within these families (4, 17), and finally our clusters with probes DSC213 and DSC441 as well as probe SRB441.
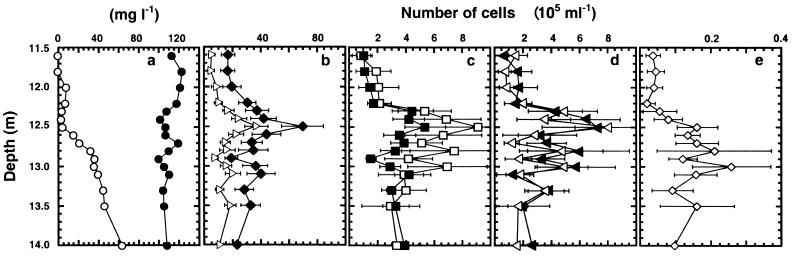
For the in situ analysis, water samples were obtained from the chemocline with a thin-layer pneumatic multisyringe sampler on 3 October, 1997 (26). The physicochemical parameters (temperature, conductivity, pH, concentration of dissolved oxygen, and turbidity) measured during sampling with a Hydropolytester HPT-C profiler displayed the characteristic stratification profile of Lake Cadagno (21, 27). Although oxygen was already depleted at a depth of 9 m, the rapid increase in sulfide concentrations (Fig. 2a) determined photometrically (26) and the turbidity profile (data not shown) indicated the formation of a condensed chemocline at a depth between 11.5 and 14 m. The turbidity profile correlated well with the number of organisms detected by DAPI staining, which ranged between (17 ± 5) × 105 and (70 ± 15) × 105 cells ml−1, showing a maximum around a depth of 12.5 m (Fig. 2b). Averaged over the whole chemocline, approximately 47% of the DAPI-stained cells were detectable by in situ hybridization with probe EUB338, again showing a maximum of cells at a depth of 12.5 m ([37 ± 8] × 105 cells ml−1) (Fig. 2b).
FIG. 2.
Vertical distribution of chemical parameters and bacteria in the chemocline of Lake Cadagno at a depth between 11.5 and 14 m. The figure graphs concentrations of sulfide (○) and sulfate (●) (a), total bacterial cells after DAPI staining (⧫) and cells detectable after in situ hybridization with probe EUB338 (▹) (b), cells detectable after in situ hybridization with probes SRB385Db (■) and SRB385 (□) (c), cells attached to aggregates of small-celled phototrophic bacteria and detectable after in situ hybridization with probe SRB385 (◂) and cells attached to aggregates of small-celled phototrophic bacteria and detectable after in situ hybridization with probes DSC213 and DSC441 (◃) (d), and cells detectable after in situ hybridization with probe SRB441 (◊) (e). Numbers determined in 40 microscopic fields of three samples are expressed as means ± standard errors (shown by error bars).
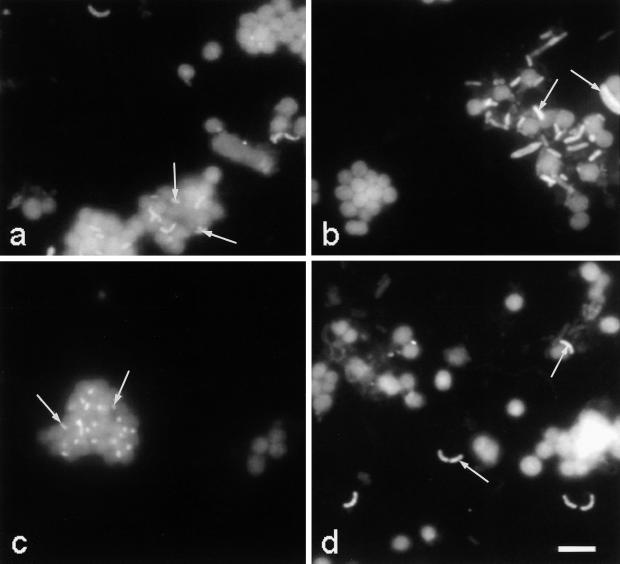
The vertical distribution profile of cells detected with probe SRB385 corresponded roughly to that of cells detected with probe SRB385Db, though at higher numbers. Probes SRB385 and SRB385Db displayed a maximum again at a depth of 12.5 m, with (9 ± 4) × 105 and (5 ± 1) × 105 cells ml−1 detected, respectively (Fig. 2c). Averaged over the whole chemocline, approximately 75% of the cells detected with probe SRB385 represented only one morphotype and were obviously associated with agglomerates of small-celled phototrophic sulfur bacteria (Fig. 2d). Small-celled phototrophic sulfur bacteria could be easily detected due to their comparatively large cell size and their pronounced autofluorescence (Fig. 3a). The remaining 25% of the cells detected with probe SRB385 were free or loosely attached to other cells or cell debris which was similar to all cells detected with probe SRB385Db. In contrast to cells detected with probe SRB385, cells detected with probe SRB385Db represented many different morphotypes (Fig. 3b).
FIG. 3.
In situ detection of sulfate-reducing bacteria with Cy3-labeled probes SRB385 targeting members of the family Desulfovibrionaceae (a), SRB385Db targeting members of the family Desulfobacteriaceae (b), a combination of probes DSC213 and DSC441 targeting a cluster of six clones of a 16S rRNA gene clone library from the chemocline of Lake Cadagno and D. thiozymogenes (c), and SRB441 targeting a second cluster with two clones of the same 16S rRNA gene clone library (d). Cells in the background mainly represent small-celled phototrophic sulfur bacteria exhibiting strong autofluorescent signals. Arrows show hybridizing cells. Bar represents 10 μm.
Averaged over the whole chemocline, probes SRB385 and SRB385Db allowed us to detect 24% (15 and 9%, respectively) of the DAPI-stained cells, which shows that sulfate-reducing bacteria make up a significant part of the bacterial population in the chemocline of Lake Cadagno. However, one can only speculate about absolute numbers, since the probes used might also detect non-sulfate-reducing members of the δ subdivision of Proteobacteria as well as several members of the α subdivision of the Proteobacteria (30). The vertical distribution profiles of sulfate-reducing bacteria detected with probes SRB385 and SRB385Db are similar to those of phototrophic sulfur bacteria that make up, on average, 33% of the DAPI-stained bacteria in the chemocline (26, 27). The high abundance of both sulfate-reducing bacteria and phototrophic sulfur bacteria supports assumptions on the dominance of these groups of bacteria in the chemocline of lakes such as Lake Cadagno (9, 10, 18, 20).
Only very few of the free or loosely attached cells hybridized with probes commonly used to analyze populations of sulfate-reducing bacteria (4, 17). None of the associated cells hybridized to these probes (results not shown). A combination of probes DSC213 and DSC441, targeting a cluster of six clones and the closely related D. thiozymogenes DSM7269, however, detected cells only associated to small-celled phototrophic sulfur bacteria (Fig. 3c). A vertical distribution profile of numbers of associated cells was obtained with values from (1 ± 1) × 105 to (8 ± 2) × 105 cells ml−1 which were not significantly different from those obtained with probe SRB385 (Fig. 2d). Probe SRB441 designed to specifically bind to sequences of two other clones only hybridized to loosely attached or free cells ([2 ± 2] × 104 to [25 ± 11] × 104 cells ml−1) (Fig. 2d and e). Averaged over the whole chemocline, numbers of cells detected with probes DSC213, DSC441, and SRB441 comprise about 64% of those obtained by hybridization with probe SRB385. This percentage indicates that the numbers of cells detected with probe SRB385 might be too high due to the detection of non-sulfate-reducing members of the δ and α subdivisions of the Proteobacteria. Another reason might be that the number of clones analyzed in our library was not sufficient to obtain all bacteria detectable with probe SRB385. Nevertheless, our results demonstrate that a numerically prominent population of bacteria closely related to D. thiozymogenes, and therefore most likely sulfate-reducing, is associated with agglomerates of small-celled phototrophic sulfur bacteria.
Although the phylogenetic relationship of our clones to the closest cultured relative, D. thiozymogenes DSM7269, does not necessarily reflect physiological relationships, the physiological traits of D. thiozymogenes might be used in speculation about the nature of the association. D. thiozymogenes was described as sulfate-reducing bacterium growing under strictly anaerobic conditions by disproportionation of thiosulfate, sulfite, or elemental sulfur to sulfate and sulfide. It was also able to grow by the oxidation of a limited range of organic compounds coupled to sulfate reduction (13). Similar to observations with D. sulfoexigens DSM10523 (6) and Desulfobulbus propionicus DSM2032 (15), disproportionation of sulfur to sulfate and sulfide was enhanced in the presence of sulfide scavengers such as amorphous ferric hydroxide (13), FeCO3, or MnO2, generally resulting in the formation of sulfate along with iron and manganese sulfides (15, 25). In the chemocline of Lake Cadagno, small-celled phototrophic sulfur bacteria that oxidize sulfide to sulfur and further to sulfate might act as alternative sulfide scavengers to ferric and manganic oxides, creating a sink for sulfide produced by sulfur disproportionation of the sulfate-reducing bacteria represented by our clones. Such speculations, however, require further investigations and can probably be supported if bacteria represented by our clones can be obtained in pure culture by using the conditions for the isolation of D. thiozymogenes (13) or D. sulfoexigens (6).
Nucleotide sequence accession numbers.
Sequence data were deposited in the EMBL and GenBank databases with accession no. AJ389622 to AJ389629.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation (NF31-46855.96) and from the canton of Ticino (Switzerland).
The authors are indebted to N. Ruggeri and A. Caminada for technical support.
REFERENCES
- 1.Amann R I, Binder B J, Olsen R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brosius J, Dull T J, Sleeter D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 3.Demarta A, Tonolla M, Caminda A-P, Ruggeri N, Peduzzi R. Phylogenetic diversity of the bacterial community from the anoxic layer of the meromictic Lake Cadagno. Doc Ist Ital Idrobiol. 1998;63:19–30. [Google Scholar]
- 4.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 5.Felsenstein J. PHYLIP manual, version 3.5C. Seattle: University of Washington; 1990. [Google Scholar]
- 6.Finster K, Liesack W, Thamdrup B. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol. 1998;64:119–125. doi: 10.1128/aem.64.1.119-125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer C, Wiggli M, Schanz F, Hanselmann K W, Bachofen R. Light environment and synthesis of bacteriochlorophyll by populations of Chromatium okenii under natural environmental conditions. FEMS Microbiol Ecol. 1996;21:1–9. [Google Scholar]
- 8.Glöckner F-O, Amann R I, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An optimized in situ hybridization protocol for planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 9.Guerrero R, Abella C, Miracle M R. Spatial and temporal distribution of bacteria in a meromictic karsic lake basin: relationships with physicochemical parameters and zooplankton. Verh Int Ver Limnol. 1978;29:2264–2271. [Google Scholar]
- 10.Guerrero R, Montesinos E, Pedros-Alio C, Esteve I, Mas J, van Gemerden H, Hofman P A G, Bakker J F. Phototrophic sulfur bacteria in two Spanish Lakes: vertical distribution and limiting factors. Limnol Oceanogr. 1985;30:919–931. [Google Scholar]
- 11.Hanselmann K, Hutter R. Geomicrobiological coupling of sulfur and iron cycling in anoxic sediments of a meromictic lake: sulfate reduction and sulfide sources and sinks in Lake Cadagno. Doc Ist Ital Idrobiol. 1998;63:85–98. [Google Scholar]
- 12.Higgins D, Thompson J, Gibson T, Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen P H, Schuhmann A, Bak F, Liesack W. Disproportionation of inorganic sulfur compounds by the sulfate-reducing bacterium Desulfocapsa thiozymogenes gen. nov., sp. nov. Arch Microbiol. 1996;166:184–192. [Google Scholar]
- 14.Lehmann C, Luehty L, Bachofen R. Tools for the evaluation of sources and sinks of sulfide in Lake Cadagno. Doc Ist Ital Idrobiol. 1998;63:99–104. [Google Scholar]
- 15.Lovely D R, Phillips E J P. Novel processes for anaerobic sulfate reduction from elemental sulfur by sulfate-reducing bacteria. Appl Environ Microbiol. 1994;60:2394–2399. doi: 10.1128/aem.60.7.2394-2399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 18.Overmann J, Beatty T, Hall K J, Pfennig N, Northcote T G. Characterization of a dense, purple sulfur bacterial layer in a meromictic lake. Limnol Oceanogr. 1991;36:846–859. [Google Scholar]
- 19.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pedros-Alio C, Montesinos E, Guerrero R. Factors determining annual changes in bacterial photosynthetic pigments in holomictic Lake Ciso, Spain. Appl Environ Microbiol. 1983;46:999–1006. doi: 10.1128/aem.46.5.999-1006.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peduzzi R, Demarta A, Tonolla M. Dynamics of the autochthonous and contaminant bacterial colonization of lakes (Lake of Cadagno and Lake of Lugano as model systems) Stud Environ Sci. 1993;55:323–335. [Google Scholar]
- 22.Rabus R, Fukui M, Wilkes H, Widdel F. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl Environ Microbiol. 1996;62:3605–3613. doi: 10.1128/aem.62.10.3605-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strunk O, Ludwig W. ARB. Munich, Germany: Computer program distributed by the Technical University Munich; 1996. [Google Scholar]
- 24.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thamdrup B, Finster K, Hansen J W, Bak F. Bacterial disproportionation of elemental sulfur coupled to chemical reduction of iron or manganese. Appl Environ Microbiol. 1993;59:101–108. doi: 10.1128/aem.59.1.101-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tonolla M, Demarta A, Hahn D, Peduzzi R. Microscopic and molecular in situ characterization of bacterial populations in the meromictic Lake Cadagno. Doc Ist Ital Idrobiol. 1999;63:31–44. [Google Scholar]
- 27.Tonolla M, Demarta A, Peduzzi R, Hahn D. In situ analysis of phototrophic sulfur bacteria in the chemocline of meromictic Lake Cadagno (Switzerland) Appl Environ Microbiol. 1998;65:1325–1330. doi: 10.1128/aem.65.3.1325-1330.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 29.Wagener S, Schulz S, Hanselmann K. Abundance and distribution of anaerobic protozoa and their contribution to methane production in Lake Cadagno (Switzerland) FEMS Microbiol Ecol. 1990;74:39–48. [Google Scholar]
- 30.Zarda B, Hahn D, Chatzinotas A, Schönhuber W, Neef A, Amann R I, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]