Abstract
BACKGROUND
Leprosy, caused by Mycobacterium leprae, is a public health problem in Brazil that affects peripheral nerves, resulting in physical disabilities. During host-pathogen interactions, the immune response determines leprosy outcomes from a localised (paucibacillary) form to a disseminated (multibacillary) form. The recognition of M. leprae involves the DC-SIGN receptor, which is present on the dendritic cells (DCs) and participates in immune activation.
OBJECTIVES
To evaluate the association of polymorphisms in the promoter region of the gene encoding DC-SIGN (CD209) and the clinical form of leprosy, and to investigate its functional effects.
METHODS
The study population included 406 leprosy patients from an endemic area in Brazil [310 multibacillary (MB); 96 paucibacillary (PB)]. A functional evaluation based on the effects of the single nucleotide variant (SNV) associated with PB leprosy on the specific immune response was also performed.
RESULTS
The GA genotype and the presence of the A allele of rs735240 (-939G>A) were associated with PB leprosy [OR: 2.09 (1.18-3.69) and 1.84 (1.07-3.14), respectively]. Carriers of the A allele showed reduced expression of CD209 and TGF-β1 in leprosy lesions in comparison with individuals with GG genotype, in addition to a higher response to the Mitsuda test.
CONCLUSION
These data suggest that rs735240 influences the immune response against M. leprae and clinical presentation of leprosy.
Key words: DC-SIGN, Mycobacterium leprae, leprosy, single nucleotide polymorphism
Leprosy is considered a public health problem in Brazil, which registered around 28,000 new cases among a total of 202,185 cases detected worldwide in 2019. 1 Although the introduction of multidrug therapy has reduced the prevalence of the disease, epidemiological indices still show active transmission of Mycobacterium leprae. 2 Besides, leprosy presents important morbidity associated with peripheral nerve damage. 3
Following the initial host-pathogen interaction, elimination of the bacillus or clinical manifestation of leprosy in several forms may occur. 4 The diversity of the possible outcomes reflects the host immune response. 5 , 6 Two polar and stable forms of leprosy are known: the tuberculoid form (TT), considered restrictive, which presents as a localised disease with few lesions and rare bacilli due to the intense cellular immune response, and the lepromatous form (LL) with disseminated lesions and a large number of bacilli that result from an inefficient immune response characterised by the inhibition of macrophage antimicrobial activity, high levels of antibodies, and activation of regulatory T cells. 7 Three intermediate and unstable forms exist between these poles: borderline tuberculoid (BT), borderline borderline (BB), and borderline lepromatous (BL), among which the cellular response gradually decreases from the tuberculoid to the lepromatous pole. 5 , 8 For treatment purposes, leprosy patients are classified as paucibacillary (PB) or multibacillary (MB), according to the number of lesions or the bacillary index, when available. 9
Mycobacterium leprae has a compact genome 10 and exhibits low genetic diversity. 11 Thus, the complexity of clinical and biological phenotypes in leprosy points to the major role of the host genetic background in determining the course of the disease. 12 , 13 This fact has been evidenced by classical and molecular genetic studies demonstrating that disease susceptibility is influenced by genes that also regulate the progression of leprosy. 14 Some genes, such as NOD2, CCDC122-LACC1, TNF, TLR1, IFNG, and IL10, have already been associated with leprosy susceptibility after being consistently replicated in different populations. 15 , 16 , 17 , 18 , 19 However, genes may also be involved in the clinical manifestations of leprosy. 20
The CD209 gene encodes DC-SIGN (CD209), a C-type lectin receptor expressed on the surface of dendritic cells (DCs), that is involved in mycobacterial recognition, 21 , 22 promoting antigen internalisation, and presentation. 23 DC-SIGN binding by mycobacteria impairs DC maturation and promotes pathogen persistence. 24 Previous studies have shown that polymorphisms in the promoter region of CD209 are associated with susceptibility to several infectious diseases 25 , 26 , 27 including tuberculosis. 28 , 29 In addition, other studies associated these polymorphisms with the severity of tuberculosis, 30 hepatitis C, 31 severe acute respiratory syndrome 32 and dengue fever. 33 , 34
Considering that the recognition of M. leprae involves DC-SIGN, but polymorphisms in this gene were not associated with leprosy susceptibility, 22 we investigated the association of single nucleotide variants (SNVs) in the promoter region of the CD209 gene with clinical forms of leprosy in the Brazilian population. In addition, we investigated the functional effects of these polymorphisms on the response to M. leprae.
SUBJECTS AND METHODS
Participants and study design - The association of rs2287886, rs4804803, rs735239, and rs735240 SNVs in the CD209 gene promoter region with clinical forms of leprosy was tested in a Middle Eastern Brazilian population of 406 leprosy patients from Rondonópolis, Mato Grosso state. This is an endemic area in Brazil, with a prevalence of 15.52 leprosy cases per 10,000 inhabitants in 2018. 35 Leprosy diagnosis was confirmed by clinical and laboratory tests performed at the outpatient service in Rondonópolis and Lauro de Souza Lima Institute, respectively. The general characteristics of the participants are presented in Table I. The classification of patients with leprosy was based on the Ridley and Jopling (R&J) criteria, 5 which considers clinical, histopathological, immunological, and bacilloscopic characteristics. Bacilloscopic evaluation was performed on the histological sections of the skin samples using the Fite-Faraco technique and corresponded to the bacilloscopic index (BI, 0-6+) associated with the evaluation of the presence of bacilli in different skin tissues (mainly in granulomas). This information (clinical, histopathological, immunological, and bacilloscopic characteristics) allows for classification in the R&J spectrum (TT, BT, BB, BL, and LL). For genetic and functional analysis, samples with BI “0” or 1+ were considered paucibacillary, which represented patients I, TT, and some BT (PB, n = 96). Samples with BI ≥ 2+ were considered multibacillary and represented some BT patients and all BB, BL, and LL patients (MB, n = 310). This modified version of the operational classification recommended by the WHO 36 , 37 was adopted since it reflects the ability of patients to contain bacillary multiplication. 38 , 39 To better characterise the clinical condition of leprosy patients, other clinical and laboratory data from the patients, such as disability grade, bacterial index, antibody response against PGL-I antigen from M. leprae, and skin response in the Mitsuda reaction were obtained from medical reports.
TABLE I. Characteristics of leprosy patients from Rondonópolis - Mato Grosso state enrolled in the genetic study (n = 406).
| Characteristics | Categories | MB (n = 310) | PB (n = 96) |
| Age (mean ± SD) | 42.9 ± 16.1 | 39.1 ± 16.1 | |
| Sex (n/%) | Male Female | 201 (64.8%) 109 (35.2%) | 45 (46.9%) 51 (53.1%) |
| Ridley and Jopling classification (n/%) | LL BL BB BT TT I | 21 (6.8%) 63 (20.3%) 79 (25.5%) 147 (47.4%) --- --- | --- --- --- 9 (9.4%) 60 (62.5%) 27 (28.1%) |
SD: standard deviation; LL: lepromatous-lepromatous leprosy; BL: borderline-lepromatous leprosy; BB: borderline-borderline leprosy; BT: borderline-tuberculoid leprosy; TT: tuberculoid-tuberculoid leprosy; I: indeterminate leprosy.
For ethnicity control, molecular ancestry was defined using 46 ancestry-informative indels, as previously described. 40 , 41 The estimates of individual ancestry (European, African, and Native American) were analysed using the ADMIXTURE software. 42
SNVs selection - The candidate SNVs rs2287886 (-139A>G), rs4804803 (-336A>G), rs735239 (-871A>G), and rs735240 (-939G>A) were chosen considering the previous association of the CD209 promoter region with the severity of infectious diseases. 31 , 43 , 44 Moreover, the CD209 promoter region has different binding sites for transcription factors, such as AP-1, Sp-1, Ets-1, and NF-kb, and the proximity of these sites has the potential to affect the transcriptional levels. 45
DNA extraction and SNVs genotyping - For genetic analysis, genomic DNA was extracted from the peripheral blood leukocytes using the salting-out method. 46 For the functional analysis, DNA was obtained from skin lesion biopsies embedded in paraffin. These samples were processed using the Qiagen DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). Genotyping was performed by allelic discrimination using fluorogenic probes (TaqMan assay numbers: C_1999341_10 (rs735240), C_1999340_10 (rs4804803), C_989421_10 (rs735239), and C_11515683_1 (rs2287886) using Viia 7 equipment (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions.
Functional evaluation - To evaluate the functional effects of the rs735240 SNV on the immune response against M. leprae, we analysed the expression levels of DC-SIGN, cytokines, and other accessory molecules using data from two previous and independent studies from our group. 47 , 48 First, we examined the mRNA expression levels in 28 leprosy patients [Supplementary data (116.3KB, pdf) (Table I)] according to the presence or absence of allele A of rs735240 using the dataset GSE74481. 47 In brief, biopsies from leprosy skin lesions were collected and stored in RNAlater solution (Ambion). RNA extraction was performed using a Precellys24 apparatus (Bertin Technologies), and RNA integrity was evaluated using a 2100 Bioanalyzer electrophoresis apparatus (GE Healthcare Bio-Sciences). Total RNA was reverse transcribed and amplified, and gene expression was evaluated using the 8X60K cDNA microarray G4858A platform (GE Healthcare Bio-Sciences) containing 60,000 probes representing the entire human genome. The microarray data were analysed using Gene Spring GX version 12.1 software (Agilent Technologies).
We also explored the expression of DC-SIGN, cytokines, and other surface markers by monocyte-derived DCs from leprosy patients primed with M. leprae antigen 48 [Supplementary data (116.3KB, pdf) (Table II)], taking into account the genotype of rs735240. In this study, monocytes from leprosy patients were purified from peripheral blood and treated with GM-CSF and IL-4 to differentiate into DCs that were stimulated by M. leprae sonicated antigen or standard maturation cocktail (IL-1β, IL-6, tumor necrosis factor, and prostaglandin E2), or remained unstimulated. After 48 h, the supernatant was collected and stored at -80ºC for cytokine measurement using commercial enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, USA). DCs were harvested for analysis of surface marker expression by flow cytometry, as previously described. 48
Furthermore, we analysed the influence of this variant on clinical and laboratory parameters in patients with leprosy recruited for the genetic study.
Data analysis - The genotypes, alleles, and carrier frequencies were compared using a univariate logistic regression model. These analyses were also performed with adjustments for covariates of sex and molecular ancestry. The last variable was used for continuous adjustment because there is no consensus regarding the use of these continuous variables to classify ethnicity as a categorical variable. Statistical software R version 2.5.1 for Windows and the Genetics package were used. A p-value < 0.05 was adopted as the cut-off for statistical significance.
For functional analyses, data were grouped into carriers and non-carriers of allele A from rs735240 SNV (AA+AG vs GG). Statistical analysis was performed using the non-parametric Mann-Whitney test, and significance was set at p-value < 0.05. All analyses were performed using the GraphPad Prism software version 7.04 (GraphPad Software Inc., La Jolla, CA, USA, 2017).
Ethics - This study was approved by the Research Ethics Committee of the Lauro de Souza Lima Institute (CAAE 68433217.0.0000.5475), and all protocols were carried out in accordance with the Helsinki Declaration of 1975, as revised in 1983, as well as Brazilian law concerning ethics in biological research.
RESULTS
rs735240 SNV was associated with the clinical form of leprosy - Genotyping was successful in more than 90% of the samples for all four SNVs studied. The heterozygous GA genotype of rs735240 SNV was significantly associated with paucibacillary leprosy (OR 2.02; CI 1.10-3-73; p = 0.0240). In addition, an association between A allele carriers and paucibacillary leprosy was noted (OR 1.85; CI 1.04-3.31; p = 0.0350) (Table II).
TABLE II. Alelle, genotype and carrier frequencies of the rs735240, rs4804803, rs735239 and rs2287886, polymorphisms in CD209 promoter region of leprosy patients. Logistic regression data for association with clinical forms of leprosy at the sample from Rondonópolis, MT, Brazil.
| Polymorphisms | Categories | PB | MB | OR (95% CI) p-value | OR (95% CI) p-value a |
| rs735240 -939G>A | G | 94 (0.52) | 318 (0.57) | * | * |
| A | 88 (0.48) | 240 (0.43) | 1.2 (0.8-2.0) 0.37 | 1.31 (0.8-2.2) 0.30 | |
| GG | 22 (0.24) | 103 (0.37) | * | * | |
| GA | 50 (0.55) | 112 (0.40) | 2.1 (1.2-3.7) 0.01 | 2.0 (1.1-3.7) 0.02 | |
| AA | 19 (0.21) | 64 (0.23) | 1.4 (0.7-2.8) 0.35 | 1.6 (0.8-3.2) 0.22 | |
| A carrier | 69 (0.66) | 176 (0.63) | 1.8 (1.1-3.1) 0.03 | 1.8 (1.0-3.3) 0.04 | |
| Total | 91 | 279 | |||
| rs4804803 -336A>G | A | 146 (0.77) | 445 (0.73) | * | * |
| G | 44 (0.23) | 165 (0.27) | 0.8 (0.5-1.4) 0,45 | 0.8 (0.4-1.4) 0.47 | |
| AA | 53 (0.56) | 165 (0.54) | * | * | |
| AG | 40 (0.42) | 115 (0.38) | 1.1 (0.7-1.7) 0.74 | 1.1 (0.6-1.8) 0.74 | |
| GG | 2 (0.02) | 25 (0.08) | 0.2 (0.1-1.1) 0.06 | 0.2 (0.1-1.1) 0.07 | |
| G carrier | 42 (0.44) | 140 (0.46) | 0.9 (0.6-1.5) 0.77 | 0.9 (0.6-1.6) 0.80 | |
| Total | 95 | 305 | |||
| rs735239 -871A>G | A | 121 (0.67) | 377 (0.68) | * | * |
| G | 59 (0.33) | 177 (0.32) | 1.0 (0.6-1.7) 0.88 | 1.1 (0.6-1.9) 0.69 | |
| AA | 39 (0.43) | 133 (0.48) | * | * | |
| AG | 43 (0.48) | 111 (0.40) | 1.3 (0.8-2.2) 0.28 | 1.4 (0.8-2.4) 0.21 | |
| GG | 8 (0.09) | 33 (0.12) | 0.8 (0.3-1.9) 0.66 | 1.0 (0.4-2.3) 0.95 | |
| G carrier | 51 (0.57) | 144 (0.52) | 1.2 (0.7-1.9) 0.44 | 1.3 (0.8-2.2) 0.31 | |
| Total | 90 | 277 | |||
| rs2287886 -139A>G | G | 130 (0.69) | 406 (0.70) | * | * |
| A | 58 (0.31) | 170 (0.30) | 1.1 (0.6-1.8) 0.81 | 1.00 (0.6-1.7) 0.97 | |
| GG | 43 (0.46) | 147 (0.51) | * | * | |
| GA | 44 (0.47) | 112 (0.39) | 1.3 (0.8-2.2) 0.23 | 1.2 (0.7-2.0) 0.48 | |
| AA | 7 (0.07) | 29 (0.10) | 0.8 (0.3-2.0) 0.67 | 0.7 (0.3-1.9) 0.54 | |
| A carrier | 51 (0.54) | 141 (0.49) | 1.2 (0.8-2.0) 0.37 | 1.1 (0.7-1.8) 0.68 | |
| Total | 94 | 288 |
a: adjusted regression values using sex and molecular ancestry covariates. CI: confidence interval; MB: multibacillary leprosy patients; OR: odds ratio; PB: paucibacillary leprosy patients.
The rs2287886, rs4804803, and rs735239 SNVs were not associated with clinical forms of leprosy, as summarised in Table II.
rs735240 SNV presented functional effects in the immune response against M. leprae - Considering the association of rs735240 with PB leprosy as well as the relevance of DC-SIGN in the recognition of mycobacteria by DCs, we analysed the influence of this variant on the immune response against M. leprae by evaluating the expression of CD209 and other molecules involved in DC activation. Allele A carriers showed lower expression of DC-SIGN in both leprosy skin lesions and monocyte-derived DCs stimulated with M. leprae, with marginal significance (Figure A-B) in comparison with non-carriers (GG genotype). In addition, the levels of TGF-β1, a cytokine induced after DC-SIGN activation, were smaller in allele A carriers under the same conditions (Figure C-D). The expression levels of CD40, ITGAX (CD11c), CCL17, CD83, CIITA, IL1B, IL6, ISG15, OAS1, RNaseL, TLR3, and TNF genes in skin lesions, as well as CD11c, CD40, CD80, CD86, HLA-DR, ICAM-1, IL-10, IL-12-23p40, IL-12p70, and TNF, which are expressed after DC activation, were also evaluated according to the rs735240 SNV, but no significant differences were found (data not shown). Although the findings for the expression levels of CD209 and TGF-β1 in leprosy lesions and DC cultures are in accordance, the data obtained in vitro included few patients and only two individuals with the GG genotype for rs735240, therefore requiring further confirmation.
Functional effects of rs735240 SNV at CD209 gene in the immune response against Mycobacterium leprae. The expression of CD209 (DC-SIGN) and TGF-β1 according to the presence or absence of the allele A was evaluated in leprosy skin lesions (A and C) and monocyte-derived dendritic cells stimulated with M. leprae antigens (B and D). Besides, the response to the Mitsuda test in leprosy patients was analyzed in allele A carriers (n = 98) and non-carriers (n = 73). MC: DCs stimulated for 48 h with a specific maturation cocktail IL-1β (25 ng/mL), IL-6 (1,000 U/mL), tumor necrosis factor (50 ng/mL) and prostaglandin E2 (10-6 M); ML: DCs stimulated with sonicated antigen of M. leprae (10 μg/mL); US: unstimulated immature DCs. Mann-Whitney test was used to statistical analysis.
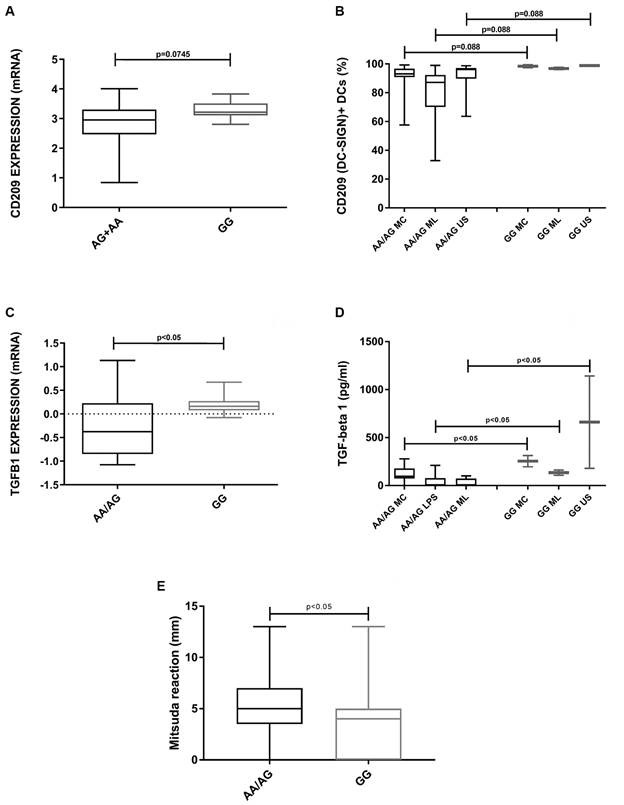
Furthermore, we also observed that the rs735240 allele A has some influence on the response to the Mitsuda test in leprosy patients. The Mitsuda test is a skin reaction based on a delayed cell-mediated immune response against mycobacteria; it is used for the prognostic and clinical classification of leprosy. 49 We observed that allele A carriers exhibited a higher Mitsuda response than non-carriers (Figure E). The response to the Mitsuda test observed in AA allele carriers was higher than that observed in GG individuals for both PB (5,7 mm x 4,8 mm) and MB patients (4,2 mm and 3,1 mm). Disability grade, bacterial index, and antibody response against the PGL-I antigen from M. leprae showed no difference related to the rs735240 SNV (data not shown).
DISCUSSION
Although polymorphisms located in the promoter region of CD209 are widely associated with the severity of various infectious diseases, 31 , 32 , 44 , 50 , 51 including tuberculosis, 30 such association has not yet been reported in leprosy. In view of the relevance of DC-SIGN in the recognition of antigens and activation of the immune response, and considering that the variations in genes that potentially influence the course of infections are largely population-dependent 52 we chose to investigate the association of candidate variants in the CD209 promoter region with the clinical manifestation of leprosy in a Brazilian population. We observed an association between the A allele and AG genotype of SNV rs735240 (-939G>A) with paucibacillary leprosy, a localised form of the disease. No other studies in the Brazilian population have investigated the association between CD209 SNVs and the clinical forms of leprosy. Worldwide, only two studies have investigated this association: one in a Japanese population evaluating the -336 SNV (rs4804803) 53 and the other investigating seven SNVs in a Pakistani population; 22 both being non-significant associations. However, it is important to mention that these studies were performed with a limited casuistic comparing 60 and 109 MB to 33 and 85 PB patients in Japan and Pakistan, respectively, while our study included 310 MB and 96 PB patients, rendering robust the data presented here.
Silva et al. 54 observed that the rs735240 SNV was associated with extrapulmonary tuberculosis susceptibility, rs2287886 showed a significant association with pulmonary tuberculosis, and rs4804803 and rs735239 provided protection against the disease, confirming the association of the promoter region in CD209 with mycobacterial diseases. rs4804803 also contributes towards genetic susceptibility to ulcerative colitis 55 in which the colonisation of the intestinal tract by mycobacteria has been described.
Considering other infectious diseases, the A allele of rs735240 is associated with the development of fungal keratitis in the Han Chinese population. 56 In addition, a higher incidence of cytomegalovirus (CMV) infection among kidney transplant recipients not receiving prophylaxis was associated with the GG genotype of rs735240, 57 and CMV reactivation after allogeneic stem cell transplantation was associated with the G allele of rs735240. 58
Furthermore, the A allele of rs735240 was associated with protection against Kawasaki disease, a systemic vasculitis of unknown etiology, 59 indicating that this SNV can modulate the immune response in different scenarios. In fact, the promoter region of the CD209 gene contains binding sites for the transcription factors AP-1, Sp-1, Ets-1, and NF-κB that participate in the activation of the immune system. 45
Our results showed lower expression of CD209 mRNA related to the A allele of rs735240 SNV in skin lesions from leprosy patients, as well as a tendency to decrease DC-SIGN expression levels in monocyte-derived DCs, even if this needs to be considered with caution because of the low number of patients evaluated in vitro. M. leprae is recognised via DC-SIGN, 22 , 60 the activation of which triggers pathways associated with IL-10 secretion and results in an immunosuppressive response that contributes to pathogen persistence. 24 Thus, the lower expression levels of DC-SIGN in A allele carriers could imply a low production of IL-10, concurring with the development of an efficient cell-mediated immune response observed in PB leprosy patients. Similarly, Li et al. 61 proposed that the AA genotype of rs735240 can protect against nasopharyngeal carcinoma by reducing DC-SIGN expression and consequently decreasing the infection of DCs and nasopharyngeal epithelial cells by the Epstein-Barr virus. In addition, Vannberg et al. observed that the GG genotype of the rs4804803 SNV is involved in the downregulation of CD209 mRNA expression, which protects against a severe form of tuberculosis characterised by pulmonary cavitation. 30
Our study also showed that carriers of the A allele presented lower TGB-β1 expression in skin lesions and reduced levels of this cytokine in cultures of monocyte-derived DCs stimulated with M. leprae antigens. This cytokine is involved in the development and maintenance of suppressor and regulatory cells in the immune response. 62 In leprosy, the TGF-β expression levels were higher in the skin lesions from LL and BL patients with a high bacilloscopic index than in paucibacillary patients, 8 , 63 as well as in PBMCs of these individuals stimulated with M. leprae antigen. 64 Lower production of TGB-β1 by A allele carriers may avoid bacillary persistence and multiplication, favoring the control of infection and the development of PB leprosy, the most restrictive form of the disease.
Another important finding in our study is the association of the A allele of rs735240 with a greater response to the Mitsuda test, an intradermic reaction based on the injection of heat-killed M. leprae. The test is considered positive when a granulomatous response with a size >5 mm occurs, which reflects a cell-mediated immune response that is able to control mycobacterial infection. The reaction is usually positive in patients with PB with localised diseases. Although, based on our data, it is not possible to determine the mechanisms involved in this association, it is reasonable to suggest that rs735240 could influence the polarisation of the immune response and favor the development of PB leprosy.
A limitation of our study is that both genetic and functional analyses involved multiple tests and would require statistical corrections. However, we did not apply these extremely conservative corrections methods as they could over-correct the data. The non-correction decision for genetic study considered the strong linkage disequilibrium between the variants and that some authors discuss the marker interdependence as a parameter for adjusting correction methods. 65 Furthermore, the functional findings were replicated in two distinct sets of data and agree with the observed genetic association, reinforcing that our findings were not by chance.
Although the influence of genetic background on leprosy development has been extensively investigated, few studies have evaluated the role of genes in the progression towards different clinical forms. This is especially important for identifying markers of disseminated disease associated with leprosy transmission. In summary, our results indicate, for the first time, an association between rs735240 and the CD209 gene and leprosy immunological dichotomy, as has been observed in other infectious diseases. In addition, this variant had a functional influence on the immune response against M. leprae.
ACKNOWLEDGEMENTS
To all the staff of ILSL that contributes to development of this study, in special Adriana Sierra Assêncio Almeida Barbosa for technical assistance.
Footnotes
Financial support: FAPESP (Grant #2009/01436-1). GG, AFB and RMC were supported by scholarships from CAPES.
REFERENCES
- 1.WHO Global leprosy (Hansen disease) update, 2019: time to step-up prevention initiatives. Wkly Epidemiol Rec. 2020;95 [Google Scholar]
- 2.Blok DJ, De Vlas SJ, Richardus JH. Global elimination of leprosy by 2020: are we on track? Parasit Vectors. 2015 doi: 10.1186/s13071-015-1143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Global leprosy update, 2018: moving towards a leprosy free world. Wkly Epidemiol Rec. 2019 [Google Scholar]
- 4.Cardoso CC, Pereira AC, Sales-Marques C, Moraes MO. Leprosy susceptibility genetic variations regulate innate and adaptive immunity, and disease outcome. Future Microbiol. 2011;6(5):533–549. doi: 10.2217/fmb.11.39. [DOI] [PubMed] [Google Scholar]
- 5.Ridley DS, Jopling WH. Classification of leprosy according to immunity A five-group system. Int J Lepr Other Mycobact Dis. 1966;34(3):255–273. [PubMed] [Google Scholar]
- 6.Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL. The continuing challenges of leprosy. Clin Microbiol Rev. 2006;19(2):338–381. doi: 10.1128/CMR.19.2.338-381.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saini C, Tarique M, Rai R, Siddiqui A, Khanna N, Sharma A. T helper cells in leprosy An update. Immunol Lett. 2017;184:61–66. doi: 10.1016/j.imlet.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Venturini J, Soares CT, Belone AF, Barreto JA, Ura S, Lauris JR. In vitro and skin lesion cytokine profile in Brazilian patients with borderline tuberculoid and borderline lepromatous leprosy. Lepr Rev. 2011;82(1):25–35. [PubMed] [Google Scholar]
- 9.WHO - World Health Organization WHO Expert Committee on Leprosy. World Health Organization technical report series. 1998;874:1–43. [PubMed] [Google Scholar]
- 10.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR. Massive gene decay in the leprosy bacillus. Nature. 2001;409(6823):1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 11.Benjak A, Avanzi C, Benito Y, Breysse F, Chartier C, Boschiroli ML. Highly reduced genome of the new species. Mycobacterium uberis, the causative agent of nodular thelitis and tuberculoid scrotitis in livestock and a close relative of the leprosy bacilli. mSphere. 2018;3(5):e00405–e00418. doi: 10.1128/mSphere.00405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prevedello FC, Mira MT. Hanseníase uma doença genética? An Bras Dermatol. 2007;82(5):451–459. [Google Scholar]
- 13.Fava VM, Dallmann-Sauer M, Schurr E. Genetics of leprosy today and beyond. Hum Genet. 2020;139(6-7):835–846. doi: 10.1007/s00439-019-02087-5. [DOI] [PubMed] [Google Scholar]
- 14.Lázaro FP, Werneck RI, Mackert CCO, Cobat A, Prevedello FC, Pimentel RP. A major gene controls leprosy susceptibility in a hyperendemic isolated population from north of Brazil. J Infect Dis. 2010;201(10):1598–1605. doi: 10.1086/652007. [DOI] [PubMed] [Google Scholar]
- 15.Sales-Marques C, Salomão H, Fava VM, Alvarado-Arnez LE, Amaral EP, Cardoso CC. NOD2 and CCDC122-LACC1 genes are associated with leprosy susceptibility in Brazilians. Hum Genet. 2014;133(12):1525–1532. doi: 10.1007/s00439-014-1502-9. [DOI] [PubMed] [Google Scholar]
- 16.Cardoso CC, Pereira AC, Brito-de-Souza VN, Duraes SMB, Ribeiro-Alves M, Nery JAC. TNF -308G>A single nucleotide polymorphism is associated with leprosy among Brazilians a genetic epidemiology assessment, meta-analysis, and functional study. J Infect Dis. 2011;204(8):1256–1263. doi: 10.1093/infdis/jir521. [DOI] [PubMed] [Google Scholar]
- 17.Sales-Marques C, Brito-De-Souza VN, Guerreiro LTA, Martins JH, Amaral EP, Cardoso CC. Toll-like receptor 1 N248s single-nucleotide polymorphism is associated with leprosy risk and regulates immune activation during mycobacterial infection. J Infect Dis. 2013;208(1):120–129. doi: 10.1093/infdis/jit133. [DOI] [PubMed] [Google Scholar]
- 18.Cardoso CC, Pereira AC, Brito-De-Souza VN, Dias-Baptista IM, Maniero VC, Venturini J. IFNG +874 T>A single nucleotide polymorphism is associated with leprosy among Brazilians. Hum Gen. 2010;128(5):481–490. doi: 10.1007/s00439-010-0872-x. [DOI] [PubMed] [Google Scholar]
- 19.Pereira AC, Brito-de-Souza VN, Cardoso CC, Dias-Baptista IMF, Parelli FPC, Venturini J. Genetic, epidemiological and biological analysis of interleukin-10 promoter single-nucleotide polymorphisms suggests a definitive role for -819C/T in leprosy susceptibility. Genes Immun. 2009;10(2):174–180. doi: 10.1038/gene.2008.97. [DOI] [PubMed] [Google Scholar]
- 20.Alter A, Grant A, Abel L, Alcaïs A, Schurr E. Leprosy as a genetic disease. Mamm Genome. 2011;22(1-2):19–31. doi: 10.1007/s00335-010-9287-1. [DOI] [PubMed] [Google Scholar]
- 21.Tailleux L, Schwartz O, Herrmann J-L , Pivert E, Jackson M, Amara A. DC-SIGN is the major Mycobacterium tuberculosis receptor on human dendritic cells. J Exp Med. 2003;197(1):121–127. doi: 10.1084/jem.20021468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barreiro LB, Quach H, Krahenbuhl J, Khaliq S, Mohyuddin A, Mehdi SQ. DC-SIGN interacts with Mycobacterium leprae but sequence variation in this lectin is not associated with leprosy in the Pakistani population. Hum Immunol. 2006;67(1-2):102–107. doi: 10.1016/j.humimm.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geijtenbeek TBH, Engering A, Van Kooyk Y. DC-SIGN, a C-type lectin on dendritic cells that unveils many aspects of dendritic cell biology. J Leukoc Biol. 2002;71(June):921–931. [PubMed] [Google Scholar]
- 24.Geijtenbeek TB, Van Vliet SJ, Koppel EA, Sanchez-Hernandez M, Vandenbroucke-Grauls CM, Appelmelk B. Mycobacteria target DC-SIGN to suppress dendritic cell function. J Exp Med. 2003;197(1):7–17. doi: 10.1084/jem.20021229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaaithanya IK, Muruganandam N, Surya P, Anwesh M, Alagarasu K, Vijayachari P. Association of oligoadenylate synthetase gene cluster and DC-SIGN (CD209) gene polymorphisms with clinical symptoms in Chikungunya virus infection. DNA Cell Biol. 2015;35(1):44–50. doi: 10.1089/dna.2015.2819. [DOI] [PubMed] [Google Scholar]
- 26.Sakuntabhai A, Turbpaiboon C, Casademont I, Lowhnoo T, Kajaste-Rudnitski A, Kalayanarooj SM. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37(5):507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zupin L, Polesello V, Alberi G, Moratelli G, Crocè SL, Masutti F. CD209 promoter polymorphisms associate with HCV infection and pegylated-interferon plus ribavirin treatment response. Mol Immunol. 2016;76:49–54. doi: 10.1016/j.molimm.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Barreiro LB, Neyrolles O, Babb CL, Tailleux L, Quach H, McElreavey K. Promoter variation in the DC-SIGN-encoding gene CD209 is associated with tuberculosis. PLoS Med. 2006;3(2):e20. doi: 10.1371/journal.pmed.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi L, Zhang K, Mo Y, Zhen G, Zhao J. The association between CD209 gene polymorphisms and pulmonary tuberculosis susceptibility a meta-analysis. Int J Clin Exp Pathol. 2015;8(10):12437–12445. [PMC free article] [PubMed] [Google Scholar]
- 30.Vannberg FO, Chapman SJ, Khor CC, Tosh K, Floyd S, Jackson-Sillah D. CD209 genetic polymorphism and tuberculosis disease. PLoS One. 2008;3(1):e1388. doi: 10.1371/journal.pone.0001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan EJ, Dring M, Ryan CM, McNulty C, Stevenson NJ, Lawless MW. Variant in CD209 promoter is associated with severity of liver disease in chronic hepatitis C virus infection. Hum Immunol. 2010;71(8):829–832. doi: 10.1016/j.humimm.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Chan KYK, Xu MS, Ching JCY, So TMK, Lai ST, Chu CM. CD209 (DC-SIGN) -336A>G promoter polymorphism and severe acute respiratory syndrome in Hong Kong Chinese. Hum Immunol. 2010;71(7):702–707. doi: 10.1016/j.humimm.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren J, Wang Z, Chen E. Different associations between DC-SIGN promoter-336G/A (rs4804803) polymorphism with severe dengue in Asians and south-central Americans a meta-analysis. Int J Environ Res Public Health. 2019;16(8):1475–1475. doi: 10.3390/ijerph16081475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakuntabhai A, Turbpaiboon C, Casadémont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37(5):507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brazilian Health Ministry Leprosy Epidemiological Record 2020. Brasilia. 2020 [Google Scholar]
- 36.WHO - World Health Organization Chemotherapy of leprosy for control programmes. World Health Organization technical report series. 1982;675:1–33. [PubMed] [Google Scholar]
- 37.WHO - World Health Organization WHO Expert Committee on Leprosy. World Health Organization technical report series. 1988;768:1–51. [PubMed] [Google Scholar]
- 38.Fleury RN. Dificuldades no emprego da classificação de Ridley e Jopling uma análise morfológica. Hansen Int. 1989;14(2):101–106. [PubMed] [Google Scholar]
- 39.Rao PN, Pratap DVS, Reddy AVR, Sujai S. Evaluation of leprosy patients with 1 to 5 skin lesions with relevance to their grouping into paucibacillary or multibacillary disease. Indian J Dermatol Venereol Leprol. 2006;72(3):207–210. doi: 10.4103/0378-6323.25781. [DOI] [PubMed] [Google Scholar]
- 40.Pereira R, Phillips C, Pinto N, Santos C, Santos SEB, Amorim A. Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS One. 2012;7(1):e29684. doi: 10.1371/journal.pone.0029684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camargo RM, Silva WL, Medeiros P, Belone AFF, Latini ACP. Polymorphisms in the TGFB1 and IL2RA genes are associated with clinical forms of leprosy in Brazilian population. Mem Inst Oswaldo Cruz. 2018;113(12):e180274. doi: 10.1590/0074-02760180274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barkhash AV, Kochneva GV, Chub EV, Mikhailova SV, Romaschenko AG. Association between polymorphisms in OAS2 and CD209 genes and predisposition to chronic hepatitis C in Russian population. Microbes Infect. 2014;16(5):445–449. doi: 10.1016/j.micinf.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Chen RF, Liu JW, Lee IK, Lee CP, Kuo HC. DC-SIGN (CD209) promoter -336 A/G polymorphism is associated with dengue hemorrhagic fever and correlated to DC-SIGN expression and immune augmentation. PLoS Negl Trop Dis. 2011;5(1):e934. doi: 10.1371/journal.pntd.0000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Yu W, Liou LY, Rice AP. Isolation and characterization of the human DC-SIGN and DC-SIGNR promoters. Gene. 2003;313(1-2):149–159. doi: 10.1016/s0378-1119(03)00674-7. [DOI] [PubMed] [Google Scholar]
- 46.John SWM, Weitzner G, Rozen R, Scriver CR. A rapid procedure for extracting genomic DNA from leukocytes. Nucleic Acids Res. 1991;19(2):408–408. doi: 10.1093/nar/19.2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belone AFF, Rosa PS, Trombone APF, Fachin LRV, Guidella CC, Ura S. Genome-wide screening of mRNA expression in leprosy patients. Front Genet. 2015;6:334–334. doi: 10.3389/fgene.2015.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braga AF, Moretto DF, Gigliotti P, Peruchi M, Vilani-Moreno FR, Campanelli AP. Activation and cytokine profile of monocyte derived dendritic cells in leprosy in vitro stimulation by sonicated Mycobacterium leprae induces decreased level of IL-12p70 in lepromatous leprosy. Mem Inst Oswaldo Cruz. 2015;110(5):655–661. doi: 10.1590/0074-02760140230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Alecrim ES, Chaves AT, Pôrto LAB, Grossi MAF, Lyon S, Rocha MOC. Reading of the mitsuda test comparison between diameter and total area by means of a computerized method. Rev Inst Med Trop São Paulo. 2019;61:e5. doi: 10.1590/S1678-9946201961005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morenikeji OB, Metelski JL, Hawkes ME, Capria AL, Seamans BN, Falade CO. CD209 and not CD28 or STAT6 polymorphism mediates clinical malaria and parasitemia among children from Nigeria. Microorganisms. 2020;8(2):158–158. doi: 10.3390/microorganisms8020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barkhash AV, Perelygin AA, Babenko VN, Brinton MA, Voevoda MI. Single nucleotide polymorphism in the promoter region of the CD209 gene is associated with human predisposition to severe forms of tick-borne encephalitis. Antiviral Res. 2012;93(1):64–68. doi: 10.1016/j.antiviral.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 52.Smatti MK, Al-Sarraj YA, Albagha O, Yassine HM. Host genetic variants potentially associated with SARS-CoV-2 a multi-population analysis. Front Genet. 2020;11:578523–578523. doi: 10.3389/fgene.2020.578523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanazawa N, Mikita N, Li HJ, Nakatani Y, Ozaki M, Kosaka M. Genetic involvement of bacterial sensor molecules in Japanese leprosy. Nihon Hansenbyo Gakkai Zasshi. 2009;78(3):255–261. doi: 10.5025/hansen.78.255. [DOI] [PubMed] [Google Scholar]
- 54.Da Silva RC, Segat L, Da Cruz HLA, Schindler HC, Montenegro LML, Crovella S. Association of CD209 and CD209L polymorphisms with tuberculosis infection in a Northeastern Brazilian population. Mol Biol Rep. 2014;41(8):5449–5457. doi: 10.1007/s11033-014-3416-y. [DOI] [PubMed] [Google Scholar]
- 55.Núñez C, Oliver J, Mendoza JL, Gómez-García M, Taxonera C, Gómez LM. CD209 in inflammatory bowel disease a case-control study in the Spanish population. BMC Med Genet. 2007;8:75–75. doi: 10.1186/1471-2350-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qu X, Che C, Gao A, Lin J, Wang N, Du X. Association of Dectin-1 and DC-SIGN gene single nucleotide polymorphisms with fungal keratitis in the northern Han Chinese population. Mol Vis. 2015;21:391–402. [PMC free article] [PubMed] [Google Scholar]
- 57.Fernández-Ruiz M, Corrales I, Arias M, Campistol JM, Giménez E, Crespo J. Association between individual and combined SNPs in genes related to innate immunity and incidence of CMV infection in seropositive kidney transplant recipients. Am J Transplant. 2015;15(5):1323–1335. doi: 10.1111/ajt.13107. [DOI] [PubMed] [Google Scholar]
- 58.Mezger M, Steffens M, Semmler C, Arlt E-M , Zimmer M. Kristjanson G-I Investigation of promoter variations in dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN) (CD209) and their relevance for human cytomegalovirus reactivation and disease after allogeneic stem-cell transplantation. Clin Microbiol Infect. 2008;14:228–234. doi: 10.1111/j.1469-0691.2007.01902.x. [DOI] [PubMed] [Google Scholar]
- 59.Kuo HC, Huang YH, Chien SC, Yu HR, Hsieh KS, Hsu YW. Genetic variants of CD209 associated with Kawasaki disease susceptibility. PLoS One. 2014;9(8):e105236. doi: 10.1371/journal.pone.0105236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soilleux EJ, Sarno EN, Hernandez MO, Moseley E, Horsley J, Lopes UG. DC-SIGN association with the Th2 environment of lepromatous lesions cause or effect? J Pathol. 2006;209(2):182–189. doi: 10.1002/path.1972. [DOI] [PubMed] [Google Scholar]
- 61.Li S, Lu Z, Yao M, Ning S, Wu Y, Zhou X. Association of single-nucleotide polymorphisms in dc-sign with nasopharyngeal carcinoma susceptibility. Dis Markers. 2017;2017:6309754–6309754. doi: 10.1155/2017/6309754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruegemer JJ, Ho SN, Augustine JA, Schlager JW, Bell MP, McKean DJ. Regulatory effects of transforming growth factor-beta on IL-2- and IL-4-dependent T cell-cycle progression. J Immunol. 1990;144:1767–1776. [PubMed] [Google Scholar]
- 63.Trombone AP, Belone A, Guidella C, Fachin L, Ramuno N, Soriani M, et al. T helper cytokines expression in leprosy forms and reactional states: serum and in situ analysis. J Immunol. 2012;188 [Google Scholar]
- 64.Saini C, Ramesh V, Nath I. Increase in TGF-ß secreting CD4+CD25+ FOXP3+ T regulatory cells in anergic lepromatous leprosy patients. PLoS Negl Trop Dis. 2014;8(1):e2639. doi: 10.1371/journal.pntd.0002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics. 2008;9:516–516. doi: 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]


