Abstract
Cellobiosidase (CbsA) is an important secreted virulence factor of Xanthomonas oryzae pv. oryzae (Xoo), which causes bacterial blight of rice. CbsA is one of several cell wall‐degrading enzymes secreted by Xoo via the type II secretion system (T2SS). CbsA is considered a fundamental virulence factor for vascular pathogenesis. CbsA has an N‐terminal glycosyl hydrolase domain and a C‐terminal fibronectin type III (FnIII) domain. Interestingly, the secreted form of CbsA lacks the FnIII domain during in planta growth. Here we show that the presence of the FnIII domain inhibits the enzyme activity of CbsA on polysaccharide substrates like carboxymethylcellulose. The FnIII domain is required for the interaction of CbsA with SecB chaperone, and this interaction is crucial for the stability and efficient transport of CbsA across the inner membrane. Deletion of the FnIII domain reduced virulence similar to ΔcbsA Xoo, which corroborates the importance of the FnIII domain in CbsA. Our work elucidates a hitherto unknown function of the FnIII domain in enabling the virulence‐promoting activity of CbsA.
Keywords: exoglucanase, FnIII domain, rice, secretion, type II secretion system, virulence, Xanthomonas
The fibronectin type III domain associated with the cellobiosidase enzyme of Xanthomonas oryzae pv. oryzae is required for efficient secretion of the protein and virulence on rice.

1. INTRODUCTION
When a phytopathogen encounters a potential host, the plant cell wall serves as a physical barrier to limit access of the pathogen to cellular contents (Kubicek et al., 2014). The plant cell wall is a recalcitrant exoskeleton that surrounds the cell protoplast and is composed of a complex network of polysaccharides including cellulose, hemicellulose, and pectin (Cosgrove, 2005). Successful plant pathogens possess essential virulence attributes such as cell wall‐degrading enzymes (CWDEs) that degrade different components of the plant cell wall (Kubicek et al., 2014).
The gram‐negative bacterial genus Xanthomonas includes species that cause diseases in almost 400 plants, including agronomically important crops (Jacques et al., 2016; Ryan et al., 2011). One member of this genus, Xanthomonas oryzae pv. oryzae (Xoo), causes bacterial blight disease of rice. Xoo secretes a battery of CWDEs including cellulase (ClsA), cellobiosidase (CbsA), xylanase (XynB), lipase (LipA), and pectinase (PglA) through the type II secretion system (T2SS) (Jha et al., 2007; Rajeshwari et al., 2005; Tayi et al., 2016). Disruption of genes encoding these enzymes in Xoo leads to either partial virulence deficiency, as in the case of ΔclsA and ΔlipA, or a more severe virulence deficiency, as seen in the case of ΔcbsA (Jha et al., 2007). The CWDEs are double‐edged swords as the damage that they cause serves as a mark of infection and results in induction of plant immune responses such as callose deposition and programmed cell death responses (Jha et al., 2007).
CbsA is an extracellular exoglucanase and its biochemical activity is critical for virulence (Tayi et al., 2018). Recently, the presence and absence of the cbsA gene in Xanthomonas was shown to be central to the switch between vascular and nonvascular pathogenesis, respectively (Gluck‐Thaler et al., 2020). Genomic comparison between pathogenic and nonpathogenic Xanthomonas sp., which are inhabitants of the same host, shows an association of the cbsA gene with pathogenicity (Bansal et al., 2019). Interestingly, CbsA is among the highest expressed proteins during the growth of Xoo in the vascular tissue (González et al., 2012; Jacobs et al., 2012). In addition, the LuxR (quorum sensing‐related transcription regulator) subfamily member of Xoo, OryR, was shown to induce expression of CbsA on treatment with macerated rice tissue (Ferluga et al., 2007; Gonzalez & Venturi, 2013). Collectively, the above reports suggest that CbsA is expressed in planta and is an important virulence factor of Xoo.
The 566‐amino acid CbsA protein has an N‐terminal glycosyl hydrolase domain and a C‐terminal fibronectin type III (FnIII) domain (Figure 1a). The FnIII domain is one of the three types of internal repeats (FnI‐FnII‐FnIII) originally identified in its eponym fibronectin, a eukaryotic plasma protein (Main et al., 1992; Ruoslahti, 1988). The FnIII domain is present in almost 2% of animal proteins with a few instances in yeast and plants as well (Bateman & Chothia, 1996; Bork & Doolittle, 1992; Tsyguelnaia & Doolittle, 1998). In the case of animals, the FnIII domain is mostly present in extracellular matrix proteins, where it is involved in protein–protein interaction. The FnIII domain also acts as a spacer between domains to facilitate unhindered biological action of the respective domains (Bork et al., 1996; Campbell & Spitzfaden, 1994).
FIGURE 1.
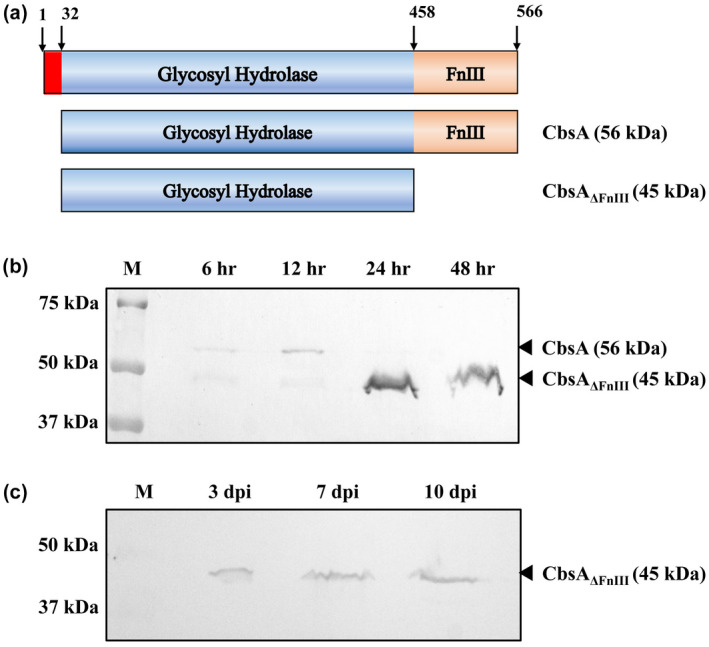
Secreted CbsA lacks the FnIII domain. (a) Schematic representation of domain organization of Xanthomonas oryzae pv. oryzae (Xoo) CbsA. CbsA has an N‐terminal signal sequence followed by the catalytic domain (glycosyl hydrolase‐6 family) and a C‐terminal fibronectin type III (FnIII) domain. (b) Western blot analysis of secreted Xoo CbsA at different time points, using anti‐CbsA antibody, showing absence of FnIII domain in the secreted CbsA at later stages of growth. (c) Western blot analysis of Xoo‐infected rice leaf exudate at 3, 7, and 10 days postinoculation, showing absence of the FnIII domain in the secreted CbsA during in planta growth
In prokaryotes, the FnIII domain was first identified in a chitinase enzyme of Bacillus circulans (Watanabe et al., 1990). Most of the initially identified FnIII domains were found to be associated with carbohydrate‐hydrolysing enzymes, but later they were also identified in the fibronectin‐binding proteins and other prokaryotic protein types (Henderson et al., 2011; Konkel et al., 2010). FnIII domains associated with bacterial hydrolases have been shown to have different roles, which include increasing thermostability of the associated protein (Brunecky et al., 2012) and acting as a spacer between the hydrolysing domain and the substrate‐binding domain for enhanced catalysis (Kataeva et al., 2002). In the case of Dickeya dadantii (a plant pathogen), genetic and biochemical studies showed that the FnIII domain associated with a pectin‐hydrolysing enzyme pectate lyase harbours a loop that acts as a type II secretion signal for the transport of the protein across the outer membrane (Pineau et al., 2014).
In Xoo, the secreted CbsA lacks the FnIII domain (Figure 1a) (Kumar et al., 2012), and this secreted form is biochemically active on soluble oligosaccharides and the polysaccharide substrate carboxymethylcellulose (CMC) (Tayi et al., 2018). In addition, it can also induce immune responses in rice tissues (Jha et al., 2007). However, the role of the FnIII domain in CbsA is not known. In the current study, we have used genetic and biochemical approaches to understand the role of the FnIII domain in CbsA function.
2. RESULTS
2.1. Secreted CbsA lacks the FnIII domain during in planta growth of Xoo in rice
To understand the possible reason for the removal of the FnIII domain, wild‐type Xoo strain BXO43 was grown and the extracellular fraction was analysed using anti‐CbsA antibody at different time points. Up to 12 h of growth, the full‐length form of the protein was present in the extracellular fraction (Figure 1b). From 24 h onwards, the major form present in the extracellular fraction was the cleaved form of CbsA lacking the FnIII domain (CbsAΔFnIII). This observation led us to check the form of CbsA that is present during Xoo infection in rice. For this purpose, wild‐type Xoo was inoculated on the susceptible rice variety Taichung Native‐1 (TN‐1), and leaf exudate was collected 3, 7, and 10 days postinoculation (dpi). Western blot analysis of the leaf exudate showed the presence of a single band corresponding to the cleaved form of CbsA (45 kDa) (Figure 1c). These observations indicate that during infection in rice, the secreted CbsA protein lacks the FnIII domain.
2.2. Presence of the FnIII domain modulates the biochemical activity of CbsA
CbsAΔFnIII is known to hydrolyse soluble oligosaccharides and polysaccharide substrate CMC in in vitro assays (Tayi et al., 2018). To assess the effect of the FnIII domain on the biochemical activity of the protein, CbsA and CbsAΔFnIII were purified from Escherichia coli (Figure S1a–c). The biochemical activity was assessed on cellotetraose, cellopentaose, cellohexaose, and CMC. CbsA showed reduced activity on cellotetraose and cellohexaose compared to CbsAΔFnIII as shown by the presence of unhydrolysed substrates (Figure 2a). CbsA did not hydrolyse CMC, but CbsAΔFnIII showed the release of cellobiose, cellotriose, and cellotetraose. This difference in activity was also observed in a CMC‐agarose plate assay where detection of enzyme activity is achieved by staining with Congo red solution and the presence of a halo indicates hydrolysis. The CbsA protein showed absence of a halo in the assay while CbsAΔFnIII showed presence of a halo (Figure 2b). Intriguingly, purified CbsA (c.3‐day‐old protein) from E. coli is spontaneously cleaved into a smaller protein that corresponds to CbsAΔFnIII in size (Figure S1d). The cleaved form showed a gain of activity on CMC (Figure 2b), indicating that it is indeed the presence of the FnIII domain that inhibits the activity on the polysaccharide substrate. In conclusion, the enzymatic activity of CbsA protein is negatively modulated by the presence of the FnIII domain.
FIGURE 2.
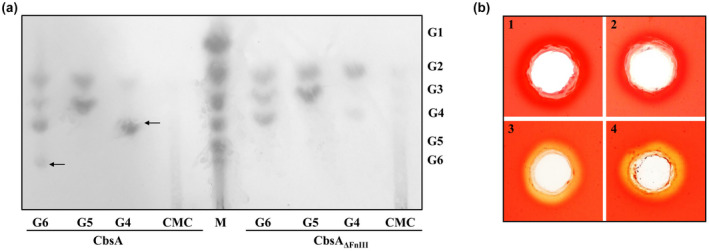
FnIII domain has an inhibitory effect on the biochemical activity of CbsA. (a) The activity of the catalytic domain of CbsA in the presence and absence of the FnIII domain was assessed on oligosaccharides and carboxymethylcellulose (CMC). CbsA and CbsAΔFnIII were incubated overnight with oligosaccharide substrates G6‐cellohexaose, G5‐cellopentaose, G4‐celloteraose, and polysaccharide substrate CMC. The hydrolysed products were separated and detected using thin‐layer chromatography as described in the experimental procedures. A mix of oligosaccharides (G1–G6) is shown in the lane labelled M. The unhydrolysed substrate is marked by a black arrow. (b) CMC‐agarose plate assay showing hydrolysis by (1) buffer, (2) CbsA, (3) CbsAΔFnIII, and (4) CbsA after cleavage of FnIII domain. The presence of a halo around the well after Congo red staining indicates the activity of the enzyme on CMC
2.3. Deletion of the FnIII domain affects the levels of CbsA protein in the extracellular space and within the cell
To investigate the requirement for the FnIII domain in CbsA, an in‐frame deletion of the FnIII domain‐coding region was made in the cbsA gene of Xoo, and the CbsA protein levels were analysed in the extracellular fraction and the whole‐cell lysate using anti‐CbsA antibody. The cbsA ΔFnIII mutant showed more than 95% reduction in the secreted CbsA level as compared to the wild type (Figure 3a,b). Additionally, in the whole‐cell lysate, the protein level showed more than 60% reduction in the mutant as compared to the wild type (Figure 3c). Although the mRNA level of cbsA was same as the wild type (Figure S2b), the total amount of CbsA protein was drastically reduced on deletion of the FnIII domain (Figure 3). These results illustrate that in absence of the FnIII domain, secretion of CbsA is heavily compromised and the level of CbsA protein within the cell is reduced.
FIGURE 3.
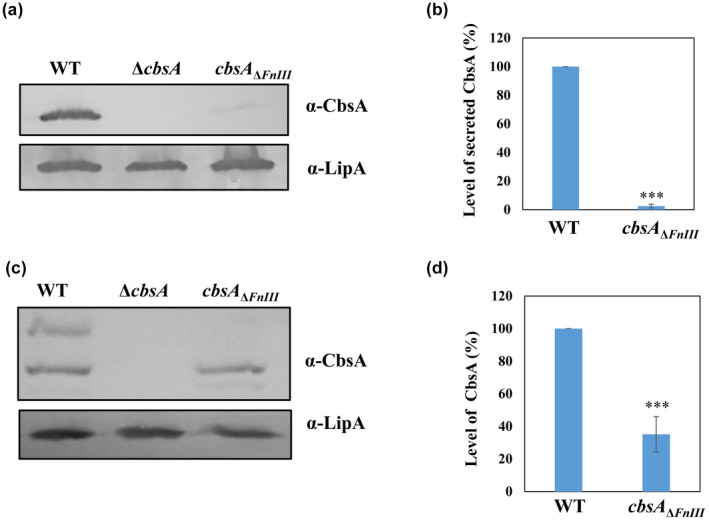
Deletion of the FnIII domain leads to a reduction in the level of CbsA protein in the extracellular space as well as within the cell. (a) Western blot analysis of the secreted CbsA protein in the culture supernatant of wild type (WT), ΔcbsA, and cbsA ΔFnIII Xanthomonas oryzae pv. oryzae (Xoo). The data indicates that the deletion of the FnIII domain results in more than 95% reduction in the level of the secreted CbsA protein. LipA protein was used as a loading control and ΔcbsA Xoo was used as a negative control. (b, d) Densitometry analysis of western blots of three independent experiments using ImageJ software. Student's two‐tailed t test was performed and compared groups were significantly different at p < 0.001 (marked with ***). (c) Western blot analysis of CbsA protein in the cell lysate of WT, ΔcbsA, and cbsA Δ FnIII Xoo indicates that deletion of the FnIII domain results in more than 60% reduction in the level of the CbsA protein
2.4. CbsA interacts with SecB chaperone through the FnIII domain
Type II secretion of proteins takes place in two steps. In the first step, the protein is transported across the cytoplasmic membrane via either the Sec system or the TAT system, depending on the signal sequence present in the secreted protein (Korotkov et al., 2012). From the periplasm, the protein is secreted into the extracellular milieu by the T2SS. Analysis of the CbsA protein sequence, using SignalP v. 5.0 (Armenteros et al., 2019), indicated the presence of a Sec‐dependent signal sequence at its N‐terminal region (Figure 4a). Proteins that are transported across the cytoplasmic membrane via the Sec system are known to remain loosely folded in the cytoplasm after their translation and are protected from aggregation and cellular protein degradation machinery by binding to the SecB chaperone (Randall & Hardy, 1995; Sakr et al., 2010). Classically, SecB recognizes its substrates via a signal sequence; however, in addition to the signal sequence, other regions of the protein may also participate in this recognition (Huang et al., 2016; Mallik et al., 2002). SecB recognition sites are known to be enriched in hydrophobic residues and the presence of acidic residues is disfavoured (Bechtluft et al., 2010; Huang et al., 2016; Knoblauch et al., 1999). Analysis of the CbsA protein sequence for hydrophobicity shows a patch of hydrophobic residues in the FnIII domain sequence (Figure 4b). Based on these observations and earlier results that deletion of the FnIII domain affects the level of CbsA protein and its secretion, we hypothesized that the FnIII domain could be the region through which CbsA binds to the SecB chaperone.
FIGURE 4.
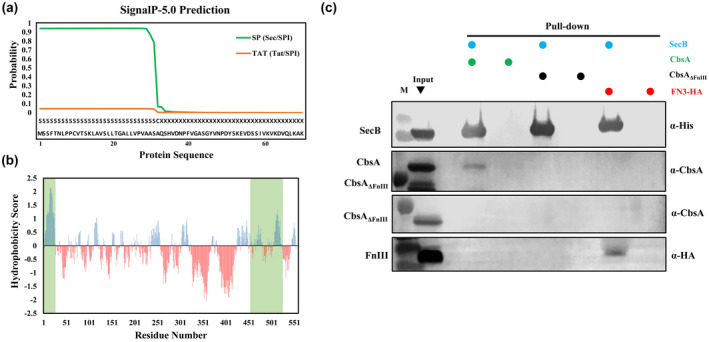
CbsA interacts with the SecB chaperone via the FnIII domain. (a) Sequence analysis of Xanthomonas oryzae pv. oryzae (Xoo) CbsA using SignalP v. 5.0 indicating the probability of secretion via the Sec or TAT pathways. (b) Hydrophobicity plot as a function of the primary sequence of CbsA protein. Positive hydrophobicity score (ProtScale‐ Kyte and Doolittle algorithm, window = 13) denotes increased hydrophobicity. The favourable sites for SecB binding in the CbsA sequence are highlighted in green. (c) Affinity pull‐down of His‐tagged SecB with CbsA/CbsAΔFnIII/FnIII in Escherichia coli followed by western blot analysis of the eluted samples using the indicated antibodies show SecB interacts with CbsA protein and the FnIII domain but not with CbsAΔFnIII protein
To investigate this, an in vitro affinity pull‐down experiment was done. For this purpose, SecB‐His, FnIII‐HA, CbsA, and CbsAΔFnIII proteins were expressed in E. coli using the isopropyl‐β‐D‐1‐thiogalactopyranoside (IPTG)‐inducible pET28b vector. Pull‐down followed by western blotting showed that SecB interacted with CbsA but not with CbsAΔFnIII. The FnIII domain alone showed binding with SecB (Figure 4c). These results suggest that CbsA interacts with SecB via the FnIII domain, which in turn facilitates stability and efficient transport of the protein across the inner membrane.
2.5. The FnIII domain of CbsA is essential for virulence of Xoo on rice
Our results indicate that the FnIII domain influences catalytic activity as well as secretion of the CbsA protein. We further investigated the virulence of cbsA ΔFnIII Xoo in comparison to wild‐type Xoo. For this purpose, the Xoo wild‐type strain BXO43, ΔcbsA, and cbsA ΔFnIII mutants were inoculated onto rice leaves and lesion lengths were measured 15 dpi. The cbsA ΔFnIII Xoo strain exhibited reduced virulence and the level of reduction was comparable to the ΔcbsA Xoo strain (Figure 5a,b). This observation confirms that the presence of the FnIII domain in the CbsA protein is critical for the optimal virulence of Xoo on rice. Furthermore, we complemented the cbsA ΔFnIII Xoo strain with cbsA or cbsA ΔFnIII, expressed via a pHM1 plasmid. On inoculation on rice, cbsA complemented the virulence phenotype of cbsA ΔFnIII Xoo, which was comparable to the wild‐type level whereas complementation with the cbsA ΔFnIII coding region showed a partial recovery (Figure 5). These results further corroborate that presence of the FnIII domain in the CbsA protein is important for the virulence of Xoo on rice.
FIGURE 5.
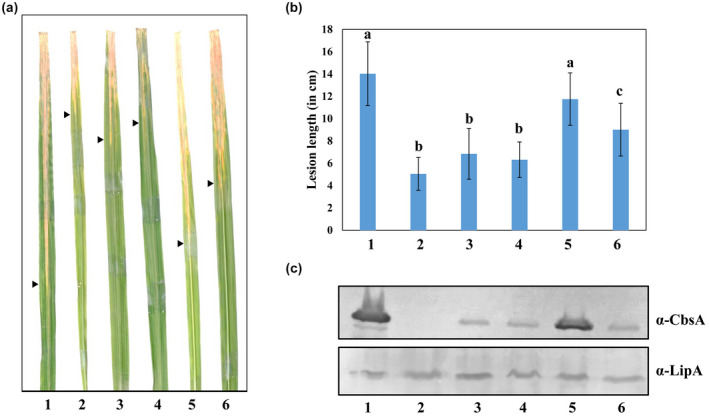
The FnIII domain of CbsA is critical for virulence of Xanthomonas oryzae pv. oryzae (Xoo) on rice. (a) Rice leaves showing lesions caused by (1) wild type, (2) ΔcbsA, (3) cbsA ΔFnIII, (4) cbsA ΔFnIII + pHM1 (empty vector), (5) cbsA ΔFnIII + pHM1‐cbsA, and (6) cbsA ΔFnIII + pHM1‐cbsA ΔFnIII Xoo strains 15 days postinoculation. Black arrowheads mark the leading edge of the lesion. (b) Columns and vertical bars indicate the mean lesion length caused by the above strains and standard deviation of at least 20 rice leaves, respectively. Student's two‐tailed t test was performed, groups with different letters were significantly different at p < 0.001. Similar results were obtained in three independent experiments. (c) Western blot analysis showing the level of secreted CbsA in the above‐mentioned strains. LipA protein was used as a loading control
2.6. Presence of the FnIII domain is associated with Sec‐dependent secretion of Xanthomonas CbsA
Domain analysis of the CbsA sequence using the InterPro tool (Blum et al., 2021) showed that the catalytic domain of CbsA in all the Xanthomonas species is linked with an FnIII domain at its C‐terminus, except in X. translucens pv. translucens (Xtt) and X. albilineans (Figure 6). In the case of X. albilineans and Xylella fastidiosa (a plant pathogen that is a close relative of Xanthomonas genus), the CbsA catalytic domain is associated with a carbohydrate‐binding domain via a long (80–150 amino acids) Gly‐Ser repeat linker. Carbohydrate‐binding domains are known to enhance the hydrolysis efficiency of the catalytic domain by facilitating prolonged substrate association (Shoseyov et al., 2006). Analysis of the CbsA protein sequence with SignalP v. 5.0 (Armenteros et al., 2019) showed the presence of an exclusively Sec‐dependent signal sequence in all the Xanthomonas spp., except Xtt (Table S1). In case of Xtt CbsA, the signal sequence showed a higher probability for TAT‐dependent secretion than Sec‐dependent secretion (Figure S3). Concomitantly, the Xtt CbsA protein lacks the FnIII domain, which is indicative of an association between Sec‐dependent secretion and presence of the FnIII domain in Xanthomonas CbsA protein. The FnIII domain is proposed to be horizontally transferred in bacterial hydrolases (Bork & Doolittle, 1992; Little et al., 1994). However, GC content and codon usage analysis of the FnIII domain in Xoo indicate that this transfer must have happened a long time ago and the codon usage pattern has been homogenized to the Xoo genome (Figure S4a,b). In conclusion, the FnIII domain appended to CbsA is a conserved feature across the Xanthomonas genus, possibly employed for efficient secretion of CbsA via the Sec pathway during pathogenesis in plants.
FIGURE 6.
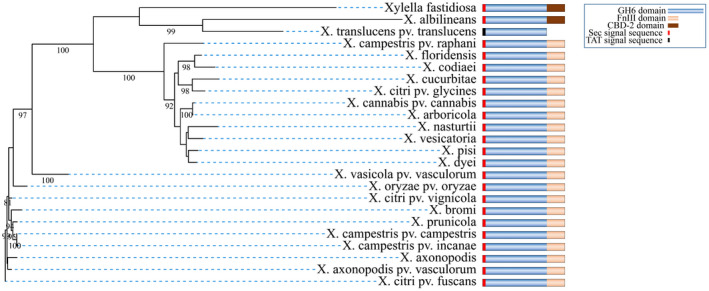
Association of Sec‐dependent secretion of CbsA and the FnIII domain in Xanthomonas. Maximum‐likelihood phylogenetic tree of CbsA protein from different Xanthomonas species and their close relative Xylella fastidiosa. Bootstrap values (>80) are shown at the branches. Domain organization and type of signal sequence present in CbsA protein sequence are shown schematically for the respective organism. Analysis of CbsA protein sequence from different Xanthomonas species showed association of the FnIII domain with the presence of a Sec‐dependent signal sequence. Xanthomonas translucens pv. translucens CbsA does not have an FnIII domain and shows a higher probability for the presence of a TAT‐dependent signal sequence over a Sec‐dependent signal sequence
3. DISCUSSION
Vascular plant pathogens cause severe systemic infections by moving long distances in the veinal system of host plants. The CWDE CbsA has been shown to be a fundamental factor in the evolution of Xanthomonas vascular pathogenicity (Gluck‐Thaler et al., 2020). CbsA is an exoglucanase that has an FnIII domain at its C‐terminus. In bacterial hydrolases, the presence of the FnIII domain is associated with diverse functions. CbsA is an example of such association with the unique feature that the secreted form of CbsA lacks the FnIII domain. In the present study, we have identified a unique function of the FnIII domain where it is required for the optimal level of CbsA secretion by Xoo, which is critical for pathogenesis in rice.
Xoo CbsA contains a Sec‐dependent secretion signal at its N‐terminus. Sec‐secreted proteins are known to remain loosely folded in the cytoplasm, which is their transport‐competent form (Cranford‐Smith & Huber, 2018; Hardy & Randall, 1991). SecB is a cytoplasmic chaperone that has an antifolding activity (Huang et al., 2016). The binding of a secretory protein with SecB serves three purposes: first, it keeps the secretory protein in the partly unfolded form; second, it protects this partly unfolded secretory protein from aggregation and cellular degradation machinery; and third, SecB hands over the bound secretory protein to SecA, which is part of the Sec translocon (Cranford‐Smith & Huber, 2018; Hardy & Randall, 1991; Sakr et al., 2010). In the case of Xoo, deletion of the FnIII domain of CbsA led to impaired secretion and a reduction in the total amount of CbsA protein, which is indicative of insufficient SecB binding. Our in vitro pull‐down experiment showed that SecB bound to CbsA but not to CbsAΔFnIII. This finding elucidates a unique function of the FnIII domain, wherein it mediates interaction with the SecB chaperone to achieve efficient transport across the cytoplasmic membrane as well as stability of the CbsA protein. It is evident that an optimal level of CbsA expression is critical for pathogenesis, which warrants an efficient transport mechanism, and it appears that it may be achieved in this case by appending an FnIII domain to this virulence factor.
Pathogens are under continuous selection pressure, imposed by their host environment, which leads to the evolution of virulence factors that facilitate colonization within their hosts (Goldstein, 2008; Thornton et al., 1999). Despite low sequence identity among different FnIII domains, the structural fold remains common, which is a Greek key β‐sandwich fold made up of seven or eight strands (Banner et al., 1996; de Pereda et al., 1999; Sharma et al., 1999; Zhao et al., 2017). This feature of the FnIII domain allows it to be highly mutated and repurposed to confer novel functions. In particular, one study has shown that the FnIII domain can accommodate multiple mutations within the same fold to become a novel binding protein (Koide et al., 1998). Possibly, the FnIII domain of CbsA has also evolved in a similar way to bind to SecB for proficient secretion.
Intriguingly, the FnIII domain of Xoo CbsA is cleaved from the protein in the later stages of laboratory‐grown cultures and during in planta infection. Similar observations have been made in Trichoderma ressei, where cellobiohydrolase I and cellobiohydrolase II undergo postsecretion proteolysis at the later growth stages to yield multiple forms of cellulases (Hagspiel et al., 1989). However, in both cases, it is not clear whether this cleavage takes place by the action of a specific protease or by spontaneous cleavage. The whole‐cell lysates of Xoo (Figure 3c) and E. coli expressing CbsA (Figure 4c) show the presence of both full‐length and cleaved forms of CbsA, which would argue for a spontaneous cleavage. Presumably, the FnIII domain has to be removed because its presence inhibits the activity of CbsA on polymeric substrates, making it less efficient for cell wall degradation. CMC is cellulose with carboxymethyl modifications. The action of exoglucanases on such substrates is facilitated by the flexibility of the substrate‐binding tunnel‐enclosing loops (Varrot et al., 1999; Zou et al., 1999). It is possible that in the presence of the FnIII domain, the movement of these loops is restricted and thus the enzyme is inactive on CMC. Furthermore, it is tempting to propose that the inhibition of the enzyme activity of the CbsA catalytic domain by the FnIII domain serves the purpose of reducing detection by the host immune system during the early phase of infection.
What is the fate of the FnIII domain after cleavage? Does it have any other function in the extracellular milieu when it is attached to CbsA in the initial growth phase? Some of these questions need further investigation. In the case of Vibrio cholerae, RbmA is one of the proteins required for biofilm formation. Structural analysis of this protein showed that it has two tandem FnIII domains that participate in biofilm formation (Giglio et al., 2013). For Xanthomonas spp. that dwell in the xylem, biofilm formation during the initial stages and dispersal in the later stages is critical for pathogenesis (Crossman & Dow, 2004; Dow et al., 2003). In addition, factors involved in biofilm formation and dispersion are often regulated by quorum sensing. Interestingly, one such factor regulates CbsA expression in the presence of macerated rice tissues (Degrassi et al., 2007; Ferluga et al., 2007). Given this information and the fact that CbsA is critical for vascular pathogenesis, it would be worthwhile to assess the role of CbsA in the context of biofilm formation during the growth of Xoo in rice.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains and growth conditions
The primers, bacterial strains, and plasmids used in this study are listed in Tables S2 and S3. Xoo strains were grown at 28°C in peptone sucrose (PS) medium and E. coli strains were grown in Luria Bertani (LB) medium at 37°C unless mentioned otherwise. The concentrations of antibiotics used were rifampicin (Rf) 50 µg/ml, spectinomycin (Sp) 50 µg/ml, ampicillin (Ap) 50 µg/ml, and kanamycin (Km) 50 µg/ml for E. coli and 15 µg/ml for Xoo.
4.2. Construction of cbsA ΔFnIII mutant in Xoo
Based on the structure of Xoo‐secreted CbsA (Kumar et al., 2012; Tayi et al., 2018), the C‐terminal region that is missing in the secreted CbsA was deleted in the Xoo genome. Genomic DNA of Xoo wild‐type strain BXO43 was used as a template and two primer pairs, fragment A‐FP/ fragment A‐RP and fragment C‐FP/ fragment C‐RP (Table S2), were used to amplify fragments A and C (fragments A and C include the flanking regions of the FnIII domain). The A + C fragment was cloned in the HindIII restriction site of pK18mobsacB plasmid (Schäfer et al., 1994). The recombinant plasmid containing the fragment A + C was transformed into E. coli S17‐1 (Simon et al., 1983). This recombinant plasmid was transferred from E. coli S17‐1 to the wild‐type Xoo BXO43 by biparental mating. Xoo cells were selected for kanamycin resistance and sucrose sensitivity. Integration of the plasmid at the expected locus was confirmed by PCR. The PCR‐confirmed single recombinants were subcultured twice in liquid medium (0.3% beef extract, 0.5% peptone) without kanamycin and plated on PS agar plates. Putative double recombinants, which include both wild‐type and mutant cells (kanamycin sensitive and sucrose resistant), were isolated. The double recombinants were screened for the desired deletion by PCR and sequencing.
4.3. Antiserum preparation
The catalytic domain of CbsA was purified from Xoo culture supernatant as described earlier (Kumar et al., 2012; Tayi et al., 2018). The purified protein was used to generate anti‐CbsA antibodies in rabbit following the method described (Aparna et al., 2009). CbsA‐specific antibodies were purified from the serum using affinity purification as reported earlier (Burbelo et al., 2002).
4.4. Protein sample preparation for western blotting
Leaf tips of 40‐day‐old Taichung Native‐1 (TN‐1) rice plants were clip inoculated with Xoo strains. At different time points, infected leaves were cut at a point 2 cm below the visible lesion, dipped into water, and incubated at 4°C overnight to collect the leaf exudate. The exudate was centrifuged at 6700 × g for 10 min to remove cells. The supernatant was concentrated and subjected to western blotting analysis.
Secreted proteins and whole‐cell lysates from Xoo cells were prepared as reported earlier (Gautam & Sharma, 2002) with some modifications. Xoo strains were grown to saturation, centrifuged, and the supernatant was taken as the extracellular fraction (EF). The EF was concentrated and an equal amount of the protein was loaded on a gel for western blotting analysis. The cell pellet from the above‐mentioned sample was washed and resuspended in sterile Milli‐Q water. Cell number was normalized based on OD600 for different strains to obtain similar protein concentrations. Cells were lysed by heating at 95°C for 10 min with 2× protein loading buffer. After the heating step, samples were chilled on ice for 5 min and centrifuged at 9700 × g for 10 min. An equal amount of supernatant was separated by SDS‐PAGE for western blotting analysis.
4.5. Western blotting
Samples were run on 12% SDS‐PAGE followed by transfer onto a nitrocellulose membrane. The membrane was subjected to 2 h of blocking with 5% Blotto powder (Bio‐Rad). After this, the membrane was washed with phosphate‐buffered saline containing 0.05% Tween (PBST) and incubated with primary antibody (α‐CbsA antibody, 1:1000; α‐LipA antibody, 1:2000; α‐HA antibody, 1:4000; abcam) or α‐polyHis antibody (1:2000; Sigma Aldrich) at room temperature for 2 h. After thorough washing with PBST, the membrane was incubated with alkaline phosphatase (ALP)‐conjugated secondary antibody (1:30,000 dilution; Sigma Aldrich) for 1 h at room temperature. This was followed by washing and incubation with ALP substrate (33 µg/ml nitroblue tetrazolium, 17 µg/ml 5‐bromo‐4‐chloro‐3‐indolyl phosphate; Roche) in ALP buffer (100 mM NaCl, 5 mM MgCl2, 100 mM Tris pH 9.5). Once the colour developed, the reaction was stopped by adding 0.5 M EDTA solution.
4.6. Purification of recombinant CbsA and CbsAΔFnIII protein and enzyme activity assay
Sequences encoding CbsA and CbsAΔFnIII protein without the signal sequence were amplified from Xoo genomic DNA using specific primer pairs (Table S2). The amplified sequence was cloned into a pETM40 expression vector (Invitrogen) between NcoI and XhoI restriction sites to add an N‐terminal maltose‐binding protein tag (MBP, to improve solubility of the recombinant protein) and a C‐terminal 6× histidine tag. A tobacco etch virus (TEV) protease cleavage site was present between the MBP and the cloned protein sequence. These constructs were overexpressed in E. coli BL‐21 codon plus cells under IPTG induction. The proteins were purified using Ni‐NTA (nitrilotriacetic acid) affinity chromatography followed by TEV protease cleavage to remove MBP. For this purpose, TEV protease and the MBP‐tagged protein were mixed in a 1:100 ratio (wt/wt) and incubated at 4°C overnight. The protein of interest was further purified from this reaction mixture by Ni‐NTA affinity chromatography followed by gel filtration. The purity of the protein was checked using SDS‐PAGE. Enzyme activity assays were performed on soluble oligosaccharides and CMC as mentioned by Tayi et al. (2018).
4.7. Reverse transcription quantitative real‐time PCR
Total RNA was isolated from Xoo wild‐type and cbsA ΔFnIII cells using TRIzol reagent (Invitrogen) as per the manufacturer's instructions. The RNA quality was assessed by agarose gel electrophoresis followed by DNase I (NEB) treatment and quantification. Primer pairs were checked for amplification efficiency using 10‐fold dilutions of genomic DNA and the pair that was closest to 100% efficiency (slope of 3.3 for C t versus log DNA copy number) was picked for further experiments. cDNA was synthesized from 5 µg of total RNA with random hexamers primers using RNA to cDNA EcoDry Premix kit (Clontech). The cDNA was diluted 10 times, and 1 µl of it was subjected to quantitative PCR (qPCR) using a DyNAmo Color Flash SYBR green qPCR kit (Thermo Scientific). The experiment was performed on a ViiA 7 real‐time PCR system (Applied Biosystems). ΔC t was calculated for Xoo wild type and cbsA Δ FnIII by subtracting the C t mean of respective samples from the C t mean of 16S rRNA, which was used as an endogenous control. The experiment was done for three independent biological replicates. Mean ΔC t and standard error of a representative experiment were plotted.
4.8. Pull‐down assay
CbsA, CbsAΔFnIII, HA‐tagged FnIII domain, and polyhistidine‐tagged SecB proteins were overexpressed in E. coli using pET28b (Novagen) expression vector. The strains were grown at 37°C until OD600 0.6 and induced with 0.1 mM IPTG. After induction, cells were grown at 18°C overnight. Cells were harvested by centrifugation and resuspended in a lysis buffer (50 mM Tris‐Cl pH 8.0, 150 mM NaCl). CbsA/CbsAΔFnIII/FnIII‐expressing cell lysates were incubated overnight with TALON beads with and without SecB‐expressing cell lysate. After binding to the cell lysate, beads were washed successively with lysis buffer and lysis buffer containing 20 mM imidazole. Finally, the bound protein fraction was eluted using elution buffer (50 mM Tris‐Cl pH 8.0, 150 mM NaCl, 250 mM imidazole). The eluted proteins were probed by western blotting.
4.9. Virulence assay
The following Xoo strains were used for virulence analysis: BXO43, ΔcbsA, cbsA ΔFnIII, cbsA ΔFnIII complemented with wild‐type cbsA, and cbsA without FnIII domain‐encoding region on pHM1 plasmid (Innes et al., 1988). Strains were grown to saturation followed by centrifugation at room temperature. The cell pellet was washed and resuspended in Milli‐Q water to achieve OD600 = 1.0. The leaf tips of 40‐day‐old TN‐1 rice plants were cut with surgical scissors dipped in the bacterial culture. Lesion lengths were measured 15 dpi.
4.10. Bioinformatics analysis
CbsA sequences from different Xanthomonas species were retrieved from the NCBI database. A multiple sequence alignment was generated using Clustal Omega on the EMBL‐EBI server (Sievers et al., 2011). The alignment was submitted to the IQ‐TREE web server (Trifinopoulos et al., 2016) to generate a maximum‐likelihood phylogenetic tree. Domain identification was done in different CbsA sequences using the InterPro online tool (Blum et al., 2021). The SignalP v. 5.0 web tool (Armenteros et al., 2019) was used to predict the type of signal sequence in CbsA proteins from different Xanthomonas species/pathovars. The hydrophobicity score of the Xoo CbsA protein sequence was calculated using the ProtScale web tool (Kyte & Doolittle, 1982).
4.11. Codon usage pattern
The codon usage pattern was analysed as described earlier (Patil & Sonti, 2004) using “The Sequence Manipulation Suite” web tool (Stothard, 2000.). The first group was chosen containing the following housekeeping genes encoding tonB‐dependent siderophore receptor (BXO1_013815): Xanthomonas adhesin‐like protein (BXO1_013910), rpfF (BXO1_008735), phytase (BXO1_006505), shikimate dehydrogenase (BXO1_016165), and secreted xylanase (BXO1_019245). The second group contained genes from the lipopolysaccharide cluster, which was previously shown to be horizontally transferred into Xoo (Patil & Sonti, 2004). This group included smtA (BXO1_014260), wxoA (BXO1_014255), wxoB (BXO1_014250), wxoC (BXO1_014240), and wxoD (BXO1_014235). The average GC content for the Xoo genome was taken based on the genome sequence of BXO1 (Kaur et al., 2019).
CONFLICT OF INTEREST
The authors declare that no conflict of interest exists.
AUTHOR CONTRIBUTIONS
R.N., R.V.S., and R.S. conceived the project and designed the experiments. R.N. and R.V.M. performed the experiments. R.N., H.K.P., R.S., and R.V.S. analysed the data, and finalized the manuscript, which was approved by all the authors. H.K.P., R.S., and R.V.S. contributed reagents/materials.
Supporting information
FIGURE S1 Expression and purification of CbsA and CbsAΔFnIII in Escherichia coli. (a) 8% SDS‐PAGE showing purified CbsA and CbsAΔFnIII tagged with maltose‐binding protein (MBP) and 6×histidine. Proteins were purified using Ni‐NTA affinity chromatography. (b) 12% SDS‐PAGE showing CbsA and CbsAΔFnIII cleaved from MBP after TEV protease digestion. The lowest band shows TEV protease, which was used in the digestion reaction. (c) 8% SDS‐PAGE gel showing CbsA and CbsAΔFnIII purified from TEV protease digestion reaction by affinity chromatography followed by gel filtration. (d) Spontaneous cleavage of CbsA into CbsAΔFnIII in a c.3‐day‐old protein preparation
FIGURE S2 Reverse transcription quantitative PCR analysis of cbsA expression in wild‐type and cbsA Δ FnIII Xanthomonas oryzae pv. oryzae (Xoo) strains. (a) Schematic of primers used for cbsA expression analysis (b) cbsA mRNA level measured in Xoo wild type and cbsA Δ FnIII mutant shows no significant difference. 16S rRNA gene was used as endogenous control. Average ΔC t (C t cbsA − C t 16s rRNA) was plotted for both strains. Student’s two‐tailed t test was performed and the difference was not significant (p = 0.329)
FIGURE S3 Sequence analysis of Xanthomonas translucens pv. translucens CbsA using SignalP v. 5.0 indicating the probability of the presence of Sec‐ or TAT‐dependent signal sequence
FIGURE S4 (a) Average GC content (%) of the Xanthomonas oryzae pv. oryzae (Xoo) genome, the catalytic domain of CbsA (CD‐CbsA), and the FnIII domain. (b) Codon usage pattern of housekeeping (HK) genes, lipopolysaccharide (LPS) cluster genes, CD‐CbsA, and FnIII domain
TABLE S1 Analysis of CbsA protein sequence from different Xanthomonas species using SignalP v. 5.0. Columns labelled as Sec and TAT represent the probability of the presence of Sec‐ and TAT‐dependent secretory signal in the sequence, respectively. Values for Xanthomonas translucens pv. translucens are highlighted in red. Accession numbers for the sequence used are given in parentheses
TABLE S2 List of primers used in the study
TABLE S3 List of bacterial strains and plasmids used in the study
ACKNOWLEDGEMENTS
R.N. acknowledges the Council of Scientific and Industrial Research (CSIR), Government of India for PhD fellowship. This work was supported by J. C. Bose fellowships to R.V.S. and R.S. from the Science and Engineering Research Board, Government of India. R.V.S. and H.K.P. acknowledge CSIR, Government of India for Plant‐Microbe and Soil Interaction and Focused basic research project grants.
Nathawat, R. , Maku, R.V. , Patel, H.K. , Sankaranarayanan, R. & Sonti, R.V. (2022) Role of the FnIII domain associated with a cell wall‐degrading enzyme cellobiosidase of Xanthomonas oryzae pv. oryzae . Molecular Plant Pathology, 23, 1011–1021. 10.1111/mpp.13205
Contributor Information
Rajan Sankaranarayanan, Email: sankar@ccmb.res.in.
Ramesh V. Sonti, Email: sonti@ccmb.res.in, Email: rameshvsonti@gmail.com.
DATA AVAILABILITY STATEMENT
All data has been included in the manuscript.
REFERENCES
- Almagro Armenteros, J.J. , Tsirigos, K.D. , Sønderby, C.K. , Petersen, T.N. , Winther, O. , Brunak, S. et al. (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature Biotechnology, 37, 420–423. [DOI] [PubMed] [Google Scholar]
- Aparna, G. , Chatterjee, A. , Sonti, R.V. & Sankaranarayanan, R. (2009) A cell wall–degrading esterase of Xanthomonas oryzae requires a unique substrate recognition module for pathogenesis on rice. The Plant Cell, 21, 1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banner, D.W. , D’Arcy, A. , Chène, C. , Winkler, F.K. , Guha, A. , Konigsberg, W.H. et al. (1996) The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature, 380, 41–46. [DOI] [PubMed] [Google Scholar]
- Bansal, K. , Midha, S. , Kumar, S. , Kaur, A. , Sonti, R.V. & Patil, P. (2019) Ecological and evolutionary insights into pathogenic and non‐pathogenic rice associated Xanthomonas . bioRxiv. 10.1101/453373 [DOI] [Google Scholar]
- Bateman, A. & Chothia, C. (1996) Fibronectin type III domains in yeast detected by a hidden Markov model. Current Biology, 6, 1544–1547. [DOI] [PubMed] [Google Scholar]
- Bechtluft, P. , Kedrov, A. , Slotboom, D.J. , Nouwen, N. , Tans, S.J. & Driessen, A.J.M. (2010) Tight hydrophobic contacts with the SecB chaperone prevent folding of substrate proteins. Biochemistry, 49, 2380–2388. [DOI] [PubMed] [Google Scholar]
- Blum, M. , Chang, H.Y. , Chuguransky, S. , Grego, T. , Kandasaamy, S. , Mitchell, A. et al. (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Research, 49, D344–D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork, P. & Doolittle, R.F. (1992) Proposed acquisition of an animal protein domain by bacteria. Proceedings of the National Academy of Sciences of the United States of America, 89, 8990–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork, P. , Downing, A.K. , Kieffer, B. & Campbell, I.D. (1996) Structure and distribution of modules in extracellular proteins. Quarterly Reviews of Biophysics, 29, 119–167. [DOI] [PubMed] [Google Scholar]
- Brunecky, R. , Alahuhta, M. , Bomble, Y.J. , Xu, Q. , Baker, J.O. , Ding, S.Y. et al. (2012) Structure and function of the Clostridium thermocellum cellobiohydrolase A X1‐module repeat: enhancement through stabilization of the CbhA complex. Acta Crystallographica, Section D: Biological Crystallography, 68, 292–299. [DOI] [PubMed] [Google Scholar]
- Burbelo, P.D. , Kisailus, E. & Peck, W. (2002) Negative purification method for the selection of specific antibodies from polyclonal antisera. BioTechniques, 33, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Campbell, I.D. & Spitzfaden, C. (1994) Building proteins with fibronectin type III modules. Structure, 2, 333–337. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2005) Growth of the plant cell wall. Nature Reviews Molecular Cell Biology, 6, 850–861. [DOI] [PubMed] [Google Scholar]
- Cranford‐Smith, T. & Huber, D. (2018) The way is the goal: how SecA transports proteins across the cytoplasmic membrane in bacteria. FEMS Microbiology Letters, 365, fny093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossman, L. & Dow, J.M. (2004) Biofilm formation and dispersal in Xanthomonas campestris . Microbes and Infection, 6, 623–629. [DOI] [PubMed] [Google Scholar]
- Degrassi, G. , Devescovi, G. , Solis, R. , Steindler, L. & Venturi, V. (2007) Oryza sativa rice plants contain molecules that activate different quorum‐sensing N‐acyl homoserine lactone biosensors and are sensitive to the specific AiiA lactonase. FEMS Microbiology Letters, 269, 213–220. [DOI] [PubMed] [Google Scholar]
- Dow, J.M. , Crossman, L. , Findlay, K. , He, Y.Q. , Feng, J.X. & Tang, J.L. (2003) Biofilm dispersal in Xanthomonas campestris is controlled by cell‐cell signaling and is required for full virulence to plants. Proceedings of the National Academy of Sciences of the United States of America, 100, 10995–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga, S. , Bigirimana, J. , Höfte, M. & Venturi, V. (2007) A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Molecular Plant Pathology, 8, 529–538. [DOI] [PubMed] [Google Scholar]
- Gautam, S. & Sharma, A. (2002) Involvement of Caspase‐3‐like protein in rapid cell death of Xanthomonas . Molecular Microbiology, 44, 393–401. [DOI] [PubMed] [Google Scholar]
- Giglio, K.M. , Fong, J.C. , Yildiz, F.H. & Sondermann, H. (2013) Structural basis for biofilm formation via the Vibrio cholerae matrix protein RbmA. Journal of Bacteriology, 195, 3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck‐Thaler, E. , Cerutti, A. , Perez‐Quintero, A.L. , Butchacas, J. , Roman‐Reyna, V. , Madhavan, V.N. et al. (2020) Repeated gain and loss of a single gene modulates the evolution of vascular plant pathogen lifestyles. Science Advances, 6, eabc4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, R.A. (2008) The structure of protein evolution and the evolution of protein structure. Current Opinion in Structural Biology, 18, 170–177. [DOI] [PubMed] [Google Scholar]
- González, J.F. , Degrassi, G. , Devescovi, G. , De Vleesschauwer, D. , Höfte, M. , Myers, M.P. et al. (2012) A proteomic study of Xanthomonas oryzae pv. oryzae in rice xylem sap. Journal of Proteomics, 75, 5911–5919. [DOI] [PubMed] [Google Scholar]
- González, J.F. & Venturi, V. (2013) A novel widespread interkingdom signaling circuit. Trends in Plant Science, 18, 167–174. [DOI] [PubMed] [Google Scholar]
- Hagspiel, K. , Haab, D. & Kubicek, C.P. (1989) Protease activity and proteolytic modification of cellulases from a Trichoderma reesei QM 9414 selectant. Applied Microbiology and Biotechnology, 32, 61–67. [Google Scholar]
- Hardy, S.J.S. & Randall, L.L. (1991) A kinetic partitioning model of selective binding of non‐native proteins by the bacterial chaperone SecB. Science, 251, 439–443. [DOI] [PubMed] [Google Scholar]
- Henderson, B. , Nair, S. , Pallas, J. & Williams, M.A. (2011) Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin‐binding proteins. FEMS Microbiology Reviews, 35, 147–200. [DOI] [PubMed] [Google Scholar]
- Huang, C. , Rossi, P. , Saio, T. & Kalodimos, C.G. (2016) Structural basis for the antifolding activity of a molecular chaperone. Nature, 537, 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes, R.W. , Hirose, M.A. & Kuempel, P.L. (1988) Induction of nitrogen‐fixing nodules on clover requires only 32 kilobase pairs of DNA from the Rhizobium trifolii symbiosis plasmid. Journal of Bacteriology, 170, 3793–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, J.M. , Babujee, L. , Meng, F. , Milling, A. & Allen, C. (2012) The in‐planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato, mBio, 3, e00114‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques, M.A. , Arlat, M. , Boulanger, A. , Boureau, T. , Carrère, S. , Cesbron, S. et al. (2016) Using ecology, physiology, and genomics to understand host specificity in Xanthomonas . Annual Review of Phytopathology, 54, 163–187. [DOI] [PubMed] [Google Scholar]
- Jha, G. , Rajeshwari, R. & Sonti, R.V. (2007) Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Molecular Plant‐Microbe Interactions, 20, 31–40. [DOI] [PubMed] [Google Scholar]
- Kataeva, I.A. , Seidel, R.D. , Shah, A. , West, L.T. , Li, X.L. & Ljungdahl, L.G. (2002) The fibronectin type 3‐like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Applied and Environmental Microbiology, 68, 4292–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, A. , Bansal, K. , Kumar, S. , Sonti, R.V. & Patil, P.B. (2019) Complete genome dynamics of a dominant‐lineage strain of Xanthomonas oryzae pv. oryzae harbouring a novel plasmid encoding a type IV secretion system. Access Microbiology, 1, e000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch, N.T.M. , Rüdiger, S. , Schönfeld, H.J. , Driessen, A.J.M. , Schneider‐Mergener, J. & Bukau, B. (1999) Substrate specificity of the SecB chaperone. Journal of Biological Chemistry, 274, 34219–34225. [DOI] [PubMed] [Google Scholar]
- Koide, A. , Bailey, C.W. , Huang, X. & Koide, S. (1998) The fibronectin type III domain as a scaffold for novel binding proteins. Journal of Molecular Biology, 284, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Konkel, M.E. , Larson, C.L. & Flanagan, R.C. (2010) Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. Journal of Bacteriology, 192, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov, K.V. , Sandkvist, M. & Hol, W.G.J. (2012) The type II secretion system: biogenesis, molecular architecture and mechanism. Nature Reviews Microbiology, 10, 336–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek, C.P. , Starr, T.L. & Glass, N.L. (2014) Plant cell wall‐degrading enzymes and their secretion in plant‐pathogenic fungi. Annual Review of Phytopathology, 52, 427–451. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Haque, A.S. , Jha, G. , Sonti, R.V. & Sankaranarayanan, R. (2012) Crystallization and preliminary crystallographic studies of CbsA, a secretory exoglucanase from Xanthomonas oryzae pv. oryzae . Acta Crystallographica, Section F: Structural Biology and Crystallization Communications, 68, 1191–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J. & Doolittle, R.F. (1982) A simple method for displaying the hydropathic character of a protein. Journal of Molecular Biology, 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Little, E. , Bork, P. & Doolittle, R.F. (1994) Tracing the spread of fibronectin type III domains in bacterial glycohydrolases. Journal of Molecular Evolution, 39, 631–643. [DOI] [PubMed] [Google Scholar]
- Main, A.L. , Harvey, T.S. , Baron, M. , Boyd, J. & Campbell, I.D. (1992) The three‐dimensional structure of the tenth type III module of fibronectin: an insight into RGD‐mediated interactions. Cell, 71, 671–678. [DOI] [PubMed] [Google Scholar]
- Mallik, I. , Smith, M.A. & Flower, A.M. (2002) Recognition of secretory proteins in Escherichia coli requires signals in addition to the signal sequence and slow folding. BMC Microbiology, 2, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, P.B. & Sonti, R.V. (2004) Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice. BMC Microbiology, 4, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pereda, J.M. , Wiche, G. & Liddington, R.C. (1999) Crystal structure of a tandem pair of fibronectin type III domains from the cytoplasmic tail of integrin α6β4. EMBO Journal, 18, 4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau, C. , Guschinskaya, N. , Robert, X. , Gouet, P. , Ballut, L. & Shevchik, V.E. (2014) Substrate recognition by the bacterial type II secretion system: more than a simple interaction. Molecular Microbiology, 94, 126–140. [DOI] [PubMed] [Google Scholar]
- Rajeshwari, R. , Jha, G. & Sonti, R.V. (2005) Role of an in planta‐expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Molecular Plant‐Microbe Interactions, 18, 830–837. [DOI] [PubMed] [Google Scholar]
- Randall, L.L. & Hardy, S.J.S. (1995) High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends in Biochemical Sciences, 20, 65–69. [DOI] [PubMed] [Google Scholar]
- Ruoslahti, E. (1988) Fibronectin and its receptors. Annual Review of Biochemistry, 57, 375–413. [DOI] [PubMed] [Google Scholar]
- Ryan, R.P. , Vorhölter, F.J. , Potnis, N. , Jones, J.B. , Van Sluys, M.A. , Bogdanove, A.J. et al. (2011) Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nature Reviews Microbiology, 9, 344–355. [DOI] [PubMed] [Google Scholar]
- Sakr, S. , Cirinesi, A.M. , Ullers, R.S. , Schwager, F. , Georgopoulos, C. & Genevaux, P. (2010) Lon protease quality control of presecretory proteins in Escherichia coli and its dependence on the SecB and DnaJ (Hsp40) chaperones. Journal of Biological Chemistry, 285, 23506–23514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer, A. , Tauch, A. , Jsger, W. , Kalinowski, J. , Thierbachb, G. & Pühler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutumicum . Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- Sharma, A. , Askari, J.A. , Humphries, M.J. , Jones, E.Y. & Stuart, D.I. (1999) Crystal structure of a heparin‐and integrin‐binding segment of human fibronectin. EMBO Journal, 18, 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov, O. , Shani, Z. & Levy, I. (2006) Carbohydrate binding modules: biochemical properties and novel applications. Microbiology and Molecular Biology Reviews, 70, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T.J. , Karplus, K. , Li, W. et al. (2011) Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. & Pühler, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology, 1, 784–791. [Google Scholar]
- Stothard, P. (2000) The sequence manipulation suite: javascript programs for analyzing and formatting protein and DNA sequences. BioTechniques, 28, 1102–1104. [DOI] [PubMed] [Google Scholar]
- Tayi, L. , Kumar, S. , Nathawat, R. , Haque, A.S. , Maku, R.V. , Patel, H.K. et al. (2018) A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice. Molecular Plant Pathology, 19, 1364–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayi, L. , Maku, R. , Patel, H.K. & Sonti, R.V. (2016) Action of multiple cell wall‐degrading enzymes is required for elicitation of innate immune responses during Xanthomonas oryzae pv. oryzae infection in rice. Molecular Plant‐Microbe Interactions, 29, 599–608. [DOI] [PubMed] [Google Scholar]
- Thornton, J.M. , Orengo, C.A. , Todd, A.E. & Pearl, F.M.G. (1999) Protein folds, functions and evolution. Journal of Molecular Biology, 293, 333–342. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos, J. , Nguyen, L.T. , von Haeseler, A. & Minh, B.Q. (2016) W‐IQ‐TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research, 44, W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsyguelnaia, I. & Doolittle, R.F. (1998) Presence of a fibronectin type III domain in a plant protein. Journal of Molecular Evolution, 46, 612–614. [DOI] [PubMed] [Google Scholar]
- Varrot, A. , Schülein, M. & Davies, G.J. (1999) Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding. Biochemistry, 38, 8884–8891. [DOI] [PubMed] [Google Scholar]
- Watanabe, T. , Suzuki, K. , Oyanagi, W. , Ohnishi, K. & Tanaka, H. (1990) Gene cloning of chitinase A1 from Bacillus circulans WL‐12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. Journal of Biological Chemistry, 265, 15659–15665. [PubMed] [Google Scholar]
- Zhao, J. , Ren, J. , Wang, N. , Cheng, Z. , Yang, R. , Lin, G. et al. (2017) Crystal structure of the second fibronectin type III (FN3) domain from human collagen α1 type XX. Acta Crystallographica Section F Structural Biology Communications, 73, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J.Y. , Kleywegt, G.J. , Ståhlberg, J. , Driguez, H. , Nerinckx, W. , Claeyssens, M. et al. (1999) Crystallographic evidence for substrate ring distortion and protein conformational changes during catalysis in cellobiohydrolase Cel6A from Trichoderma reesei . Structure, 7, 1035–1045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Expression and purification of CbsA and CbsAΔFnIII in Escherichia coli. (a) 8% SDS‐PAGE showing purified CbsA and CbsAΔFnIII tagged with maltose‐binding protein (MBP) and 6×histidine. Proteins were purified using Ni‐NTA affinity chromatography. (b) 12% SDS‐PAGE showing CbsA and CbsAΔFnIII cleaved from MBP after TEV protease digestion. The lowest band shows TEV protease, which was used in the digestion reaction. (c) 8% SDS‐PAGE gel showing CbsA and CbsAΔFnIII purified from TEV protease digestion reaction by affinity chromatography followed by gel filtration. (d) Spontaneous cleavage of CbsA into CbsAΔFnIII in a c.3‐day‐old protein preparation
FIGURE S2 Reverse transcription quantitative PCR analysis of cbsA expression in wild‐type and cbsA Δ FnIII Xanthomonas oryzae pv. oryzae (Xoo) strains. (a) Schematic of primers used for cbsA expression analysis (b) cbsA mRNA level measured in Xoo wild type and cbsA Δ FnIII mutant shows no significant difference. 16S rRNA gene was used as endogenous control. Average ΔC t (C t cbsA − C t 16s rRNA) was plotted for both strains. Student’s two‐tailed t test was performed and the difference was not significant (p = 0.329)
FIGURE S3 Sequence analysis of Xanthomonas translucens pv. translucens CbsA using SignalP v. 5.0 indicating the probability of the presence of Sec‐ or TAT‐dependent signal sequence
FIGURE S4 (a) Average GC content (%) of the Xanthomonas oryzae pv. oryzae (Xoo) genome, the catalytic domain of CbsA (CD‐CbsA), and the FnIII domain. (b) Codon usage pattern of housekeeping (HK) genes, lipopolysaccharide (LPS) cluster genes, CD‐CbsA, and FnIII domain
TABLE S1 Analysis of CbsA protein sequence from different Xanthomonas species using SignalP v. 5.0. Columns labelled as Sec and TAT represent the probability of the presence of Sec‐ and TAT‐dependent secretory signal in the sequence, respectively. Values for Xanthomonas translucens pv. translucens are highlighted in red. Accession numbers for the sequence used are given in parentheses
TABLE S2 List of primers used in the study
TABLE S3 List of bacterial strains and plasmids used in the study
Data Availability Statement
All data has been included in the manuscript.


