Abstract
Motivation
The analysis of cancer genomes provides fundamental information about its etiology, the processes driving cell transformation or potential treatments. While researchers and clinicians are often only interested in the identification of oncogenic mutations, actionable variants or mutational signatures, the first crucial step in the analysis of any tumor genome is the identification of somatic variants in cancer cells (i.e. those that have been acquired during their evolution). For that purpose, a wide range of computational tools have been developed in recent years to detect somatic mutations in sequencing data from tumor samples. While there have been some efforts to benchmark somatic variant calling tools and strategies, the extent to which variant calling decisions impact the results of downstream analyses of tumor genomes remains unknown.
Results
Here, we quantify the impact of variant calling decisions by comparing the results obtained in three important analyses of cancer genomics data (identification of cancer driver genes, quantification of mutational signatures and detection of clinically actionable variants) when changing the somatic variant caller (MuSE, MuTect2, SomaticSniper and VarScan2) or the strategy to combine them (Consensus of two, Consensus of three and Union) across all 33 cancer types from The Cancer Genome Atlas. Our results show that variant calling decisions have a significant impact on these analyses, creating important differences that could even impact treatment decisions for some patients. Moreover, the Consensus of three calling strategy to combine the output of multiple variant calling tools, a very widely used strategy by the research community, can lead to the loss of some cancer driver genes and actionable mutations. Overall, our results highlight the limitations of widespread practices within the cancer genomics community and point to important differences in critical analyses of tumor sequencing data depending on variant calling, affecting even the identification of clinically actionable variants.
Availability and implementation
Code is available at https://github.com/carlosgarciaprieto/VariantCallingClinicalBenchmark.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The exponential growth in both, the generation and access to genomic data from tumor samples and cancer patients, is transforming all aspects of this disease, from basic research to its clinical care (Hyman et al., 2017). For example, thanks to sequencing data, we are beginning to understand the etiology of the mutational processes that affect cancer cells (Alexandrov et al., 2013). Furthermore, we are now able to track and reconstruct the phylogenetic tree of tumor evolution (Nik-Zainal et al., 2012). Similarly, the large cohorts of cancer patients that have been sequenced so far, have helped us identify germline and somatic mutations that predispose or drive carcinogenesis, respectively (Bailey et al., 2018; Huang et al., 2018), laying the foundations of personalized cancer care.
The first crucial step in analyzing cancer sequencing data is the identification of genetic variants, particularly those of somatic origin. In that sense, the research community has made great efforts to assess the performance of the many different somatic variant callers available (Alioto et al., 2015; Cai et al., 2016; Sandmann et al., 2017; Wang et al., 2013; Xiao et al., 2021; Xu, 2018). However, so far, there has been no agreement on which variant caller, nor strategy to combine them, is the most suitable. For instance, The Cancer Genome Atlas (TCGA) implemented different variant callers on multiple papers throughout its history (Abeshouse et al., 2017; Ciriello et al., 2015; Robertson et al., 2017). This eventually led to the Multi-Center Mutation Calling in Multiple Cancers (MC3) project (Ellrott et al., 2018) to address standardization and reproducibility issues at the end of TCGA. During MC3, many groups worked together to define a clear and unique strategy to combine the output of multiple variant calling tools. Other groups have explored the use of machine-learning approaches to combine the output of different variant calling tools (Anzar et al., 2019; Wood et al., 2018). However, despite all these efforts, it is still unclear which variant calling tool, or combination of tools, is optimal to analyze cancer genomics data.
The biggest challenge in determining the optimal variant calling tool or strategy is the lack of gold standard sets of somatic variants. Another likely important reason is that it is difficult to define a metric in cancer genomics. At the end of the day somatic variant calling is a means to an end, as researchers and oncologists are interested not in the variant calling itself, but rather on the results of downstream analyses. Sequencing data from tumors can be used for many different secondary analyses, from finding cancer driver genes and mutations to determining the presence of clinically actionable mutations or quantifying the effects of mutational signatures. Since none of the somatic variant callers or strategies is perfect, it is possible that the answer to all these secondary analyses differs depending on which somatic variant calling tool or strategy is used.
While there have been benchmarking studies comparing how mutation callers find somatic mutations, to the best of our knowledge there has been no systematic study of the impact on variant calling tools in secondary analyses. In this article, we studied how decisions at the somatic variant calling stage of cancer genomics data affect the results of three different secondary analyses: detection of cancer driver genes and mutations, quantification of mutational signatures and identification of clinically actionable variants.
2 Materials and methods
2.1 Variant calling datasets
To compare the effects of different mutation calling approaches in secondary analyses, we analyzed the entire set of TCGA somatic mutation files comprising 10 189 patients from 33 different cancer types and spanning more than 3 500 000 unique somatic variants. We aimed to explore the impact of different somatic variant calling strategies in downstream analyses of cohorts with different sizes, mutational signatures and mutational burdens. The Genomic Data Commons (GDC) portal (https://portal.gdc.cancer.gov) gives access to all the processed whole-exome sequencing (WXS) data for all the TCGA projects. In particular, the GDC created the DNA-Seq pipeline (https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/DNA_Seq_Variant_Calling_Pipeline/) to process all TCGA samples in a uniform way (Grossman et al., 2016). Briefly, this pipeline includes sample preprocessing, alignment to the human reference genome GRCh38.d1.vd1 followed by BAM cleaning and somatic variant calling with variant annotation and aggregation. Somatic variants were identified in WXS data by comparing allele frequencies in matched tumor-normal samples. The GDC used four different variant calling tools to identify somatic mutations: MuSE (Fan et al., 2016), MuTect2 (Cibulskis et al., 2013), SomaticSniper (Larson et al., 2012) and VarScan2 (Koboldt et al., 2012). After analyzing the WXS data for each individual sample, the GDC pipeline includes an aggregation step that combines variants from all cases of a cancer cohort into a single TCGA project mutation annotation format (MAF) file. For a detailed explanation of the GDC DNA-Seq pipeline see Supplementary Methods.
Therefore, for each of the 33 TCGA cancer types, we downloaded the four different Somatic aggregated MAF files with all the somatic mutations for each variant caller (MuSE, MuTect2, SomaticSniper and VarScan2). Additionally, we computed three extra mutation call sets per TCGA project: a Consensus of two variant callers (Consensus2) file with those variants that were called by at least two out of the four aforementioned variant callers, a Consensus of three variant callers (Consensus3) file with those variants that were called by at least three out of the four variant callers and a Union file with every somatic variant called by any variant caller.
2.2 Detecting cancer driver genes
To identify cancer driver genes, we used the IntOGen pipeline (https://bitbucket.org/intogen/intogen-plus/src/master/, March 20, 2020) (Gonzalez-Perez et al., 2013). Specifically, we analyzed every somatic variant file (MuSE, MuTect2, SomaticSniper, VarScan2, Consensus2, Consensus3 and Union) of each of the 33 TCGA projects separately. We did not run IntOGen using PanCancer approaches on all samples combined. IntOGen integrates the result of seven driver discovery methods: OncodriveFML (Mularoni et al., 2016), OncodriveCLUSTL (Arnedo-Pac et al., 2019), dNdScv (Martincorena et al., 2017), CBaSE (Weghorn and Sunyaev, 2017), HotMAPS (Tokheim et al., 2016), smRegions (Martínez-Jiménez et al., 2020a) and MutPanning (Dietlein et al., 2020). The driver discovery methods integrated in IntOGen explore different signals of positive selection, such as clustering of mutations in protein structures or mutational functional bias, to pinpoint which driver genes deviate from the estimated neutral mutation rate using the set of input somatic mutations. The results of these tools are then combined by accounting each method credibility—the relative credibility for each method is based on the ability of the method to give precedence to well-known genes already collected in the Cancer Gene Census (Sondka et al., 2018) catalogue of driver genes—to produce a consensus ranking of genes using a TIER based classification. Finally, IntOGen also provides a weighted combined P-value for each ranked gene. For the purpose of our analysis, we only considered true driver genes those within TIER 1 and TIER 2 (q-value <0.05). We, therefore, discarded genes classified in TIER 3 and TIER 4.
2.3 Benchmarking variant calling strategies with driver genes
We considered the curated set of known driver genes from IntOGen (https://www.intogen.org/download, release date February 1, 2020) as our reference set to benchmark how the different mutation call sets can be used to detect cancer driver genes. This set encompasses both, newly detected and previously annotated cancer driver genes in the Cancer Gene Census (https://cancer.sanger.ac.uk/census) of Catalogue Of Somatic Mutations In Cancer (COSMIC) (Forbes et al., 2017). To further assess and compare our results, we also benchmarked against a second reference set of cancer driver genes published by the PanCancerAtlas-MC3-project (Bailey et al., 2018). We restricted our benchmarking analysis to only those genes annotated as known cancer driver genes in the 33 cancer types we analyzed (MC3 cancer driver genes uniquely identified using PanCancer approaches on all samples combined were not considered). Furthermore, in the case of IntOGen reference set, we only considered those driver genes identified within TCGA cohorts (i.e. driver genes uniquely identified by IntOGen in non-TCGA cohorts, such as ICGC or PCAWG cohorts were filtered out).
We used multiple metrics (Table 1) to assess the performance of the different variant calling strategies when detecting driver genes with IntOGen in downstream analyses. We defined our true positives (TP), false positives (FP) and false negatives (FN) as follows:
Table 1.
Benchmarking metrics
| Metric | Definition |
|---|---|
| Precision=TP/(TP+FP) | Also known as positive predictive value. It is the ratio of correctly detected driver genes among all driver genes detected by IntOGen with a given somatic variant call set. |
| Recall=TP/(TP+FN) | Also known as sensitivity. It is the ratio of correctly detected driver genes by IntOGen among all driver genes within the reference set. |
| F1-score =(2×Precision×Recall)/(Precision+Recall) | Harmonic average of precision and recall. The best value is 1 and the worst is 0. |
TP: those driver genes detected by IntOGen with a given variant call set that are within the reference set.
FP: those driver genes detected by IntOGen with a given variant call set that are outside the reference set.
FN: those driver genes within the reference set not identified by IntOGen with a given variant call set.
2.4 Mutational signature analysis
We used deconstructSigs (Rosenthal et al., 2016) 1.8.0 R package to quantify the presence of different mutational signatures in the different mutation call sets. In brief, deconstructSigs accounts for the trinucleotide context of each mutation to classify the six different base substitutions (C > A, C > G, C > T, T > A, T > C and T > G) into 96 possible mutation types (Alexandrov et al., 2013). The signature matrix with the number of times a mutation was found within each trinucleotide context was compared against COSMIC Single Base Substitution (SBS) signatures (available at https://cancer.sanger.ac.uk/signatures/sbs) (Alexandrov et al., 2020).
Finally, deconstructSigs uses an iterative approach to assign different weights to each signature and estimate their contribution to the mutational profile of the tumor sample. We filtered out those samples with <50 mutations. Since we analyzed WXS samples, the signature matrix was normalized to reflect the absolute frequency of each trinucleotide context as it would have taken place in the whole genome. This way we adjusted for differences in trinucleotide abundances between exome and whole genome in order to compare our signatures to the ones extracted from whole genomes (available in synapse.org, ID syn12009743).
2.5 Clinically actionable variants analysis
We used the Molecular Oncology Almanac (Reardon et al., 2021) (https://github.com/vanallenlab/moalmanac, November 4, 2021) (MOAlmanac) to detect alterations that might be therapeutically actionable. Briefly, MOAlmanac is a clinical interpretation algorithm paired with an underlying knowledge base for precision oncology to enable integrative interpretation of multimodal genomic data for point-of-care decision making and translational-hypothesis generation. The primary objective of MOAlmanac is to identify and associate molecular alterations with therapeutic sensitivity and resistance as well as disease prognosis. This is done for ‘first-order’ genomic alterations (individual events, such as somatic variants) as well as ‘second-order’ events [those that may be descriptive of global processes in the tumor, such as tumor mutational burden or microsatellite instability (MSI)]. In addition to clinical insights, MOAlmanac annotates and evaluates first-order events based on their presence in numerous other well established datasources as well as highlights connections between them. Overall, MOAlmanac is an open-source computational method for integrative clinical interpretation of individualized molecular profiles.
Since this method is currently geared toward hg19/b37 reference files, we needed to liftover genome coordinates between assemblies for all the Somatic MAFs using CrossMap (Zhao et al., 2014) version 0.3.4 (99.99% of variants were successfully remapped).
2.6 Purity and ploidy dataset
We used purity and ploidy ABSOLUTE annotations (Hoadley et al., 2018) for all TCGA samples available at https://gdc.cancer.gov/about-data/publications/pancanatlas. These annotations were used to adjust the variant allele frequencies (VAFs) by cancer DNA fraction and ploidy to use them in all the analyses.
Almost 97% of TCGA mutation call set cases (9871/10 189 samples) present purity and ploidy information. However, 85% of cases (8673/10 189 samples) match both mutation and purity/ploidy information at the TCGA analyte level (meaning both sources of information come from the same TCGA analyte). Thus, to ensure that the adjusted VAF information presented in our study was sufficiently accurate, we decided to report the adjusted VAF information for this 85% cases. However, when adjusting VAF information at the TCGA analyte level, 1% of variants ended up with adjusted VAFs >1. Therefore, we only used the unadjusted VAFs in our analyses for this 1% of variants and for the variants of the 15% aforementioned cases.
2.7 Clinical metadata
We retrieved tumor stage information from the TCGA-Clinical Data Resource (Liu et al., 2018) file available at https://gdc.cancer.gov/about-data/publications/pancanatlas.
3 Results
3.1 Effects of variant calling in the detection of cancer driver genes
One of the most widespread uses of somatic mutation data from cohorts of cancer patients is the identification of cancer driver genes (Bailey et al., 2018; Martínez-Jiménez et al., 2020b). The tools to detect these genes are sensitive to which somatic mutations are included in the final analysis, as they can bias some aspects of the randomization in which most cancer driver detection tools rely (Arnedo-Pac et al., 2019; Dietlein et al., 2020; Martincorena et al., 2017; Martínez-Jiménez et al., 2020a; Mularoni et al., 2016; Tokheim et al., 2016; Weghorn and Sunyaev, 2017).
To assess to what extent variant calling affects the detection of cancer driver genes, we used IntOGen (Gonzalez-Perez et al., 2013) to find driver genes in 231 different mutation call sets for the 33 different cancer types from TCGA. The seven mutation call sets of each cancer type are distributed as follows: one mutation set with all the calls from one of the four variant calling tools [MuSe (Fan et al., 2016), MuTect2 (Cibulskis et al., 2013), SomaticSniper (Larson et al., 2012) and VarScan2 (Koboldt et al., 2012)], another mutation set—Consensus2—with all those mutations found by, at least, two of the four variant callers, another consensus mutation set—Consensus3—with all those mutations found by, at least, three of the four variant callers and a final mutation set with all the mutations found by any mutation caller—Union (Fig. 1).
Fig. 1.
Intersection of mutation calls across all variant calling strategies for the 33 TCGA cancer types. This UpSetR plot shows the number of variants uniquely identified by one variant calling tool (single point) and variants called by different tools (linked points). Bar-plot indicates intersection size and colors indicate the number of variants present in the PanCancerAtlas MC3 project. Violin plots represent VAF distribution adjusted by cancer DNA fraction and ploidy; colors indicate total coverage (read depth) across loci. Bottom left plot indicates variant call set size
One of the main concerns while determining the optimal variant calling tool or strategy is the difficulty to classify mutation calls as TP due to the lack of gold standard sets of somatic variants. The best way to tackle this issue is by experimentally validating the mutation calls with an orthogonal technology. However, in the case of the TCGA somatic call set only 3% of unique somatic variants (110263/3592923) have been validated according to the information in ‘GDC_Validation_Status’ from the TCGA Somatic MAFs. Therefore, we considered including ‘MC3_Overlap’ information indicating whether a particular somatic variant overlaps with an MC3 variant for the same sample pair as proxy for bona fide calls. The 87% of unique somatic variants (3127800/3592923) in the TCGA call set are included in the MC3 project (Ellrott et al., 2018). Furthermore, we included VAF information adjusted by cancer DNA fraction and ploidy to better assess variant calling results. Variant callers tend to perform better when detecting clonal mutations (VAF =0.5) whereas they struggle to call subclonal ones (VAF <0.5).
The variant calling results (Fig. 1) show that the somatic mutation call sets from SomaticSniper and MuTect2 were, respectively, the smallest and largest from the individual variant callers. More importantly, 53% of somatic variants were shared among all variant calling strategies spanning a median VAF range around 0.5. Interestingly, MuTect2 uniquely identified 11.7% of all somatic variants, most of them with a very low VAF range. Thus, many of these variants are not included in the MC3 project call set. However, the very high coverage (read depth) across these loci prevents us from discarding these calls as TP and suggests that MuTect2 has high sensitivity to identify subclonal somatic variants.
We wondered whether the different capabilities of the variant calling strategy tools to detect mutations according to their VAF ranges may be clinically related to tumor stage as more advanced tumors tend to be more heterogeneous. However, we were not able to find any correlation in this regard in part due to the high rate of samples with missing American Joint Committee on Cancer (AJCC) stage information.
Having assessed the influence of various tumor properties in the number of mutations called by each tool and combination strategy, we next quantified the effect that they have when detecting cancer driver genes. To that end, we used IntOGen to detect cancer driver genes in the 231 somatic mutation call sets (Fig. 2A and B and Supplementary File 1).
Fig. 2.
Performance of different variant calling strategies when detecting cancer driver genes with IntOGen. (A) Correlation between cancer driver genes and median number of mutations per megabase. (B) Correlation between cancer driver genes and cohort sample size. The number of cancer driver genes detected by IntOGen with different call sets in each cancer type positively correlates with median number of mutations per megabase (A) and sample size (B). Shaded area indicates 95% bootstrapped confidence interval. (C) Boxplots represent the different performance metrics scores of the variant calling strategies when detecting cancer driver genes with IntOGen for the 33 TCGA cancer types. Boxplots are sorted by mean metric score. Metric scores when benchmarking against IntOGen (left panel) and PanCancerAtlas MC3 project (right panel) reference sets of known cancer driver genes are shown. (D) Alluvial plot indicating best performing variant calling strategy according to F1-score for each cancer type when benchmarking against IntOGen (left panel) and PanCancerAtlas MC3 project (right panel) reference sets of known cancer driver genes. Y-axis indicates number of cancers in each group
Overall, we found that there are wide differences in the number of detected cancer driver genes in each cohort depending on which somatic variant calling tool or strategy we used. For example, in the case of prostate cancer [prostate adenocarcinoma (PRAD)], the set of mutations from MuTect2 leads to the detection of 33 cancer driver genes, whereas the set from VarScan2 leads to 62 driver genes. Similarly, in the case of bladder cancer [bladder urothelial carcinoma (BLCA)], the Union leads to the detection of 54 cancer driver genes, whereas the set of mutations from MuSE leads to 86 driver genes. Interestingly, the number of cancer driver genes detected in each mutation call set has a positive correlation with the median number of mutations per megabase (spearman rho = 0.56, P-value <2.2e-16), as already described in the final driver analysis of TCGA (Bailey et al., 2018). Additionally, the number of cancer driver genes detected in each mutation call set positively correlates with the number of samples in each cohort (spearman rho = 0.36, P-value <2.1e-08).
To further assess the possible effects that different sample sizes may have on the performance of specific variant call sets upon detection of cancer driver genes, we conducted a downsampling experiment using the largest TCGA cohort available, the breast invasive carcinoma (BRCA) cohort with 986 samples (Supplementary Fig. S1). To this end, we created three new BRCA cohorts with different sample sizes by subsetting the 25%, 50% and 75% of all BRCA samples, respectively. Furthermore, to select the samples comprising each one of these three newly created BRCA cohorts, we conducted three iterations by selecting different samples for each cohort, obtaining a total of nine different cohorts (three with 25% samples, three with 50% samples and three with 75% samples) to better assess the robustness of the results. While conducting the three different iterations to select the samples, we adjusted for AJCC tumor stage to avoid any confounding effect this variable may have on the results. This analysis confirmed that the number of cancer driver genes detected positively correlates with the number of samples in each cohort (spearman rho = 0.38, P-value = 0.0012). Surprisingly, the Consensus3 proved to be the less robust of all strategies with very important differences in the number of cancer driver genes detected within each cohort. For example, in the 50% BRCA cohort (n = 496), 62 cancer driver genes were detected with the Consensus3 second iteration call set, whereas only 29 cancer driver genes were detected with the Consensus3 first iteration call set.
Next, we benchmarked our results against a reference set of known cancer driver genes from IntOGen. We also considered the set of cancer driver genes published by the PanCancerAtlas-MC3-project (Bailey et al., 2018) as a second reference set to further assess our results. In both cases, we restricted our reference sets to only those genes annotated as cancer driver genes in the 33 tumor types we analyzed. For the IntOGen reference set, we only considered those cancer driver genes identified within TCGA cohorts. For the MC3 reference set we removed those cancer driver genes uniquely identified by PanCancer approaches on all samples combined.
The benchmarking results of the 33 cancer types showed, to our surprise, that the Union variant calling strategy is the best one when detecting cancer driver genes with IntOGen and benchmarking against IntOGen reference set (Fig. 2C left panel and Supplementary File 2). Also, when benchmarked against MC3 reference set (Fig. 2C right panel and Supplementary File 2), the Union call set remains the top performer according to recall score, being outperformed by MuSE, VarScan2 and SomaticSniper when looking at F1-score and precision results. Interestingly, Consensus3 proved to be amongst the lower performance strategies across all metrics when compared to both reference sets. Consensus2 showed to be pretty robust, being the second-best method when comparing against IntOGen reference set. However, it was outperformed by the Union in all cases. Regarding the four single variant caller performances, it is quite difficult to decide which one is the best one, as their performance depends on the metric and reference set used.
To further assess our results, and considering that Consensus2 performance seemed to be pretty robust, we benchmarked all possible two-caller intersections in a subset of five cancer types: adrenocortical carcinoma (ACC), BLCA, BRCA, PRAD and uterine corpus endometrial carcinoma (UCEC) (Supplementary Fig. S2 and Supplementary Files 3 and 4). According to F1-score metric, Consensus2 outperformed all other possible two-caller intersections when compared against both reference sets. Likewise, SomaticSniper and VarScan2 intersection proved to be the second-best two-caller intersection method.
We next wondered whether certain variant calling strategies are more suitable for specific cancer types. From a clinical point of view, knowing beforehand which variant caller is the best one for a particular cancer or group of cancers would be very helpful and could help inform patient treatment improving the clinical outcome. To this end, we classified all the 33 TCGA cancer types into different groups (Hoadley et al., 2018): hematologic and lymphatic cancers include acute myeloid leukemia (LAML), lymphoid neoplasm diffuse large B cell lymphoma (DLBC) and thymoma (THYM); urologic cancers contain BLCA, PRAD, testicular germ cell tumors, kidney renal cell carcinoma, kidney chromophobe and kidney renal papillary cell carcinoma; gynecologic tumors comprise ovarian (OV), UCEC, cervical squamous cell carcinoma and endocervical adenocarcinoma and BRCA; endocrine cancers include thyroid carcinoma and ACC; central nervous system malignancies contain glioblastoma multiforme and brain lower-grade glioma; gastrointestinal tumors include esophageal carcinoma (ESCA), stomach adenocarcinoma (STAD), colon adenocarcinoma (COAD), rectum adenocarcinoma (READ), liver hepatocellular carcinoma, cholangiocarcinoma and pancreatic adenocarcinoma; thoracic tumors contain lung adenocarcinoma, lung squamous cell carcinoma (LUSC) and mesothelioma; soft tissue cancers include sarcoma and uterine carcinosarcoma; finally the remaining cancer types were classified as ‘other’ including head and neck squamous cell carcinoma, pheochromocytoma and paraganglioma, skin cutaneous melanoma (SKCM) and uveal melanoma.
When analyzing the best variant calling strategy for each cancer type (Fig. 2D and Supplementary Fig. S3 and Supplementary File 2) we observed that the Union is still the best variant calling strategy for the majority of cancer types, especially for gastrointestinal tumors according to F1-score. Interestingly, MuTect2 showed very good results being the best variant caller in a variety of cancer types and being the best strategy alongside the Union when considering precision as the metric of interest. Surprisingly, SomaticSniper proved to be the best variant caller for hematologic and lymphatic malignancies, specifically for DLBC and THYM cancer types, but not for LAML malignancies where it was outperformed by other strategies. Consensus2 was the best strategy in the majority of cancer types when considering recall as the metric of interest.
Focusing on the total number of cancer driver genes detected by IntOGen with the different variant calling strategies across the different groups of cancer types (Supplementary Fig. S4 and Supplementary Files 1 and 2), we observed that in most of the cancers (gastrointestinal, gynecologic, urologic and ‘other’ cancer types) the majority of cancer driver genes detected were shared among all the variant calling strategies. Nevertheless, we found some exceptions in specific cancer types, such in the case of thoracic and hematologic and lymphatic malignancies where SomaticSniper uniquely identified 36 and 28 cancer driver genes respectively, in the latter case most of them from LAML malignancies. Furthermore, Consensus3 was the call set with the largest number of cancer driver genes identified by IntOGen in central nervous system and gastrointestinal cancers, including 41 and 43 uniquely identified cancer driver genes respectively. Overall, our results show important differences in the number and identity of the cancer driver genes detected in a cohort of patients depending on which tool is used to identify somatic variants.
3.2 Somatic mutations in cancer driver genes
Even if one can identify a gene as a driver in a cohort using a variant call set, it is possible that the variant caller misses some individual mutations of that gene in some samples. This could have important implications for patients, as the presence or absence of mutations in cancer driver genes can determine whether patients will receive certain treatments or not (Hyman et al., 2017). To evaluate the impact of variant calling when finding mutations in cancer driver genes, we calculated the number of patients harboring missense and/or nonsense mutations in cancer driver genes depending on the mutation set used (Fig. 3 and Supplementary Fig. S5).
Fig. 3.
Detection of somatic mutations in cancer driver genes. This UpSetR plot shows the number of somatic missense and nonsense variants in cancer driver genes uniquely identified by one tool (single point) and by different tools (linked points). Bar-plot indicates intersection size and colors indicate the cancer driver gene role. Violin plots represent VAF distribution adjusted by cancer DNA fraction and ploidy; colors indicate total coverage (read depth) across loci. Top bar-plot indicates the ratio of missense and nonsense mutations. Bottom left plot indicates variant call set size
As expected, there is great variability in the detection of somatic mutations in cancer driver genes depending on the variant calling strategy used. Overall, there is a correlation between the total number of mutations called by each method and the number of mutations identified in cancer driver genes (Fig. 3). MuTect2, VarScan2 and, specially, Consensus2 detected more mutations in cancer driver genes than Consensus3 and SomaticSniper. Interestingly, we found that 61% of all missense and nonsense mutations in cancer driver genes were called by all variant callers. Furthermore, 56.5% of all missense and nonsense mutations were found in tumor suppressor genes with 30% of them being nonsense mutations. On the other hand, 37.5% of all missense and nonsense mutations in cancer driver genes were found in oncogenes with 96.5% of them being missense mutations. The remaining 6% of all the mutations affected genes with unknown roles. Importantly, none of the somatic variant call sets (except the Union) had all the mutations in all cancer driver genes, suggesting that we need to use multiple variant callers to ensure that we are detecting all missense and nonsense mutations in cancer driver genes.
We also found important differences when looking at the number of patients bearing at least one missense and/or nonsense mutation in specific cancer driver genes. Specifically, we quantified the number of missed mutations by each variant caller tool or strategy in the four most mutated cancer driver genes (TP53, KRAS, PTEN and PIK3CA) across the 33 cancer types (Supplementary Fig. S5). For example, depending on the variant caller used, up to 22% of UCEC patients (196 patients) differ their PTEN mutational status depending on the variant call set. Similarly, 6% of UCEC patients (32 patients) vary their PIK3CA mutational status when comparing Consensus3 and Union call sets. Importantly, up to 27% of PADD patients (49 patients) carrying a mutation in KRAS could be missing depending on the variant calling strategy used. Finally, regarding samples harboring TP53 mutations, up to 19% of ESCA patients (35 patients), 26% of LUSC patients (128 patients), 35% of OV patients (153 patients) and 20% of READ patients (27 patients) could be missing depending on the variant call set used.
3.3 Mutational signatures
The analysis of mutational signatures is important to understand the biological mechanisms underlying somatic mutations, such as defective DNA repair, mutagenic exposures, DNA replication infidelity or enzymatic DNA modifications. These mutational processes have implications in the understanding of cancer etiology and may inform patient treatment.
We analyzed the mutational signatures of five cancer types—ACC, BLCA, BRCA, PRAD and UCEC—so that they spanned a variety of mutational processes, ranges of purity, mutation rates and cohort sizes within TCGA. For example, ACC is one of the smallest cohorts within TCGA (n = 92), as well amongst those with the highest tumor purity (average purity 80%) (Aran et al., 2015). On the other hand, BRCA is the largest cohort in TCGA (n = 986). Another factor that can alter the efficiency of tools to detect cancer driver genes is the mutation rate of the cohort, hence why we included UCEC, which is amongst the cancer types with highest mutation rates (Bailey et al., 2018). Finally, BLCA and PRAD are amongst the cohorts that are closest to the TCGA average in all these aspects, making them good representatives of the average tumor sample.
We focused the mutational signatures analysis on those signatures that have been proved to contribute mutations to the corresponding cancer types (Alexandrov et al., 2020) (Fig. 4 and Supplementary File 5). We detected all the expected mutational signatures in all cancer types regardless of the variant calling tool or strategy used. As expected, the mutational signatures contributing the most mutations to individual tumor genomes were SBS1, SBS2, SBS5, SBS13 and SBS40.
Fig. 4.
The percentage of mutations contributed by each mutational signature to individual tumor genomes. The size of each dot represents the proportion of samples of each tumor type that shows the mutational signature. The color of each dot represents the median signature contribution per individual tumor genome in samples that show the signature. Tumors that had few mutations (<50) or that were poorly reconstructed by the signature assignment were excluded. ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; PRAD, prostate adenocarcinoma; UCEC, uterine corpus endometrial carcinoma
We observed SBS5 and SBS40 as flat signatures contributing to multiple types of cancer, although their proposed etiology remains unknown. Furthermore, SBS5, SBS40 and SBS1 mutations have been proved to correlate with age. Specifically, SBS1 may reflect the number of cell divisions a cell has undergone. On the other hand, cancers with high APOBEC activity, specially BLCA and to a lesser extent BRCA, show an increase in the mutational burden of SBS2 and SBS13, both of them related to the APOBEC family of cytidine deaminases activity.
We found no differences in the quantification of mutational signatures regardless of the variant call set used in any of the five cancer cohorts analyzed. We would like to emphasize that one of the main sources of FP callings are germline mutations in CpG sites that are miscalled as somatic. Hence, the lack of significant differences in SBS1 (characterized by C > T mutations at NCG trinucleotides; N being any base) results is relevant. Overall, it seems that mutational signatures are pretty robust to variant calling decisions.
3.4 Differences in clinically actionable mutations depending on the variant calling strategy
Another important goal of the analysis of somatic cancer genomes is the identification of clinically actionable variants (CAVs). These are somatic variants that help oncologists and physicians decide whether they should give a treatment to a cancer patient, as they are associated with sensitivity, resistance or disease prognosis. Therefore, properly assessing the presence of such variants in the genome of cancer cells is of ultimate clinical importance. To that end, we used the Molecular Oncology Almanac (Reardon et al., 2021) (https://github.com/vanallenlab/moalmanac, November 4, 2021) (MOAlmanac) to identify and associate somatic variants with therapeutic sensitivity and resistance as well as disease prognosis.
We found 36 874 CAVs (Supplementary Fig. S6 and Supplementary File 6) described as biomarkers for a selected tumor type, meaning that the disease for which the association has been reported coincides with the cancer type of the tumor under analysis. These somatic variants are classified according to different levels of clinical actionability or biological relevance depending on how closely they match an alteration–action relationship, as given by catalogued assertions. In total, 6% (2182/36 874) are putatively actionable variants (i.e. exact match between gene, variant classification and protein change with a catalogued variant), 71% (26 214/36 874) are classified as investigate actionability variants (i.e. gene and feature type—somatic variant—match but not either the variant classification or specific protein alteration) and 23% (8478/36 874) are classified as biologically relevant (i.e. gene match only).
Only a little over half of all CAVs were detected by all variant calling strategies (21 198 out of 36 874, 58%). Amongst variant callers, MuTect2 and VarScan2 identified 11% (4084/36 874) of CAVs that were missed by SomaticSniper and MuSE. Moreover, Mutect2 identified an additional 3536 CAVs (10% of all of CAVs). Importantly, all variant callers had some unique CAVs, highlighting the importance of using more than one variant caller when analyzing WXS data to ensure that no CAVs are missed.
MOAlmanac further classifies putatively actionable and investigate actionability somatic variants according to a predictive implication that describes the strength of clinical evidence for a given relationship between a somatic variant and a clinical action. Thus, these variants were matched independently on catalogued events associated with therapeutic sensitivity, therapeutic resistance and disease prognosis with different evidence levels: Food and Drug Association (FDA)-approved (the FDA recognizes an association between the alteration and recommend clinical action); Guideline (this relationship is catalogued as a guideline for standard of care treatment); Clinical trial (the alteration is or has been used as an eligibility criterion for a clinical trial); Clinical evidence (the relationship is reported in a clinical study that did not directly involve a clinical trial); Preclinical evidence (this relationship is reported in a study involving mice, cell line or patient derived models); Inferential evidence (the relationship is inferred as a result of mathematical modeling or an association between molecular features).
In total, 5354 variants were associated with therapeutic sensitivity and 514 were associated with therapeutic resistance (Fig. 5 and Supplementary Files 7 and 8). Importantly, 55% (2935/5354) of variants associated with therapeutic sensitivity and 59% (304/514) of variants associated with therapeutic resistance were detected by all variant calling strategies, respectively.
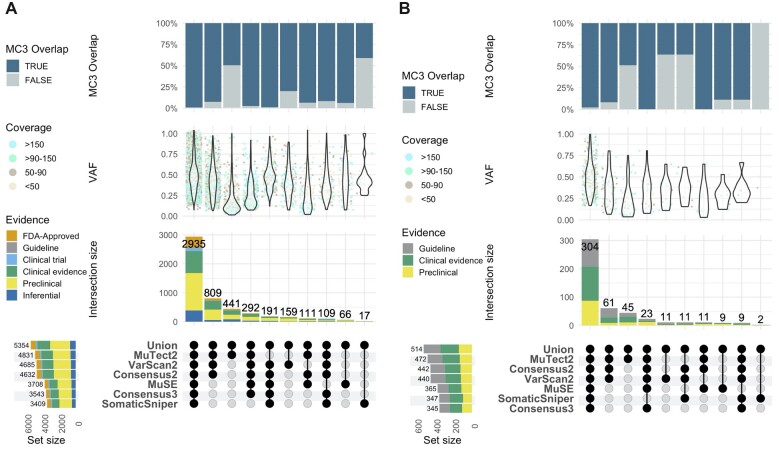
Fig. 5.
Clinically actionable somatic mutations associated to therapeutic sensitivity and resistance. This UpSetR plot shows the number of clinically actionable somatic mutations associated to therapeutic sensitivity (A) and therapeutic resistance (B) detected by the Molecular Oncology Almanac with the different variant calling strategies in the complete set of TCGA projects. Single points indicate those variants uniquely identified by one variant call set. Linked points indicate those variants identified by multiple variant call sets. These clinically actionable somatic variants are classified according to different evidence levels. Bar-plot indicates intersection size and colors indicate the association evidence level. Violin plots represent VAF distribution adjusted by cancer DNA fraction and ploidy; colors indicate total coverage (read depth) across loci. Top bar-plot indicates the ratio of variants presents in the PanCancerAtlas MC3 project. Bottom left plot indicates variant call set size. Only those clinically actionable somatic variants in which the disease for which the association has been reported coincides with the cancer type of the tumor under analysis are shown
Interestingly, 11.3% (607/5354) of the variants associated with therapeutic sensitivity were found to have FDA-Approved evidence level associations and 27.7% (1483/5354) have a Clinical evidence level. Most of the variants, 44.8% (2396/5354), have a Preclinical evidence level and finally 12.6% (675/5354) have an Inferential evidence level. More importantly, 15.1% (809/5354) were uniquely detected by MuTect2 and VarScan2 (and Consensus2), comprising 12.7% (77/607) of all the variants with FDA-Approved evidence level association. Likewise, MuTect2 uniquely identified 6.3% (38/607) of all FDA-Approved evidence level variants. Finally, very important differences in the detection of clinically actionable variants associated with therapeutic sensitivity were found across variant call sets, with MuTect2, VarScan2 and Consensus2 detecting 21.7% (1163/5354) more variants on average than MuSE, Consensus3 and SomaticSniper.
Furthermore, 869 variants were found to have an association with disease prognosis (Supplementary Fig. S7 and Supplementary File 9) and 71% (617/869) were detected by all variant callers. About 52.2% (454/869) were associated with a favorable prognosis and 47.8% (415/869) with an unfavorable one.
Finally, we looked for clinically actionable variants associated with MSI. This phenotype, MSI, is a hypermutation pattern that occurs at genomic microsatellites caused by impaired DNA mismatch repair. Mismatch repair deficiency that leads to MSI has been described more frequently in colorectal (COAD and READ), endometrial (UCEC) and gastric (STAD) adenocarcinomas (Bonneville et al., 2017; Cortes-Ciriano et al., 2017). Furthermore, it is known that colorectal patients with DNA mismatch repair deficiency have been shown to be more susceptible to immunotherapies, such as programmed cell death (PD-1) immune blockade. Thus, accurate identification of variants associated with MSI is of therapeutic importance.
We found a total of 1276 variants associated with MSI (Supplementary Fig. S8A and Supplementary File 10). In this case, the effect of variant calling strategy is even more significant than for the rest of CAVs, as only 19.5% of all variants (249/1276) where detected by all variant calling strategies. To further assess these important findings, we compared the performance of the different variant calling strategies to identify patients harboring at least one variant associated to MSI. To this end, we selected the four cancer types where MSI has been described more frequently (UCEC, COAD, STAD and READ) and created a reference set of MSI-High (MSI-H) samples described in the literature (Bonneville et al., 2017; Cortes-Ciriano et al., 2017). As expected from previous results, MuTect2, VarScan2 and Consensus2 uniquely identified 69.7% (191/274) of patients with MSI associated variants that were indeed classified as MSI-H samples in the literature (Bonneville et al., 2017; Cortes-Ciriano et al., 2017) (Supplementary Fig. S8B). Only 20% (55/274) of MSI-H patients were detected to bear at least one MSI associated variant with all variant calling approaches. Finally, it is worth mentioning the 49 patients detected by all variant callers that were not classified as MSI-H samples. This is likely due to the fact that we only consider those samples bearing at least one MSI associated variant, which is different from the MSI-H status. For the purpose of the analysis, we considered that MSI-H samples were expected to bear at least one MSI associated variant but not the other way around.
4 Discussion
The analysis of sequencing data from cancer genomes is critical, among others, to understand cancer etiology, identify the events driving the transformation of healthy cells into cancerous ones or guide the treatment of cancer patients (Alexandrov et al., 2013; Bailey et al., 2018; Huang et al., 2018; Hyman et al., 2017; Nik-Zainal et al., 2012). Each of these analyses relies on the proper identification of true somatic variants in the cancer genome, which can be done with many different computational tools. However, we currently do not understand how variant calling approaches impact the final results of cancer sequencing data.
Here, we have quantified the impact of changing variant calling tools or strategies in three different secondary analyses across 33 different cancer types. We have shown that variant calling decisions have no impact on mutational signatures results but, importantly, may lead to significant differences in the identification of cancer driver genes and clinically actionable variants.
While we found no magic recipe, the single recommendation that we believe can be applied in all circumstances is to use, at least, more than one variant calling tool and test the results of any secondary analysis in the different variant call sets. This would give researchers a sense of how much their results might vary depending on the variant calling and whether additional efforts into running other variant calling tools are necessary or not. A useful rule of thumb is to run as many variant callers as possible using the mutations from the Union of all variant calling tools. Taking the mutations from the Consensus of two or more variant callers is the second-best alternative when running multiple variant callers. In the case of running only one variant caller, MuTect2 would be the preferred option in general, albeit we also hope that the detailed results that we provide for the different cancer types in Figure 2D help researchers in deciding which variant caller to use.
Regarding cancer driver genes, while the performance of each variant calling tool or strategy can vary depending on the cancer type, the overall results suggest that one will get the best results using the mutations from the Union of all variant calling tools. The result of the Union variant call set was a surprise, because we initially expected that the likely high number of FP somatic mutations in the Union call set would lower the predictive power of the cancer driver detection tools in IntOGen, but this was not the case. We believe that this likely reflects the robustness of IntOGen to the presence of FP in the somatic mutation set. Another unexpected finding was that one of the most common approaches to combine somatic variant call sets, Consensus3 (Bailey et al., 2018), had some of the worse overall results when detecting cancer driver genes. On the other hand, Consensus2 showed very robust results overall, being the second-best strategy when considering recall as the metric of interest. Thus, very restrictive methods, such as Consensus3, seemed to badly penalized IntOGen cancer driver genes detection tools. Nevertheless, considering the specific cancer type is important, such is the case of hematologic and lymphatic malignancies like DLBC and THYM, where SomaticSniper proved to be the best caller.
Importantly, we have also found differences in the detection of somatic missense and nonsense mutations in cancer driver genes. In some cases, a specific cancer driver gene mutation status (i.e. PTEN in UCEC) could differ in more than 20% of patients depending on the variant call set used. This result suggests that it is important to use, at least, more than one variant calling tool to analyze cancer genomes. Otherwise, a significant number of mutations in cancer driver genes can be missed. Specially considering that Consensus2 was the strategy that detected more missense and nonsense mutations in cancer driver genes.
Mutational signatures analysis is pretty robust to variant calling decisions. We found no differences in the quantification of mutational signatures across the five cancer types analyzed.
However, if the goal of the analysis of the somatic genome is to find clinically actionable mutations, we need to be aware that there are considerable differences depending on the somatic mutation calling used. Only half (57.5%) of all clinically actionable variants were detected by all variant calling strategies. On average, MuTect2, VarScan2 and Consensus2 detect 20% more clinically actionable variants than MuSE, Consensus3 and SomaticSniper. This trend remains when looking at variants associated to therapeutic sensitivity. Importantly, we found greater differences when detecting of MSI associated variants, with MuTect2, VarScan2 and Consensus2 uniquely identifying 70% of MSI-H samples. Accurately identifying these variants is of therapeutic importance considering their relevance for immunotherapy treatments.
Finally, one of the main sources of variation between variant calling strategies is the identification of subclonal mutations. Here, we included VAF information adjusted by cancer DNA fraction and ploidy, observing that MuTect2 has high sensitivity to identify subclonal somatic variants. However, intra-tumor heterogeneity would be another important factor to consider (Dentro et al., 2021) since the analysis of heterogeneous cancers (i.e. PRAD) would yield more variable results compared to those of homogeneous cancers (i.e. SKCM) (Supplementary Fig. S4).
We acknowledge several limitations in our study. For example, we are not considering results for copy number and structural variants in the mutation call sets. We also have not explored the impact of other important variables, such as sequencing coverage. It is possible that with deeper coverages, such as those provided by targeted sequencing of gene panels, the differences we observed here for the variant callers are smaller.
Overall, we hope this study will help researchers understand how variant calling decisions might impact their results. It is important to account for the clinical implications that variant calling decisions have on different downstream analyses, especially in such important aspects of cancer genomics like driver genes and the identification of actionable variants. Moreover, we hope that this study will help guide variant calling design while considering the needs and goals of the different research projects.
Supplementary Material
Acknowledgements
We would like to thank the patients that donated the samples for The Cancer Genome Atlas, without them this work would not be possible. We would also like to thank Abel González-Pérez, Collin Tokheim, Brendan Reardon and Eliezer M. Van Allen for their valuable discussions and insights.
Funding
This work was supported by the BSC-Lenovo Master Collaboration Agreement (2015) and the IBM-BSC Joint Study Agreement (JSA) on Precision Medicine under the IBM-BSC Deep Learning Center Agreement (to C.A.G.-P.). F.M.-J. was supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme [Grant Agreement No. 682398]. A.V. received support from Institució Catalana de Recerca i Estudis Avançats (ICREA). E.P.-P. received support by a La Caixa Junior Leader Fellowship [LCF/BQ/PI18/11630003] from Fundación La Caixa and a Ramon y Cajal fellowship from the Spanish Ministry of Science [RYC2019-026415-I]. The Barcelona Supercomputing Center and IRB Barcelona are recipients of a Severo Ochoa Centre of Excellence Award from Spanish Ministry of Science, Innovation and Universities (MICINN; Government of Spain). The Josep Carreras Leukaemia Research Institute and IRB Barcelona are supported by CERCA (Generalitat de Catalunya). E.P-P. is supported by the Spanish Science Ministry (PID2019-107043R1-I00).
Conflict of Interest: The authors declare that they do not have any conflict of interest.
Contributor Information
Carlos A Garcia-Prieto, Josep Carreras Leukaemia Research Institute (IJC), Badalona, Spain; Barcelona Supercomputing Center (BSC), Barcelona, Spain.
Francisco Martínez-Jiménez, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain.
Alfonso Valencia, Josep Carreras Leukaemia Research Institute (IJC), Badalona, Spain; Institució Catalana de Recerca i Estudis Avançats (ICREA), Barcelona, Spain.
Eduard Porta-Pardo, Josep Carreras Leukaemia Research Institute (IJC), Badalona, Spain; Barcelona Supercomputing Center (BSC), Barcelona, Spain.
References
- Abeshouse A. et al. (2017) Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell, 171, 950–965.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L.B. et al. ; Australian Pancreatic Cancer Genome Initiative. (2013) Signatures of mutational processes in human cancer. Nature, 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L.B. et al. ; PCAWG Consortium. (2020) The repertoire of mutational signatures in human cancer. Nature, 578, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alioto T.S. et al. (2015) A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat. Commun., 6, 10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzar I. et al. (2019) NeoMutate: an ensemble machine learning framework for the prediction of somatic mutations in cancer. BMC Med. Genomics, 12, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D. et al. (2015) Systematic pan-cancer analysis of tumour purity. Nat. Commun., 6, 8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnedo-Pac C. et al. (2019) OncodriveCLUSTL: a sequence-based clustering method to identify cancer drivers. Bioinformatics, 35, 4788–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M.H. et al. ; Cancer Genome Atlas Research Network. (2018) Comprehensive characterization of cancer driver genes and mutations. Cell, 173, 371–385.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville R. et al. (2017) Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol., 1. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L. et al. (2016) In-depth comparison of somatic point mutation callers based on different tumor next-generation sequencing depth data. Sci. Rep., 6, 36540–36549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K. et al. (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol., 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G. et al. ; TCGA Research Network. (2015) Comprehensive molecular portraits of invasive lobular breast cancer. Cell, 163, 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ciriano I. et al. (2017) A molecular portrait of microsatellite instability across multiple cancers. Nat. Commun., 8, 15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentro S.C. et al. ; PCAWG Evolution and Heterogeneity Working Group and the PCAWG Consortium. (2021) Characterizing genetic intra-tumor heterogeneity across 2,658 human cancer genomes. Cell, 184, 2239–2254.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietlein F. et al. (2020) Identification of cancer driver genes based on nucleotide context. Nat. Genet., 52, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrott K. et al. (2018) Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst., 6, 271–281.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y. et al. (2016) MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol., 17, 178. https://doi.org/10.1186/s13059-016-1029-6. [DOI] [PMC free article] [PubMed]
- Forbes S.A. et al. (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res., 45, D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A. et al. (2013) IntOGen-mutations identifies cancer drivers across tumor types. Nat. Methods, 10, 1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman R.L. et al. (2016) Toward a shared vision for cancer genomic data. N. Engl. J. Med., 375, 1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoadley K.A. et al. (2018) Cell-of-Origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell, 173, 291–304.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. et al. (2018) Pathogenic germline variants in 10,389 adult cancers. Cell, 173355–173370.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman D.M. et al. (2017) Implementing genome-driven oncology. Cell, 168, 584–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D.C. et al. (2012) VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res., 22, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D.E. et al. (2012) SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics, 28, 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. (2018) An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell, 173, 400–416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I. et al. (2017) Universal patterns of selection in cancer and somatic tissues. Cell, 171, 1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Jiménez F. et al. (2020a) Systematic analysis of alterations in the ubiquitin proteolysis system reveals its contribution to driver mutations in cancer. Nat. Cancer, 1, 122–135. [DOI] [PubMed] [Google Scholar]
- Martínez-Jiménez F. et al. (2020b) A compendium of mutational cancer driver genes. Nat. Rev. Cancer, 20, 555–572. [DOI] [PubMed] [Google Scholar]
- Mularoni L. et al. (2016) OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol., 17, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S. et al. ; Breast Cancer Working Group of the International Cancer Genome Consortium. (2012) The life history of 21 breast cancers. Cell, 149, 994–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon B. et al. (2021) Integrating molecular profiles into clinical frameworks through the molecular oncology almanac to prospectively guide precision oncology. Nat. Cancer, 2, 1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A.G. et al. (2017) Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell, 171, 540–556.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A.G. et al. ; TCGA Research Network. (2017) Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell., 32, 204–220.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal R. et al. (2016) deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol., 17, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann S. et al. (2017) Evaluating variant calling tools for Non-Matched Next-Generation sequencing data. Sci. Rep., 7, 43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondka Z. et al. (2018) The COSMIC cancer gene census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer, 18, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokheim C. et al. (2016) Exome-scale discovery of hotspot mutation regions in human cancer using 3D protein structure. Cancer Res., 76, 3719–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. et al. (2013) Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med., 5, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weghorn D., Sunyaev S. (2017) Bayesian inference of negative and positive selection in human cancers. Nat. Genet., 49, 1785–1788. [DOI] [PubMed] [Google Scholar]
- Wood D.E. et al. (2018) A machine learning approach for somatic mutation discovery. Sci. Transl. Med., 10, eaar7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W. et al. (2021) Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat. Biotechnol., 39, 1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. (2018) A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput. Struct. Biotechnol. J., 16, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H. et al. (2014) CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics, 30, 1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.