Significance
Although turnover of sex chromosomes is very common in many vertebrate lineages, the transition process is still elusive. We studied the sex-determining region (SDR) of 12 congeneric fish species of Takifugu fish that compose an important model for the study of genomics and sex determination. We found that while nine species retained their ancestral SDR, three species had acquired derived SDRs. Although the derived SDRs resided in three different chromosomes, they harbored a shared supergene flanked by two putative transposable elements. The results highlight the underestimated role of a mobile supergene in turnover of sex chromosomes in vertebrates.
Keywords: sex chromosome evolution, sex-determining gene, structural variants, transposon
Abstract
Recent studies have revealed a surprising diversity of sex chromosomes in vertebrates. However, the detailed mechanism of their turnover is still elusive. To understand this process, it is necessary to compare closely related species in terms of sex-determining genes and the chromosomes harboring them. Here, we explored the genus Takifugu, in which one strong candidate sex-determining gene, Amhr2, has been identified. To trace the processes involved in transitions in the sex-determination system in this genus, we studied 12 species and found that while the Amhr2 locus likely determines sex in the majority of Takifugu species, three species have acquired sex-determining loci at different chromosomal locations. Nevertheless, the generation of genome assemblies for the three species revealed that they share a portion of the male-specific supergene that contains a candidate sex-determining gene, GsdfY, along with genes that potentially play a role in male fitness. The shared supergene spans ∼100 kb and is flanked by two duplicated regions characterized by CACTA transposable elements. These results suggest that the shared supergene has taken over the role of sex-determining locus from Amhr2 in lineages leading to the three species, and repeated translocations of the supergene underlie the turnover of sex chromosomes in these lineages. These findings highlight the underestimated role of a mobile supergene in the turnover of sex chromosomes in vertebrates.
Sex-determining genes and the chromosomes harboring them have been maintained in therian mammals and birds for more than 100 million years (1). However, such stability and conservation of the sex-determination system are not universal in vertebrates. Indeed, it has been shown that sex chromosomes have changed (“turned over”) in many poikilothermic vertebrate lineages, such as fishes, amphibians, and nonavian reptiles (2–7).
Among them, fishes are a particularly attractive group of animals in which to investigate the rapid turnover of sex chromosomes and/or sex-determining genes, since different sex-determination mechanisms often exist within a group of closely related species, and the sex-determining genes have been identified in some of these groups. However, in the majority of cases, distinct sex-determining genes were only identified in phylogenetically distant species. Therefore, it is challenging to illuminate key genomic changes associated with the evolutionary transition between the ancestral and derived sex-determination systems, and hence evolutionary drivers facilitating the transition remain largely vague. Although extensive comparative studies have been ongoing in some genera, each of which includes a relatively well-studied species such as medaka, guppy, pejerrey, Nile tilapia, threespine stickleback, rainbow trout, and northern pike (8–17), the transition between the ancestral and derived sex-determining genes has only been traced at the molecular level in medaka and its relatively distant species that diverged more than 20 million years ago (18, 19).
Takifugu is a genus of fish that includes fugu (T. rubripes), a model species for comparative genomics (20), and ∼20 closely related species (21) (Fig. 1A). It has been known that sex chromosomes in the lineage leading to fugu have remained undifferentiated for more than 4 million years (1, 15). The anti-Müllerian hormone receptor type 2 (Amhr2) is most likely the sex-determining gene in fugu, T. pardalis, and T. poecilonotus, since the single nucleotide polymorphism (SNP; SNP7271) on exon 9 of the gene was the sole polymorphism associated with phenotypic sex (G/C in male) and there is no sign of sequence differentiation beyond this SNP between the X and Y chromosomes (15, 16). However, a recent study of this genus showed a possible transition of the sex-determining genes in one of the congeneric species, T. niphobles, in which the sex-determining locus mapped to a location distinct from that of Amhr2 (15, 16). Moreover, the strong suppression of recombination around the sex-determining locus in T. niphobles suggested that this species potentially has sex chromosomes with an early stage of sequence differentiation. Since the ancestor of the Takifugu genus underwent rapid speciation 2–5 million years ago, the genus will provide a unique opportunity for understanding the process of rapid sex chromosome turnover and the initiation of sex chromosome differentiation, as well as the genomic changes associated with each of these.
Fig. 1.
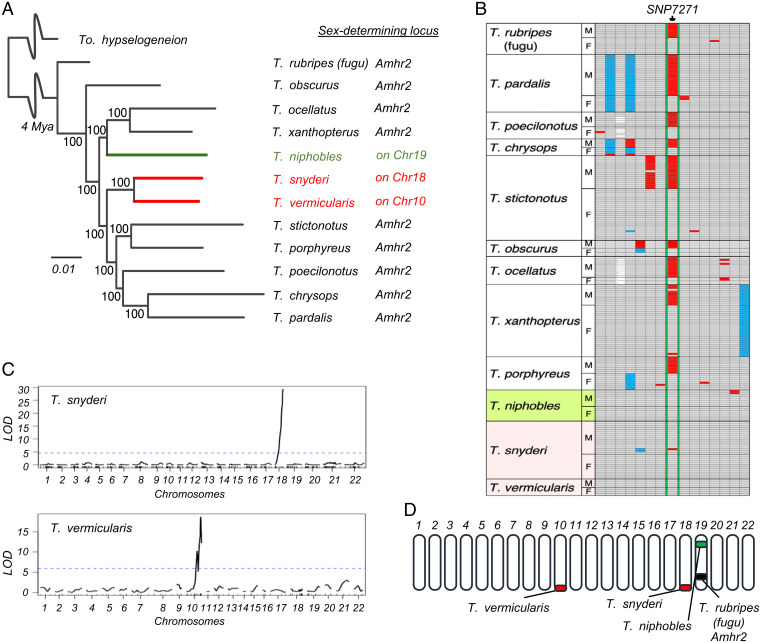
The genus Takifugu contains at least three species with sex chromosome turnover. (A) Phylogenetic relationship of 12 Takifugu species and their sex-determining loci. The green and red lines indicate lineages with a distinct sex-determining locus from Amhr2. Bootstrap values are shown at the nodes. Trees were rooted with Torquigener hypselogeneion as an outgroup. Divergence times at the crown age of the genus are based on fossil records and mitochondrial and transcriptome divergence (22). (B) A comparison of single nucleotide polymorphisms (SNPs) in Amhr2 among 12 Takifugu species suggests that sex is not determined by the SNP7271 in the three species. The data for four fugu species (T. rubripes [fugu], T. pardalis, T. poecilonotus, and T. niphobles) are taken from ref. 16 for comparison. SNP7271 of Amhr2 and seven SNPs on either side of it are shown. SNP7271 of Amhr2 is flanked by thick green lines. Gray cells indicate that an individual is homozygous for the reference allele, blue cells indicate that an individual is homozygous for the alternative alleles, and red cells indicate that an individual is heterozygous. White cells refer to deletions. (C) Linkage analyses map the sex-determining locus to Chr18 in T. snyderi and to Chr10 in T. vermicularis. QTL analysis was conducted using 1,225 markers for 98 full-sibling progenies in T. snyderi family A, and using 139 markers for 62 full-sibling progenies in T. vermicularis family D. The logarithm of the odds (LOD) score is on the y-axis, and linkage groups (chromosomes) are on the x-axis. Chromosome identity is based on conserved synteny with the reference genome of fugu (T. rubripes) defined in ref. 23. The marker position is indicated on the x-axis. The significance level (P = 0.05) is indicated by a dashed line. (D) Map location of the sex-determining locus in genus Takifugu. There are 22 chromosomes in the haploid set of Takifugu. The different sex-determining loci in this genus are schematically shown. The sex-determining loci of T. rubripes (black) and T. niphobles (green) have been mapped to distinct locations on Chr19 in previous studies (15, 16). F, female; M, male.
To gain insight into the transition process between the sex-determination systems, we expanded our previous work to include 12 Takifugu species and found that sex chromosome replacements have occurred in three species. We then generated a genome assembly for each of the three species and unexpectedly found that despite the fact that the chromosomal location of the sex-determining locus is distinct among the three species, these loci share a very similar male-specific region. We speculated that repeated translocation of a preexisting sex-determining locus may be more prevalent in vertebrates than previously thought and may have caused rapid turnover of sex chromosomes.
Results
The Sex-Determining SNP in Amhr2 Is Absent in 3 of 12 species.
Previous studies suggested that the sex-determining role of SNP7271 in Amhr2 is not conserved in one of four Takifugu species, namely T. niphobles (16) (shown in green in Fig. 1A). To gain a more comprehensive view of the transition of sex-determining loci in this genus, we sequenced the genomic segment harboring the SNP7271 site from females and males of eight Takifugu species that had not been examined previously (Fig. 1B and SI Appendix, Table S1). We found that while the association between SNP7271 polymorphisms and gonadal sex was conserved among six species, this was not the case in the other two; the SNP locus was homozygous for the C allele (the female allele) in T. snyderi and T. vermicularis, regardless of their sex, except in one T. snyderi male individual (Fig. 1B and SI Appendix, Table S1, shown in red in Fig. 1A). We performed additional genotyping of the SNP site and found that the G allele (the male allele) was absent in wild populations of the two species examined (n = 50 for each sex of each species) (SI Appendix, Table S1).
Linkage Mapping Identifies Sex-Determining Loci.
The genotyping results implied that the sex-determining loci have turned over in lineages leading to T. snyderi and T. vermicularis. Alternatively, the role of master sex determiner in these species may have been taken over by environmental factors. To test these hypotheses, we produced 98 full-sibling progenies (Family A) in T. snyderi and 62 full-sibling progenies (Family D) in T. vermicularis from wild-caught parents (SI Appendix, Table S2), and we performed genome-wide linkage mapping of the sex-determining locus. We found a strong linkage between gonadal sex and paternally inherited alleles at loci near the distal end of Chr18 in T. snyderi (logarithm of the odds [LOD] = 29.3; P = 1.8 × 10−23) and Chr10 in T. vermicularis (LOD = 18.7; P = 4.3 × 10−15) (Fig. 1C). Indeed, the markers f1659 on Chr18 and sca541-1 on Chr10 exhibited perfect segregation with phenotypic sex among the 83 progeny of T. snyderi and the 62 progeny of T. vermicularis (SI Appendix, Table S3). We further confirmed this result in additional families of T. snyderi (P < 1.0 × 10−16) and T. vermicularis (P = 1.8 × 10−39) by marker association (Fisher exact test) (SI Appendix, Table S3). The linkage analysis also revealed that the marker loci and the synteny of these markers are largely conserved between the species (SI Appendix, Fig. S1).
A Phylogenetic Framework Suggests Three Independent Turnovers.
Summing up the results we have reported here thus far and those of previous studies (15, 16), it is now evident that while the role of the Amhr2 locus in sex determination is largely conserved among Takifugu species, at least three turnover events have occurred in this genus, resulting in four distinct sex-determining loci (Fig. 1D). To infer the ancestral and derived states of the four sex-determining loci, we generated a phylogenetic tree using whole-genome resequencing data of 12 Takifugu species and one outgroup species (Fig. 1A and SI Appendix, Table S4). According to the most parsimonious interpretation of the phylogeny, the results corroborate the previous hypothesis that T. niphobles has obtained a derived sex-determining locus on Chr19, while having lost the ancestral male-determining allele (the G allele) at the Amhr2 locus on the same chromosome (16) (green in Fig. 1A). Moreover, the phylogenetic tree indicates that sex chromosomes are likely to have turned over at least twice in the lineage leading to the sister species, T. snyderi and T. vermicularis (red in Fig. 1A).
Having detected the three turnover events related to sex chromosomes within Takifugu species, we turned to the detailed characterization of the three sex-determining loci that have likely arisen recently (shown in green and red in Fig. 1A).
k-mer Analysis Identifies Male-Specific Sequences in T. niphobles.
Although we previously showed that the sex-determining locus in T. niphobles maps to Chr19 with an XX/XY system (16) (shown in green in Fig. 1A), no male-specific sequences have been found. To identify male-specific sequences in T. niphobles, we first searched for the male-specific k-mers in the resequencing data of six females and five males (SI Appendix, Table S5) and obtained the 35-mer sequences unique to males. Using these sequences, we extracted the pair-end reads from the same resequencing data and assembled them into 551 contigs with a total of 218-kb sequences (SI Appendix, Table S6). We then masked repetitive sequences in the contigs and used the Basic Local Alignment Search Tool (BLAST) to search against a fugu reference genome sequence, FUGU5 (23). This revealed that similar sequences exist on multiple chromosomes in T. rubripes, including Chr6, Chr12, and Chr22 (SI Appendix, Table S7).
Construction of a Chromosomal-Scale Genome Assembly of a T. niphobles YY Male.
As the k-mer approach suggested that the male-specific region of T. niphobles is likely absent in the fugu reference genome, we attempted to generate a reference genome sequence for T. niphobles, including its Y chromosome and the male-specific region. Complete sequence assemblies of Y chromosomes have not been generated for many organisms (but see some exceptions in refs. 24–28) because of technical difficulties related to the discrimination of X and Y in the male genome and/or the assembly of regions containing high levels of repetitive sequences. To alleviate these problems, we sequenced the genome of a male T. niphobles carrying two Y chromosomes (instead of X and Y) and generated a draft genome sequence using a hybrid assembly strategy with PacBio long reads and Illumina short reads (SI Appendix, Fig. S2 and Tables S8 and S9). The assembly consists of 728 contigs with an N50 of 6.08 Mb and a total size of ∼396 Mb. Further scaffolding with Hi-C data placed 288 contigs into 22 superscaffolds (pseudochromosomes) that had chromosomal-level length (longer than 10 Mb) (SI Appendix, Fig. S3B). The size of these pseudochromosomes covered 91.2% of the total genome and their number was consistent with the karyotype of this species (29) (SI Appendix, Fig. S3B and Tables S10 and S11). The total number of scaffolds in the Hi-C–based, proximity-guided assembly was 424. The scaffolding increased N50 from 6.1 Mb to 16.2 Mb and that of the longest scaffold from 15 Mb to 29.8 Mb (Table 1 and SI Appendix, Fig. S3B). A Benchmarking Universal Single-Copy Orthologs (BUSCO) search against the 4,584 single-copy orthologs for Actinopterygii suggested that the assembly was missing only 2.3% of the core genes (SI Appendix, Table S12). Furthermore, ∼95% of the short reads from multiple male individuals were uniquely mapped to the assembled genome with a mapping quality above 20. These metrics indicated the high quality of the chromosomal scale assembly. The assembly showed highly conserved synteny with the fugu reference genome (SI Appendix, Fig. S4) and, therefore, chromosome identities for the T. niphobles were assigned based on those of T. rubripes. Based on the conserved synteny shown here and the genomic positions of the sex-linked markers reported previously (16), we identified the pseudochromosome corresponding to the T. niphobles sex chromosome (Chr19), which comprises 23 contigs totaling 17 Mb in length (SI Appendix, Fig. S5 and Table S11). Because this pseudochromosome was assembled from the YY male, we denoted it as Chr19Y.
Table 1.
Assembly statistics for the genomes of Takifugu niphobles, T. snyderi, and T. vermicularis
| T. niphobles | T. snyderi | T. vermicularis | |
|---|---|---|---|
| Genotype | YY | XY | XY |
| Sequencing and scaffolding strategies | PacBio, Illumina, Hi-C | Nanopore, stLF, Illumina | PacBio, Illumina |
| No. of scaffolds/contigs | 424 | 1,097 | 1,756 |
| Total length, Mb | 396 | 399 | 429 |
| Average length, kb | 935 | 363 | 244 |
| Longest scaffold/contig | 29.8 Mb | 22.3 Mb | 8,669 kb |
| Shortest scaffold/contig, bp | 1,000 | 162 | 2,979 |
| Scaffold/contig N50, Mb | 16 | 6 | 1.6 |
stLF, single-tube long-fragment read.
The Male-Specific Region Is Flanked by Pseudoautosomal Regions in T. niphobles.
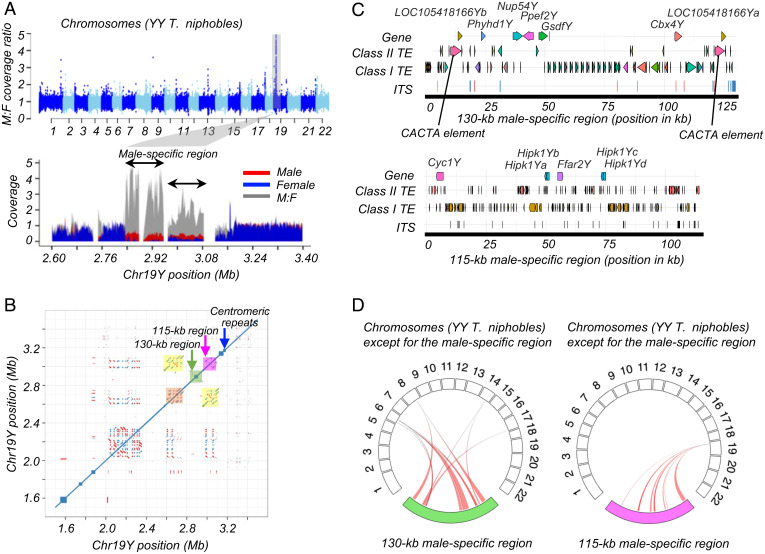
To determine the male-specific region in this assembly, we aligned resequencing reads from 8 females and 10 males against the assembly and compared the relative depth of coverage between males and females. We detected a clear difference in the coverage in the region (∼245 kb) on Chr19Y (Fig. 2A), suggesting that the region may be specific to males. The male-specific region is composed of two subregions with different relative coverage; the left-half region (130 kb) showed a relatively higher male to female ratio than the right-half region (115 kb) (Fig. 2A). The absence of differences in coverage depth between sexes along the flanking regions indicates that these regions are shared between Y and X chromosomes.
Fig. 2.
The male-specific region in Takifugu niphobles. (A) The male-specific region in T. niphobles was revealed by differences in the depth of coverage between males and females. We aligned resequencing reads from 10 males and 8 females against the assembled genome of a T. niphobles YY male and calculated the relative depth of coverage between sexes at a window size of 1 kb (y-axis). In the upper plot for the genome-wide analysis, chromosomes are on the x-axis. In the lower plot, the male-specific region and part of the pseudoautosomal region on Chr19Y are shown. The normalized coverage of males, females, and their ratio are shown in red, blue, and gray, respectively. (B) The self-comparison dot plot of the genomic region containing the male-specific region (the positions from 1.5 to 3.5 Mb). The left half (130 kb) and right half (115 kb) of the male-specific region are shaded in green and dark pink, respectively. Marked sharing of the repetitive sequence between the right-half region (dark pink) and a pseudoautosomal region (orange) was evident by dense dots at the areas in light yellow, suggesting the accumulation of large and common repetitive sequences in the two regions. Direct (blue) and inverted (red) repeats are visualized as the accumulation of dots off the central diagonal line. The blue arrow indicates the accumulation of a centromeric repeat. (C) Schematic representations of the repeat annotation in the male-specific region in T. niphobles. Each haplotype is annotated with triangles to depict annotated full-length genes, class I transposable elements (TEs), and class II TEs. Rectangles represent interstitial telomeric sequences (ITSs) (TTAGGG)n ≥ 2. (D) Circos plots showing the syntenic relationships between genes in the male-specific region and other parts of the genome. The reference chromosomes (scaled in megabases) are shown from 1 to 22 in clockwise orientation (the male-specific regions were masked from Chr19). The male-specific regions shown in the lower part of plot are scaled in kilobases. Genic sequences exhibiting a high similarity between the male-specific regions and other parts of the genome are connected by colored curves. The left-side plot suggests that autosome to Y transposition is the main source of the expansion of the 130-kb male-specific region, whereas the right-side plot suggests that the 115-kb region originated from segmental duplications on a pseudoautosomal region. F, female; M, male.
Accumulation of Large Repetitive Sequences Is Heterogeneous in the Male-Specific Region of T. niphobles.
Accumulations of large repetitive sequences have been observed in the nonrecombining regions of sex chromosomes across diverse taxa (30, 31). To determine if this is the case in T. niphobles, we aligned Chr19Y to itself and found that the repeat accumulation was still modest in about half of the male-specific region (130 kb) (Fig. 2B; shaded in green), while the other 115 kb contained many large, repetitive sequences (Fig. 2B; shaded in pink). Furthermore, the self-alignment plot implied that the 115-kb region is likely derived from the duplication of a region residing on the pseudoautosomal region near the male-specific 130-kb region (Fig. 2B; shaded in orange).
Repetitive Sequences Are Accumulated in the Male-Specific Region of T. niphobles.
The repeat sequence analysis revealed accumulation of repeat sequences within the 245-kb male-specific regions; repeat sequences occupied 42.5% and 62.2% of the 130-kb and 115-kb regions, respectively (Fig. 2C and SI Appendix, Table S13). While these values are greater than that of the whole Chr19Y (14.9%), the pseudoautosomal regions near the male-specific region also harbor repeat-rich sequences (SI Appendix, Fig. S5). For example, repeat sequences occupied 46.3% of the ∼1-Mb pseudoautosomal region (ranging from the position 2 Mb to the proximal boundary to the male-specific region).
A Candidate Sex-Determining Gene in T. niphobles Resides in the Male-Specific Region Flanked by Putative Transposable Elements.
Since the k-mer analysis suggested that part of the male-specific region may have been derived from an autosomal region, we tested this possibility by performing BLAST searches of the male-specific sequences (130 kb + 115 kb) against the reference genome sequences of T. niphobles and T. rubripes (FUGU5).
We found that the majority of the 130-kb male-specific region was composed of small segments that showed high sequence similarity to portions of other chromosomes, suggesting that the transposed sequences were the main source of the male-specific region (Fig. 2D). Furthermore, we annotated seven protein-coding genes that are paralogous to the genes on three different autosomes (Fig. 2C): GsdfY, Ppef2Y, Nup54Y, Phyhd1, Cbx4Y, and two LOC105418166Y (uncharacterized protein-coding genes that appear to be conserved in teleost fish). Of these, GsdfY is worth noting since the ortholog of this gene has been reported as the master sex-determining gene in a teleost species, Oryzias luzonensis (18). Furthermore, loss of function of this gene was found to result in female-to-male sex reversal in two teleost species, Nile tilapia and medaka (32, 33). Our RNA-seq analysis confirmed the paralog-specific expression from this locus in differentiating male gonads, along with the expressions of Ppef2Y and Nup54Y (SI Appendix, Fig. S6 and Table S14). As for Phyhd1Y, Cbx4Y, and LOC105418166Ys, their expression of either the male-specific or autosomal paralogs was observed, but paralog-specific expression could not be ensured, due to the lack of diagnostic nucleotide sites that can distinguish the transcripts of these paralogs.
Intriguingly, the sequence annotation also revealed that the male-specific region is flanked at both ends by two CACTA transposable elements containing the catalytic center of the transposase (Fig. 2C). CACTA elements, also described as EnSpm elements, belong to the class II transposons that utilize a cut-and-paste mechanism for their transposition (34–36). A full-length CACTA element contains two open reading frames, one encoding a transposase and the other a protein of unknown function. Although we were unable to show solid evidence that the two CACTA elements of T. niphobles are structurally intact, we found that the catalytic center of the transposase called the DDD/E domain is conserved (SI Appendix, Fig. S7).
The search involving the other half of the male-specific region (115 kb) identified six protein-coding genes (Cyc1Y, Hipk1Ya, Hipk1Yb, Hipk1Yc, Hipk1Yd, and Ffar2Y), though their roles in sex determination have not been reported (Fig. 2C). Each gene had more than one untruncated or truncated paralog on Chr19Y at the outside of the male-specific region (Fig. 2D), corroborating the hypothesis that the 115-kb region originated from duplication events on the pseudoautosomal regions (Fig. 2B). Among these paralogs, the expression of either the male-specific or pseudoautosomal paralogs of Cyc1Y was observed in the developing gonads. However, the paralog-specific expression was not assessed, due to the lack of diagnostic nucleotide sites.
Verification of the Male-Specific Region of T. niphobles.
To verify the overall accuracy of the assembled sequences of the 130-kb male-specific region and its adjacent pseudoautosomal regions, we designed 12- and 5-primer pairs targeting each of the two regions, respectively (SI Appendix, Table S15), and successfully obtained all amplicons. Sequence alignment revealed that the amplicon contigs were largely concordant with the target region of the Chr19Y assembly (SI Appendix, Fig. S8). Discordant regions comprised only 1.2% of the amplicon contigs and 4.5% of the target region of the Chr19Y assembly. The mismatch may have been due to misassembly, misamplification of amplicons, or true polymorphisms between individuals used for the tiling polymerase chain reaction (PCR) and the Chr19Y construction. We also confirmed the male specificity of the region using a wild population (n = 20 for each sex; P = 3.0 × 10−10) with a diagnostic primer pair that could yield amplicons of different size from the male-specific region (352 bp) and its paralogous region (408 bp) (SI Appendix, Fig. S9). Although one potential sex-reversed fish was observed, similar incomplete penetrance of the sex-determining locus has been reported in experimental families of this species (16). Thus, the reliability of the assembly results of the 130-kb male-specific region and one of its adjacent region in Chr19Y was confirmed. Note that we were not able to apply this approach to the 115-kb region, due to its highly repetitive nature.
k-mer Analysis Identifies Male-Specific Sequences in T. snyderi and T. vermicularis.
We used similar strategies to characterize the sex-determining locus of T. snyderi on Chr18 and that of T. vermicularis on Chr10 (shown in red in Fig. 1A). First, we searched for the male-specific 35-mers in the resequencing data of 24 individuals for each sex of T. snyderi (three pools of both females and males, each pool containing eight individuals) and four individuals for each sex of T. vermicularis (SI Appendix, Table S5), and assembled them into contigs (SI Appendix, Table S6). BLAST searches of the contigs against the T. niphobles assembly revealed that many of the male-specific contigs in both species showed high similarity to part of the male-specific sequence in T. niphobles. For example, the male-specific contigs in both species contain a coding sequence of GsdfY (SI Appendix, Table S7). This result was unexpected, as the chromosomal locations of the sex-determining loci in the three species are distinct from each other, raising the possibility that the male-specific region is shared among the three species, at least in part.
Construction of the Genome Assemblies of T. snyderi and T. vermicularis.
We then generated a genome assembly for T. snyderi and T. vermicularis, including their Y chromosomes. Because individuals with the YY genotype were not available, the genome of an XY male was sequenced for each species. We used Oxford Nanopore long reads and MGI single-tube long-fragment reads for T. snyderi, and PacBio long reads and Illumina short reads for T. vermicularis (SI Appendix, Tables S16–S18). The male assembly for T. snyderi consisted of 1,097 scaffolds with an N50 of 6.0 Mb and a total size of ∼399 Mb, whereas the male assembly for T. vermicularis comprised 1,756 contigs with an N50 of 1.57 Mb and a total size of ∼428 Mb (Table 1). A BUSCO search against the 4,584 single-copy orthologs for Actinopterygii suggested that only 1.2% and 2.1% of the core genes were undetected in the genome assemblies of T. snyderi and T. vermicularis, respectively (SI Appendix, Table S12). Approximately 95% of the short reads from male individuals of both T. snyderi and T. vermicularis mapped uniquely to the respective assembled genomes, with a mapping quality above 20.
The Assembled Genomes for T. snyderi and T. vermicularis Contain Their Male-Specific Regions.
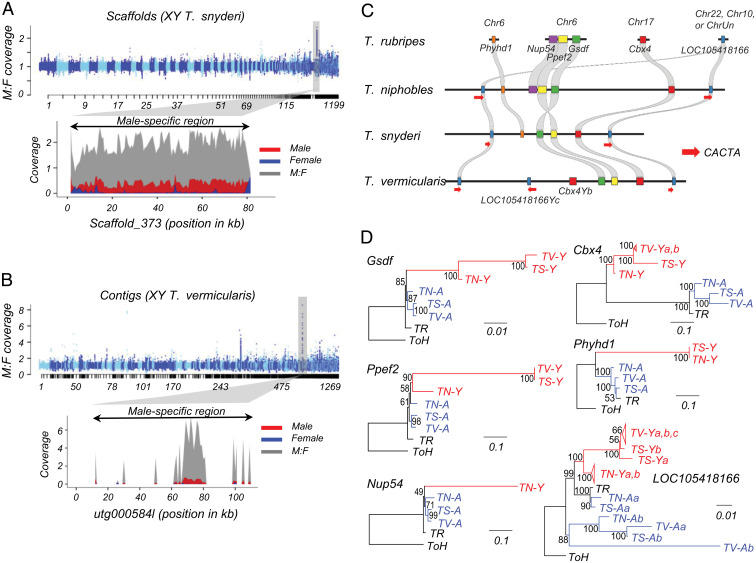
To determine the male-specific region in the assembly for T. snyderi, we aligned resequencing reads from females and males and compared the relative depth of coverage between them. This analysis revealed a clear difference in coverage between sexes on scaffold_373 (79 kb) (Fig. 3A). A BLAST search revealed conserved synteny between this region and the 130-kb male-specific region harboring GsdfY in T. niphobles (Fig. 3C and SI Appendix, Fig. S10A).
Fig. 3.
Translocation of the shared male-specific region drove the turnover of sex chromosomes in Takifugu species. (A) The male-specific region in T. snyderi. We aligned resequencing reads from 24 males and 24 females against the assembled genome of a T. snyderi XY male and calculated the relative depth of coverage between sexes at a sliding-window size of 1 kb. In the upper plot showing the genome-wide analysis, scaffolds are on the x-axis. The lower plot shows the male-specific region on scaffold_373. The normalized coverage of males, females, and their ratio are shown in red, blue, and gray, respectively. (B) The male-specific region in T. vermicularis. We aligned resequencing reads from 7 males and 12 females against the assembled genome of a T. vermicularis XY male. In the upper plot showing the genome-wide analysis, contigs are on the x-axis. The lower plot shows the male-specific region found on contig utg000584l. Note that this contig contains large inverted duplications and a repeat-rich region (SI Appendix, Fig. S10B) in which paired-end reads cannot be mapped except in several segments. The contiguous sequence spanning all the male-specific segments was defined as the male-specific region. (C) Syntenic relationship between the male-specific genes of T. niphobles, T. snyderi, and T. vermicularis, and their homologs of T. rubripes. Each haplotype is annotated with pentagons to depict annotated full-length genes. Arrows indicate CACTA transposable elements containing the catalytic center of a transposase. (D) Maximum-likelihood clustering of the male-specific and autosomal paralogs in T. niphobles, T. snyderi, and T. vermicularis. Red and blue colors represent the male-specific (-Y) and autosomal (-A) paralogs, respectively. TN, T. niphobles; TR, T. rubripes; TS, T. snyderi; TV, T. vermicularis. The autosomal orthologs in Torquigener hypselogeneion (ToH) were used as outgroups. Bootstrap values are shown at the nodes of each tree.
We used the same approach for T. vermicularis, and found coverage differences between sexes on contig utg000584l (97 kb) (Fig. 3B). As in the case of T. snyderi, sequence similarity was observed between this contig and the 130-kb male-specific region in T. niphobles. In addition, this contig contained large inverted duplications and a repeat-rich region where paired-end reads could not be properly mapped (SI Appendix, Fig. S10B).
The association of the male-specific region with phenotypic sex in a wild population of both species (n = 19 and 20 for each sex, respectively, of both T. snyderi and T. vermicularis) was supported using a diagnostic primer pair that could produce amplicons of different sizes from the male-specific region and its paralogous region (P = 5.7 × 10−11 and P = 3.3 × 10−9, respectively) (SI Appendix, Fig. S9).
Male-Specific Regions Are Largely Conserved among the Three Species.
Manual annotation of the male-specific regions in T. niphobles (130 kb), T. snyderi (79 kb), and T. vermicularis (105 kb) revealed similar characteristics of these regions; they harbor GsdfY and some other shared genes, and are flanked by two CACTA transposons (Fig. 3C). The sizes of the segments are also similar: 115 kb in T. niphobles, 61 kb in T. snyderi, and 105 kb in T. vermicularis. The gene contents in these regions were found to be similar but distinct. For example, while GsdfY, Ppef2Y, Cbx4Y, LOC105418166Ya, and LOC105418166Yb were shared among all three species, Nup54Y was observed only in T. niphobles. Moreover, T. vermicularis appeared to have lost Phyhd1Y but acquired an additional copy of Cbx4Y and LOC105418166Y (Fig. 3C and SI Appendix, Fig. S10).
To understand the history of the male-specific region that appeared to be conserved but located on different chromosomes in the three species, we constructed phylogenic trees for each gene in the male-specific region and its autosomal paralog(s) across the three species (Fig. 3D). This phylogenetic analysis identified three main branching patterns (Fig. 3D and SI Appendix, Figs. S11 and S12). The pattern of Gsdf indicated that the male-specific region (red in Fig. 3D) and the autosomal paralogs (blue in Fig. 3D) in the three species diverged after their split with the lineage leading to T. rubripes, but before the diversification of the three species. The second pattern, including Ppef2 and Nup54, suggests that the topology at the crown age of the genus is ambiguous. The third pattern, including Cbx4, Phyhd1, and LOC105418166, suggested that these paralog pairs diverged before the split of the three species with T. rubripes. These results indicate that the male-specific region may have been formed due to capture of GsdfY by the preexisting segment containing other paralogs. However, the presence of a single homolog for these genes (excluding LOC105418166) in T. rubripes (Fig. 3 C and D) raises questions about this theory, although lineage-specific gene loss in T. rubripes is one possible explanation. Alternatively, an elevated rate of evolution due to the reduced efficacy of purifying selection in the nonrecombining region may underlie the gene topology in which gene trees are discordant with the species phylogeny (37). For example, the rapid evolution of the male-specific paralogs (e.g., Phyhd1Y) may result in “long branch attraction” in which these paralogs are clustered with an outgroup homolog (38). Furthermore, positive selection in the male-specific paralogs after the gene duplication but before speciation would give rise to the third pattern (39).
Despite the heterogeneous topologies observed in the gene trees, all paralog pairs in the three species were clustered not according to species genealogy but rather to paralog groups (red in Fig. 3D and SI Appendix, Figs. S11–S13), suggesting that the male-specific genes were likely transposed or translocated as a unit into the current genomic positions after the initial formation of the supergene. When focusing on the male-specific paralogs for each species, duplicates of the gene flanking the male-specific region, LOC105418166Ys, were clustered according to species group (Fig. 3C). Duplicates of Cbx4Y in T. vermicularis were also clustered according to species group. Thus, it is likely that the linage-specific duplication of genes occurred in the gene cassette of the male-specific region.
Although the male-specific supergene likely evolved once in the common ancestor of the three species, the three species are polyphyletic in the inferred phylogeny (Fig. 1A). This could be due to incomplete lineage sorting (40) or introgression of the supergene (41). However, the topology of the gene trees (Fig. 3D) (the male-specific paralogs are not nested within the clade of the autosomal paralogs) excludes a simple introgression as an explanation of the discordance. To examine the degree to which ancestral polymorphisms have affected the divergence of Takifugu, we used topology weighting by iterative sampling of sub-trees analysis and the f4 test (SI Appendix, Figs. S14 and S15). Although the distinction between the two scenarios (i.e., incomplete lineage sorting and ancient introgression of the supergene) was unclear, these analyses suggested that a substantial amount of shared polymorphism has shaped the speciation of Takifugu, with the evidence of gene flow between T. niphobles and the last common ancestor of T. snyderi and T. vermicularis (SI Appendix, Fig. S15).
Discussion
Based on a combination of extensive genetic and genomic data, this study documents evidence that the evolution of the male-specific supergene underlies the replacement of the sex-determining gene in an ancestor of a subset of Takifugu fishes and that subsequent translocation of the supergene drove a rapid sex chromosome turnover without a simultaneous change in the sex-determining gene. Presumably, the accumulation of genes in the male-specific region conferred a selective advantage of this region over the ancestral sex-determining gene, Amhr2. However, the insertion of the supergene could have caused sex chromosome differentiation that resulted in a selective disadvantage, namely the accumulation of repeats (toxic Y hypothesis) (42), which may, in turn, have facilitated translocation of the core part of the supergene at relatively short intervals.
Formation of the Male-Specific Supergene.
Supergenes are sets of tightly linked genes that contribute to a complex phenotype (43–48). Sex-determination regions can be regarded as supergenes because suppressed recombination, if present, causes cosegregation of genes linked to the sex-determining gene (49).
Although direct evidence is lacking in Takifugu fish, the genes linked to GsdfY in the core male-specific region may play a role in testis differentiation, because either they or their paralogs are expressed in the male gonad during gonadal differentiation (SI Appendix, Fig. S6). Moreover, studies in other animals have suggested that some genes in the male-specific region of the three Takifugu fishes are involved in gonadal differentiation and/or sexual behavior. For example, Nup54 regulates nucleocytoplasmic transport, whose possible role in gonadal differentiation has been proposed based on its conserved expression in teleost fish (50). In Drosophila, this gene is essential in transposon silencing in the ovary (51), and there is evidence suggesting that it plays a role in sexual conflict by influencing female behavior (52). Moreover, this gene is contained in a supergene under sexual antagonistic selection in rainbow trout (53). Ppef2 is predominantly expressed in photoreceptors and the pineal gland, and it participates in phototransduction in mammals. This gene is also contained in the rainbow trout supergene under sexual antagonistic selection (53). Cbx4 codes for a member of the Polycomb-group proteins that control cell identity and development (54). Although its role in sexual dimorphism has not been reported, Cbx2 codes for another member of the group and is required to stabilize the testis pathway in mice (55).
The Evolutionary Development of the Core Male-Specific Region.
Although supergenes contribute to many fascinating phenotypes (11–14, 44–48), their evolutionary development is often only incompletely understood. In particular, the analysis of sex-determining regions has been difficult due to the extensive accumulation of repetitive sequences (see some exceptions in refs. 24–28). Our study provided a unique opportunity to infer the process involved in assembling the male-specific supergene and its subsequent modifications during repeated translocation.
It is likely that the core male-specific region evolved in the common ancestor of the three species after diverging from the lineages leading to T. rubripes and T. obscurus through the combination of at least two processes. One includes segmental duplication encompassing Nup54, Ppef2, and Gsdf on Chr6 and the translocation of this region to the future sex chromosome (Fig. 3C and SI Appendix, Figs. S13 and S16), given that the genomic arrangement of the three genes adjacent to each other is a conserved feature among teleost fish (50). The other process is the gathering of other unlinked genes (Phyhd1, Cbx4, and LOC105418166) through independent duplications and translocations (Fig. 3C and SI Appendix, Fig. S16). It is possible that the latter process partly predates the former, as the phylogenetic analysis in this study implied that the emergence of Phyhd1Y and LOC105418166Y occurred prior to that of GsdfY. The original segment, if this is the case, was probably not male specific until it captured GsdfY.
Based on the core structure formation, it is parsimonious to consider that Nup54Y was lost before the divergence of T. snyderi and T. vermicularis, while the gene content was retained in the lineage leading to T. niphobles (Fig. 3C and SI Appendix, Fig. S16). Then, in the T. vermicularis lineage, while Phyhd1Y was lost, the segmental duplication of part of the male-specific region resulted in the emergence of LOC105418166Yc and Cbx4Yb (Fig. 3C and SI Appendix, Figs. S10 and S16). The variations in gene content in the shared male-specific region suggest that adaptive replacements of genes may have taken place in these species and could be ongoing.
The Jumping Sex-Determining Locus.
In vertebrates, transposition or translocation of a preexisting sex-determining locus has only been reported in salmonids (56). Comparative analyses of the sex-determining loci in the three salmonid species (57) suggested that an ∼4.1-kb sequence containing the sex-determining gene sdY is conserved along with transposable elements. Although an RNA-mediated mechanism has been proposed for the transposition of the sdY locus in salmonids, different mechanisms likely played a role in the movement of the male-specific region in Takifugu, as the size of the supergene spans ∼100 kb. A notable feature of the gene arrangement of the conserved male-specific region is that the region is flanked by two duplicated regions harboring CACTA transposable element and LOC105418166Y (Fig. 3C). Therefore, the flanking transposons are candidate drivers for relocating the male-specific region. However, we were unable to find the terminal inverted repeats that are expected to reside at the ends of transposons if a cut-and-paste process is involved in transposition. Although it is possible that the terminal inverted repeats have degenerated, the same orientation of the two transposons is incompatible with the standard model of cut-and-paste transposition (34). Based on the orientation of the two flanking regions, we prefer the hypothesis that nonallelic homologous recombination played a role in the translocation. This process is thought to contribute to duplication and translocation events in animals and plants (58–60) and suggests that recombination between two directly oriented homologous sequences (typically, repeats or transposable elements) can lead to deletion and insertion of the genomic region flanked by these sequences (61). As a result, duplicated and translocated fragments are frequently bordered by transposable elements (62). Of note, this model is only speculative in Takifugu, and the mechanism underlying the translocation of supergene is still undetermined.
Roles of Translocations in Accumulation and Purging of Deleterious Sequences.
It is hypothesized that sex chromosomes start to differentiate when recombination around the sex-determining region stops. While the accumulation of deleterious mutations on the differentiating sex chromosomes will eventually reduce fitness (6, 42, 63, 64), the “jumping” ability of the sex-determining locus is a way to purge the deleterious mutations (56). Consistent with these hypotheses, our analysis of k-mers and read coverage suggested that half of the male-specific region (115 kb), which contains many large repetitive sequences in T. niphobles, appeared to be purged from T. snyderi and T. vermicularis after the translocations. The 115-kb region may have arisen due to the reduced recombination triggered by the insertion of the 130-kb region containing GsdfY in an ancestor of the three species. However, it is possible that the 115-kb region has recently emerged in T. niphobles in a species-specific manner. The distinction of the two scenarios was challenging due to the presence of multiple duplicated loci corresponding to genes in the T. niphobles 115-kb region in the genomes of Takifugu fishes, which made it difficult to distinguish orthologs and paralogs. To fully evaluate the consequence of repeated translocation of the sex-determining supergene in the three species, more contiguous sequences of the genomes for T. snyderi and T. vermicularis are needed.
Remarks
This study uncovered a process involved in the replacement of both sex chromosomes and sex-determining genes in closely related species and highlighted the importance of repeated translocation of the sex-determining region. Moreover, the construction of contigs for the jumping sex-determining supergene revealed that it has a unique structure flanked by two putative transposable elements. Outside of vertebrates, the translocation or transposition of a preexisting sex-determining gene was recently characterized in insects (65) and plants (66, 67). The repeated translocation of a sex-determining locus may be more prevalent in vertebrates than previously thought and may have caused rapid turnover of sex chromosomes.
Materials and Methods
Conservation of the Sex-Determining SNP in Amhr2.
The genotype at SNP7271 on Amhr2 is associated with phenotypic sex in fugu (15, 16). We previously examined whether the association is conserved in T. pardalis, T. poecilonotus, and T. niphobles, and found that while it was identified in T. pardalis and T. poecilonotus, the locus was fixed with the female allele (C allele) in T. niphobles (15, 16). In this study, we extended the analysis to eight Takifugu species, namely T. chrysops, T. stictonotus, T. obscurus, T. ocellatus, T. xanthopterus, T. porphyreus, T. snyderi, and T. vermicularis, and genotyped a total of 342 wild-caught and 15 aquacultured fish by a direct-sequencing approach. The genomic region containing exon9 of Amhr2 and its adjacent regions was PCR amplified and directly sequenced, as previously reported (16). The sequence data are provided in SI Appendix (Fasta1_amhr2exon9.fas). Additional genotyping of SNP7271 of T. snyderi (n = 100) and T. vermicularis (n = 100) was performed by high-resolution melting analysis using a Lightscanner (Idaho Technology) following the method described in ref. 68. Information on the sampling locations, sex, and association analysis (Fisher exact test) results of the fish of each species is provided in SI Appendix, Table S1.
Linkage Mapping.
Experimental families of T. snyderi.
Three families (families A, B, and C) of T. snyderi were produced between three wild males and a single wild female caught at Suruga Bay, Shizuoka Prefecture, Japan (SI Appendix, Table S2). Procedures for artificial fertilization and rearing conditions were defined as previously reported for T. niphobles (69). Fish at 122 and 174 d postfertilization (dpf) were combined and used for the following analyses. Phenotypic sex was visually determined under a microscope.
Genotyping of SNP markers in T. snyderi.
Genotyping by random amplicon sequencing–direct technology (70, 71) was used to obtain the genome-wide genotype data from 98 fish in Family A. Genomic DNA was extracted from the caudal fin using the Gentra Puregene tissue kit (Qiagen). Library construction and sequencing on an Illumina HiSeq 2500 platform (paired end 100 bp) were conducted by Eurofin Genomics as described in ref. 71. Genotyping procedures are described in the SI Appendix, Methods.
Linkage map construction and mapping of the sex-determining locus in T. snyderi.
The sex-determining locus was analyzed by the interval mapping implemented in the R/qtl package (version 1.41) (72) as described in ref. 16. A detailed description of the procedures is provided in the SI Appendix, Methods.
Fine mapping of the sex-determining locus using microsatellite markers in T. snyderi.
Although interval mapping is a powerful tool to identify loci underlying differences in phenotypes, it is possible that the genotype–phenotype association observed at the distal end of Chr18 is family specific. To test this possibility, we used 83, 25, and 62 fish from families A, B, and C, respectively, and genotyped markers f1618 and f1659, which were previously mapped near the distal end of Chr18 in the fugu genome. The association between gonadal sex and genotype was assessed by Fisher exact test (SI Appendix, Table S3). To increase the resolution, we genotyped two marker loci (f1335 and f1433) on Chr18 for 765 fish from the three families (342, 158, and 265 fish in families A, B, and C, respectively). After identifying 30 recombinants, 26 microsatellite markers flanked by the two markers on Chr18 were genotyped for these individuals (SI Appendix, Fig. S1). Genotyping procedures are described in ref. 23. Information on the markers is provided in SI Appendix, Table S19.
Experimental families of T. vermicularis.
For T. vermicularis, two pairs of wild-caught parents from Ariake Sea (Nagasaki Prefecture, Japan) were mated independently to produce two families, families D and E (SI Appendix, Table S2). Fish production and phenotyping were performed as for T. snyderi.
Linkage map construction and mapping of the sex-determining locus in T. vermicularis.
We first conducted a genome-wide linkage analysis of the sex-determining locus using 62 fish at 127 dpf in family D with 138 genome-wide microsatellite markers (SI Appendix, Table S2). The linkage between phenotype and genotype observed at the distal end of Chr10 in family D was confirmed using 164 fish in family E sampled at 131 to 161 dpf by genotyping 35 markers anchored on Chr10 in T. rubripes. The genotype information is provided in SI Appendix (mapping genotype.xlsx).
The phylogenetic framework of Takifugu.
To generate a species phylogenetic tree, we first obtained whole-genome resequencing data from 12 Takifugu species and one outgroup (one individual per species): T. rubripes, T. snyderi, T. vermicularis, T. niphobles, T. pardalis, T. poecilonotus, T. chrysops, T. stictonotus, T. obscurus, T. ocellatus, T. xanthopterus, T. porphyreus, and Torquigener hypselogeneion (outgroup). We then used RaxML (version 0.8) to construct the phylogenetic tree (see details in the SI Appendix, Methods).
k-mer Analysis to identify male-specific sequences.
A detailed description of k-mer analyses of T. niphobles, T. snyderi, and T. vermicularis is provided in the SI Appendix, Methods.
Genome Assembly in a T. niphobles YY Male.
Long- and short-read sequencing.
We sequenced the genome of a YY individual on a PacBio Sequel platform. The production of the YY fish was reported previously (16). In brief, we first identified an XY sex-reversed female using sex-linked microsatellite markers (f2003 and f2006) to cross with an XY male. The offspring consisted of individuals carrying XX, XY, or YY sex chromosome sets. Then the YY fish was selected based on the sex-linked markers f2003 and f2006. Genomic DNA of the YY fish was isolated from the caudal fin by the conventional phenol/chloroform/isoamyl-alcohol method. Size selection, library preparation, and sequencing were conducted at the National Institute of Genetics. Approximately 56.6-Gb sequences with total reads of 5,445,262 (N50 = 17 kb) were derived from eight PacBio SMRT cells (SI Appendix, Table S8). Genomic DNA from the YY fish was also used for whole-genome Illumina resequencing. A TruSeq DNA PCR-Free library was prepared and sequenced (2 × 245 bp) on the HiSeq 2500 platform at the National Institute of Genetics. The data have been registered in the DNA Data Bank of Japan Sequence Read Archive (accession no. DRA012992).
Hybrid genome assembly, Hi-C scaffolding, and gene annotation.
Detailed procedures are provided in the SI Appendix, Methods. In brief, Illumina reads were first trimmed and assembled into contigs. We then obtained an initial hybrid assembly by anchoring the contigs to the PacBio long reads. We corrected potential sequencing errors in the initial assembly by realigning the long reads and the Illumina contigs and polished it. For long-read-only assembly, a Pairwise mApping Format file was generated using Minimap2 (73) and converted into an assembly graph using Miniasm (74). Then, a de novo assembly sequence (unitig) was extracted from the assembly graph file. We polished the two assemblies and merged them. Furthermore, Hi-C data from an XY male individual were used to scaffold this merged genome. An overview of the genome assembly is shown in SI Appendix, Fig. S2. Gene annotation was conducted using MAKER (version 3.01.02) (75), which utilizes both evidence-based methods (RNA-seq data) and ab initio predictions. RNA-seq data were obtained from the differentiating gonads of T. niphobles at 90 dpf.
Characterization of the male-specific region.
For a comparison of the relative depth of read coverage between males and females, we sequenced an additional two females and four males of T. niphobles from Lake Hamana (SI Appendix, Methods and Table S5). The data were registered in the DNA Data Bank of Japan Sequence Read Archive (accession no. DRA012877). We then mapped the resequencing data of 10 males and 8 females (SI Appendix, Table S5) onto the genome assembly of the T. niphobles YY male using the Burrows-Wheeler Aligner Maximal Exact Match (BWA-MEM) algorithm (76) (SI Appendix, Methods). To test if the male-specific region comprised segments that were duplicated and translocated from other regions of the genome, we first masked repetitive sequences from the two male-specific regions using the repeat database of T. niphobles YY genome assembly. We then aligned the male-specific regions to the genome assembly using the nucmer algorithm with option -l 50 -c 65 implemented in MUMmer (version 4.0) (77). We restricted our analyses to the genic regions and visualized synteny blocks longer than 300 bp with 90% sequence identity. We verified the contiguity of the assembled sequence by tiling PCR. Moreover, the presence of the male-specific region in the wild population of T. niphobles was confirmed by a diagnostic primer. A detailed description of these strategies is presented in the SI Appendix, Methods.
Genome assembly in a T. snyderi XY male and a T. vermicularis XY male.
The whole-genome sequence of a T. snyderi XY individual from Lake Hamana was obtained using PromethION long reads (Nanopore Technologies) and MGI single-tube long-fragment reads (MGISEQ-2000RS) (SI Appendix, Tables S16 and S17), whereas that of a T. vermicularis XY individual from Ariake Sea was obtained using PacBio long reads (SI Appendix, Table S18) and Illumina paired-end short reads (Sample identifier: S9; SI Appendix, Table S5). Detailed procedures for DNA extraction, sample preparation, sequencing, genome assembly, genome annotation, and identification and characterization of the male-specific region are provided in the SI Appendix, Methods.
Phylogenetic analysis of male-specific genes and their autosomal paralogs.
To determine the phylogenetic relationships of the male-specific genes and their autosomal paralog(s), we used RAxML (version 0.8) (SI Appendix, Methods).
Admixture analysis.
To investigate the traces of incomplete lineage sorting and/or past introgression in Takifugu species, we used resequencing data from four T. rubripes individuals, five T. niphobles individuals, five T. synderi individuals, three T. vermicularis individuals, and a single individual for T. chrysops, T. poecilonotus, and T. pardalis. A single individual of Torquigener hypselogeneion was used as outgroup for all analyses. We quantified the genealogic relationships throughout the genome of Takifugu using topology weighting by iterative sampling of sub-trees (78). We also investigated admixture/introgression across the genomes using Patterson D statistics (79, 80), f4-ratio statistics (81), and f-branch statistics (82) (SI Appendix, Methods).
Supplementary Material
Acknowledgments
This work was in part supported by Grants-in-Aid for Scientific Research (grants 22H00377, 221S0002, 24380102, 15H04542, 18H02277, 16K14966, 18K05815, and 17H06425). This work was also supported by the Cooperative Research Grant of the Genome Research for BioResource, NODAI Genome Research Center, Tokyo University of Agriculture, and JST-Mirai Program (grants JPMJMI18CH and JPMJMI21C1). The authors are grateful to Shuya Kato for helping with the data analysis and anonymous reviewers for their valuable comments and suggestions that improved this article.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.E.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121469119/-/DCSupplemental.
Data Availability
Next-generation sequencing (NGS) reads data have been deposited in DNA Data Bank of Japan Sequence Read Archive (https://www.ddbj.nig.ac.jp/dra/index-e.html) (DRA012890, DRA012891, DRA012877, DRA012889, DRA012878, DRA012992, DRA012876, DRA012888, DRA014181, and DRA014182). All study data, other than the NGS data, are included in the article and/or supporting information.
References
- 1.Bachtrog D., et al. ; Tree of Sex Consortium, Sex determination: Why so many ways of doing it? PLoS Biol. 12, e1001899 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charlesworth D., Mank J. E., The birds and the bees and the flowers and the trees: Lessons from genetic mapping of sex determination in plants and animals. Genetics 186, 9–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall Graves J. A., Peichel C. L., Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 11, 205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miura I., An evolutionary witness: The frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex Dev. 1, 323–331 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Ezaz T., Sarre S. D., O’Meally D., Graves J. A., Georges A., Sex chromosome evolution in lizards: Independent origins and rapid transitions. Cytogenet. Genome Res. 127, 249–260 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi K., Hamaguchi S., Novel sex-determining genes in fish and sex chromosome evolution. Dev. Dyn. 242, 339–353 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Furman B. L. S., Evans B. J., Sequential turnovers of sex chromosomes in African clawed frogs (Xenopus) suggest some genomic regions are good at sex determination. G3 (Bethesda) 6, 3625–3633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myosho T., Takehana Y., Hamaguchi S., Sakaizumi M., Turnover of sex chromosomes in Celebensis group medaka fishes. G3 (Bethesda) 5, 2685–2691 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darolti I., et al. , Extreme heterogeneity in sex chromosome differentiation and dosage compensation in livebearers. Proc. Natl. Acad. Sci. U.S.A. 116, 19031–19036 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Q., et al. , The rise and fall of the ancient northern pike master sex-determining gene. eLife 10, e62858 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlesworth D., Bergero R., Graham C., Gardner J., Keegan K., How did the guppy Y chromosome evolve? PLoS Genet. 17, e1009704 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hattori R. S., et al. , A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. U.S.A. 109, 2955–2959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böhne A., et al. , Repeated evolution versus common ancestry: Sex chromosome evolution in the haplochromine cichlidx Pseudocrenilabrus philander. Genome Biol. Evol. 11, 439–458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross J. A., Urton J. R., Boland J., Shapiro M. D., Peichel C. L., Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5, e1000391 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamiya T., et al. , A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8, e1002798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ieda R., et al. , Identification of the sex-determining locus in grass puffer (Takifugu niphobles) provides evidence for sex-chromosome turnover in a subset of Takifugu species. PLoS One 13, e0190635 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yano A., et al. , The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 6, 486–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myosho T., et al. , Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191, 163–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamahira K., et al. , Mesozoic origin and ‘out-of-India’ radiation of ricefishes (Adrianichthyidae). Biol. Lett. 17, 20210212 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aparicio S., et al. , Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297, 1301–1310 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Yamanoue Y., et al. , Explosive speciation of Takifugu: Another use of fugu as a model system for evolutionary biology. Mol. Biol. Evol. 26, 623–629 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Hedges S. B., Marin J., Suleski M., Paymer M., Kumar S., Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kai W., et al. , Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol. Evol. 3, 424–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skaletsky H., et al. , The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Hughes J. F., et al. , Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483, 82–86 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peichel C. L., et al. , Assembly of the threespine stickleback Y chromosome reveals convergent signatures of sex chromosome evolution. Genome Biol. 21, 177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao L., et al. , The Y chromosome sequence of the channel catfish suggests novel sex determination mechanisms in teleost fish. BMC Biol. 17, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafati N., et al. , Reconstruction of the birth of a male sex chromosome present in Atlantic herring. Proc. Natl. Acad. Sci. U.S.A. 117, 24359–24368 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyaki K., Tabeta O., Kayano H., Karyotypes in six species of pufferfishes genus Takifugu (Tetraodontidae, Tetraodontiformes). Fish. Sci. 61, 594–598 (1995). [Google Scholar]
- 30.Charlesworth B., The evolution of sex chromosomes. Science 251, 1030–1033 (1991). [DOI] [PubMed] [Google Scholar]
- 31.Charlesworth D., Charlesworth B., Marais G., Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118–128 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Jiang D.-N., et al. , gsdf Is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 83, 497–508 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Zhang X., et al. , Autosomal gsdf acts as a male sex initiator in the fish medaka. Sci. Rep. 6, 19738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wicker T., et al. , A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Yuan Y.-W., Wessler S. R., The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. U.S.A. 108, 7884–7889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Howe K., et al. , The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerrard D. T., Filatov D. A., Positive and negative selection on mammalian Y chromosomes. Mol. Biol. Evol. 22, 1423–1432 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Siddall M., Long-branch abstractions. Cladistics 15, 9–24 (1999). [Google Scholar]
- 39.Mawaribuchi S., Yoshimoto S., Ohashi S., Takamatsu N., Ito M., Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 20, 139–151 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pease J. B., Haak D. C., Hahn M. W., Moyle L. C., Phylogenomics reveals three sources of adaptive variation during a rapid radiation. PLoS Biol. 14, e1002379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon G., Kitano J., Kirkpatrick M., The origin of a new sex chromosome by introgression between two stickleback fishes. Mol. Biol. Evol. 36, 28–38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen A. H., Bachtrog D., Toxic Y chromosome: Increased repeat expression and age-associated heterochromatin loss in male Drosophila with a young Y chromosome. PLoS Genet. 17, e1009438 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwander T., Libbrecht R., Keller L., Supergenes and complex phenotypes. Curr. Biol. 24, R288–R294 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Joron M., et al. , Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477, 203–206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., et al. , A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493, 664–668 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Kess T., et al. , A migration-associated supergene reveals loss of biocomplexity in Atlantic cod. Sci. Adv. 5, eaav2461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joron M., et al. , A conserved supergene locus controls colour pattern diversity in Heliconius butterflies. PLoS Biol. 4, e303 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Labonne J. D. J., Tamari F., Shore J. S., Characterization of X-ray-generated floral mutants carrying deletions at the S-locus of distylous Turnera subulata. Heredity 105, 235–243 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Charlesworth D., The status of supergenes in the 21st century: Recombination suppression in Batesian mimicry and sex chromosomes and other complex adaptations. Evol. Appl. 9, 74–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gautier A., Le Gac F., Lareyre J.-J., The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes. Gene 472, 7–17 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Munafò M., et al. , Channel nuclear pore complex subunits are required for transposon silencing in Drosophila. eLife 10, e66321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nallasivan M. P., Haussmann I. U., Civetta A., Soller M., Channel nuclear pore protein 54 directs sexual differentiation and neuronal wiring of female reproductive behaviors in Drosophila. BMC Biol. 19, 226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pearse D. E., et al. , Sex-dependent dominance maintains migration supergene in rainbow trout. Nat. Ecol. Evol. 3, 1731–1742 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Luis N. M., et al. , Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell 9, 233–246 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Moreno S. A., et al. , CBX2 is required to stabilize the testis pathway by repressing Wnt signaling. PLoS Genet. 15, e1007895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertho S., Herpin A., Schartl M., Guiguen Y., Lessons from an unusual vertebrate sex-determining gene. Philos. Trans. R. Soc. Lond. B Biol. Sci. 376, 20200092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faber-Hammond J. J., Phillips R. B., Brown K. H., Comparative analysis of the shared sex-determination region (SDR) among salmonid fishes. Genome Biol. Evol. 7, 1972–1987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuzminov A., Homologous recombination—Experimental systems, analysis, and significance. Ecosal Plus 4(2) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang S., et al. , Repetitive element-mediated recombination as a mechanism for new gene origination in Drosophila. PLoS Genet. 4, e3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parks M. M., Lawrence C. E., Raphael B. J., Detecting non-allelic homologous recombination from high-throughput sequencing data. Genome Biol. 16, 72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woodhouse M. R., Pedersen B., Freeling M., Transposed genes in Arabidopsis are often associated with flanking repeats. PLoS Genet. 6, e1000949 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wicker T., Buchmann J. P., Keller B., Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. 20, 1229–1237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blaser O., Grossen C., Neuenschwander S., Perrin N., Sex-chromosome turnovers induced by deleterious mutation load. Evolution 67, 635–645 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Charlesworth B., Charlesworth D., The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 1563–1572 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma A., et al. , Male sex in houseflies is determined by Mdmd, a paralog of the generic splice factor gene CWC22. Science 356, 642–645 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Tennessen J. A., et al. , Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 16, e2006062 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang W., et al. , A general model to explain repeated turnovers of sex determination in the Salicaceae. Mol. Biol. Evol. 38, 968–980 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsunaga T., et al. , An efficient molecular technique for sexing tiger pufferfish (fugu) and the occurrence of sex reversal in a hatchery population. Fish. Sci. 80, 933–942 (2014). [Google Scholar]
- 69.Hosoya S., et al. , The genetic architecture of growth rate in juvenile Takifugu species. Evolution 67, 590–598 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Enoki H., “The construction of pseudomolecules of a commercial strawberry by DeNovoMAGIC and new genotyping technology, GRAS-Di” in Proceedings of the Plant and Animal Genome Conference XXVII (San Diego, CA, 2019), pp. 37002
- 71.Hosoya S., et al. , Random PCR-based genotyping by sequencing technology GRAS-Di (genotyping by random amplicon sequencing, direct) reveals genetic structure of mangrove fishes. Mol. Ecol. Resour. 19, 1153–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Broman K. W., Wu H., Sen S., Churchill G. A., R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H., Minimap and miniasm: Fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cantarel B. L., et al. , MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marçais G., et al. , MUMmer4: A fast and versatile genome alignment system. PLOS Comput. Biol. 14, e1005944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin S. H., Van Belleghem S. M., Exploring evolutionary relationships across the genome using topology weighting. Genetics 206, 429–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Green R. E., et al. , A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Durand E. Y., Patterson N., Reich D., Slatkin M., Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Patterson N., et al. , Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malinsky M., et al. , Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2, 1940–1955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Next-generation sequencing (NGS) reads data have been deposited in DNA Data Bank of Japan Sequence Read Archive (https://www.ddbj.nig.ac.jp/dra/index-e.html) (DRA012890, DRA012891, DRA012877, DRA012889, DRA012878, DRA012992, DRA012876, DRA012888, DRA014181, and DRA014182). All study data, other than the NGS data, are included in the article and/or supporting information.