Significance
The paradigmatic example of factor-dependent pausing in transcription elongation is σ-dependent pausing, in which sequence-specific σ-DNA interaction with a − 10 element-like sequence in a transcribed region results in pausing of a σ-containing transcription elongation complex. It has been proposed that σ-dependent pausing involves DNA scrunching, and that sequences downstream of the −10 element-like sequence modulate DNA scrunching. Here, using site-specific protein–DNA photocrosslinking, high-throughput sequencing, and cryoelectron microscopy structure determination, we show directly that σ-dependent pausing involves DNA scrunching, we define a consensus sequence for formation of a stable scrunched paused complex that is identical to the consensus sequence for pausing in initial transcription, and we identify positions of DNA scrunching on DNA nontemplate and template strands. Our results illuminate the structural and mechanistic basis of σ-dependent transcriptional pausing.
Keywords: RNA polymerase, sigma, transcription elongation, pausing, DNA scrunching
Abstract
In σ-dependent transcriptional pausing, the transcription initiation factor σ, translocating with RNA polymerase (RNAP), makes sequence-specific protein–DNA interactions with a promoter-like sequence element in the transcribed region, inducing pausing. It has been proposed that, in σ-dependent pausing, the RNAP active center can access off-pathway “backtracked” states that are substrates for the transcript-cleavage factors of the Gre family and on-pathway “scrunched” states that mediate pause escape. Here, using site-specific protein–DNA photocrosslinking to define positions of the RNAP trailing and leading edges and of σ relative to DNA at the λPR′ promoter, we show directly that σ-dependent pausing in the absence of GreB in vitro predominantly involves a state backtracked by 2–4 bp, and σ-dependent pausing in the presence of GreB in vitro and in vivo predominantly involves a state scrunched by 2–3 bp. Analogous experiments with a library of 47 (∼16,000) transcribed-region sequences show that the state scrunched by 2–3 bp—and only that state—is associated with the consensus sequence, T−3N−2Y−1G+1, (where −1 corresponds to the position of the RNA 3′ end), which is identical to the consensus for pausing in initial transcription and which is related to the consensus for pausing in transcription elongation. Experiments with heteroduplex templates show that sequence information at position T−3 resides in the DNA nontemplate strand. A cryoelectron microscopy structure of a complex engaged in σ-dependent pausing reveals positions of DNA scrunching on the DNA nontemplate and template strands and suggests that position T−3 of the consensus sequence exerts its effects by facilitating scrunching.
The RNA polymerase (RNAP) holoenzyme initiates transcription by binding double-stranded promoter DNA, unwinding a turn of promoter DNA to yield an RNAP-promoter open complex (RPo) containing an ∼13-bp unwound “transcription bubble” and selecting a transcription start site (1–4). In bacteria, promoter binding and promoter unwinding are mediated by the transcription initiation factor σ, which, in the context of the RNAP holoenzyme, participates in sequence-specific protein–DNA interactions with the promoter −35 element, recognized by σ region 4 (σR4), and the promoter −10 element, recognized by σ region 2 (σR2) (5, 6).
The first ∼11 nt of an RNA product are synthesized as an RNAP-promoter initial transcribing complex (RPitc) in which RNAP remains anchored on promoter DNA through sequence-specific interactions with σ (1, 3). In initial transcription, RNAP uses a “scrunching” mechanism of RNAP-active-center translocation, in which, in each nucleotide-addition cycle, RNAP remains stationary on DNA and unwinds one base pair of DNA downstream of the RNAP active center, pulls the unwound single-stranded DNA (ssDNA) into and past the RNAP active center, and accommodates the additional unwound ssDNA as bulges in the transcription bubble (1, 7–10). Scrunching enables the capture of free energy from multiple nucleotide additions and the stepwise storage of captured free energy in the form of stepwise increases in the amount of DNA unwinding (RPitc,2 to RPitc,11). Thus, scrunching provides the mechanism to capture and store the free energy required to break RNAP-promoter interactions in subsequent promoter escape (1, 7, 9, 10).
Following synthesis of an RNA product of ∼11 nt, promoter escape occurs. Promoter escape entails 1) entry of the RNA 5′ end into the RNAP RNA-exit channel; 2) displacement of σ from the RNAP RNA-exit channel, driven by steric clash with the RNA 5′ end (11–14); 3) disruption of protein-DNA interaction between σ and the promoter −35 element; and 4) rewinding of the upstream half of the transcription bubble, from the −10 element through the transcription start site, a process termed “unscrunching” (9, 14, 15). The product of this series of reactions is a transcription elongation complex (TEC) containing a threshold length of ∼11 nt of RNA and having an altered RNAP-σ interface in which a subset of the interactions previously made between RNAP and σ are lost (16–19). Because of the partial loss of RNAP σ interactions, the affinity of RNAP for σ is decreased, and σ typically dissociates in a time-dependent fashion (16, 17, 20–28).
In contrast to initial transcription, which proceeds through a scrunching mechanism, transcription elongation proceeds through a “stepping” mechanism, in which RNAP steps forward by 1 bp relative to DNA for each nucleotide added to the RNA product (29). Each nucleotide-addition cycle of transcription requires translocation of the RNAP active center relative to DNA and RNA, starting from a “pre-translocated” state and yielding a “post-translocated” state. Translocation of the RNAP active center repositions the RNA 3′ end from the RNAP addition site (A site) to the RNAP product site (P site), rendering the A site available to bind the next extending nucleoside triphosphate (NTP) (30–33).
The rate of RNA synthesis is not uniform across the DNA template. At certain template positions, transcription is interrupted by “pausing”—i.e., nucleotide-addition cycles that occur on the second or longer timescale (3, 30, 32, 34). Pausing can often involve entry of the transcription complex into an off pathway, backtracked state where the RNAP active center has reverse translocated relative to DNA and RNA, rendering the active center unable to add NTPs (35–38). Pausing can impact gene expression by reducing the rate of RNA synthesis, facilitating engagement of regulatory factors with RNAP, modulating formation of RNA secondary structures, or enabling synchronization of transcription and translation (39). RNAP can be induced to pause by DNA sequences (sequence-dependent pausing) or by interacting proteins (factor-dependent pausing) (34).
The first identified and still paradigmatic example of factor-dependent pausing is σ-dependent pausing (16, 40–43). A σ-containing TEC in a promoter-proximal transcribed region (prior to σ dissociation), or, to a lesser degree, in a promoter-distal transcribed region (following σ dissociation and σ reassociation), can recognize and engage, through sequence-specific σ-DNA interactions, a transcribed-region sequence that resembles a promoter element (16, 21, 22, 41–48). These sequence-specific σ-DNA interactions anchor the σ-containing TEC at the sequence resembling a promoter element, resulting in σ-dependent transcriptional pausing. Typically, a σ-dependent pause element (SDPE) is a sequence that resembles a consensus promoter −10 element, often supplemented by a sequence that resembles a consensus discriminator element. σ-Dependent pausing, particularly σ-dependent pausing in promoter-proximal regions, enables coordination of the timing of transcription elongation with the timing of other biological processes. In the best-characterized example, σ-dependent pausing 16–17 bp downstream of the transcription start site of the bacteriophage λ PR′ promoter (λPR′; Fig. 1A) coordinates transcription elongation and regulation of transcription termination by providing time for loading of the transcription antitermination factor λQ (40, 49–51). In other well-characterized examples, σ-dependent pausing 18 bp and 25 bp downstream of the bacteriophage 21 and 82 PR′ promoters coordinates transcription elongation and regulation of transcription termination in a similar manner (41, 49, 52, 53). Genome-wide analyses suggest that σ-dependent pausing occurs in as many as 20% of transcription units in Escherichia coli (45, 54) and is functionally linked to expression levels of stress-related genes (54).
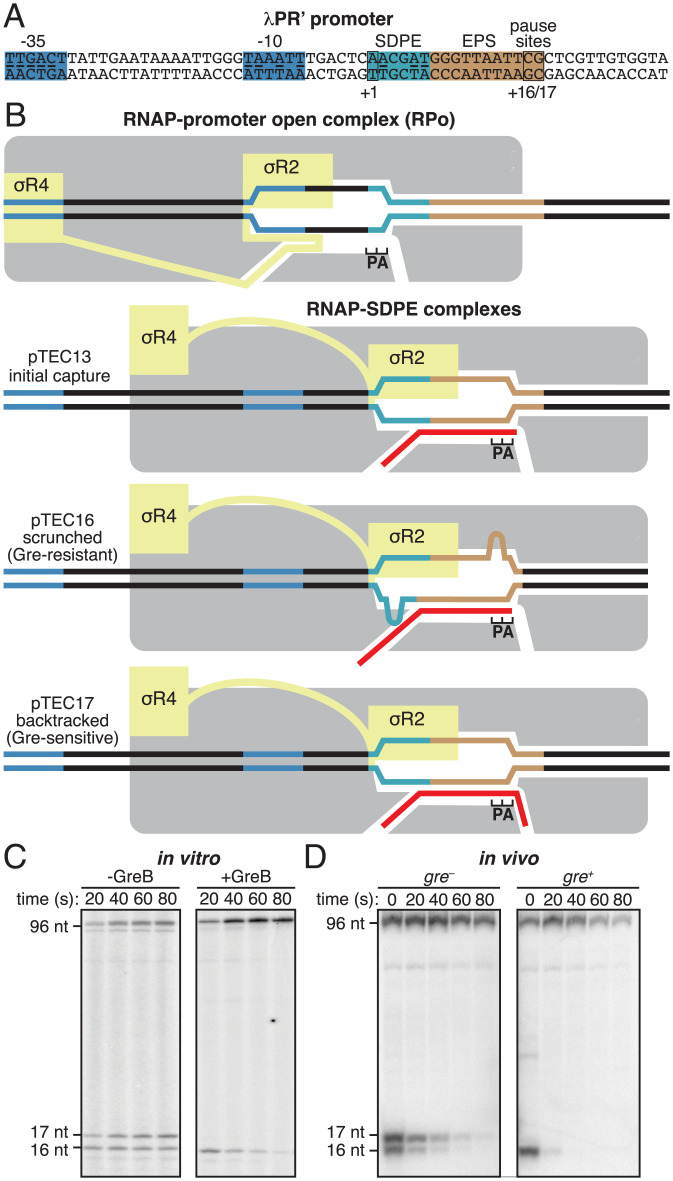
Fig. 1.
σ-Dependent pausing at λPR’: scrunched and backtracked states. (A) λPR′ promoter. Blue, –35 element and –10 element; light blue, SDPE; brown; elemental pause site (EPS); black rectangles, transcription start site (+1) and pause sites (+16/+17); underlining, consensus nucleotides of sequence elements. (B) Initiation complexes and paused complexes at λPR′. Four complexes are shown: 1) initiation complex, RPo; 2) initial-capture σ-dependent paused complex, pTEC13 (where “pTEC” denotes paused TEC and “13” denotes 13 nt RNA product); 3) scrunched σ-dependent paused complex, pTEC16; and 4) backtracked σ-dependent paused complex, pTEC17. Gray, RNAP core; yellow, σ; red, RNA product; P and A, RNAP active-center product and addition sites; blue, –35 element and –10 element; light blue, SDPE; brown, EPS; black, other DNA (nontemplate-strand above template-strand). Scrunching of DNA strands is indicated by bulges in DNA strands. (C and D) RNA product length and Gre-factor sensitivity in pTECs. (C) RNA product distributions in vitro for transcription reactions in absence or presence of GreB at indicated times after addition of NTPs. (D) RNA product distributions in vivo for gre− or gre+ cells at indicated times after addition of rifampin (Rif). Positions of pTEC-associated RNA products (16 nt and 17 nt) and full-length RNA product (96 nt) are indicated.
It has been proposed that in σ-dependent pausing, following the initial engagement of the SDPE (“pause capture”; Fig. 1B, line 2), the paused TEC (pTEC) can extend RNA for several nucleotides, using a scrunching mechanism (Fig. 1B, line 3), and the resulting pTECs with extended RNA equilibrate between scrunched states and backtracked states (Fig. 1B, lines 3–4) (10, 41, 55–59). It further has been proposed that the scrunched states are intermediates on the pathway to pause escape, the backtracked states are off the pathway to pause escape, and DNA sequences downstream of the SDPE, including sequences related to the consensus sequence for elemental pausing in transcription elongation, modulate the duration of the pause (“pause lifetime”) through effects on the relative occupancies of scrunched and backtracked states (41, 55–59). Evidence in support of these proposals comes from measurement of RNA product lengths, DNA footprinting (40, 41, 50, 55–58), and analysis of sensitivity of complexes to transcript cleavage factors of the Gre family (45–47, 55), which promote transcript cleavage in backtracked states but not in other states (Fig. 1C, ref. 60). However, the evidence is not definitive.
Here, we use in vitro and in vivo site-specific protein–DNA photocrosslinking, high-throughput sequencing, heteroduplex-template transcription experiments, and cryoelectron microscopy (cryo-EM) structure determination, to define the mechanistic and structural basis of σ-dependent pausing at λPR′.
Results
σ-Dependent Pausing at λPR′: pTEC RNA-Product Length and Gre-Factor Sensitivity.
It previously has been shown that at λPR′, in vitro, in the absence of Gre factors, the pTEC is present both in a state with 16 nt of RNA (pTEC16) and a state with 17 nt of RNA (pTEC17) (40, 51, 55). It also previously has been shown that at λPR′, in vitro, in the presence of Gre factors, the pTEC is present predominantly as pTEC16 (55). The data in Fig. 1C confirm these results, and the data in Fig. 1D show that the same pattern is obtained in vivo. Thus, in gre− cells, the pTEC is present both as pTEC16 and as pTEC17, whereas in gre+ cells, the pTEC is present essentially exclusively as pTEC16.
Use of Site-Specific Protein–DNA Photocrosslinking to Define Positions of RNAP Trailing and Leading Edges and of σ Relative to DNA at λPR′: Approach.
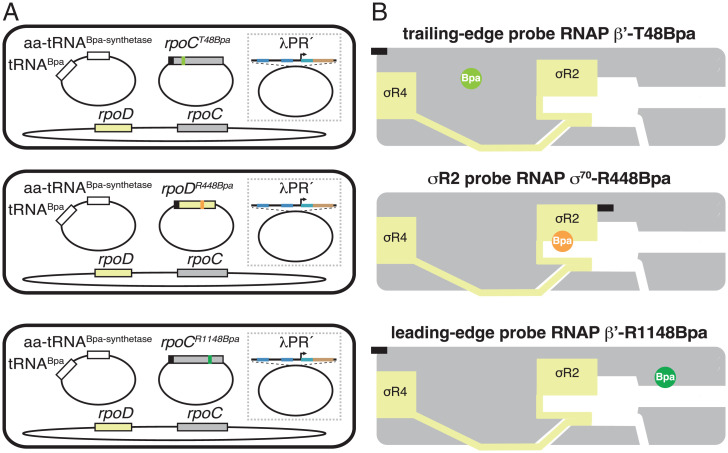
We used unnatural amino acid mutagenesis (61) to incorporate the photoactivatable crosslinking agent p-benzoyl-L-phenylalanine (Bpa) into RNAP holoenzyme at specific positions on the RNAP trailing edge (TE), the RNAP leading edge (LE), and σR2 (β′ residue 48, β′ residue 1148, and σ70 residue 448; Fig. 2). The resulting RNAP holoenzyme derivatives behaved indistinguishably from unmodified wild-type RNAP in terms of RNA-product-length distributions of paused complexes and Gre sensitivities of paused complexes (SI Appendix, Fig. S1). Using the resulting RNAP holoenzyme derivatives, in purified form for experiments in vitro, and in situ, inside living cells, for experiments in vivo, we performed site-specific protein–DNA photocrosslinking (62–65) to define the positions of the RNAP TE, the RNAP LE, and σR2 relative to DNA in RNAP-promoter complexes (RPo) and in RNAP-SDPE complexes (pTEC) at λPR′ (Fig. 3 and SI Appendix, Figs. S2–S4). From the observed positions of the RNAP LE relative to DNA, we then inferred the position of the RNAP-active-center nucleotide-addition site (A site) by subtracting 5 nt, as described previously (65).
Fig. 2.
Use of site-specific protein–DNA photocrosslinking to define positions of RNAP TEs and LEs and of σ relative to DNA at λPR′: approach. (A) Two-plasmid (for in vitro studies) or three-plasmid (for in vivo studies) merodiploid system for coproduction, in E. coli cells, of decahistidine-tagged RNAP-β′ T48Bpa, RNAP-β′ R1148Bpa, or RNAP-σ70 R448Bpa in the presence of untagged wild-type RNAP holoenzyme. First plasmid carries gene for decahistidine-tagged RNAP β′ subunit (gray rectangle) with nonsense codon at position 48 (olive green; Top row); decahistidine-tagged σ70 (light yellow rectangle) with nonsense codon at position 448 (orange; Center row); or gene for decahistidine-tagged RNAP β′ subunit with nonsense codon at position 1148 (forest green; Bottom row). Second plasmid carries genes for engineered Bpa-specific nonsense-suppressor tRNA and aminoacyl-tRNA synthetase (white rectangles). Third plasmid (shown inside dashed box), when present, carries λPR′ promoter or λPR′ promoter derivative. Chromosome (shown below plasmids) carries genes for wild-type RNAP β′ subunit and σ70. Black rectangles, decahistidine-tag coding sequence. (B) Bpa-modified RNAPs. Olive green circle, TE Bpa; orange circle, σR2 Bpa; forest green circle, LE Bpa. Black rectangles, decahistidine-tag. Other colors and symbols as in Fig. 1 A and B.
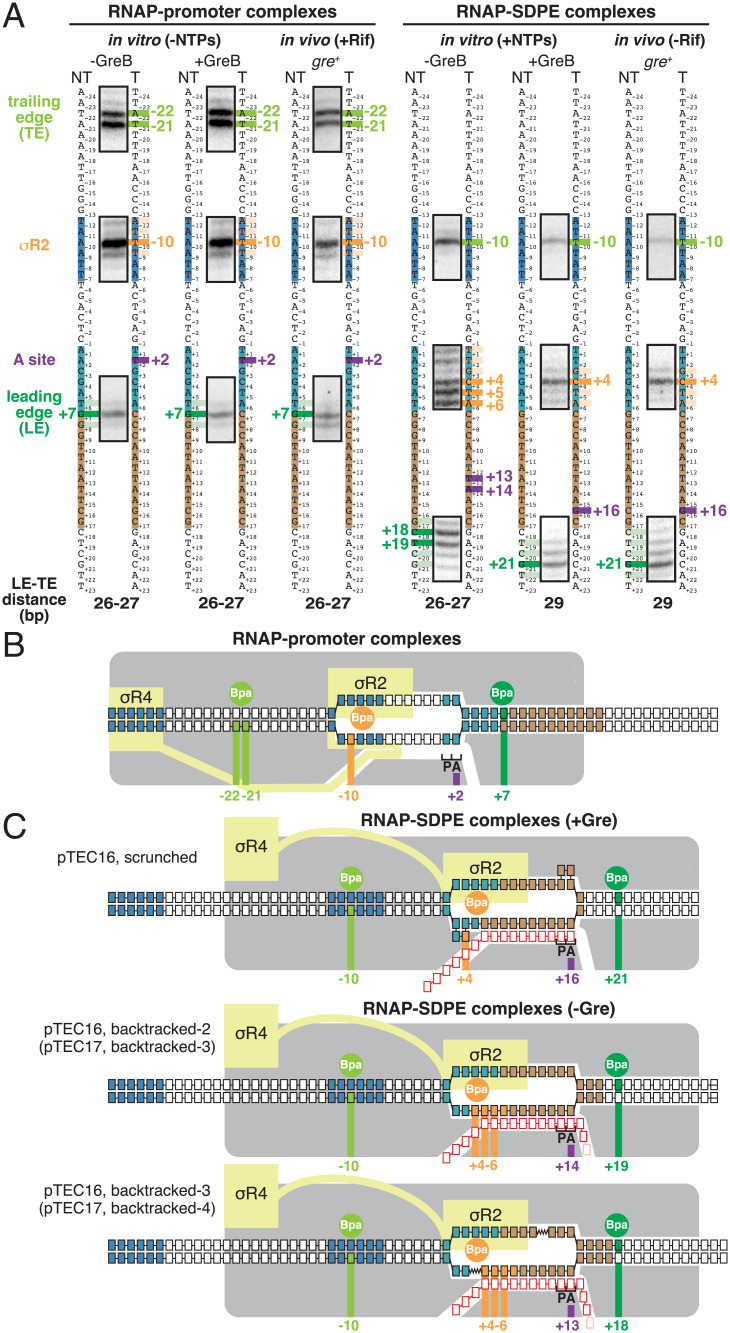
Fig. 3.
Use of site-specific protein–DNA photocrosslinking to define positions of RNAP TEs and LEs and of σ relative to DNA at λPR′: results. (A) Positions of RNAP TEs and LEs and of σR2 in RNAP-promoter complexes (Left) or RNAP-SDPE complexes (pTECs) (Right) at λPR′. For each experimental condition in vitro and in vivo, identified at top, figure shows segments of gel images for primer-extension mapping of crosslinking sites (full gel images in SI Appendix, Figs. S2–S4), nontemplate- and template-strand sequences of λPR′ (to Left and Right, respectively, of gel images; –35 element, –10 element, SDPE, and EPS colored as in Fig. 1A), observed crosslinking sites (olive green for RNAP TE, forest green for RNAP LE, and orange for σR2), inferred positions of RNAP-active-center A site (violet), and inferred modal TE/LE distances. (B) Mechanistic interpretation of data for RNAP-promoter complexes at λPR′ (A, Left). Olive green circle and olive green vertical lines denote Bpa site at RNAP TE and observed crosslinking sites in DNA for Bpa at RNAP TE, forest green circle and forest green vertical line denote Bpa site at RNAP LE and observed crosslinking site in DNA for Bpa at RNAP LE, and orange circle and orange vertical line denote Bpa site in σR2 and observed crosslinking site in DNA for Bpa in σR2. Violet vertical line denotes inferred position of RNAP-active-center A site. Gray, RNAP core; yellow, σ; P and A, RNAP active-center product and addition sites; black boxes with blue fill, –35 element and –10 element nucleotides; black boxes with light blue fill, SDPE nucleotides; black boxes with brown fill, EPS nucleotides; other black boxes, other DNA nucleotides (nontemplate-strand nucleotides above template-strand nucleotides). (C) Mechanistic interpretation of data for RNAP-SDPE complexes at λPR′ (pTEC; A, Right). Three complexes are shown: (1) a scrunched σ-dependent paused complex with 16 nt RNA product and RNAP-active-center A-site at position +16 (pTEC16 scrunched; Top row); (2) a backtracked σ-dependent paused complex with 16- or 17-nt RNA product and RNAP-active-center at position +14 (pTEC16, backtracked-2 or pTEC17, backtracked-3; Center row); and (3) a backtracked σ-dependent paused complex with 16 or 17 nt RNA product and RNAP-active-center at position +13 (pTEC16, backtracked-3 or pTEC17, backtracked-4; Bottom row). Red boxes, RNA nucleotides; pink boxes, additional RNA nucleotide present in 17 nt RNA product. Other colors as in B. Scrunched segments of nontemplate and template DNA strands in pTEC16 shown as bulges.
To assess RPo and pTEC at λPR′ in vitro, we formed transcription complexes in the absence and presence, respectively, of NTPs (Fig. 3A and SI Appendix, Figs. S2–S4). To assess RPo and pTEC at λPR′ in vivo, we performed experiments in the presence and absence, respectively, of the transcription inhibitor rifampin, which blocks extension of RNA beyond a length of 2–3 nt (66, 67) and thus prevents formation of TEC (Fig. 3A and SI Appendix, Figs. S2–S4).
Use of Site-Specific Protein–DNA Photocrosslinking to Define Positions of RNAP TE and LE and of σ Relative to DNA at λPR′: Results.
The crosslinking results for RPo at λPR′ show that the RNAP LE/TE distance is 26–27 bp, consistent with an unscrunched transcription complex (62–64, 68), that the RNAP active-center A site interacts with promoter position +2, and that σR2 interacts with the promoter −10 element (promoter position −10) (Fig. 3 A, Left, and B). The crosslinking pattern for RPo at λPR′ is identical for experiments performed in vitro in the absence of GreB, in vitro in the presence of GreB, and in vivo in gre+ cells (Fig. 3 A, Left, and B).
The crosslinking results for pTEC at λPR′ in vitro in the presence of GreB and in vivo in gre+ cells—where, as shown above, the pTEC is present exclusively as pTEC16 (Fig. 1 C and D)—show that the LE–TE distance is 29 bp, show that the RNAP active-center A site interacts with position +16, and show that σR2 interacts with the SDPE (position +4) (Fig. 3A, Right, columns 2 and 3). The LE–TE distance in pTEC16 in the presence of Gre indicates that the complex contains 2–3 bp of DNA scrunching (LE–TE distance of 29 bp for pTEC16 vs. LE–TE distance of 26–27 bp for RPo). The crosslinking results for the RNAP LE in pTEC16 in the presence of Gre indicate that the RNAP active-center A site has translocated forward by 14 steps, from position +2 in RPo to position +16. The crosslinking results for σR2 in pTEC16 in the presence of Gre indicate that σR2 has disengaged from the promoter –10 element and has engaged with the SDPE. Taken together, the crosslinking results establish that pTEC16 in the presence of Gre is a σ-dependent paused TEC having 2–3 bp of DNA scrunching (Fig. 3C, Top).
The crosslinking results for pTEC at λPR′ in vitro in the absence of Gre—where, as shown above, the pTEC is present as both pTEC16 and pTEC17 (Fig. 1C)—show that the LE–TE distance is 26–27 bp, the RNAP active-center A site interacts with position +13 or +14, and σR2 interacts with the SDPE (positions +4 to +6) (Fig. 3A, Right, column 1). The LE–TE distances in pTEC16 and pTEC17 in the absence of Gre indicate that the complexes are unscrunched (LE–TE distance of 26–27 bp for pTEC16 and pTEC17 in the absence of Gre vs. LE–TE distance of 26–27 bp for RPo). The crosslinking results for the RNAP LE in pTEC16 and pTEC17 in the absence of Gre together with the 16 and 17 nt lengths of the RNA products in those complexes indicate that the RNAP active-center A site first translocated forward by 14 or 15 steps, from position +2 in RPo to position +16 or +17, and then reverse translocated–backtracked by 2–4 bp. The crosslinking results for σR2 in pTEC16 and pTEC17 in the absence of Gre indicate that σR2 has disengaged from the promoter –10 element and has engaged with the SDPE. Taken together, the crosslinking results establish that pTEC16 and pTEC17 in the absence of Gre are σ-dependent paused TECs backtracked by 2–4 bp (Fig. 3C, Center and Bottom).
The RNAP TE position in pTEC16 in the presence of Gre is the same as the RNAP TE positions in pTEC16 and pTEC17 in the absence of Gre (Fig. 3A, Right, columns 2–3 vs. column 1). In contrast, the RNAP LE position in pTEC16 in the presence of Gre differs by 2–3 bp, in a downstream direction, from the RNAP TE positions in pTEC16 and pTEC17 in the absence of Gre (Fig. 3A, Right, columns 2–3 vs. column 1).
A defining hallmark of DNA scrunching is a change in the position of RNAP LE relative to DNA without a corresponding change in the position of the RNAP TE relative to DNA, resulting in a change in LE–TE distance (7, 8, 63, 68). The results in Fig. 3 establish that σ-dependent pTECs formed in vitro or in vivo in the presence of Gre exhibit this defining hallmark of DNA scrunching. Thus, the RNAP LE position in pTEC16 in the presence of Gre is different, by 2–3 bp, from the RNAP LE position in pTEC16 in the absence of Gre (Fig. 3A, Right, columns 2–3 vs. column 1), whereas, in contrast, the RNAP TE position in pTEC16 in the presence of Gre is identical to the RNAP TE position in pTEC16 in the absence of Gre (Fig. 3A, Right, columns 2–3 vs. column 1).
Sequence Determinants for Scrunching in σ-Dependent Pausing.
It has been hypothesized that scrunching occurs in σ-dependent pausing and has been further hypothesized that DNA sequences downstream of the SDPE modulate pause lifetime through effects on DNA scrunching (10, 55–59). The results in Fig. 3 establish that scrunching occurs in σ-dependent pausing at λPR′ and provide an experimental approach—namely, crosslinking of the RNAP LE to DNA—that enables scrunched states and unscrunched states in σ-dependent pausing to be distinguished for any DNA sequence containing an SDPE.
In a next set of experiments, we applied this experimental approach to a library of DNA sequences containing the λPR′ SDPE and all ∼16,000 possible 7-bp DNA sequences spanning the λPR′ pause site (λPR′ positions +14 to +20; Fig. 4A) to assess whether DNA sequences downstream of the SDPE determine scrunching in σ-dependent pausing and, if so, to define the sequence determinants (Fig. 4).
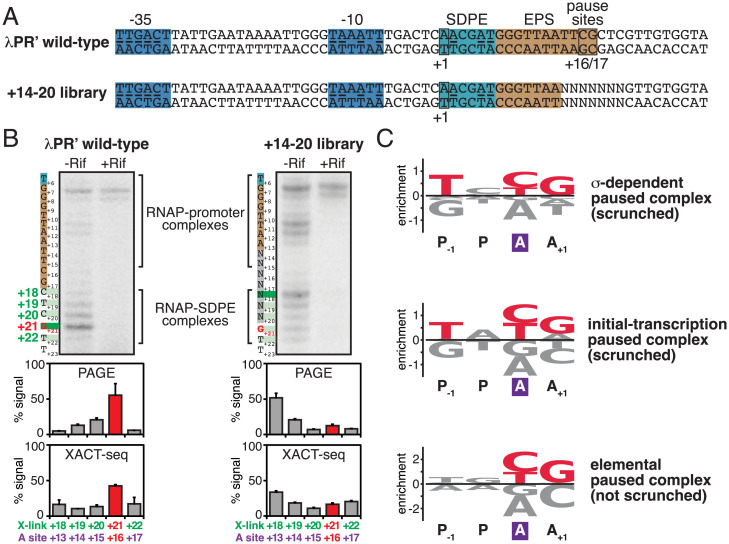
Fig. 4.
Sequence determinants for scrunching in σ-dependent pausing. (A) DNA templates containing wild-type λPR′ or +14 to +20 library. NNNNNNN, randomized nucleotides of +14 to +20 library. Other colors as in Fig. 1A. (B) Positions of RNAP LE in wild-type λPR’ and +14 to +20 library in vivo. Top, PAGE analysis of crosslinking. For each experimental condition, identified at top, figure shows gel image for primer-extension mapping of crosslinking sites, nontemplate-strand sequence (to left of gel image; SDPE and EPS colored as in Fig. 1A), and observed crosslinking sites (forest green). Center, quantitation (mean ± SD) of PAGE analysis of crosslinking. Bottom, quantitation (mean ± SD) of XACT-seq analysis of crosslinking. In all subpanels, the observed major crosslinking site for pTEC at λPR′ (position +21) and inferred major RNAP-active-center A-site position for pTEC at λPR′ (position +16) are highlighted in red. (C) Sequence logos quantifying formation and/or stability of scrunched σ-dependent paused complex (Top; this work); formation and/or stability of scrunched initial-transcription paused complex (Center) (65); and elemental pausing in transcription elongation (Bottom) (69–72). Positions are labeled relative to RNAP-active-center A site (violet rectangle) and P-site. Red, most highly preferred DNA nucleotides. Logos were generated using Logomaker (93) as described in SI Appendix, Materials and Methods.
First, using procedures analogous to those of the previous section, we transformed gre+ cells producing RNAP holoenzyme having Bpa incorporated at the RNAP LE (β′ residue 1148) with a plasmid carrying λPR′ or, in parallel, plasmids carrying sequences from the “+14 to +20 library,” and we then ultraviolet-irradiated cells to initiate RNAP-DNA photocrosslinking, lysed cells, isolated crosslinked material, and mapped crosslinks by primer extension and urea-PAGE (Fig. 4B). For λPR′, the results show that the RNAP LE crosslinks predominantly at position +21, indicating that the RNAP active-center A site interacts predominantly with positions +16, as expected, based on the results in the preceding section, for the scrunched σ-dependent paused complex at λPR′ (Figs. 3 and 4B, Left). In contrast, for the +14 to +20 library, the RNAP LE crosslinks predominantly at position +18, indicating that the RNAP active-center A site interacts predominantly with position +13, as expected for unscrunched complexes (such as, for example, the backtracked σ-dependent paused complexes of Fig. 3C). We conclude that positions +14 to +20 of λPR′ contains sequence information crucial for formation and/or stability of the scrunched state during σ-dependent pausing, consistent with previous proposals (57, 58). We further conclude, from comparison of the yield of the scrunched state for the +14 to +20 library vs. for λPR′ (∼10% vs. ∼50%; Fig. 4B, Center), that only a fraction, ∼1/5, of the ∼16,000 possible sequences at positions +14 to +20 support formation and/or stability of the scrunched state during σ-dependent pausing.
Second, in order to identify sequence determinants that influence formation and/or stability of the scrunched state during σ-dependent pausing, we performed deep sequencing of the primer-extension products of the preceding paragraph, following the xlinking-of-active-center-to-template-sequencing (XACT-seq) procedure of Winkelman et al. (65) (Fig. 4B, Bottom). Consistent with the PAGE analysis of the preceding paragraph, the XACT-seq analysis shows that ∼10% of sequence reads corresponded to the scrunched σ-dependent paused complex (i.e., sequence reads for which RNAP LE crosslinking occurred at position +21, indicating interaction of the RNAP-active-center A site with position +16) (Fig. 4B, Bottom). Analysis of this subset of sequence reads showed clear sequence preferences, yielding the consensus sequence T+14N+15Y+16G+17, or, expressed in terms of RNAP-active-center A-site and P-site positions, TP−1NPYAGA+1 (where N is any nucleotide and Y is pyrimidine) (Fig. 4C, Top, and SI Appendix, Fig. S5).
λPR′ contains a perfect match to the consensus sequence T+14N+15Y+16G+17 (Fig. 4A). λPR′ positions +16 and +17 have been shown to affect pause capture efficiency and pause lifetime (58). Our results indicate that the sequence at positions +16 and +17 affects formation and/or stability of the scrunched state of the σ-dependent paused complex at λPR′, and are consistent with the view that the effects of sequence on pause capture efficiency and pause lifetime are consequences of the effects of sequence on formation and/or stability of the scrunched state.
This consensus sequence obtained in this work for formation and/or stability of a scrunched σ-dependent paused complex (TP–1NPYAGA+1; Fig. 4C, Top) is identical to the consensus sequence obtained in previous work for formation and/or stability of a scrunched initial-transcription paused complex (TP–1NPYAGA+1; Fig. 4C, Middle) (65). The consensus sequence also is similar to the downstream, most highly conserved portion of the consensus sequence obtained in previous work for elemental pausing in transcription elongation (YAGA+1; Fig. 4C, Bottom) (69–72); however, the consensus sequences for the scrunched σ-dependent paused complex and scrunched initial transcription paused complex show a substantially stronger conservation of TP–1 than the consensus sequence for elemental pausing in transcription elongation (Fig. 4C).
Strand-Dependence of Sequence Determinants for Scrunching in σ-Dependent Pausing.
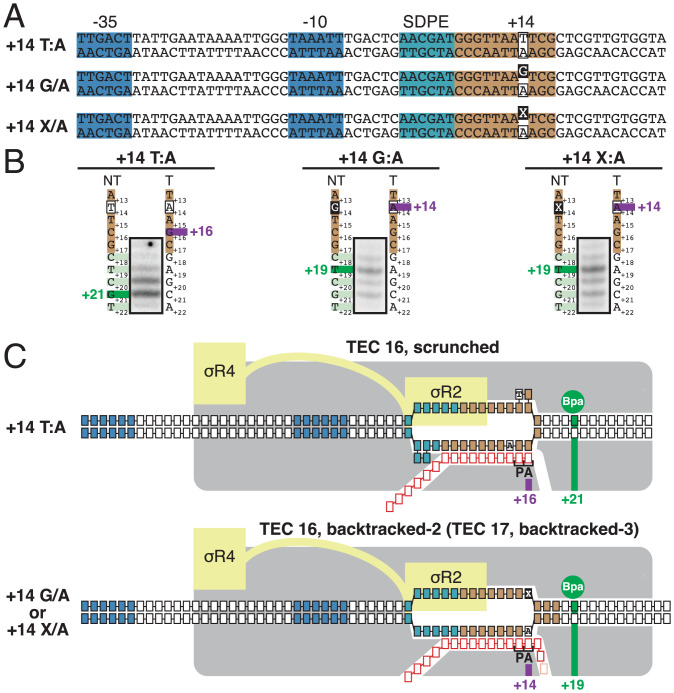
As described in the preceding section, the consensus sequence for formation of a scrunched σ-dependent paused complex contains a strongly conserved T:A base pair at position P-1 (T on the nontemplate stand; A on the template strand) that also is strongly conserved in the consensus sequence for formation of a scrunched initial-transcription complex but that is not strongly conserved in the consensus sequence for elemental pausing in transcription elongation (Fig. 4C). To determine whether specificity at this position resides in the nontemplate-strand T or the template-strand A, we analyzed formation of the scrunched state in σ-dependent pausing, in vitro, in the presence of GreB, using heteroduplex templates in which the nontemplate-strand T was replaced by a nonconsensus nucleotide or an abasic site, and the template-strand A was unchanged (Fig. 5A). The results show that the presence of a nonconsensus nucleotide or an abasic site on the nontemplate strand at position P-1 abrogates formation of the scrunched state in σ-dependent pausing (Fig. 5B). We conclude that the sequence information responsible for the preference for T:A at position P-1 resides, at least in part, in the DNA nontemplate strand (Fig. 5C).
Fig. 5.
Strand-dependence of sequence determinants for scrunching in σ-dependent pausing. (A) λPR′ promoter derivatives containing consensus nucleotide T at nontemplate strand position +14 (Top row; +14 T/A), nonconsensus nucleotide G at nontemplate strand position +14 (Center row; +14 G/A), or abasic site at nontemplate strand position +14 (Bottom row; +14 X/A). Raised black-filled box, nonconsensus nucleotide, or abasic site. Other colors as in Fig. 1A. (B) Positions of RNAP LE on λPR′ promoter derivatives of A. For each promoter derivative, figure shows gel image for primer-extension mapping of crosslinking sites, nontemplate- and template-strand sequences (to left and right of gel image; EPS colored as in Fig. 1A), observed crosslinking sites (forest green), and inferred RNAP-active-center A-site position. (C) Mechanistic interpretation of data in B. Colors as in Fig. 3 B and C.
Structural Basis of Scrunching in σ-Dependent Pausing.
Structures of σ-containing TECs have been reported previously (18, 19), but a structure of a scrunched, paused, σ-containing transcription elongation complex, such as the species defined in results in Figs. 3–5, has not been reported previously. To determine the structural basis of σ-dependent pausing, we performed cryo-EM structure determination, analyzing a σ-dependent pTEC prepared in solution. We incubated a synthetic nucleic-acid scaffold containing the λPR′ promoter, a consensus SDPE positioned as in λPR′, and a 15-bp noncomplementary region corresponding to the transcription bubble of the σ-dependent pTEC at λPR′ (Fig. 6A) with E. coli RNAP σ70 holoenzyme, and we applied samples to grids, flash-froze samples, and performed single-particle-reconstruction cryo-EM (Fig. 6 and SI Appendix, Fig. S6). Reconstitution of complexes from synthetic nucleic-acid scaffolds containing a noncomplementary region corresponding to the transcription bubble and a preannealed RNA oligomer corresponding to the RNA product enables preparation of homogeneous, defined TECs, even TECs that are formed at low abundance or that have low stabilities or short lifetimes, when formed by promoter-dependent transcription initiation on fully complementary DNA (18, 19, 38, 73–79).
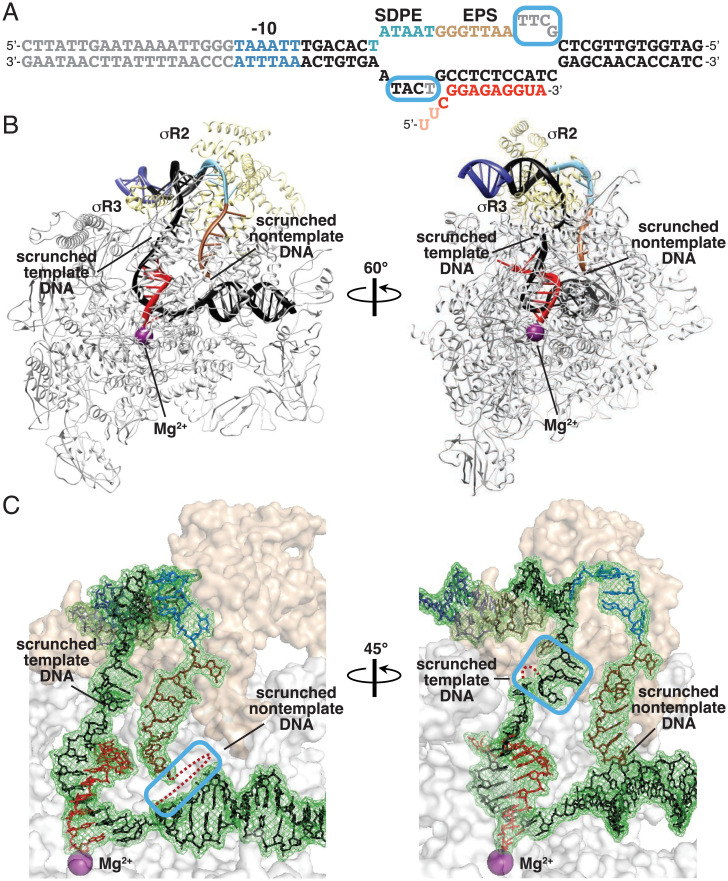
Fig. 6.
Structural basis for scrunching in σ-dependent pausing. (A) Nucleic-acid scaffold. DNA, black (–10 element, SDPE, EPS, and disordered nucleotides in blue, light blue, brown, and gray, respectively; noncomplementary region corresponding to unwound transcription bubble indicated by raised and lowered letters); RNA, red (disordered nucleotides in pink); cyan boxes, nontemplate- and template-strand DNA nucleotides disordered or repositioned due to DNA scrunching. (B) Cryo-EM structure of scrunched σ-dependent paused TEC (pTEC; two orthogonal view orientations). Violet sphere, RNAP-active-center catalytic Mg2+. Other colors as in A. (C) Cryo-EM density and atomic model, showing interactions of RNAP and σ with DNA and RNA. Cyan boxes, nontemplate-strand (Left) and template-strand (Right) DNA nucleotides disordered or repositioned due to DNA scrunching; red dots, DNA nucleotides disordered due to DNA scrunching. Other colors as in A.
The cryo-EM structure of pTEC has an overall resolution of 3.8 Å (SI Appendix, Fig. S6). Map quality is high, with ordered, traceable, density for the following: RNAP, σR2, σR3, 13 bp of upstream double-stranded (dsDNA), all except 4 nt of the nontemplate strand of the transcription bubble, all except 1 nt of the template strand of the transcription bubble, 13 bp of downstream dsDNA, and 12 nt of RNA, corresponding to the 12-nt segment closest to the RNA 3′ end (Fig. 6 B and C).
The cryo-EM structure of pTEC shows a σ-containing transcription elongation complex in an RNAP posttranslocated state (Fig. 6 B and C). σR2 interacts with the SDPE in pTEC, making the same sequence-specific protein–DNA interactions as made by σR2 with the −10 element in RPo and RPitc, and σR3 interacts with the DNA segment immediately upstream of the SDPE in pTEC, making the same protein–DNA interactions as made by σR3 in RPo and RPitc (Fig. 6B) (11–13, 80, 81). The transcription-bubble length is 16 bp, corresponding to the 15-bp noncomplementary region and one additional melted base pair immediately downstream of the noncomplementary region (Fig. 6 B and C). The structure of pTEC shows 3 bp of DNA scrunching as compared to the structure of RPo (16 bp unwound vs. 13 bp unwound) (12, 80–82) (SI Appendix, Fig. S7A). Scrunching in pTEC results in disorder of four nontemplate-strand nucleotides at the downstream edge of the transcription bubble (λPR′ positions +14 through +17; Fig. 6C and SI Appendix, Fig. S7A, Left). Scrunching in pTEC also results in disorder of one template-strand nucleotide and repositioning of three template-strand nucleotides in the upstream part of the transcription bubble, between upstream dsDNA and the RNA-DNA hybrid (λPR′ positions +5 and +2 through +4) (Fig. 6C and SI Appendix, Fig. S7A, Right).
In RPo, σR2 having Bpa incorporated at position 448 crosslinks more strongly to the template-stand nucleotide at the third position of the –10 element than to the template-stand nucleotide at the fourth position of the –10 element (Fig. 3 A and B and SI Appendix, Fig. S3), whereas, in pTEC, σR2 having Bpa incorporated at position 448 crosslinks less strongly to the template-stand nucleotide at the third position of the SDPE than to the template-stand nucleotide at the fourth position of the SDPE (Fig. 3 A and B). Comparison of the structures of RPo and pTEC explains this difference in σR2-DNA crosslinking in RPo and pTEC (SI Appendix, Fig. S7B). Namely, in RPo, residue 448 of σR2 is closer to the nucleotide at the third position of the –10 element (∼12 Å vs. ∼19 Å; SI Appendix, Fig. S7B, Left), whereas, as a result of DNA scrunching and repositioning of template-strand nucleotides in pTEC, residue 448 of σR2 is closer to the fourth position of the SDPE (∼13 Å vs. ∼15 Å; SI Appendix, Fig. S7B, Right).
The nontemplate strand of the nucleic-acid scaffold used to obtain the structure of pTEC contained a perfect match to the consensus sequence for formation of scrunched σ-dependent paused complexes and the consensus sequence for formation of scrunched initial-transcription paused complexes (positions +14 to +17; Figs. 4C, Top and Middle, and 6A). The structure of pTEC shows that the nontemplate-strand nucleotide that is strongly specified in the consensus sequences for scrunched σ-dependent paused complexes and scrunched initial-transcription paused complexes (Fig. 4C, Top and Middle)—but that is not strongly specified in the consensus sequence for elemental pausing in transcription elongation (Fig. 4C, Bottom)—is the first nucleotide of the 4-nt nontemplate-strand segment that is disordered due to DNA scrunching (position +14; Fig. 6 B and C and SI Appendix, Fig. S7A, Left). The fact that this nucleotide is strongly specified in the consensus sequences for scrunched σ-dependent paused complexes and scrunched initial-transcription paused complexes (Fig. 4C, Top and Middle)—but is not strongly specified in the consensus sequence for elemental pausing in transcription elongation, which does not involve DNA scrunching (Fig. 4C, Bottom) (75)—together with the observation that this nucleotide is the first nucleotide in the 4-nt nontemplate-strand segment repositioned due to DNA scrunching, suggests that specificity at this position is associated with DNA scrunching and reflects sequence-dependent differences in the ability to accommodate DNA scrunching. Confirming the hypothesis that specificity at this position reflects sequence-dependent differences in the ability to accommodate DNA scrunching will require additional structural or biochemical information.
The structure of the σ-containing pTEC shows RNAP in a posttranslocated state (Fig. 6 B and C), rather than in the half-translocated state that has been observed in structures of a non–σ-containing, RNA-hairpin–containing pTEC (75, 79). The difference in observed translocational states may reflect a real difference in the predominant translocational state for σ-dependent pausing vs. non–σ-dependent pausing, or may reflect only an apparent difference, arising from the presence of a posttranslocation-favoring, RNA-DNA hybrid sequence in the σ-containing pTEC structure, arising from different selections among translocational states present in the equilibrium population (34), or arising from different sensitivities of σ-containing and non–σ-containing pTECs to a noncomplementary transcription-bubble region.
Discussion
Our results 1) establish, through mapping of positions of the RNAP LEs and TEs relative to DNA, that σ-dependent pausing at λPR′ involves a scrunched state with 2–3 bp of DNA scrunching and an unscrunched, backtracked state (Fig. 3); 2) define a consensus sequence for formation and/or stability of a scrunched σ-dependent paused complex (TP–1NPYAGA+1) that is identical to the consensus sequence for formation and/or stability of a scrunched initial-transcription paused complex (TP–1NPYAGA+1) and similar to the consensus sequence for elemental pausing in transcription elongation (YAGA+1) (Fig. 4); 3); show the position that is strongly specified in the consensus sequences for the scrunched σ-dependent paused complex and the scrunched initial-transcription paused complex, but not in the consensus sequence for elemental pausing in transcription elongation (TP–1), is recognized through the nontemplate DNA strand (Fig. 5); and 4) provide an atomic structure of a scrunched σ-dependent paused complex that suggests specificity at position TP–1 is associated with DNA scrunching and reflects sequence-dependent differences in the ability to accommodate DNA scrunching (Fig. 6).
Our results provide direct evidence for the hypothesis that, in σ-dependent pausing at λPR′, following pause capture, the pTEC extends RNA by 3–4 nt using a scrunching mechanism, and for the hypothesis that the resulting pTECs with extended RNA can collapse to yield a backtracked state (10, 41, 55–59).
DNA scrunching has been shown to mediate translocation of the RNAP active center relative to DNA in transcription-start-site selection, initial transcription, and promoter escape during transcription initiation (7–9, 63, 64, 68, 83). DNA stepping has been thought to mediate translocation of the RNAP active center relative to DNA during transcription elongation (29). Our results establish that this distinction between the mechanisms of RNAP-active-center translocation during transcription initiation and transcription elongation is not absolute, showing that DNA scrunching mediates translocation of the RNAP active center under certain circumstances in pausing and pause escape during transcription elongation. The shared feature of the transcription complexes that engage in DNA scrunching during transcription initiation and the transcription complexes shown here to engage in DNA scrunching in pausing and pause escape during transcription elongation is the presence of sequence-specific σ-DNA interactions that anchor the TE of RNAP relative to DNA, preventing RNA extension through a DNA stepping mechanism, and thereby necessitating RNA extension through a DNA scrunching mechanism.
Generalizing from this observation, we propose that DNA scrunching occurs in pausing and pause escape during transcription elongation whenever sequence-specific σ-DNA interaction anchors the TE of RNAP relative to DNA, including, for example, whenever a σ-containing TEC encounters an SDPE in a transcribed region (e.g., σ-dependent pausing at sequences other than λPR′ SDPE) (41, 44–48, 54, 84, 85), or whenever a sequence-specific DNA-binding protein that interacts with RNAP engages a σ-containing TEC in a transcribed region [e.g., transcription antitermination factor Q at a Q-binding element upstream of an SDPE (57), or a transcription activator protein, such as catabolite activator protein (CAP), able to interact with RNAP from an appropriately positioned DNA site upstream of an SDPE].
Generalizing further, we suggest that DNA scrunching occurs in pausing and pause escape during transcription elongation in any circumstance in which any sequence-specific protein–DNA interaction anchors the TE of RNAP relative to DNA. Examples potentially include 1) pausing induced by RfaH, which binds to a σ-free TEC and makes sequence-specific protein–DNA interactions with the transcription-bubble non–template-DNA strand similar to the sequence-specific interactions between σ and an SDPE (76, 86); 2) pausing induced by other NusG/RfaH-family transcription factors (87), such as Bacillus subtilis NusG, that bind to a σ-free TEC and make sequence-specific protein–DNA interactions with the transcription-bubble nontemplate-DNA strand similar to the sequence-specific interactions between σ and an SDPE (88, 89); 3) pausing induced by other factors that bind to a TEC and make sequence-specific protein–DNA interactions with an appropriately positioned upstream DNA site; and 4) pausing induced by sequence-specific protein–DNA interaction between RNAP α subunit C-terminal domain (α-CTD) and an appropriately positioned upstream DNA site.
We suggest that the DNA scrunching that occurs in pausing and pause escape during transcription elongation—like the DNA scrunching that occurs in initial transcription and promoter escape during transcription initiation (1, 7–10)—serves as the mechanism to capture and store the free energy required to break the sequence-specific protein–DNA interactions that anchor RNAP on DNA. DNA scrunching during pausing enables capture of free energy from multiple nucleotide additions and stepwise storage of the captured free energy in the form of stepwise increases in the amount of DNA unwinding. Upon rewinding of the upstream part of the unwound DNA (e.g., the SDPE in σ-dependent pausing), the free energy captured and stored during scrunching is accessed to drive pause escape.
We note that the approaches of this work—specifically, mapping of positions of the RNAP LEs and TEs relative to DNA using Bpa-modified RNAP—will enable direct determination whether, and if so how, DNA scrunching occurs in each of the above circumstances.
Materials and Methods
Protein–DNA photocrosslinking was performed as in Winkelman et al. (63) and Yu et al. (64). XACT-seq was performed as in Winkelman et al. (65), and the resulting data were analyzed using custom Python scripts. The cryo-EM structure was determined using single-particle reconstruction. Full details of methods are presented in the SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
Cryo-EM data were collected at the Rutgers University Cryo-EM and Nanoimaging Facility and the University of Michigan Life Sciences Institute Cryo-EM Facility. Cryo-EM data analysis were performed with assistance from Chengyuan Wang. We thank Jeremy Bird for providing GreB. Work was supported by National Institutes of Health (ES030791 to M.S., GM133777 to J.B.K., GM041376 to R.H.E., and GM118059 to B.E.N.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201301119/-/DCSupplemental.
Data Availability
Sequencing reads, atomic model, and map data have been deposited in NIH/NCBI Sequence Read Archive, Protein Data Bank, and Electron Microscopy Data Bank (90–92).
References
- 1.Winkelman J. T., Nickels B. E., Ebright R. H., “The transition from transcription initiation to transcription elongation: Start-site selection, initial transcription, and promoter escape” in RNA Polymerase as a Molecular Motor, Landick R., Wang J., Strick T. R., Eds. (RSC Publishing, Cambridge, UK, ed. 2, 2021). [Google Scholar]
- 2.Chen J., Boyaci H., Campbell E. A., Diverse and unified mechanisms of transcription initiation in bacteria. Nat. Rev. Microbiol. 19, 95–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazumder A., Kapanidis A. N., Recent advances in understanding σ70-dependent transcription initiation mechanisms. J. Mol. Biol. 431, 3947–3959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruff E. F., M. T. Record, Jr, Artsimovitch I., Initial events in bacterial transcription initiation. Biomolecules 5, 1035–1062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feklístov A., Sharon B. D., Darst S. A., Gross C. A., Bacterial sigma factors: A historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Paget M. S., Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules 5, 1245–1265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapanidis A. N., et al. , Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margeat E., et al. , Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys. J. 90, 1419–1431 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Revyakin A., Liu C., Ebright R. H., Strick T. R., Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts J. W., Biochemistry. RNA polymerase, a scrunching machine. Science 314, 1097–1098 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Li L., Molodtsov V., Lin W., Ebright R. H., Zhang Y., RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription. Proc. Natl. Acad. Sci. U.S.A. 117, 5801–5809 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo Y., Steitz T. A., Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell 58, 534–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu R. S., et al. , Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 289, 24549–24559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami K. S., Masuda S., Darst S. A., Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science 296, 1280–1284 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Mekler V., et al. , Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 108, 599–614 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Mooney R. A., Darst S. A., Landick R., Sigma and RNA polymerase: An on-again, off-again relationship? Mol. Cell 20, 335–345 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Nickels B. E., et al. , The interaction between σ70 and the β-flap of Escherichia coli RNA polymerase inhibits extension of nascent RNA during early elongation. Proc. Natl. Acad. Sci. U.S.A. 102, 4488–4493 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J., et al. , Structural basis of Q-dependent transcription antitermination. Nat. Commun. 10, 2925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin Z., Kaelber J. T., Ebright R. H., Structural basis of Q-dependent antitermination. Proc. Natl. Acad. Sci. U.S.A. 116, 18384–18390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-Nahum G., Nudler E., Isolation and characterization of σ(70)-retaining transcription elongation complexes from Escherichia coli. Cell 106, 443–451 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Deighan P., Pukhrambam C., Nickels B. E., Hochschild A., Initial transcribed region sequences influence the composition and functional properties of the bacterial elongation complex. Genes Dev. 25, 77–88 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harden T. T., et al. , Bacterial RNA polymerase can retain σ70 throughout transcription. Proc. Natl. Acad. Sci. U.S.A. 113, 602–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapanidis A. N., et al. , Retention of transcription initiation factor σ70 in transcription elongation: Single-molecule analysis. Mol. Cell 20, 347–356 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Mooney R. A., et al. , Regulator trafficking on bacterial transcription units in vivo. Mol. Cell 33, 97–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay J., et al. , Translocation of σ(70) with RNA polymerase during transcription: Fluorescence resonance energy transfer assay for movement relative to DNA. Cell 106, 453–463 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Raffaelle M., Kanin E. I., Vogt J., Burgess R. R., Ansari A. Z., Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol. Cell 20, 357–366 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Reppas N. B., Wade J. T., Church G. M., Struhl K., The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell 24, 747–757 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Shimamoto N., Kamigochi T., Utiyama H., Release of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase depends mainly on time elapsed after the start of initiation, not on length of product RNA. J. Biol. Chem. 261, 11859–11865 (1986). [PubMed] [Google Scholar]
- 29.Abbondanzieri E. A., Greenleaf W. J., Shaevitz J. W., Landick R., Block S. M., Direct observation of base-pair stepping by RNA polymerase. Nature 438, 460–465 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belogurov G. A., Artsimovitch I., The mechanisms of substrate selection, catalysis, and translocation by the elongating RNA polymerase. J. Mol. Biol. 431, 3975–4006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erie D. A., Yager T. D., von Hippel P. H., The single-nucleotide addition cycle in transcription: A biophysical and biochemical perspective. Annu. Rev. Biophys. Biomol. Struct. 21, 379–415 (1992). [DOI] [PubMed] [Google Scholar]
- 32.Larson M. H., Landick R., Block S. M., Single-molecule studies of RNA polymerase: One singular sensation, every little step it takes. Mol. Cell 41, 249–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L. R., Landick R., “Substrate loading, nucleotide addition, and translocation by RNA polymerase” in RNA Polymerases as Molecular Motors, Buc H., Strick S., Eds. (Royal Society of Chemistry, London, UK, 2009). [Google Scholar]
- 34.Kang J. Y., Mishanina T. V., Landick R., Darst S. A., Mechanisms of transcriptional pausing in bacteria. J. Mol. Biol. 431, 4007–4029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nudler E., Mustaev A., Lukhtanov E., Goldfarb A., The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89, 33–41 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Lerner E., et al. , Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 113, E6562–E6571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Artsimovitch I., Landick R., Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl. Acad. Sci. U.S.A. 97, 7090–7095 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komissarova N., Kashlev M., Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. U.S.A. 94, 1755–1760 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landick R., Transcriptional pausing as a mediator of bacterial gene regulation. Annu. Rev. Microbiol. 75, 291–314 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Ring B. Z., Yarnell W. S., Roberts J. W., Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell 86, 485–493 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Perdue S. A., Roberts J. W., Σ(70)-dependent transcription pausing in Escherichia coli. J. Mol. Biol. 412, 782–792 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Artsimovitch I., Post-initiation control by the initiation factor σ. Mol. Microbiol. 68, 1–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petushkov I., Esyunina D., Kulbachinskiy A., Possible roles of σ-dependent RNA polymerase pausing in transcription regulation. RNA Biol. 14, 1678–1682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldman S. R., Nair N. U., Wells C. D., Nickels B. E., Hochschild A., The primary σ factor in Escherichia coli can access the transcription elongation complex from solution in vivo. eLife 4, e10514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nickels B. E., Mukhopadhyay J., Garrity S. J., Ebright R. H., Hochschild A., The σ 70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat. Struct. Mol. Biol. 11, 544–550 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Brodolin K., Zenkin N., Mustaev A., Mamaeva D., Heumann H., The σ 70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nat. Struct. Mol. Biol. 11, 551–557 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Hatoum A., Roberts J., Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol. Microbiol. 68, 17–28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mooney R. A., Landick R., Tethering sigma70 to RNA polymerase reveals high in vivo activity of sigma factors and sigma70-dependent pausing at promoter-distal locations. Genes Dev. 17, 2839–2851 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts J. W., et al. , Antitermination by bacteriophage lambda Q protein. Cold Spring Harb. Symp. Quant. Biol. 63, 319–325 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Yarnell W. S., Roberts J. W., The phage lambda gene Q transcription antiterminator binds DNA in the late gene promoter as it modifies RNA polymerase. Cell 69, 1181–1189 (1992). [DOI] [PubMed] [Google Scholar]
- 51.Grayhack E. J., Yang X. J., Lau L. F., Roberts J. W., Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell 42, 259–269 (1985). [DOI] [PubMed] [Google Scholar]
- 52.Guo H. C., Kainz M., Roberts J. W., Characterization of the late-gene regulatory region of phage 21. J. Bacteriol. 173, 1554–1560 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goliger J. A., Roberts J. W., Sequences required for antitermination by phage 82 Q protein. J. Mol. Biol. 210, 461–471 (1989). [DOI] [PubMed] [Google Scholar]
- 54.Sun Z., Yakhnin A. V., FitzGerald P. C., Mclntosh C. E., Kashlev M., Nascent RNA sequencing identifies a widespread σ70-dependent pausing regulated by Gre factors in bacteria. Nat. Commun. 12, 906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marr M. T., Roberts J. W., Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell 6, 1275–1285 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Zhilina E., Esyunina D., Brodolin K., Kulbachinskiy A., Structural transitions in the transcription elongation complexes of bacterial RNA polymerase during σ-dependent pausing. Nucleic Acids Res. 40, 3078–3091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strobel E. J., Roberts J. W., Regulation of promoter-proximal transcription elongation: Enhanced DNA scrunching drives λQ antiterminator-dependent escape from a σ70-dependent pause. Nucleic Acids Res. 42, 5097–5108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strobel E. J., Roberts J. W., Two transcription pause elements underlie a σ70-dependent pause cycle. Proc. Natl. Acad. Sci. U.S.A. 112, E4374–E4380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bird J. G., Strobel E. J., Roberts J. W., A universal transcription pause sequence is an element of initiation factor σ70-dependent pausing. Nucleic Acids Res. 44, 6732–6740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borukhov S., Sagitov V., Goldfarb A., Transcript cleavage factors from E. coli. Cell 72, 459–466 (1993). [DOI] [PubMed] [Google Scholar]
- 61.Chin J. W., Martin A. B., King D. S., Wang L., Schultz P. G., Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winkelman J. T., et al. , Crosslink mapping at amino acid-base resolution reveals the path of scrunched DNA in initial transcribing complexes. Mol. Cell 59, 768–780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winkelman J. T., et al. , Multiplexed protein-DNA cross-linking: Scrunching in transcription start site selection. Science 351, 1090–1093 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu L., et al. , The mechanism of variability in transcription start site selection. eLife 6, e32038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winkelman J. T., et al. , XACT-Seq comprehensively defines the promoter-position and promoter-sequence determinants for initial-transcription pausing. Mol. Cell 79, 797–811.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McClure W. R., Cech C. L., On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 253, 8949–8956 (1978). [PubMed] [Google Scholar]
- 67.Carpousis A. J., Gralla J. D., Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J. Mol. Biol. 183, 165–177 (1985). [DOI] [PubMed] [Google Scholar]
- 68.Winkelman J. T., Chandrangsu P., Ross W., Gourse R. L., Open complex scrunching before nucleotide addition accounts for the unusual transcription start site of E. coli ribosomal RNA promoters. Proc. Natl. Acad. Sci. U.S.A. 113, E1787–E1795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herbert K. M., et al. , Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell 125, 1083–1094 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larson M. H., et al. , A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vvedenskaya I. O., et al. , Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science 344, 1285–1289 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imashimizu M., et al. , Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 16, 98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sidorenkov I., Komissarova N., Kashlev M., Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol. Cell 2, 55–64 (1998). [DOI] [PubMed] [Google Scholar]
- 74.Vassylyev D. G., Vassylyeva M. N., Perederina A., Tahirov T. H., Artsimovitch I., Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Kang J. Y., et al. , RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell 69, 802–815.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang J. Y., et al. , Structural basis for transcript elongation control by NusG family universal regulators. Cell 173, 1650–1662.e1614 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang C., et al. , Structural basis of transcription-translation coupling. Science 369, 1359–1365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Webster M. W., et al. , Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Guo X., et al. , Structural basis for NusA stabilized transcriptional pausing. Mol. Cell 69, 816–827.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y., et al. , Structural basis of transcription initiation. Science 338, 1076–1080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feng Y., Zhang Y., Ebright R. H., Structural basis of transcription activation. Science 352, 1330–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bae B., Feklistov A., Lass-Napiorkowska A., Landick R., Darst S. A., Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife 4, e08504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vvedenskaya I. O., et al. , Interactions between RNA polymerase and the core recognition element are a determinant of transcription start site selection. Proc. Natl. Acad. Sci. U.S.A. 113, E2899–E2905 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shikalov A. B., Esyunina D. M., Pupov D. V., Kulbachinskiy A. V., Petushkov I. V., The σ24 subunit of Escherichia coli RNA polymerase can induce transcriptional pausing in vitro. Biochemistry (Mosc.) 84, 426–434 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Petushkov I., Esyunina D., Kulbachinskiy A., σ38-dependent promoter-proximal pausing by bacterial RNA polymerase. Nucleic Acids Res. 45, 3006–3016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Artsimovitch I., Landick R., The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203 (2002). [DOI] [PubMed] [Google Scholar]
- 87.Wang B., Artsimovitch I., Nus G., NusG, an ancient yet rapidly evolving transcription factor. Front. Microbiol. 11, 619618 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yakhnin A. V., Kashlev M., Babitzke P., NusG-dependent RNA polymerase pausing is a frequent function of this universally conserved transcription elongation factor. Crit. Rev. Biochem. Mol. Biol. 55, 716–728 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yakhnin A. V., Murakami K. S., Babitzke P., Nus G., NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J. Biol. Chem. 291, 5299–5308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.C. Pukhrambam, B. E. Nickels, Structural and mechanistic basis of sigma-dependent transcriptional pausing. NIH/NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA797396/. Deposited 14 January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.V. Molodtsov, M. Su, R. H. Ebright, Escherichia coli σ70-dependent paused transcription elongation complex. Protein Data Bank. https://www.rcsb.org/structure/7N4E. Deposited 4 June 2021. [Google Scholar]
- 92.V. Molodtsov, M. Su, R. H. Ebright, Escherichia coli σ70-dependent paused transcription elongation complex. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-24148. Deposited 4 June 2021. [Google Scholar]
- 93.Tareen A., Kinney J. B., Logomaker: Beautiful sequence logos in Python. Bioinformatics 36, 2272–2274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads, atomic model, and map data have been deposited in NIH/NCBI Sequence Read Archive, Protein Data Bank, and Electron Microscopy Data Bank (90–92).