Significance
Pneumococcal infections are major contributors to morbidity and mortality worldwide. Introduction of pneumococcal conjugated vaccines (PCVs) into the childhood vaccination program has led to a decrease in invasive pneumococcal disease (IPD) in vaccinated children but concurrently to an increase of nonvaccine-type IPD, also in nonvaccinated age groups such as the elderly. Thus, novel vaccine approaches are urgently needed, especially for the elderly, targeting all pneumococci causing IPD. Here, we show that pneumococcal membrane particles (MPs) evoke a serotype-independent cross-protection against IPD. This protection is dependent on the presence of the two conserved lipoproteins MalX and PrsA. We suggest that MPs can be used for pneumococcal vaccine development.
Keywords: Streptococcus pneumoniae, pneumococci, serotype-independent vaccine, membrane particles, membrane vesicles
Abstract
Pneumococcal conjugate vaccines (PCVs) used in childhood vaccination programs have resulted in replacement of vaccine-type with nonvaccine-type pneumococci in carriage and invasive pneumococcal disease (IPD). A vaccine based on highly conserved and protective pneumococcal antigens is urgently needed. Here, we performed intranasal immunization of mice with pneumococcal membrane particles (MPs) to mimic natural nasopharyngeal immunization. MP immunization gave excellent serotype-independent protection against IPD that was antibody dependent but independent of the cytotoxin pneumolysin. Using Western blotting, immunoprecipitation, mass spectrometry, and different bacterial mutants, we identified the conserved lipoproteins MalX and PrsA as the main antigens responsible for cross-protection. Additionally, we found that omitting the variable surface protein and vaccine candidate PspA from MPs enhanced protective immune responses to the conserved proteins. Our findings suggest that MPs containing MalX and PrsA could serve as a platform for pneumococcal vaccine development targeting the elderly and immunocompromised.
The pneumococcus (Streptococcus pneumoniae) is a major cause of morbidity and mortality globally, being the most common cause of respiratory tract infections such as otitis and sinusitis, but also a major contributor to more severe diseases such as pneumonia without or with sepsis and meningitis (i.e., invasive pneumococcal disease [IPD]) (1, 2). Risk groups for acquiring pneumococcal infections include young children, the elderly, immunocompromised individuals, and those with a prior influenza A virus infection. Current pneumococcal vaccines target a limited number of the 100 pneumococcal capsular serotypes identified so far (3–5). First, polysaccharide-based vaccines were launched (PPV23), but due to limited protection in the risk groups, so-called pneumococcal conjugate vaccines (PCVs) were introduced where the sugars are conjugated to a protein carrier to provide a stronger antibody response. Since 2010, 10-valent (PCV10) and 13-valent (PCV13) vaccines are used worldwide in childhood vaccination programs. PCV introduction has led to a significant decrease in IPD in vaccinated children and a reduction in nasopharyngeal carriage of vaccine type (VT) strains in healthy children (6–11). However, pneumococcal carriage rates in children have remained the same due to replacement of VT with nonvaccine type (NVT) strains (8, 9). This also resulted in an expansion of NVTs among IPD cases in nonvaccinated age groups, affecting the efficacy of the PCVs (12). Thus, in 2016 the IPD incidence among the elderly in Sweden did not decrease after PCV introduction, and more than 70% of cases were caused by NVTs (12). PCV13 includes an additional three serotypes not included in PCV10 (i.e., serotypes 3, 6A, and 19A). So far, its efficacy against IPD caused by serotype 3 is being debated (13), and in Sweden, serotype 3 now constitutes a major serotype among IPD cases in the elderly (8, 12). This underlines the importance of finding a new vaccine against serotype 3. Recently, a 20-valent vaccine was approved by the US Food and Drug Administration for adults, including PCV13 serotypes as well as serotypes 8, 10A, 11A, 12F, 15B, 22F, and 33F. Other vaccine approaches include protein-based vaccines. Recently, a study was published in which five pneumococcal candidates were identified and validated as immunogens. Zangari et al. (14) found protection against pneumococcal colonization using a nonencapsulated strain, but not against the encapsulated variant, suggesting that the capsule might affect mucosal protection.
Like other gram-positive bacteria, pneumococci release extracellular vesicles (EVs) derived from their cytoplasmic membrane (15, 16). It has been suggested that colonization events during childhood generate serum antibodies to pneumococcal antigens (17). Several of these antigens relate to proteins that recently were shown to be enriched in purified EVs from liquid-grown bacteria (18, 19). We argue that EVs produced during colonization represent major vehicles for immunogens that may generate protective immunity toward carriage and IPD. Furthermore, we previously showed that pneumococcal EVs can be internalized by host cells such as respiratory epithelial and dendritic cells (DCs) and can lead to DC activation and release of proinflammatory cytokines (18). Also, in vivo experiments have shown that intramuscular immunization of mice with pneumococcal EVs protect mice after homologous serotype challenge (20).
Here, we show that spontaneously produced pneumococcal membrane particles (MPs) isolated from plate-grown bacteria provide a serotype-independent cross-protection against IPD in mice that is antibody dependent but is not dependent on the pneumococcal cytotoxin pneumolysin (Ply). We identify the major protective antigens for cross-protection as the two conserved membrane-bound lipoproteins MalX and PrsA, which are both needed for full protection. We also suggest that the highly variable immunodominant surface protein PspA, which differs between pneumococcal serotypes, may mask immune responses to conserved membrane-bound proteins that could be used as antigens in vaccines.
Results
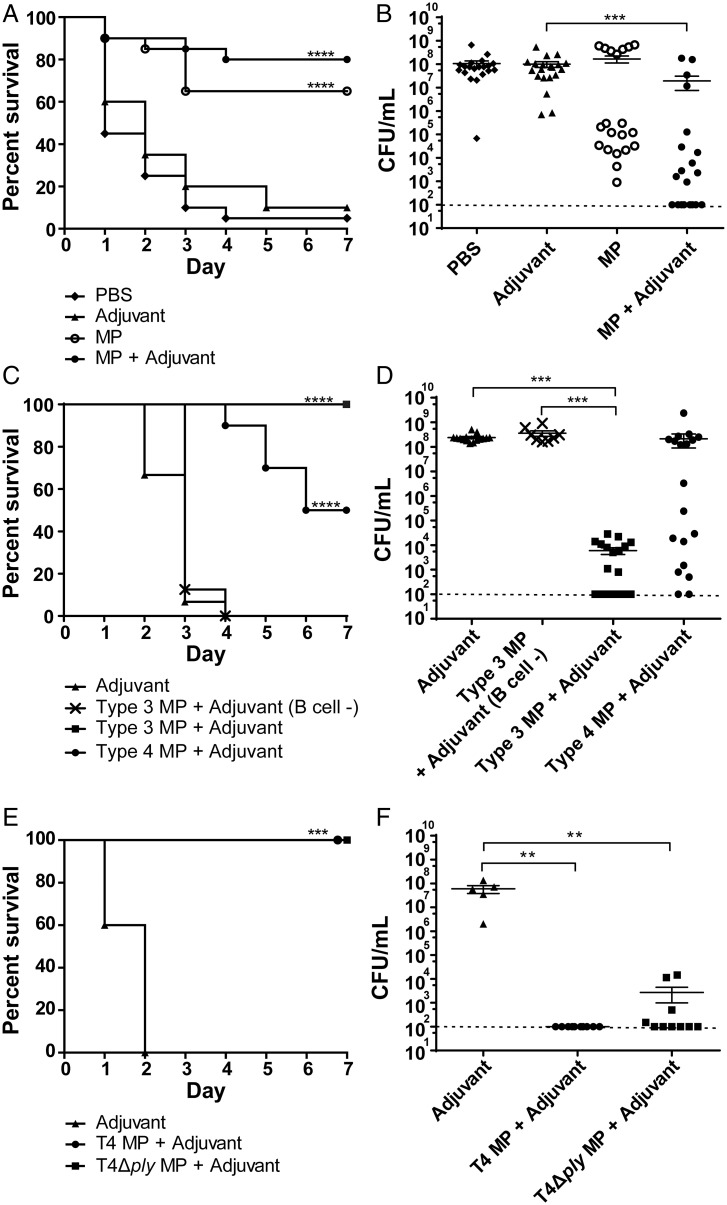
MPs from Plate-Grown Pneumococci Evoke a Serotype-Independent Cross-Protection against Invasive Pneumococcal Infection.
Vesicles were isolated from plate-grown pneumococci as a proxy for biofilm-growing pneumococci during nasopharyngeal carriage and were called MPs (SI Appendix, Fig. S1 provides characteristics). Wild-type (WT) mice (C57BL/6) were immunized twice intranasally with MPs isolated from the strain T4 (TIGR4) (21), alone or combined with the adjuvant aluminum hydroxide (used in humans, but not generally for mucosal immunization) or with phosphate-buffered saline (PBS) or adjuvant alone for the control groups. After 4 wk, mice were infected intranasally with 5 × 106 colony-forming units (CFUs) of the serotype 1 strain BHN733. Mice were followed for 7 d, and survival was recorded. Heterologous serotype challenge with immunization using MP+adjuvant from T4 and infection with serotype 1 conferred 80% survival. Also, in the absence of adjuvant, we observed significant protection (65% survival) (Fig. 1A). Bacterial loads in the lungs were consistent with the survival data, with lower bacterial numbers in immunized mice compared to control mice treated with adjuvant only (Fig. 1B). Bacterial CFUs in the blood were monitored daily until sacrifice (SI Appendix, Fig. S2 A–D). For the PBS and adjuvant groups, only mice without bacteremia survived. Seven of the mice vaccinated with MP+adjuvant cleared the bacteria from the blood, suggesting that even though the immunization was intranasal, immune clearance occurred systemically. In summary, intranasal immunization with MPs gives a serotype-independent cross-protection against IPD.
Fig. 1.
Intranasal immunization with MPs confers protection against intranasal pneumococcal infection. (A and B) T4 MPs confer cross-protection against infection with a serotype 1 strain. (A) Intranasal immunization of C57BL/6 mice with MPs from strain T4 of serotype 4 and subsequent infection with strain BHN733 of serotype 1 resulted in 80% survival; 20 mice per group. (B) Bacteria (in CFUs) in the lungs of immunized and infected mice in A after sacrifice. Each dot represents one mouse. (C and D) Immunization with MPs of serotype 3 confers antibody-dependent protection against intranasal infection with the same strain. (C) Percentage of mice that survived the intranasal infection with serotype 3 bacteria of WT C57BL/6 mice or of B cell–deficient mice (muMt knockout mice) immunized with MPs from the same serotype 3 strain BHN428 or of WT mice immunized with MPs from the serotype 4 strain T4; 20 mice per group. (D) Number of bacteria (CFUs) in the lungs of mice from C at sacrifice. Each dot represents one mouse. (E and F) Protection conferred by MPs against intranasal pneumococcal challenge is not dependent on the cholesterol-binding cytotoxin Ply. (E) Percentage survival after intranasal infection with T4 of WT C57BL/6 mice immunized with MPs from T4 or its Ply-deficient strain T4Δply; 10 mice per group, 5 mice immunized with adjuvant as control. (F) Number of bacteria (CFUs) in the lungs of mice at sacrifice. Each dot represents one mouse. Error bars represent the standard error of the mean (SEM). Dashed horizontal line in (B, D, and F) represents the limit of detection for the lung CFU counts using this method. **P < 0.01; ***P < 0.001; ****P < 0.0001.
The experiments were done using MPs (and not EVs isolated from liquid cultures) since intranasal immunization of mice with MPs from strain T4 gave higher protection and higher antibody levels after immunization than what was observed using EVs from the same strain (Fig. 1 A and B and SI Appendix, Figs. S2 A–D and S3).
MPs from Serotype 3 Pneumococci Evoke Serotype-Independent and Antibody-Dependent Protection.
Since there is an urgent need for a vaccine against serotype 3 (12), we purified and characterized MPs from the serotype 3 strain BHN428. These MPs were larger than those from T4, with diameters up to 850 nm, and they possessed more cytosolic proteins and fewer predicted cell wall–associated and secreted proteins (SI Appendix, Fig. S1). Mice were immunized intranasally with MPs of serotype 3. In the 7-d survival experiment, all mice immunized with MPs of serotype 3+adjuvant survived, while adjuvant-treated control mice all succumbed within 4 d (Fig. 1C). Immunized mice showed lower bacterial numbers in the lungs and blood compared to adjuvant-treated mice (Fig. 1D and SI Appendix, Fig. S2 E and F). Immunization using MPs from T4 and challenge with serotype 3 bacteria (BHN428) provided 50% cross-protection (Fig. 1C), and the bacterial load in the lungs of surviving mice tended to be lower than that of the adjuvant control and those that succumbed to infection (Fig. 1D). The 10 mice that showed lower lung counts had no detectable bacteria in the blood (SI Appendix, Fig. S2G). These data show that MPs of type 3 showed full protection against homologous challenge in mice. Furthermore, the data suggest that T4 MPs evoke cross-protection against genetically distant pneumococcal lineages, such as those of serotype 3.
To investigate whether protection is antibody dependent, we immunized B cell–deficient mice (muMt knockout mice) with MPs from serotype 3 and challenged them intranasally with the same strain. In the 7-d survival experiment, none of the B cell–deficient mice survived, demonstrating that protection is largely antibody dependent (Fig. 1C). Bacterial numbers in the lungs and blood were high and similar to what was detected in WT mice given adjuvant only and were significantly higher than in WT mice immunized with the same MPs (Fig. 1D and SI Appendix, Fig. S2 F–H). Thus, our data suggest that intranasal immunization with MPs provides antibody-mediated protection.
Ply Is Not Essential for the Protection Evoked by MPs.
The pneumococcal cytotoxin Ply has been discussed as a vaccine candidate because Ply-deficient strains are highly attenuated in mice infection models (22, 23). However, the potential toxic effects of Ply may be an obstacle in a protein-based pneumococcal vaccine (24). We therefore investigated whether protection is Ply dependent using MPs isolated from an isogenic strain lacking Ply (T4Δply). Mice immunized with MPs from T4Δply and challenged with T4 showed 100% protection (Fig. 1E). Also, these mice showed CFUs in the lungs similar to those of mice immunized with MPs from T4 and had no bacteria in the blood (Fig. 1F and SI Appendix, Fig. S2 I–K). We conclude that Ply is not crucial for protection against IPD mediated by MPs.
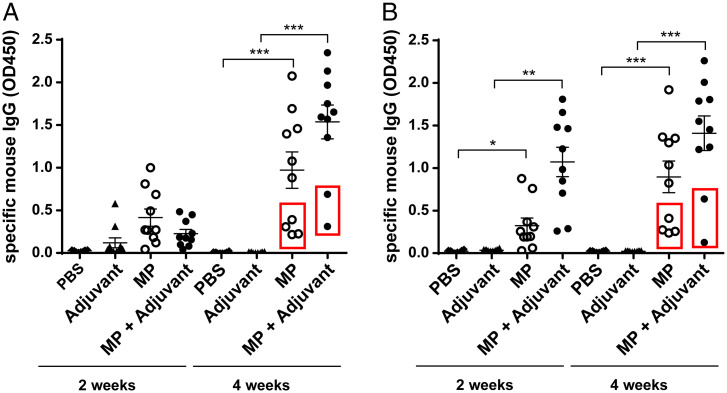
MP Immunization Leads to Pneumococcal-Specific Antibodies against Noncapsular Antigens.
Next, we analyzed the presence of pneumococcal-specific antibodies in sera of mice immunized intranasally with MPs from T4. Using enzyme-linked immunosorbent assay (ELISA), we detected immunoglobulin (Ig)G specific for T4 in sera from mice 2 wk and 4 wk after immunization, and the IgG levels were already higher in immunized mice than in adjuvant and PBS control groups 2 wk after immunization and were significantly higher after 4 wk (Fig. 2A). Furthermore, we detected IgG with affinity for the nonencapsulated mutant strain T4R, thus indicating reactivity to noncapsular antigens (Fig. 2B).
Fig. 2.
Intranasal immunization with MPs elicits production of pneumococcal-specific IgG to noncapsular antigens. Mice were immunized intranasally with MPs from strain T4. Using ELISA, pneumococcal-specific IgG was detected in wells coated with (A) WT T4 or (B) T4R (isogenic mutant in the capsule of T4 bacteria). The low responder mice in the MP+Adjuvant and MP groups are those that did not survive after serotype 1 challenge. These mice are highlighted in the graphs within red borders. Each dot represents one mouse. Error bars represent the standard error of the mean (SEM). *P < 0.05; **P < 0.01; ***P < 0.001.
The Membrane-Bound Pneumococcal Lipoproteins MalX and PrsA Are Responsible for the Cross-Protection Evoked by MPs.
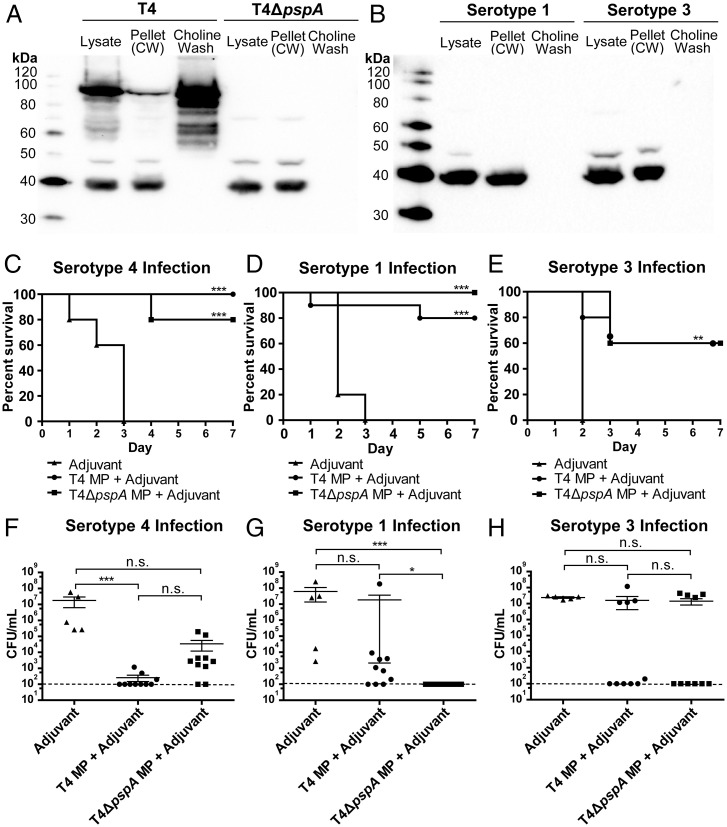
To identify which pneumococcal proteins in MPs are responsible for protection, T4 was grown in liquid medium and then washed with elevated concentrations of choline chloride to release choline-binding proteins (CBPs) that associate noncovalently with the cell surface. Western blot analyses of the cell pellets and supernatants of choline-washed cells were performed using sera from immunized mice (with T4 MPs+adjuvant) as the source of primary antibodies. A major band of ∼80 kDa was observed mainly in the supernatant fraction, which was absent in a T4ΔpspA mutant, thus identifying it as the highly immunogenic choline-binding pneumococcal surface protein PspA (Fig. 3A). PspA has been described as a putative vaccine candidate against IPD and colonization (23), but its sequence is highly variable among clinical strains, which likely explains why no PspA bands were observed in choline-washed samples from BHN733 (serotype 1) and BHN428 (serotype 3) (Fig. 3B) (25, 26). However, two additional protein bands of slightly less than 40 and 50 kDa were observed in the lysates and pellet fractions, respectively, of choline-washed cells of T4, T4ΔpspA and the serotype 1 and 3 strains (Fig. 3 A and B).
Fig. 3.
Cross-protection against IPD is not dependent on PspA. (A and B) Western blot analysis of bacterial lysates, supernatant samples from choline-washed cells (treated with 5% choline chloride to release CBPs) and from lysates of corresponding choline-washed cells (Pellet CW). Binding of sera from mice immunized with T4 MP+Adjuvant (used as primary antibody) to (A) T4 and T4ΔpspA and (B) serotype 1 and serotype 3 cells were analyzed. Sera came from mice included in experiments represented in Fig. 1 A and B. (C–E) Percentage of mice immunized with either T4 MPs or T4ΔpspA MPs that survived an intranasal infection with (C) T4, (D) serotype 3 (BHN428), or (E) serotype 1 (BHN733) bacteria; 10 mice per group (the group immunized with T4ΔpspA MPs and infected with serotype 1 included 9 mice), 5 mice/pneumococcal strain immunized with adjuvant as control. (F–H) Number of bacteria (CFUs) in the lungs of mice infected with (F) T4, (G) serotype 1 (BHN733), or (H) serotype 3 (BHN428) bacteria at sacrifice. Each dot represents one mouse. The Dashed horizontal line (F–H) represents the limit of detection for the lung CFU counts using this method. Error bars represent the standard error of the mean (SEM). *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant.
To study the role played by PspA in protection by MPs, we immunized mice with MPs from the mutant strain T4ΔpspA and challenged them with T4. Immunized mice showed 80% protection (Fig. 3C), suggesting that antigens other than PspA provide protection. Indeed, when mice immunized with T4ΔpspA MPs were challenged with the serotype 1 strain BHN733, a 100% cross-protection was observed, reinforcing that the protective antigens in MPs were neither PspA nor the capsular polysaccharide (Fig. 3D). Similar results were obtained when mice were challenged with serotype 3 pneumococci (BHN428). Here, 60% protection was observed with T4ΔpspA MPs, which was in the same range as for T4 MPs (Fig. 3E). For all three infection experiments, the bacterial loads in the lungs (Fig. 3 F–H) and blood (SI Appendix, Fig. S4A) were low in surviving mice. ELISAs and coating with serotype 1, 3, and 4 pneumococci, respectively, showed higher serotype 1– and 3–specific IgG titers in mice immunized with MPs from T4ΔpspA compared to T4 MPs (SI Appendix, Fig. S4B).
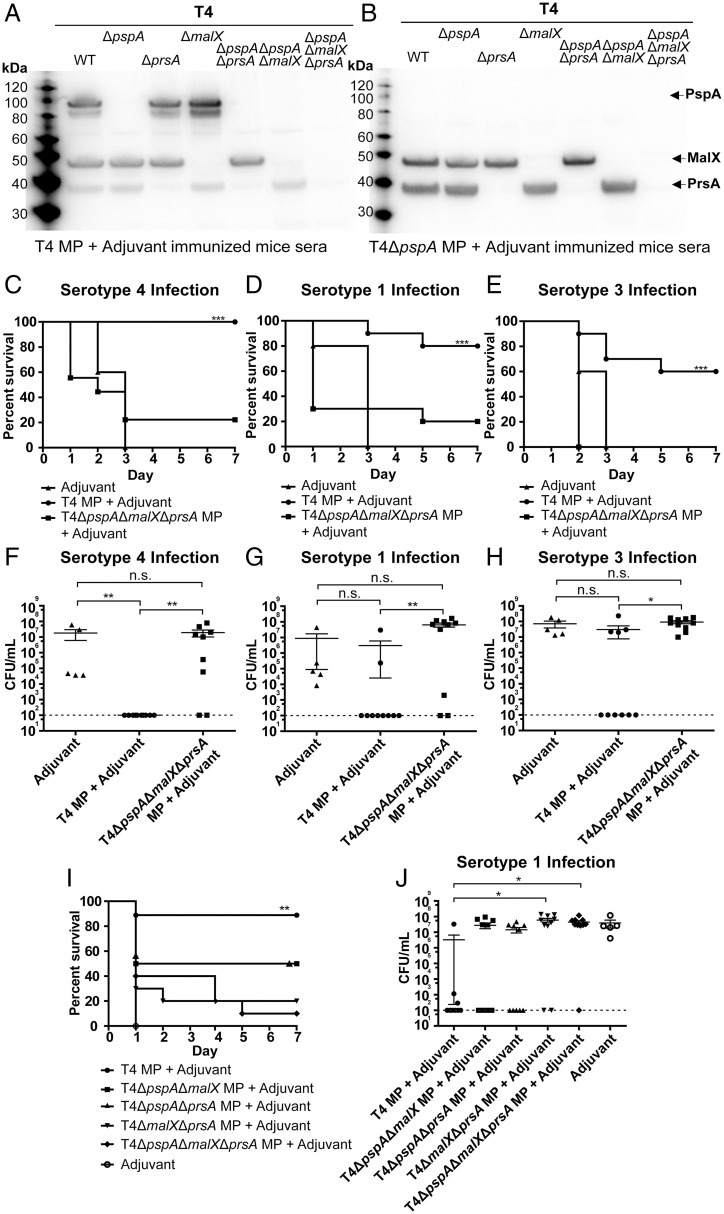
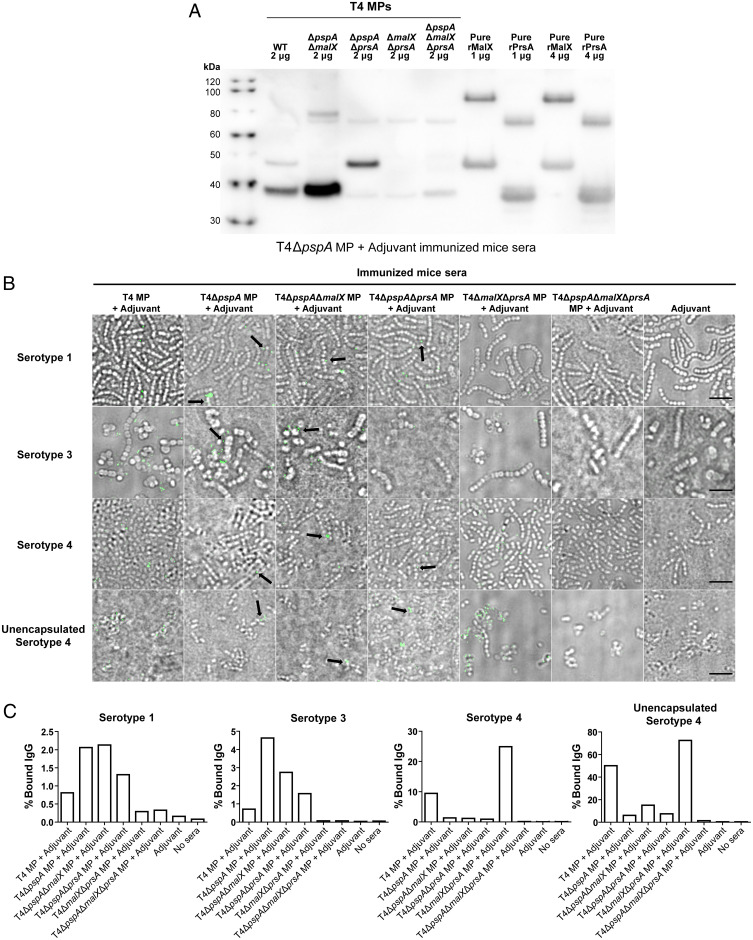
Since we found that PspA in MPs was not important for heterologous serotype protection, we next performed a series of immunoprecipitation (IP) experiments to capture additional putative antigens using sera from mice immunized with MPs from T4+adjuvant and T4ΔpspA+adjuvant or from adjuvant-treated mice as a control. Major proteins recognized by immune sera by Western blot analysis in lysates of T4 were subjected to mass spectrometry analysis for protein identification. Importantly, the hits with the highest scores from IP using sera from T4ΔpspA MP-immunized mice were two lipoproteins, PrsA and MalX. These proteins were not detected in IP with sera from the adjuvant control. As expected, PspA was identified as one of the top hits in the IP with sera from T4 MPs, but MalX was also enriched with these sera, albeit with a lower score (SI Appendix, Tables S1–S3). Additionally, the masses of MalX and PrsA corresponded to the immunoreactive bands of ∼40 and ∼50 kDa that we observed in the lysates and cell pellet fractions by Western blot analysis, as addressed earlier in this section (Fig. 3 A and B). To further validate that PspA, PrsA, and MalX were the major reactive proteins from the immune sera, we complemented the IP experiment with Western blot analyses in which lysates blotted from WT T4 and mutants in pspA, prsA, and malX were incubated with sera from mice immunized with MPs from T4 or T4ΔpspA (Fig. 4 A and B). Indeed, sera from T4 MP-immunized mice reacted with the three major proteins, and absence of these protein bands in the corresponding mutant background confirmed these proteins to be PspA, PrsA, and MalX (Fig. 4A). Thus, although PrsA was not enriched in IP with the T4 MP-immune sera, the Western blot analysis clearly showed that these sera still reacted to this protein. Likewise, using sera from mice immunized with T4ΔpspA MPs revealed two major bands of slightly less than 40 and 50 kDa, absent in mutants having a prsA or a malX deletion, thus confirming the identity of these proteins (Fig. 4B).
Fig. 4.
Cross-protection against IPD is mediated by the lipoproteins PrsA and MalX. (A and B) Validation of the results from the IP experiment and mass spectrometry analysis confirming the identity of the major immunoreactive proteins. Lysates from T4 WT; single mutants T4ΔpspA, T4ΔprsA, and T4ΔmalX; double mutants T4ΔpspAΔprsA and T4ΔpspAΔmalX; and triple mutant T4ΔpspAΔmalXΔprsA as indicated were subjected to Western blot analysis. PVDF membranes were incubated with immune sera from mice immunized with (A) T4 MP+Adjuvant or (B) T4ΔpspA MP+Adjuvant. Sera came from mice included in experiments presented in Fig. 3 C–E. A second incubation with sheep anti-mouse IgG–HRP conjugated facilitated chemiluminescent detection. (C–E) Percentage of mice immunized with either T4 MPs or T4ΔpspAΔmalXΔprsA MPs that survived an intranasal infection with (C) T4, (D) serotype 1 (BHN733), or (E) serotype 3 (BHN428) bacteria; 10 mice per group (the group immunized with T4ΔpspAΔmalXΔprsA MPs and infected with T4 included 9 mice). Five mice/pneumococcal strain were immunized with adjuvant as control. (F–H) CFUs in the lungs of mice infected with (F) T4, (G) serotype 1 (BHN733), or (H) serotype 3 (BHN428) bacteria at sacrifice. Dashed horizontal line in (F–H) represents the limit of detection for the lung CFU counts using this method. (I) Percentage of mice immunized with T4, T4ΔpspAΔmalX, T4ΔpspAΔprsA, T4ΔmalXΔprsA, orT4ΔpspAΔmalXΔprsA MPs that survived an intranasal infection with serotype 1 strain BHN733, showing both MalX and PrsA are required for high percentage of survival. (J) CFUs in the lungs of mice from the experiment in I; 10 mice per group (except for T4 MP + Adjuvant, which had 9 mice). Five mice/pneumococcal strain were immunized with adjuvant as control. Each dot represents one mouse. Error bars represent the standard error of the mean (SEM). *P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant.
To further investigate if the identified PrsA and MalX proteins were responsible for the cross-protection, we first isolated MPs from a triple mutant lacking both prsA and malX in the T4ΔpspA background, T4ΔpspAΔprsAΔmalX. Mice were immunized with either MPs from T4+adjuvant or from the triple mutant+adjuvant and then infected intranasally with either T4, BHN733 (serotype 1), or BHN428 (serotype 3). Immunization with T4 MPs showed again 100%, 80%, and 60% protection against these strains, respectively, confirming the previous results (Fig. 3 C–E). However, when MPs from T4ΔpspAΔprsAΔmalX were used for immunization, mice showed only 20% protection toward T4 and type 1 (BHN733) and no protection toward type 3 (BHN428) (Fig. 4 C–E). Lung and blood CFUs were in accordance with these findings, as were IgG titers (Fig. 4 F–H and SI Appendix, Fig. S5 A and B).
To study whether cross-protection depends on both lipoproteins, three new mutant strains were created in strain T4 where pspA and malx, or pspA and prsA, or malX and prsA were deleted (Fig. 4 I and J and SI Appendix, Fig. S6 A–F). MPs from the three strains were used to immunize mice, and then the mice were challenged with a serotype 1 strain. The data suggest that both MalX and PrsA are needed for cross-protection since mice immunized with MPs from either of the mutant strains showed lower survival than mice immunized with MPs from the WT strain (Fig. 4I). Also, the bacterial numbers in the lungs and blood were consistent with the survival data (Fig. 4J and SI Appendix, Fig. S6 A–F). We conclude that both MalX and PrsA are needed in MPs for cross-protection against a pneumococcal infection.
MP-Raised Antibodies Recognize PrsA and MalX in MPs Better Than Recombinant Mature Proteins.
Sera from mice immunized with MPs isolated from T4ΔpspA were used in Western blots after sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) separation of MP-proteins and recombinant PrsA and MalX (Fig. 5 A and B). More PrsA- and MalX-specific antibodies bound the native proteins present in MPs compared to the respective recombinant protein. This was despite the much lower content of the antigen present in MPs compared to the recombinant form of the protein lacking the amino-terminal sequence and lacking lipid modification of the terminal cysteine residue required for the formation of membrane-bound lipoproteins.
Fig. 5.
MP-raised antibodies recognize PrsA and MalX in MPs better than recombinant proteins. (A) Western blot analysis of T4 WT and mutant vesicles as well as recombinant proteins rMalX and rPrsA, using sera from mice immunized with T4ΔpspA MP+Adjuvant (from experiment in Fig. 3), showing a stronger signal of antibody binding against vesicles compared to the pure recombinant protein lacking the signal peptide. (B) Microscopy images of antibody binding against pneumococci of serotypes 1, 3, and 4 and unencapsulated serotype 4 (T4R), using sera from mice immunized with different mutant vesicles (obtained in experiments in Figs. 3 and 4), followed by incubation with anti-mouse secondary antibody labelled with Alexa Fluor 488, emitting the green fluorescence signal. While PspA antibodies represent a majority of the antibodies binding to the serotype 4 surface (since most of the signal was depleted when using T4ΔpspA MPs), this is not the case with serotypes 3 and 4, where the signal is comparable when vesicles with or without PspA were used for immunization. Black arrows highlight preferential antibody binding to division sites using sera from mice immunized with vesicles lacking PspA, indicating that MalX and PrsA might be exposed in the septum region. (Scale bar, 5 μm.) (C) Quantification of the binding of IgG from immunized mice to S. pneumoniae bacteria by flow cytometry. Bacterial cells were incubated with sera from mice immunized with MPs harvested from the indicated pneumococcal strains, cells bound to mouse IgGs stained with anti-mouse IgG antibody and counted by flow cytometry. The percentages of stained cells are plotted. At least 5,000 events (cells) were counted per condition. “No sera” indicates control samples where bacteria were not incubated in mice sera. Corresponding dot plots and gatings from the flow cytometry analyses are displayed in SI Appendix, Fig. S7.
Sera from mice immunized with MPs from WT serotype 4 and its mutant derivatives contained antibodies that bound to the surface of serotype 1, 3, and 4 bacteria if the MPs contained MalX, PrsA, or both. PspA, after T4 MP immunization, represented the main antibody target on the serotype 4 surface, as most of the signal was depleted when using T4ΔpspA MPs (Fig. 5B). Also, capsular expression decreased binding (Fig. 5 B and C and SI Appendix, Fig. S7).
To investigate the opsonophagocytic activity of the cross-reactive antibodies, serotype 1 pneumococci were incubated with sera from mice immunized with T4 MP+adjuvant and subsequently incubated with RAW mouse macrophages. The ratio of adhered bacteria was significantly higher when pneumococci were incubated with sera from mice immunized with MP+adjuvant compared to adjuvant alone (SI Appendix, Fig. S8A). Also, these pneumococci with better adherence were significantly less able to survive inside the cells compared to pneumococci incubated with control serum (SI Appendix, Fig. S8B).
MalX and PrsA Are Conserved Lipoproteins in S. pneumoniae.
To investigate the conservation of prsA and malX, we performed a BLAST search of the nucleotide sequences of these genes against the annotated PubMLST database containing 8,351 pneumococcal genomes belonging to several serotypes and sequence types (27, 28). For prsA encoding a membrane-bound lipoprotein acting as a cis-trans prolyl-isomerase, 99.5% of the pneumococcal genomes (8,325/8,351 genomes) had a prsA sequence more than 98% identical to the T4 sequence. Similarly, for malX encoding a lipoprotein acting as an ABC maltose transporter, 98.5% of the genomes (8,318/8,351) had a malX sequence more than 98% identical to the T4 sequence. This indicates a very high sequence conservation of these two genes across different strains and serotypes of pneumococci. However, single-nucleotide polymorphisms leading to amino acid replacements were found.
Discussion
At any time point, up to 30 to 60% of healthy preschool children are colonized by S. pneumoniae in the nasopharynx. It is believed that pneumococcal colonization is an immunization event, explaining why nasopharyngeal colonization decreases with age (29). Sera taken from healthy adults frequently contain antibodies directed against pneumococcal membrane proteins, many of which are highly conserved between pneumococcal strains (19). S. pneumoniae produces spontaneously, during growth on solid and in liquid media, membrane protrusions across the cell wall that pinch off from the bacterial cell without observed lysis, resulting in membrane vesicles of different sizes confined by the plasma membrane and enclosing a cytosolic cargo (SI Appendix, Fig. S1) (18). We suggest that such membrane protrusions are spontaneously produced during pneumococcal colonization. As MPs contain only low levels of capsular polysaccharide and cell wall material, their plasma membranes are readily targeted by complement C3 deposition, facilitating uptake into antigen-presenting cells (18). MPs may contain the entire set of known membrane-bound proteins, including lipoproteins, hydrophilic proteins anchored to the outer leaflet of the plasma-membrane through their N-terminal lipid moiety attached to a conserved cysteine (SI Appendix, Fig. S1) (18). In addition, many CBPs are also found on MPs, probably associated with the choline residues decorating the membrane-associated lipoteichoic acids. Many membrane-bound proteins have specialized localizations, such as those involved in septum formation and cell division (30). We hypothesize that the superior immuno-protective effects found here of MPs isolated from solid agar plates over EVs, isolated from liquid-growing cells (SI Appendix, Fig. S3), could be that the former are enriched for membrane proteins localized to the septum region, whereas the latter are also the result of vesicle formation following bacterial autolysis.
There is an urgent need for a new pneumococcal vaccine approach against IPD, especially for the elderly. It has been demonstrated that high age is a main risk factor for IPD (31). The underlying mechanisms are not known but could be due to a waning immune memory from nasopharyngeal exposure events during childhood. In the post-PCV vaccination era, the elderly are protected from IPD caused by pneumococcal strains expressing VT capsules, since VT strains have been eliminated from the nasopharynx of vaccinated children (9). However, PCV vaccination in children has resulted in replacement of VT with NVT strains in nonvaccinated age groups, such as the elderly (8). A vaccine for the elderly should ideally be protective irrespective of serotype, thereby targeting NVT strains and strains producing a serotype 3 capsule. Even though serotype 3 is included in PCV13, it is still a major serotype causing IPD, particularly among the elderly (12). Here, we show that intranasal immunization of mice with MPs from serotype 4 protects against heterologous challenge with strains of serotypes 1 and 3. Also, we find that this serotype-independent protection and bacterial clearance is largely antibody dependent but not dependent on the cytotoxin Ply, since deletion of Ply in the MPs does not affect the protective effect. Moreover, we found that although the highly diverse CBP PspA is a dominant immunogen in the MPs, it only marginally contributes to the homologous serotype protection observed, and it is not required for cross-protection.
Importantly, we identified two highly conserved pneumococcal membrane-anchored lipoproteins, MalX and PrsA, as the major protective antigens in the MPs, and they are both needed for protection. MalX is part of an ABC transporter complex and binds maltooligosaccharides of various sizes. MalX was in a signature mutagenesis screen identified as a protein required for lung infection in an intranasal challenge model (32), and serum antibodies directed against MalX have been detected in healthy adults (19). PrsA is a conserved cis-trans prolyl isomerase that facilitates protein secretion by promoting extracellular folding of several secreted proteins. PrsA has been shown to contribute to nasopharyngeal colonization in a murine model and enhances bacterial resistance to phagocytosis (33). It was suggested that these effects by PrsA may be indirect by promoting proper folding of adhesins and other virulence-associated surface proteins. We suggest that intranasal immunization with MPs elicits neutralizing antibodies to the two natively folded lipoproteins MalX and PrsA that prevent bacterial proliferation in the respiratory tract. Recombinant proteins of the respective lipoprotein lacking signal peptide and lipid modifications were significantly less recognized by antibodies present in sera of MP immunized mice compared to native lipoproteins present in the MPs. This finding indicates that the protective epitopes on MalX and PrsA may be localized close to the plasma membrane. We previously demonstrated that complement C3b gets access to the plasma membrane at division sites where the capsular layer seems thin or absent (34). Our finding that PrsA and MalX antibodies, elicited by MP immunization, bind to the bacterial cells as foci, many of which are located at or close to division sites, suggests that the PrsA and MalX lipoproteins are present at sites on the bacteria that are targetable from the outside by neutralizing antibodies.
In conclusion, we found that the main protective antigens in MPs are two conserved lipoproteins, MalX and PrsA, that are enriched when the surface protein PspA is lacking. We suggest that MPs could be used as a platform for producing a protein-based vaccine against pneumococcal infections.
Materials and Methods
Pneumococcal Strains.
The following strains of S. pneumoniae were used: T4 (TIGR4 of serotype 4) (21), isogenic mutants lacking Ply (T4Δply) (35), or the capsule (T4R) (36) or pspA, prsA, and malX (SI Appendix, Table S4), BHN733 of serotype 1, and BHN428 of serotype 3. T4, BHN733, and BHN428 were grown in C+Y-medium, pH 7.9 to 8.0, at 37 °C. Growth was followed by measuring optical density (OD) at 600 nm with a spectrophotometer (Genesys 20; Thermo Spectronic).
Construction of Pneumococcal Mutant Strains.
For generation of pneumococcal mutants using transformation, a general procedure was employed as previously described (37). To generate double and triple mutants, the same procedure was iterated using sequenced mutants that had acquired the corresponding mutations. Confirmation of mutants was done with Sanger sequencing of PCR amplicons covering the cognate loci (SI Appendix, Tables S4 and S5).
Pneumococcal mutants were created in which the open reading frames (ORFs) of pspA (SP0117), prsA (SP0981), and malX (SP2108) (gene numbers in parenthesis) were deleted and replaced with antibiotic resistance ORFs (prsA and malX) or a cassette consisting of a promoter and ORF (pspA), as indicated in SI Appendix, Table S4. Transformation constructs were made with sequential overlap PCR, where the ∼700 bp upstream and ∼700 bp downstream regions flanking the target genes were PCR amplified from genomic DNA isolated from T4 with primers given in SI Appendix, Table S4. These primers contained nonannealing overhang sequences complementary to the 5′ and 3′ ends of the antibiotic ORFs or cassette to be fused. In a second PCR, ∼700-bp up- and downstream regions were fused to the corresponding antibiotic cassette (or ORF) through thermal annealing followed by overlap extension PCR. The final PCR products that contained the joined constructs (700-bp upstream region plus the antibiotic resistance ORF or cassette plus the 700 bp-downstream region) were purified (PCR cleanup kit; Qiagen) and used to transform the T4 strain. All strains created in this study are listed in SI Appendix, Table S4 and all primers in SI Appendix, Table S5.
Preparation of Pneumococcal EVs and MPs.
EVs were isolated from liquid cultures as described before (18). Briefly, bacteria were grown in C+Y medium until OD600 nm = 0.9 and removed by centrifugation, and the supernatant was filtered and centrifuged. EVs were washed twice and resuspended in PBS. MPs were isolated from plate-grown bacteria. Briefly, bacteria were grown overnight on blood agar plates at 37 °C with 5% CO2, scraped off the plates, resuspended in PBS, and removed by centrifugation (17,000 rcf, 30 min). The supernatant was passed through a 0.2-µm filtering device (Filtropur S 0.2; Sarstedt) (except for serotype 3) and ultracentrifuged (170,000 rcf, 12 h). Pelleted vesicles were resuspended in PBS. Both EVs and MPs were further purified using Optiprep Density Gradient Medium (Sigma-Aldrich). After being resuspended in PBS, EV and MP protein concentrations were assessed by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific), and preparations were stored at −80 °C.
Animal Experiments.
All animal experiments were approved by the local ethical committee (Stockholms Norra djurförsöksetiska nämnd). For each immunization experiment, male C57BL/6 WT 5-wk-old mice were used. Before immunization, mice were anesthetized by isofluorane (Abbott) inhalation and then challenged intranasally with 50 µL/mouse (10 μg total protein content/mouse in 50 μL) EV or MP preparation, either alone or in combination with the adjuvant aluminum hydroxide (Sigma-Aldrich; 10 mg/mL in PBS) or PBS or the adjuvant alone for the control groups. Immunization was repeated after 2 wk from the first immunization, following the same procedure as described above. After 4 wk of immunization, mice were infected intranasally (50 µL/mouse) with 5 × 106 CFUs for T4 and serotype 1 or 1 × 106 CFUs for serotype 3. Mice were anesthetized by isofluorane inhalation prior to bacterial challenge. After infection, clinical symptoms of the mice were monitored multiple times daily (in accordance with the ethical permit). Blood samples (5 µL/mouse) were taken every infection day, and bacteremia levels were assessed by plating serial dilutions of blood samples onto blood agar plates. Mice that reached humane end points were killed according to ethical regulations. Lungs and spleens were collected for further analyses. Bacterial CFUs were calculated after serial dilutions of lung homogenates that were plated onto blood agar plates. MuMt knockout mice in the C57BL/6 background (The Jackson Laboratory) were used as the B cell–deficient model.
SDS-PAGE and Western Blotting.
Detection of pneumococcal proteins in EVs and MPs was performed as previously described (18, 38). Briefly, Pierce BCA Protein Assay Kit (Life Technologies) was used to determine the total protein quantity in EVs or MPs. Samples were resolved by SDS-PAGE using NuPAGE 4 to 12% Bis-Tris protein gels (Invitrogen) and electroblotted onto poly(vinylidene difluoride) (PVDF) membranes. Membranes were blocked with 5% skim milk in PBS containing 0.1% Tween-20 and incubated with sera from immunized mice (as primary antibodies, 1:5,000) and horseradish peroxidase (HRP)–conjugated secondary antibodies (1:10,000). Membranes were developed with Amersham ECL Plus Western blotting detection system (GE Healthcare Life Sciences) using a ChemiDoc XRS+ (Bio-Rad Laboratories). For Western blot detection using bacterial lysates, T4, T4ΔpspA, T4ΔprsA, T4ΔmalX, T4ΔpspAΔprsA, T4ΔpspAΔmalX, T4ΔΔprsAΔmalX, and T4ΔpspAΔprsAΔmalX, serotype 1 BHN733, and serotype 3 BHN428 were grown overnight on blood agar plates and inoculated in C+Y medium at 37 °C. These precultures were grown to mid-log phase and used to start new cultures in C+Y media at OD620 = 0.05. When the cultures reached OD620 = 0.5 (0.5 mL), each strain was centrifugated (10,000 rpm, 5 min), and the cell pellet was washed with 1 mL PBS, centrifugated (10,000 rpm, 5 min), dissolved in 250 µL 1x SDS-PAGE loading buffer including 25 mM 1,4-dithiothreitol (DTT), and then boiled for 5 min. For assessment of sera binding to pneumococcal CBPs, cells were after-growth incubated with 5% choline chloride for 10 min at 37 °C and centrifuged, and the supernatant was collected. The pellet was further washed with 5% choline chloride to remove CBPs. Pellets and supernatant fractions were mixed with loading buffer, and Western blot analysis was performed as described above.
IP.
A column-based IP protocol was set up using the Protein G HP SpinTrap/Ab Spin Trap kit (GE Healthcare). The resin from the Protein G Sepharose kit columns were washed three times with PBS and then removed from the columns and transferred to Eppendorf tubes in a 300-µL slurry in PBS. Samples (100 µL) of 10-fold diluted immune sera from mice immunized with MPs from T4 and T4ΔpspA and of sera from adjuvant challenged mice were added to the resin in separate tubes and incubated for 30 min at room temperature. Following incubation, the resins were washed twice with 400 µL PBS using centrifugation (100 rfc, 1 min). The pelleted resin fractions, containing IgG molecules from the immune sera, were resuspended in 400 µL of a pneumococcal cell lysate from T4 generated from mid-log phase cultures grown in C+Y media at 37 °C and transferred to Eppendorf tubes (500 µL per tube) and treated with 5 µL 10% Triton ×100 (0.1% end concentration) for 30 min to induce prominent autolysin-mediated lysis of the cultures. These lysates were centrifugated (14,000 rpm, 5 min) to remove cell wall debris, and 400 µL of the supernatant was used per sample. The lysate/antisera–Protein G Sepharose mixtures were incubated for 30 min at room temperature, after which the samples were transferred back to the spin columns, fitted into collection tubes, and centrifugated (100 rcf, 1 min) to remove unbound proteins. The columns were successively washed five times with 400 µL PBS with centrifugation (100 rcf, 1 min), elution was done with 400 µL 100 mM sodium citrate buffer, pH 2.5, with centrifugation (100 rcf, 1 min), and the samples were collected into tubes containing 40 µL 1 M Tris buffer, pH 8.0, to neutralize the pH. Samples were concentrated using SpedVac (Thermo Fisher Scientific) to 100 µL, frozen, and sent for mass spectrometry analysis.
Mass Spectrometry.
Analysis of EVs and MPs was performed as described previously (18). Briefly, a urea-containing buffer was used to lyse EVs and MPs. Proteins were reduced, alkylated, and digested in solution by trypsin. A Pierce C18 Spin Column (Thermo Fisher Scientific) was used to purify the sample that was dried and resolved in 0.1% formic acid. Peptides were separated in reversed phase on a C18-column and electrosprayed on-line to a Q Exactive Plus mass spectrometer (Thermo Finnigan). Tandem mass spectrometry (MS/MS) was performed applying higher-energy collisional dissociation (HCD). The Sequest algorithm in Proteome Discoverer 1.4 (Thermo Fisher Scientific) was used to search databases toward a FASTA database of TIGR4 (for serotype 4) or SP3-BS71 (for serotype 3) proteins from UniProtKB. Criteria for protein identification were at least two matching peptides of 95% confidence level. In order to avoid false-positives, only proteins with a score of 20 or above were included in the analysis. Subcellular localizations of proteins were predicted as before (18).
For analysis of the IP samples, the following mass spectrometry setup was used.
Chemicals and reagents.
Acetonitrile (ACN), formic acid (FA), and ammonium bicarbonate (NH4HCO3) were obtained from Merck. Protease inhibitor mixture and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich. For tryptic digestion, iodoacetamide (IAA), urea, and DTT were obtained from Sigma-Aldrich, and grade-modified trypsin (V5073) was from Promega. Ultrapure water was prepared by Milli-Q water purification system (Millipore).
In-solution tryptic digestion of proteins.
The whole volume of the samples (100 μL) was used for in-solution digestion. A volume of 10 μL 45 mM DTT was added, and the mixtures were incubated at 50 °C for 15 min and then cooled to ambient temperature. Then 10 μL 100 mM IAA was added, and the mixtures were incubated for 15 min at room temperature in darkness. Finally, trypsin solution was added to yield a final trypsin/protein concentration of 5% (wt/wt). The tryptic digestion was performed at 37 °C overnight in darkness. Thereafter, the samples were desalted using the SPE Pierce C18 Spin Columns (Thermo Fisher Scientific). These columns were activated by 2 × 200 μL 50% ACN and equilibrated with 2 × 200 μL 0.5% TFA. The tryptic peptides were adsorbed to the media using two repeated cycles of 40 μL sample loading, and the column was washed using 3 × 200 μL 0.5% TFA. Finally, the peptides were eluted in 3 × 50 μL 70% ACN and dried. Dried peptides were resolved in 30 μL 0.1% FA prior to nano–liquid chromatography (LC)–MS/MS.
LC-MS/MS analysis.
The nano–LC-MS/MS experiments were performed using Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen) equipped with a nano electrospray ion source. The peptides were separated by C18 reversed-phase LC using an EASY-nLC 1000 system (Thermo Fisher Scientific). A setup of precolumn and analytical column was used. The precolumn was a 2-cm EASYcolumn (ID 100 µm, 5-µm particles; Thermo Fisher Scientific), while the analytical column was a 10-cm EASY-column (ID 75 µm, 3-µm particles; Thermo Fisher Scientific). Peptides were eluted with a 90-min linear gradient from 4 to 100% ACN at 250 nL min−1. The mass spectrometer was operated in positive ion mode, acquiring a survey mass spectrum with resolving power 70,000 (full width, half maximum) m/z 400 to 1,750 using an automatic gain control (AGC) target of 3 × 106. The 10 most intense ions were selected for HCD fragmentation (25% normalized collision energy), and MS/MS spectra were generated with an AGC target of 5 × 105 at a resolution of 17,500. The mass spectrometer worked in data-dependent mode.
Data analysis.
The acquired data (.RAW-files) were processed by Proteome Discoverer software (Thermo Fisher Scientific, version [nr 1.4.1.14]) using the Sequest algorithm toward a combined database containing protein sequences from the strain TIGR4 proteome (2,178 entries) downloaded from Uniprot 2019–10. The following parameters were used for data processing: maximum 10 ppm and 0.02 13 Da error tolerances for the survey scan and MS/MS analysis, respectively; trypsin as digesting enzyme; carbamidomethylation of cysteine as fixed modification; oxidation of methionine as variable modification; and maximum of two missed cleavage sites. The target decoy PSM validator was used to calculate false discovery rate (FDR). An FDR of maximum 5% for peptide identification was accepted, and the search criteria for protein identification were set to at least two matching peptides per protein.
Mouse IgG ELISA.
To detect T4- and T4R-specific mouse IgG in sera of T4 EV, T4 MP, and T4R MP immunized mice, bacteria were grown on blood agar plates overnight at 37 °C, resuspended in PBS, and heat inactivated for 2 h at 60 °C. After the bacteria were diluted to OD600 0.6, optical plates were coated with 100 μL bacteria in 0.1 M sodium carbonate buffer, pH 9.5, overnight at 4 °C. Wells were washed three times and incubated with 200 μL PBS with 2.5% skim milk for 2 h and, after three washes, were incubated with 100 μL mice sera diluted 1:500 in PBS for 1 h at room temperature. Wells were washed three times and incubated with 100 μL anti-mouse IgG-HRP diluted 1:500 in PBS for 1 h at room temperature, and the procedure was continued as described above.
To detect T4, serotype 1 (BHN733)– and serotype 3 (BHN428)–specific mouse IgG in sera of mice immunized with T4 MP and T4ΔpspA MP, bacteria were grown on blood agar plates overnight at 37 °C, resuspended in PBS, and heat inactivated for 2 h at 60 °C. After the bacteria were diluted to OD600nm = 0.6, optical plates were coated with 100 μL bacteria in PBS with complete Protease Inhibitor Mixture (Roche) overnight at 4 °C. Wells were washed three times and incubated with 200 μL PBS with 2.5% skim milk for 2 h and, after three washes, were incubated with 100 μL mice sera diluted 1:1,000 in PBS with 1% bovine serum albumin (BSA) for 1 h in room temperature. Wells were washed three times and incubated with 100 μL anti-mouse IgG-HRP diluted 1:1,000 in PBS with 1% BSA for 1 h at room temperature, and the procedure was continued as described above.
Immunofluorescence Microscopy.
After growth, pneumococci (T4, T4R, serotype 1 BHN733, and serotype 3 BHN428) were stained using sera from mice immunized with T4 MPs+adjuvant, T4ΔpspA MPs+adjuvant, T4ΔpspAΔmalX MPs+adjuvant, T4ΔpspAΔprsA MPs+adjuvant, T4ΔmalXΔprsA MPs+adjuvant, T4ΔpspAΔmalXΔprsA MPs+adjuvant, and adjuvant only control as primary antibody (dilution 1:100 in PBS 1% BSA) followed by incubation with the secondary antibody Alexa Fluor 488 goat anti-mouse (dilution 1:500 in PBS 1% BSA). Cells were fixed with 4% paraformaldehyde (PFA) and mounted onto a slide using Vectashield anti-fade mounting medium (Vector Laboratories). Imaging was performed using a Deltavision Elite microscope (Applied Precision) and a scientific complementary metal-oxide–semiconductor camera. Images were acquired with fluorescein isothiocyanate laser intensity of 50% and exposure time of 200 ms using Softworx (Applied Precision).
Flow Cytometry of Bacteria Labeled with IgGs from Mouse Sera.
Bacterial strains were grown in C+Y medium static at 37 °C to OD600nm = 0.5. Then, 500 µL bacterial culture was centrifugated (10,000 rpm, 5 min), and the supernatant was removed. The bacteria were washed with PBS and resuspended in 100 µL of 1% BSA in PBS. After incubation at room temperature for 30 min, 1 µL of a 1:10 dilution of pooled mice sera was added to the bacteria suspensions (final serum concentration, 1:1,000). After 1-h incubation at room temperature, the bacteria were washed with PBS and resuspended in 100 µL PBS containing 1% BSA and the antibody Alexa Fluor 488 goat anti-mouse IgG (H+L) (Invitrogen, Cat. #A-11001) diluted 1:1,000. Following 1-h incubation at room temperature, the bacteria were washed with PBS and fixed with 4% PFA, resuspended in 300 µL PBS, and analyzed quantitatively by flow cytometry using a Gallios flow cytometer (Beckman Coulter). The analysis of the flow cytometry data was performed with the software FlowJo (v10.8.1). The gating was determined based on the cell distribution in the control samples where the bacteria were not incubated with sera (SI Appendix, Fig. S7).
Opsonophagocytosis Assay with RAW Cells.
RAW 264.7 murine macrophages were grown and maintained at 37 °C, 5% CO2 in RPMI medium (Gibco) supplemented with 10% (vol/vol) fetal bovine serum (HyClone). To assess the opsonophagocytic activity of antibodies in immunized mice sera, 2 × 105 RAW 264.7 cells were seeded in 24-well plates and incubated overnight at 37 °C. Serotype 1 bacteria were incubated for 30 min at 37 °C with 5% CO2 with 20% serum from mice. RAW cells were then washed with PBS and incubated for 1.5 h with 2.5 × 107/well of pretreated bacteria. Cells were washed three times with PBS to remove unattached bacteria. To measure total uptake of bacteria, cells were incubated with a 50/50 solution of 2% saponin (Sigma-Aldrich) and trypsin-ethylenediaminetetraacetic acid (trypsin-EDTA , Gibco) for 15 min at 37 °C to lyse eukaryotic cells, and total bacteria were plated for enumeration. To evaluate phagocytosis, 300 µg/mL Gentamicin (Sigma-Aldrich) and 0.12 mg/mL Penicillin G (Sigma-Aldrich) were added to separate wells and incubated 15 min at 37 °C to kill extracellular bacteria. Cells were washed three times with PBS and incubated with a 50/50 solution of 2% saponin and trypsin-EDTA for 15 min at 37 °C to lyse eukaryotic cells. To evaluate bacterial killing inside macrophages, separate wells were treated with antibiotics (as for phagocytosis), washed three times with PBS, and incubated for 1 h at 37 °C with medium. Cells were washed three times with PBS and incubated with a 50/50 solution of 2% saponin and trypsin-EDTA for 15 min at 37 °C to lyse eukaryotic cells.
Statistical Analysis.
Statistical analysis for multiple comparisons was done using a nonparametric ANOVA test, and Dunn’s test was applied to assess differences between pairs. Two-group comparisons were analyzed using a nonparametric two-tailed Mann–Whitney U test. Analysis of survival curves was done using a log-rank Mantel-Cox test. Statistically significant data were defined as *P < 0.05; **P < 0.01; ***P < 0.001; and **** = P < 0.0001.
Supplementary Material
Acknowledgments
We thank the MS-based proteomics facility at Uppsala University (Sweden) for processing and analysis of MS samples. This work was supported by grants from the Family Erling Persson Foundation, the Swedish Foundation for Strategic Research, the Swedish Research Council, the Stockholm County Council, the Torsten Söderberg foundation, and the Knut and Alice Wallenberg Foundation.
Footnotes
Reviewers: R.R., Toscana Life Sciences Foundation; and J.W., New York University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2122386119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Henriques-Normark B., Tuomanen E. I., The pneumococcus: Epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 3, a010215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iovino F., Seinen J., Henriques-Normark B., van Dijl J. M., How does Streptococcus pneumoniae invade the brain? Trends Microbiol. 24, 307–315 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Harboe Z. B., et al. , Pneumococcal serotypes and mortality following invasive pneumococcal disease: A population-based cohort study. PLoS Med. 6, e1000081 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley S. D., et al. , Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2, e31 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ganaie F., et al. , A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral Streptococcus. MBio 11, e00937-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanquet G., et al. ; SpIDnet/I-MOVE+ Pneumo Group, Effect of childhood pneumococcal conjugate vaccination on invasive disease in older adults of 10 European countries: Implications for adult vaccination. Thorax 74, 473–482 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savulescu C., et al. ; SpIDnet Group, Effect of high-valency pneumococcal conjugate vaccines on invasive pneumococcal disease in children in SpIDnet countries: An observational multicentre study. Lancet Respir. Med. 5, 648–656 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Galanis I., et al. , Effects of PCV7 and PCV13 on invasive pneumococcal disease and carriage in Stockholm, Sweden. Eur. Respir. J. 47, 1208–1218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindstrand A., et al. , Unaltered pneumococcal carriage prevalence due to expansion of non-vaccine types of low invasive potential 8 years after vaccine introduction in Stockholm, Sweden. Vaccine 34, 4565–4571 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Moore M. R., et al. , Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: Analysis of multisite, population-based surveillance. Lancet Infect. Dis. 15, 301–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitney C. G., et al. , Active Bacterial Core Surveillance of the Emerging Infections Program Network, Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348, 1737–1746 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Naucler P., et al. , Comparison of the impact of pneumococcal conjugate vaccine 10 or pneumococcal conjugate vaccine 13 on invasive pneumococcal disease in equivalent populations. Clin. Infect. Dis. 65, 1780–1789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horácio A. N., Silva-Costa C., Lopes J. P., Ramirez M., Melo-Cristino J.; Portuguese Group for the Study of Streptococcal Infections, Serotype 3 remains the leading cause of invasive pneumococcal disease in adults in Portugal (2012-2014) despite continued reductions in other 13-valent conjugate vaccine serotypes. Front. Microbiol. 7, 1616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zangari T., et al. , Pneumococcal capsule blocks protection by immunization with conserved surface proteins. NPJ Vaccines 6, 155 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown L., Wolf J. M., Prados-Rosales R., Casadevall A., Through the wall: Extracellular vesicles in gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 13, 620–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E. Y., et al. , Gram-positive bacteria produce membrane vesicles: Proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9, 5425–5436 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Lebon A., et al. , Natural antibodies against several pneumococcal virulence proteins in children during the pre-pneumococcal-vaccine era: The Generation R Study. Infect. Immun. 79, 1680–1687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Codemo M., et al. , Immunomodulatory effects of pneumococcal extracellular vesicles on cellular and humoral host defenses. MBio 9, e00559-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giefing C., et al. , Discovery of a novel class of highly conserved vaccine antigens using genomic scale antigenic fingerprinting of pneumococcus with human antibodies. J. Exp. Med. 205, 117–131 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olaya-Abril A., et al. , Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J. Proteomics 106, 46–60 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Tettelin H., et al. , Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293, 498–506 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Subramanian K., Henriques-Normark B., Normark S., Emerging concepts in the pathogenesis of the Streptococcus pneumoniae: From nasopharyngeal colonizer to intracellular pathogen. Cell. Microbiol. 21, e13077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogunniyi A. D., et al. , Contributions of pneumolysin, pneumococcal surface protein A (PspA), and PspC to pathogenicity of Streptococcus pneumoniae D39 in a mouse model. Infect. Immun. 75, 1843–1851 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian K., et al. , Pneumolysin binds to the mannose receptor C type 1 (MRC-1) leading to anti-inflammatory responses and enhanced pneumococcal survival. Nat. Microbiol. 4, 62–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchiya M., et al. , Genetic diversity of pneumococcal surface protein A (PspA) in paediatric isolates of non-conjugate vaccine serotypes in Japan. J. Med. Microbiol. 67, 1130–1138 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Ferreira D. M., et al. , Characterization of protective mucosal and systemic immune responses elicited by pneumococcal surface protein PspA and PspC nasal vaccines against a respiratory pneumococcal challenge in mice. Clin. Vaccine Immunol. 16, 636–645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolley K. A., Bray J. E., Maiden M. C. J., Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiser J. N., Ferreira D. M., Paton J. C., Streptococcus pneumoniae: Transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beilharz K., et al. , Control of cell division in Streptococcus pneumoniae by the conserved Ser/Thr protein kinase StkP. Proc. Natl. Acad. Sci. U.S.A. 109, E905–E913 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naucler P., Darenberg J., Morfeldt E., Ortqvist A., Henriques Normark B., Contribution of host, bacterial factors and antibiotic treatment to mortality in adult patients with bacteraemic pneumococcal pneumonia. Thorax 68, 571–579 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Hava D. L., Camilli A., Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 (2002). [PMC free article] [PubMed] [Google Scholar]
- 33.Cron L. E., et al. , Surface-associated lipoprotein PpmA of Streptococcus pneumoniae is involved in colonization in a strain-specific manner. Microbiology (Reading) 155, 2401–2410 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Pathak A., et al. , Factor H binding proteins protect division septa on encapsulated Streptococcus pneumoniae against complement C3b deposition and amplification. Nat. Commun. 9, 3398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Littmann M., et al. , Streptococcus pneumoniae evades human dendritic cell surveillance by pneumolysin expression. EMBO Mol. Med. 1, 211–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernebro J., et al. , Capsular expression in Streptococcus pneumoniae negatively affects spontaneous and antibiotic-induced lysis and contributes to antibiotic tolerance. J. Infect. Dis. 189, 328–338 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Balaban M., et al. , Secretion of a pneumococcal type II secretion system pilus correlates with DNA uptake during transformation. Proc. Natl. Acad. Sci. U.S.A. 111, E758–E765 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellroth P., et al. , LytA, major autolysin of Streptococcus pneumoniae, requires access to nascent peptidoglycan. J. Biol. Chem. 287, 11018–11029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.