Significance
Finding that pan-tolerance derives from defects in carbohydrate regulation connects stress-mediated lesions with metabolic change, identifies a stable type of tolerance, and demonstrates a widely shared death response. Manipulation of the response should improve antimicrobial efficacy, preserve beneficial bacteria during antimicrobial use, and protect industrial bacteria from toxic products. Mutations in many genes can interfere with stress-mediated metabolism; thus, mutation to pan-tolerance could be a high-probability event. Finding that selection of tolerance to one lethal stressor confers tolerance to many, if not all, indicates that massive disinfectant consumption potentially undermines antimicrobial efficacy and immune defenses against pathogenic bacteria. Since pan-tolerance is hidden from current surveillance of resistance, the work indicates a need for facile methods to measure tolerance.
Keywords: commonly shared death mechanism, cAMP-Crp transcription regulatory system, reactive oxygen species (ROS), stress-mediated killing, carbohydrate phosphotransferase system (PTS)
Abstract
Recent work indicates that killing of bacteria by diverse antimicrobial classes can involve reactive oxygen species (ROS), as if a common, self-destructive response to antibiotics occurs. However, the ROS-bacterial death theory has been challenged. To better understand stress-mediated bacterial death, we enriched spontaneous antideath mutants of Escherichia coli that survive treatment by diverse bactericidal agents that include antibiotics, disinfectants, and environmental stressors, without a priori consideration of ROS. The mutants retained bacteriostatic susceptibility, thereby ruling out resistance. Surprisingly, pan-tolerance arose from carbohydrate metabolism deficiencies in ptsI (phosphotransferase) and cyaA (adenyl cyclase); these genes displayed the activity of upstream regulators of a widely shared, stress-mediated death pathway. The antideath effect was reversed by genetic complementation, exogenous cAMP, or a Crp variant that bypasses cAMP binding for activation. Downstream events comprised a metabolic shift from the TCA cycle to glycolysis and to the pentose phosphate pathway, suppression of stress-mediated ATP surges, and reduced accumulation of ROS. These observations reveal how upstream signals from diverse stress-mediated lesions stimulate shared, late-stage, ROS-mediated events. Cultures of these stable, pan-tolerant mutants grew normally and were therefore distinct from tolerance derived from growth defects described previously. Pan-tolerance raises the potential for unrestricted disinfectant use to contribute to antibiotic tolerance and resistance. It also weakens host defenses, because three agents (hypochlorite, hydrogen peroxide, and low pH) affected by pan-tolerance are used by the immune system to fight infections. Understanding and manipulating the PtsI-CyaA-Crp–mediated death process can help better control pathogens and maintain beneficial microbiota during antimicrobial treatment.
Knowledge of bacterial cell death has been obtained largely by treating cultures with lethal agents and then identifying the resulting lesions, their consequences, and their repair. Such studies establish that bacterial death can derive from many distinct mechanisms, each specific to the type of primary lesion generated. However, in the last decade, work initiated by the Collins’ group (1) has reported that the lethal action of three diverse antimicrobials correlates with a metabolic shift in which an increase in respiration is followed by an accumulation of reactive oxygen species (ROS) and macromolecular damage (2–8). In this scheme, ROS appear to play a central role shared by lethal agents that generate distinct lesions. The idea that diverse primary lesions would lead to a general, ROS-based mechanism of self-destruction was counter-intuitive, because bacteria possess many genes specific for repairing diverse lethal lesions and robust systems for protecting from oxidative stress. Moreover, challenges to Collins’ initial work (9, 10) led to controversy (11–14). Although subsequent reports largely support a role for ROS in death triggered by diverse stress types, these studies have been directed at ROS, largely by perturbing intracellular ROS and then correlating the perturbation with bacterial survival. Many studies, if not all, show a correlation rather than a causal relationship between a surge in ROS levels and cell death. To address this issue in an unbiased way, we asked whether a widely shared death process exists without a priori consideration of ROS. Genetic analysis was then used to identify genes that connect various types of primary lesions with downstream lethal events.
Since distinguishing dying from dead cells is difficult and since the physiologically inactive death state is hard to interrogate, directly studying death has been challenging. Examining antideath mutants whose defects clearly interfere with the active lethal process provides an unambiguous way to identify genes involved in death. Such mutants, which are also called tolerant mutants (15–17), can be enriched by treating bacteria with a highly lethal agent and then screening for cells that neither grow nor die when exposed to a lethal stressor. The absence of a change in minimal inhibitory concentration (MIC) is crucial to the definition of tolerance (17, 18). High (increased) survival is imprecise, because it could derive from two mechanistically distinct phenomena: decreased growth inhibition or decreased killing or both. Tolerance and antideath refer only to the latter. The key to successful screening for tolerance is finding an agent that rapidly and extensively kills wild-type cells but still allows mutant recovery. The enrichment procedure must not allow resistant mutants to emerge and amplify, because resistant growth would make recovery of tolerant mutants very difficult. Several disinfectants have the properties desired for enrichment of tolerant mutants.
In the present work, we used phenol to enrich tolerant mutants of Escherichia coli; we then tested for cross-tolerance to a wide variety of lethal stressors. Several tolerant mutants were obtained that, unlike tolerant mutants studied previously (19) (reviewed in ref. 15), showed little effect on bacterial growth. Genetic analysis associated tolerance with a deficiency in ptsI, a gene in the carbohydrate uptake phosphotransferase system (PTS) (20). This deficiency conferred tolerance to a variety of lethal disinfectants, antimicrobials, and environmental stresses. The PTS phosphorylation cascade increases levels of cAMP (21), and deficiencies in adenyl cyclase also conferred pan-tolerance. The PTS-cAMP-Crp regulatory cascade defines a broadly applicable death pathway by conveying signals from diverse, stress-mediated lesions to a shared, ROS-mediated damaging of macromolecules.
The existence of a widely shared and potentially universal death pathway implies that massive consumption of disinfectants, which has been greatly exacerbated during the current COVID-19 pandemic, may be enriching mutants that are cross-tolerant to diverse antimicrobials and thus capable of reducing antimicrobial efficacy and successful treatment.
Results
Enrichment, Identification, and Characterization of Phenol-Tolerant Mutants.
To determine whether a general death pathway exists in bacteria, we enriched antideath mutants by exposing E. coli cultures to phenol, a prototype disinfectant for aseptic surgery (22). Phenol was chosen because it kills bacteria rapidly and extensively; no mechanism for resistance is known, and no phenol-resistant mutant has been reported. As expected, we failed to obtain low-susceptibility “resistant” mutants after multiple rounds of challenge, possibly because phenol simultaneously attacks multiple targets. However, we did recover several mutants that exhibited tolerance, defined as decreased phenol-mediated killing (Fig. 1 A–C) with wild-type susceptibility to growth inhibition (e.g., unaltered MIC) (SI Appendix, Table S1).
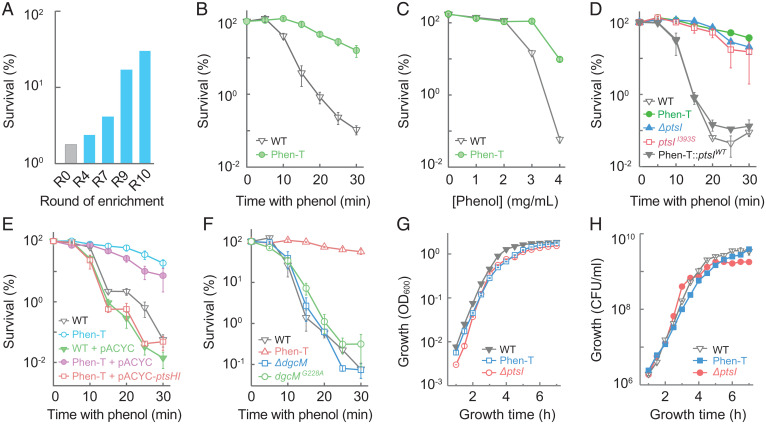
Fig. 1.
Enrichment, identification, and characterization of a mutant tolerant to killing by phenol. (A) Survival of wild-type E. coli after multiple rounds of challenge with 3.5 mg/mL phenol. (B) Reduced rate of killing from 3.5 mg/mL phenol by a phenol-tolerant mutant (Phen-T), obtained from A. (C) Elevated concentrations of phenol required to kill the Phen-T mutant during a 15-min treatment. (D) PtsI defect is responsible for phenol tolerance. E. coli cultures were treated with 3.5 mg/mL phenol prior to measurement of survival. Strains: wild-type, Phen-T mutant, ΔptsI mutant, ptsI point (I393S) mutant, Phen-T mutant back-crossed with wild-type ptsI. (E) Expression of ptsHI operon from a plasmid complemented the Phen-T tolerance phenotype. Killing measurements (as in B) were used with the following strains: wild-type, wild-type harboring pACYC184 vector plasmid, Phen-T, Phen-T harboring pACYC184, and Phen-T harboring pACYC184-ptsHI (expressing the ptsHI operon from the plasmid). (F) Mutations in dgcM failed to account for phenol tolerance of the Phen-T mutant. Cultures of wild-type cells and Phen-T, ΔdgcM::Kan, and dgcM G228A mutants were treated and processed as in B. (G and H) Phen-T and ptsI deficiencies have little effect on culture growth. Overnight cultures of wild-type cells and the Phen-T and the ΔptsI mutants were diluted 2,000-fold into fresh LB medium and grown aerobically at 37 °C; at the indicated times turbidity (OD600, G) or colony-forming units (H) were determined. Data represent average of three biological replicates; error bars indicate SEM. See SI Appendix, Tables S1 and S2 for supporting information.
Comparative whole-genome sequencing of a representative, phenol-tolerant mutant (Phen-T) with its parental, ancestral strain revealed the presence of two point mutations, one in ptsI (I393S), a gene encoding enzyme I (EI) of the carbohydrate PTS (20), and the other in dgcM (G228A), which encodes a diguanylate cyclase involved in control of curli biosynthesis (23) (SI Appendix, Table S2). Replacement of the ptsI mutant allele with the corresponding wild-type gene, using CRISPR-based allelic exchange (24), largely reversed phenol tolerance, and introduction of a ptsI (I393S) or ΔptsI allele into wild-type cells conferred phenol tolerance similar to that seen with the Phen-T mutant (Fig. 1D and SI Appendix, Table S1). Moreover, a plasmid-borne ptsHI operon reversed Phen-T–mediated phenol tolerance (Fig. 1E and SI Appendix, Table S1). In contrast, introduction of dgcM (G228A) or ΔdgcM alleles into the parental, wild-type strain conferred neither phenol tolerance nor resistance (Fig. 1F and SI Appendix, Table S1). dgcM was not studied further. Collectively, these data indicate a role for ptsI but not dgcM in phenol-mediated bacterial death.
Unlike findings with previous work on tolerance (15, 25), slow growth did not account for tolerance, as growth rates were similar for the Phen-T and ΔptsI mutants and wild-type cells (Fig. 1 G and H). Moreover, genetic defects associated with growth-defective tolerance (15, 19) do not include ΔptsI or genes identified below. Overall, the data establish that a ptsI deficiency confers a new type of tolerance to the lethal action of phenol while retaining wild-type MIC (e.g., no resistance). Since Phen-T and ΔptsI mutants exhibited a similar phenotype, subsequent work focused on the ΔptsI mutant.
We note that successful screening for tolerant mutants demonstrates that growth inhibition (measured as MIC) and killing (measured as loss of viable counts) are distinct events, because they can be experimentally separated. The former, which is associated with resistance, derives from the formation of stressor-specific primary lesions, which is affected by factors such as stressor uptake, efflux, and drug-target interactions (our experimental design eliminated such factors from consideration). In contrast, killing (death) arises from a subsequent cellular response to primary stress-mediated lesions.
A Deficiency in ptsI Confers a Pan-Tolerance (Antideath) Phenotype to Diverse Lethal Stressors and Treatments.
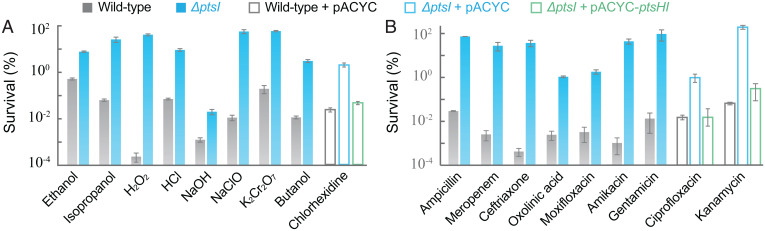
To determine whether PtsI is involved in a death process caused by stressors of diverse types, we examined the ΔptsI mutant for killing by a variety of disinfectants. The mutant displayed reduced killing by ethanol, isopropanol, chlorhexidine, hydrogen peroxide, hydrochloric acid (pH 3), sodium hydroxide (pH 10), sodium hypochlorite, potassium dichromate, and butanol (Fig. 2A and SI Appendix, Fig. S1 A–J) with little effect on MIC (SI Appendix, Table S3). Complementation with a plasmid-borne wild-type ptsI gene was achieved with two diverse, representative disinfectants: phenol and chlorhexidine (Figs. 1E and 2A and SI Appendix, Fig. S1C). Similar results were obtained with the Phen-T mutant (SI Appendix, Fig. S1 K–S and Table S3) (quantitative differences between the two mutants with some stressors may derive from the additional dgcM allele in the Phen-T mutant, but by itself the dgcM mutation has no effect). We conclude that a PTS deficiency causes tolerance to killing by diverse disinfectant types. The absence of an effect on MIC emphasizes that inhibition of growth and killing are mechanistically distinct.
Fig. 2.
ptsI deficiency protects from the lethal action of diverse stressors. (A) Killing by diverse disinfectants. Exponentially growing cultures of E. coli wild-type and ΔptsI mutant were treated with ethanol (13% for 50 min), isopropanol (9% for 30 min), chlorhexidine (12 μg/mL for 5 h), hydrogen peroxide (25 mM for 30 min), hydrochloric acid (pH 2.5 for 60 min), sodium hydroxide (pH10 for 40 min), sodium hypochlorite (0.03% for 100 min), potassium dichromate (1 mg/mL for 3 h), or butanol (2% for 25 min) prior to measurement of survival. Complementation was performed by expression of a plasmid-borne wild-type ptsHI operon (pACYC-184- ptsHI) for two representative disinfectants, chlorhexidine and phenol (also see SI Appendix, Fig. S1 C and D). (B) Killing by diverse antimicrobials. Survival of exponentially growing wild-type and ΔptsI mutant cells was measured after treatment with ampicillin (5× MIC for 4 h), meropenem (10× MIC for 8 h), ceftriaxone (20× MIC for 8 h), ciprofloxacin (5× MIC for 2.5 h), oxolinic acid (16× MIC for 5 h), moxifloxacin (10× MIC for 3 h), kanamycin (3× MIC for 2 h), amikacin (3× MIC for 2 h), or gentamicin (3× MIC for 1 h). Complementation was performed by expression of a plasmid-borne wild-type ptsHI operon (pACYC-184-ptsHI) for two representative antimicrobials, ciprofloxacin and kanamycin (also see SI Appendix, Fig. S2 D and G). Data are averages of three biological replicates; error bars indicate SEM. See SI Appendix, Figs. S1–S3 and Tables S1 and S3 for supporting information.
We next compared killing of wild-type cells and a ΔptsI mutant by a variety of lethal antimicrobials. The ptsI mutation reduced killing by all antimicrobials tested, including β-lactams (ampicillin, meropenem, ceftriaxone), quinolones (ciprofloxacin, oxolinic acid, moxifloxacin), and aminoglycosides (kanamycin, amikacin, gentamicin) (Fig. 2B and SI Appendix, Fig. S2). For drugs that kill in a concentration-dependent fashion (26), such as ciprofloxacin and kanamycin, tolerance was also observed when killing was measured using various drug concentrations for a fixed time (SI Appendix, Fig. S2 J and K). Complementation with a plasmid-borne wild-type ptsI gene was achieved with two diverse antimicrobials, ciprofloxacin and kanamycin (Fig. 2B and SI Appendix, Fig. S2 D and G). The Phen-T mutant showed results similar to those observed with the ΔptsI mutant (SI Appendix, Fig. S3). As with disinfectants, the ptsI deficiency mainly affected antimicrobial-mediated killing, not growth inhibition (SI Appendix, Tables S1 and S3).
Finally, we tested the ptsI mutant for killing by two environmental stressors, UV irradiation and high osmolarity. The ΔptsI mutant displayed the antideath phenotype for both lethal conditions (SI Appendix, Fig. S2 L and M). Thus, a ptsI deficiency confers tolerance to killing by a wide variety of lethal stressors.
PTS Phosphorelay Affects Stress-Mediated Killing through an Interplay with Adenyl Cyclase.
To examine molecular events underlying the pan-tolerance phenotype associated with ptsI deficiency, we focused on phenol and ciprofloxacin as two representative probes for diverse stressor types. We found that chemical inhibition of PtsI (EI) activity interfered with killing. For example, dimethyl 2-oxoglutarate, an inhibitor of EI (27), reduced killing by both phenol and ciprofloxacin (Fig. 3 A and B) without altering MIC (SI Appendix, Table S1) when supplied at a concentration that by itself did not inhibit bacterial growth (SI Appendix, Fig. S4A). The protective effect of dimethyl 2-oxoglutarate on killing was also concentration-dependent (SI Appendix, Fig. S4B). Moreover, blocking the PTS phosphorelay by amino acid substitutions in the phosphorylation sites of PtsI (H189Q), PtsH (H15Q), or Crr (H91Q) reduced killing by phenol and ciprofloxacin (Fig. 3 C–H) without altering MIC (SI Appendix, Table S1). Deletion of genes encoding products downstream from Crr (enzyme IIA or EIIA)—such as ptsG, malX, ascF, and glvC, which are involved in carbohydrate phosphorylation and uptake (28)—did not confer tolerance to phenol or ciprofloxacin (SI Appendix, Fig. S4 C and D). Collectively, these results indicate that early steps of PtsI-PtsH-Crr phosphorelay activity, rather than late-stage PTS carbohydrate transport functions, are required for lethality by diverse bactericidal agents.
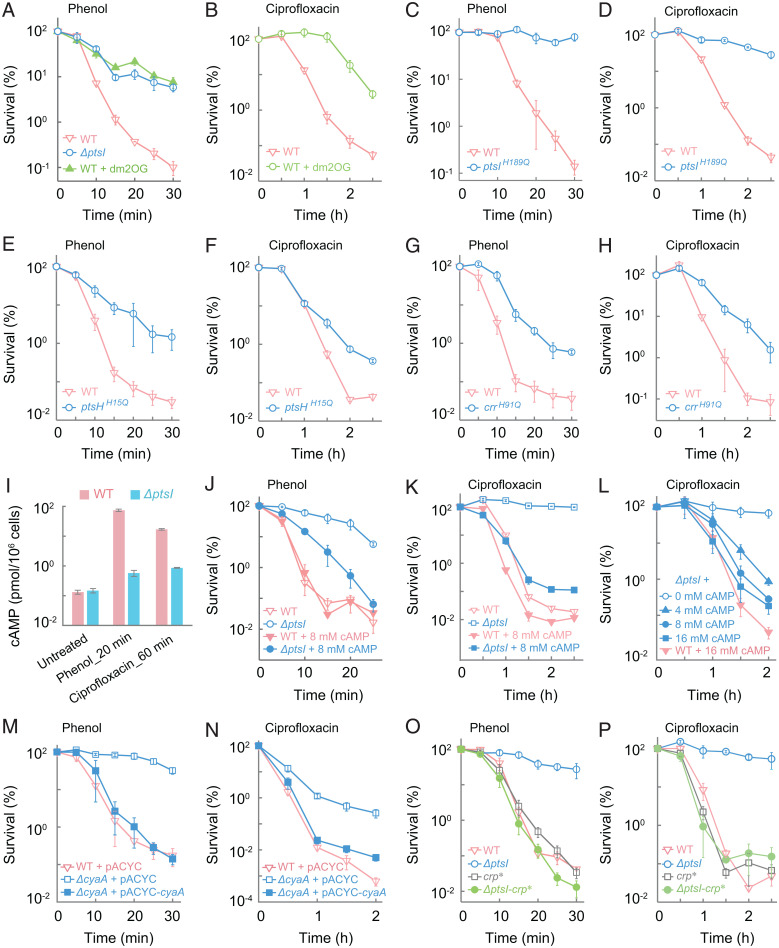
Fig. 3.
PtsI participates in phenol- and ciprofloxacin-mediated killing via early steps of the PTS phosphorelay and a subsequent interaction with the cAMP-Crp transcription regulatory axis. (A and B) Protective effect of dimethyl-2-oxoglutarate (dm2OG) on phenol- or ciprofloxacin-mediated killing. Wild-type E. coli (WT) was pretreated with 1 mg/mL dm2OG for 40 min before treatment with 3.5 mg/mL phenol (A) or 5× MIC ciprofloxacin (B) for the indicated times. As a reference, in A the ΔptsI mutant received no dm2OG pretreatment. (C–H) Protection due to abolished phosphorylation site in PtsI, PtsH, or Crr. Wild-type, a ptsI (H189Q) mutant (C and D), a ptsH (H15Q) mutant (E and F), and a crr (H91Q) mutant (G and H) were treated with 3.5 mg/mL phenol (C, E and G) or 5× MIC ciprofloxacin (D, F, and H) for the indicated times. (I) Suppression of stress-induced intracellular cAMP production by ΔptsI. Exponentially growing wild-type and ΔptsI mutant cells were treated with 3.5 mg/mL phenol or 5× MIC ciprofloxacin for the indicated times after which intracellular cAMP levels were determined. (J–L) Suppression of ptsI-mediated protection by exogenous cAMP. Wild-type and ΔptsI cells were pretreated with 8 mM cAMP for 15 min before addition of phenol (J) at 3.5 mg/mL or ciprofloxacin (K) at 5× MIC. L shows the concentration dependence of cAMP-mediated suppression of killing by 5× MIC ciprofloxacin with the ΔptsI mutant. (M and N) Deficiency of cyaA protects from phenol- and ciprofloxacin-mediated killing. Cultures of wild-type and ΔcyaA mutant cells were treated with 3.5 mg/mL phenol (M) or 10× MIC ciprofloxacin (N) for the indicated times. Complementation was observed with a plasmid-borne wild-type cyaA. (O and P) Suppression of the protective effect of ΔptsI by crp*. Exponentially growing cultures of wild-type, ΔptsI, crp*, and ΔptsI-crp* double-mutant cells were treated with 3.5 mg/mL phenol (O) or 5× MIC ciprofloxacin (P) for the indicted times. Data indicate average of three biological replicates; error bars indicate SEM. See SI Appendix, Fig. S4 and Tables S1 and S3 for supporting information.
Since phosphorylated Crr (EIIAGlc) activates adenyl cyclase (21, 29, 30), we asked whether the ptsI deficiency confers tolerance by lowering cAMP levels. Exposure to phenol or ciprofloxacin triggered a surge in cellular cAMP levels with wild-type cells; such a surge was reduced by a deficiency in ptsI (Fig. 3I). Addition of exogenous cAMP to cultured bacteria partially reversed ΔptsI-mediated protection from the lethal activity of phenol and almost completely reversed protection from ciprofloxacin-mediated killing in a concentration-dependent manner (Fig. 3 J–L). Moreover, cAMP supplementation had no effect on MIC (SI Appendix, Table S3). Furthermore, a deficiency in cyaA, which encodes adenyl cyclase, reduced killing by ciprofloxacin and phenol without affecting MIC (Fig. 3 M and N and SI Appendix, Table S1); such protection from killing was reversed when a plasmid-borne wild-type cyaA was used to complement the ΔcyaA mutation (Fig. 3 M and N). We also combined ΔptsI with a crp* allele, which encodes a constitutively active variant of the transcription regulator Crp that bypasses the requirement for cAMP binding to activate Crp (31, 32). The added crp* allele eliminated ΔptsI-mediated tolerance (Fig. 3 O and P and SI Appendix, Table S1). This observation suggests that PtsI stimulates death through cAMP activation of Crp. Finally, when we examined a deficiency of cyaA or crp for pan-tolerance, we found that both mutations protected from killing by various agents while exhibiting little effect on MIC (Fig. 4 and SI Appendix, Figs. S5 and S6 and Tables S1 and S3). Again, complementation with a wild-type copy of cyaA or crp restored the protection from killing by a deficiency in either of these two genes with four diverse stressor types (Fig. 4 and SI Appendix, Figs. S5 F and I and S6 C, F, H, and I). Overall, the data indicate that ΔptsI-mediated pan-tolerance is achieved by blocking the PTS phosphorelay, which results in loss of adenyl cyclase activation that normally occurs via phosphorylated Crr (EIIAGlc) (21, 30). Reduced cAMP production and subsequent lack of Crp activation protect from killing by diverse lethal agents.
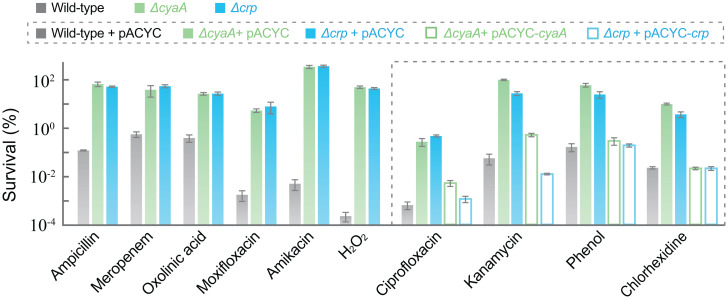
Fig. 4.
Deficiency of cyaA or crp protects from killing by diverse lethal stressors. Exponentially growing wild-type, ΔcyaA, and Δcrp cultures were treated with ampicillin (4× MIC for 3 h), meropenem (6× MIC for 8 h), oxolinic acid (10 MIC for 2 h), moxifloxacin (10× MIC for 2 h), amikacin (3× MIC for 2 h), hydrogen peroxide (25 mM for 30 min), ciprofloxacin (10× MIC for 2 h), kanamycin (5× MIC for 2 h), phenol (3.5 mg/mL for 25 min), or chlorhexidine (12 μg/mL for 4 h) prior to measurement of survival. Complementation with plasmid-borne wild-type cyaA or crp (pACYC-184-cyaA or pACYC-184-crp) was performed for four diverse stressors (ciprofloxacin, kanamycin, phenol, and chlorhexidine). (Also see SI Appendix, Figs. S5 F and I and S6 C, F, H, and I for more complementation data.) Data are averages of three biological replicates; error bars indicate SEM. See SI Appendix, Figs. S5 and S6 and Tables S1 and S3 for supporting information.
Both ptsI and cyaA mutants were also obtained in a tolerance screen using antimicrobials rather than phenol. In this set of experiments, the antimicrobials (kanamycin, ciprofloxacin, and ampicillin) were administered sequentially to E. coli cultures, with two challenge rounds per drug. Four pan-tolerant mutants were obtained. One harbored a D300E substitution in CyaA; the other three exhibited mutations in ptsI (two were premature stop-codon defects, and one was an 81-bp deletion) (SI Appendix, Table S2). The difference in pan-tolerance alleles obtained from the two different enrichment approaches may derive from the two experiments being carried out at different times using different starting cultures.
PTS-mediated modulation of CyaA (adenylate cyclase) activity by Crr phosphorylation and the subsequent connection to the cAMP-Crp master transcription regulatory system fit well with previous observations in which factors affecting cAMP levels modulate killing by β-lactams, fluoroquinolones, hydrogen peroxide, and hypochlorous acid (33–36). We conclude that E. coli evades the lethal action of antimicrobials and disinfectants through the same mechanism, with the PTS lying upstream of the cAMP-Crp regulatory cascade.
ptsI Deficiency Confers Tolerance through Down-Regulation of Genes Encoding Enzymes of the TCA Cycle, Electron Transport Chain Units, and ATP Synthases.
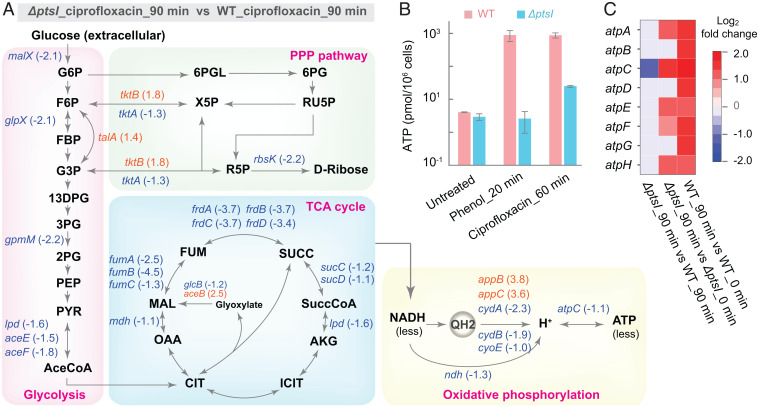
Since the cAMP-Crp system regulates carbon utilization and energy metabolism (37), we next examined the effect of a deficiency in the PTS on energy metabolism. For this experiment, we performed RNA-sequencing (RNA-seq) assays using ciprofloxacin rather than phenol as a stressor, because the latter kills so rapidly that massive killing might occur before transcription patterns change. When we compared cells treated with ciprofloxacin to those serving as untreated controls, we found that ciprofloxacin treatment elevated expression of many genes involved in the TCA cycle, glycolysis, pentose phosphate pathway (PPP), and oxidative phosphorylation with both wild-type and the ptsI mutant cells; however, such elevation was lower with the ptsI mutant than with wild-type cells (SI Appendix, Fig. S7 A and B). When ciprofloxacin-treated samples were compared, the ptsI mutant showed reduced expression of many genes involved in the TCA cycle, electron transport chain, and glycolysis, but elevated expression of several genes in the PPP relative to wild-type cells (Fig. 5A).
Fig. 5.
ptsI deficiency associated with a metabolic flux shift and down-regulation of ATP synthesis. (A) Effects of a deficiency in ptsI on ciprofloxacin-induced (5× MIC) expression of genes involved in metabolic flux (glycolysis, PPP, TCA cycle) and oxidative phosphorylation. Shown are genes up-regulated (orange; number in parenthesis represents log2 increase) or down-regulated (blue; number in parenthesis represent log2 decrease) by greater than twofold in the indicated metabolic pathways, as revealed by RNA-seq analysis. (B) Deficiency in ptsI restricts the increase in cellular ATP level stimulated by phenol or ciprofloxacin. Wild-type and ΔptsI-mutant cells were treated with 3.5 mg/mL phenol for 20 min or 5× MIC ciprofloxacin for 60 min. ATP levels were determined and expressed as pmol/106 cells. Data are averages of three biological replicates; error bars indicate SEM. (C) Suppression of ciprofloxacin-mediated up-regulation of ATP synthase genes by a deficiency in ptsI. Ciprofloxacin treatment at 5× MIC for 90 min up-regulated all eight genes encoding ATP synthases in wild-type cells; a deficiency in ptsI inhibited all up-regulation and suppressed atpC expression, as revealed by RNA-seq analysis. Panel shows comparisons; far right bar shows color scale. See SI Appendix, Fig. S7 for supporting information.
Even in the absence of stress, the ptsI deficiency reduced the expression of many genes involved in the TCA cycle, oxidative phosphorylation, and the conversion of pyruvate to acetyl-CoA, a substrate that fuels the TCA cycle. Elevated expression of many genes involved in glycolysis and PPP was seen in the same comparison (SI Appendix, Fig. S7C). Thus, a deficiency in ptsI shifts energy metabolism from the TCA cycle to glycolysis and PPP; that shift would precondition bacterial cells for becoming tolerant to lethal agents.
Shifting metabolic flux away from the TCA cycle to glycolysis and PPP was expected to reduce ATP synthesis. Indeed, when we measured intracellular ATP levels during phenol or ciprofloxacin treatment, they were 50- to 200-fold lower in the ΔptsI mutant than in the wild-type control (Fig. 5B). Similar suppression of a ciprofloxacin-induced, time-dependent ATP increase was observed with the cyaA and crp mutants (SI Appendix, Fig. S8 A–C). When the ptsI mutant was further examined by RNA-seq, samples from ciprofloxacin-treated and -untreated cells showed that ciprofloxacin increased the expression of only half of the ATP synthase genes, while with wild-type cells expression of all the genes in the set was up-regulated by two- to fourfold. Moreover, the level of up-regulation was lower in the mutant than in the corresponding wild-type cells (Fig. 5C). Comparison of the ptsI mutant and wild-type cells revealed that with ciprofloxacin treatment, the ptsI deficiency suppressed expression of all ATP synthase genes, with that of atpC suppressed the most (e.g., greater than twofold) (Fig. 5C).
Collectively, the data suggest that pan-tolerance is achieved by the diversion of carbon flux from the TCA cycle to glycolysis and the pentose phosphate pathway via defects in the PTS-cAMP regulatory system. That diversion reduces electron transfer and ATP synthesis during lethal stress. The central nature of this metabolic shift allows it to affect the lethal action of diverse stressors.
Intracellular ROS Accumulation Contributes to the Common Death Mechanism.
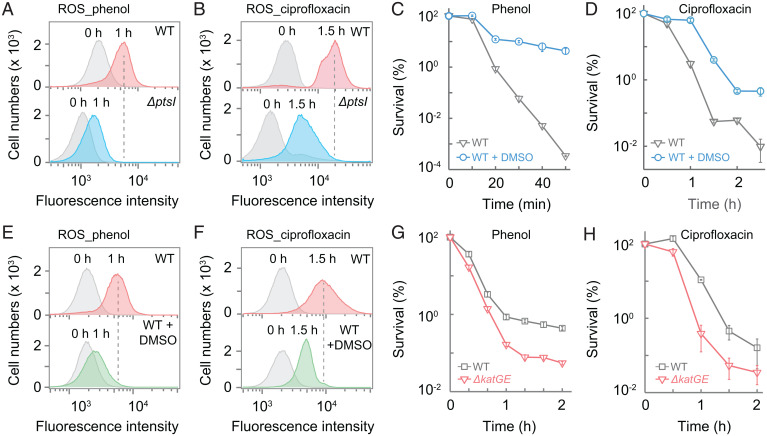
Since ROS are byproducts of electron transfer by the respiratory chain during production of ATP (38, 39) and since stress-mediated ATP synthesis is reduced in a ΔptsI mutant (Fig. 5 B and C and SI Appendix, Fig. S8 A–C), we expected stress-mediated ROS accumulation to also be lower in the mutant. Moreover, pilot work by us and others (33–35) suggested that reduction of cAMP levels correlates with suppression of stressor-mediated ROS accumulation and protection from stress-mediated killing. When ROS accumulation was measured by flow cytometry using the fluorescent dye carboxyl-H2DCFDA [5(6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate], treatment with phenol or ciprofloxacin elevated ROS levels, but to lower extents with the ptsI-deficient mutant than with wild-type cells (Fig. 6 A and B). Moreover, a deficiency in any of three key genes (ptsI, cyaA, or crr), which define the lethal pathway currently studied, suppressed a ciprofloxacin-induced ROS surge (SI Appendix, Fig. S8D).
Fig. 6.
Intracellular ROS associated with stress-mediated death. (A and B) ptsI deficiency lowers phenol- and ciprofloxacin-stimulated intracellular ROS levels. Wild-type and ΔptsI-mutant cells were pretreated with carboxy-H2DCFDA for 20 min and then treated with 3.5 mg/mL phenol (A) or with 5× MIC ciprofloxacin (B) for the indicated times. Samples were subjected to flow cytometry for analysis of intracellular ROS levels. Similar results were obtained in three replicate experiments. (C and D) Protective effect of DMSO on survival. Exponentially growing wild-type cultures were treated with 3.5 mg/mL phenol (C) or 5× MIC ciprofloxacin (D) for the indicated times in the absence or presence of 5% DMSO. (E and F) Suppression of ROS accumulation by DMSO. Wild-type cells were pretreated with carboxy-H2DCFDA for 20 min and then treated with 3.5 mg/mL phenol (E) or with 5× MIC ciprofloxacin (F) for the indicated times before intracellular ROS levels were measured by flow cytometry. (G and H) Catalase/peroxidase deficiency increases phenol- or ciprofloxacin-mediated killing. Exponentially growing cultures of wild-type and a katG-katE double mutant were treated with 3 mg/mL phenol (G) or 5× MIC ciprofloxacin (H) for the indicated times. Data are averages of three biological replicates; error bars indicate SEM. See SI Appendix, Tables S1 and S3 for supporting information.
A contribution of ROS to stress-mediated killing can be assessed by treating cells with agents that lower ROS accumulation. In the present work, addition of dimethyl sulfoxide (DMSO), which can scavenge hydroxyl radicals (40, 41), protected wild-type cells from being killed by phenol and lowered the rate of killing by ciprofloxacin (Fig. 6 C and D) without affecting MIC (SI Appendix, Table S3). DMSO also lowered the phenol- or ciprofloxacin-mediated intracellular ROS surge (Fig. 6 E and F). Moreover, a deficiency in ROS detoxification (katGE) increased killing by phenol and ciprofloxacin (Fig. 6 G and H) with no effect on MIC (SI Appendix, Table S1). Collectively, our data, plus earlier work showing that ROS contribute to killing by lethal stressors (7, 11, 12) but not to growth inhibition (42, 43), indicate that reduced ROS accumulation is responsible for the pan-tolerance phenotype of ΔptsI.
Discussion
Challenging E. coli cells with phenol or antimicrobials resulted in the recovery of pan-tolerant mutants. Characterization of these mutants revealed a death pathway shared by lethal insults of many diverse types. These data demonstrate that, unlike resistance, which is usually stressor-class-specific, tolerance may be a cellular property that is not stressor-specific and is likely to simultaneously apply to many, if not all, lethal stressors. Pan-tolerance was conferred by a genetic deficiency in ptsI that reduced cAMP synthesis, since 1) lethal stress stimulated an increase in intracellular cAMP, which was suppressed by ΔptsI, and exogenous cAMP reversed ΔptsI-mediated tolerance; 2) defects in cAMP synthase caused pan-tolerance; and 3) combining ΔptsI with crp*, an allele encoding a variant Crp that bypasses the requirement of cAMP binding for transcription activation (31, 32), eliminated ΔptsI-mediated pan-tolerance. Reduced levels of cAMP were associated with a shift in metabolic flux from the TCA cycle to the PPP and glycolysis, thereby lowering ATP production and the associated ROS accumulation. The concordance among stress-mediated cAMP level increase, reduction in stress-stimulated ATP production, and ROS accumulation in ΔptsI, ΔcyaA, and Δcrp mutants defines a PTS-cAMP-Crp regulatory cascade that controls cell death. The metabolic shift to glycolysis and PPP also produces metabolic intermediates, such as nucleotides, fatty acids, and aromatic amino acids (44, 45), that are needed for repair of stress-mediated damage (1, 3, 7). In addition, NADPH generated from the PPP provides reducing power to synthesize glutathione, which counters oxidative stress (46). Thus, the PTS-cAMP-Crp regulatory axis can regulate stress-mediated cell death in several ways.
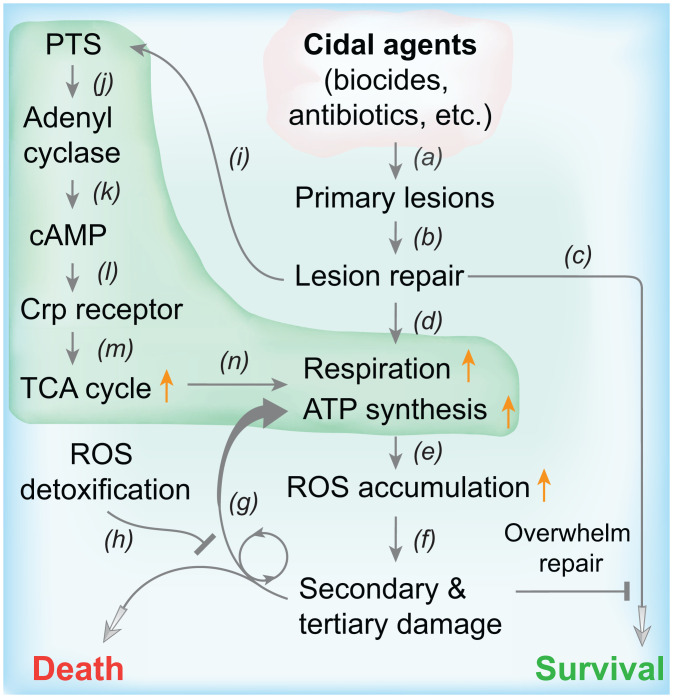
A scheme describing stress-mediated metabolic death emerges from combining the present observations with previous reports (Fig. 7). We postulate that repair of primary damage imposes a sudden demand for ATP (47, 48), which stimulates carbohydrate metabolism (via the PTS-cAMP-Crp pathways), respiration, ATP synthesis, and ROS accumulation. In turn, ROS damage macromolecules; such damage stimulates additional ROS accumulation in a self-amplifying cycle that eventually overwhelms repair and leads to cell death (7). Bacteria also possess ROS-detoxifying systems whose deficiencies do not affect normal growth or stressor resistance but render cells more vulnerable to killing by diverse stressors (3, 43). Wild-type versions likely protect cells from moderate levels of lethal stress.
Fig. 7.
Proposed relationships of PTS, cAMP-Crp, respiration, ROS accumulation, and damage repair in stress-mediated bacterial death. Bactericidal treatments produce primary lesions (a) that are recognized by repair systems (b). If repair is sufficient, cells survive (c) unless overwhelmed by downstream damage. Repair increases demand for ATP and stimulates respiration (d), resulting in elevated ATP synthesis. ROS accumulate as by-products during ATP synthesis (e); ROS damage macromolecules (f), including DNA, protein, and lipids. Macromolecular damage from ROS stimulates additional ROS accumulation and damage in a self-amplifying cycle (f and g) that leads to cellular repair being overwhelmed and ultimately to cell death unless ROS-detoxifying systems are sufficient to suppress ROS accumulation (h). The demand for ATP by repair also stimulates carbohydrate metabolism via the PTS (i), which in turn stimulates adenyl cyclase (j) to synthesize cAMP (k). cAMP binds to and activates Crp (l), which up-regulates genes involved in the TCA cycle (m), thereby producing elevated levels of NADH that fuel respiration (n), subsequent ATP synthesis, and ROS production. L-shaped green shading indicates pathways from which pan-tolerance mutants may emerge.
Since energy metabolism involves multiple steps and enzymes, pan-tolerance mutations are unlikely to be limited to the PTS or cAMP-Crp pathways. Discovery of additional pan-tolerant mutations will expand our knowledge of the general death pathway. We emphasize that the death pathway we describe refers to a cellular response to stress and not to death from physical destruction, such as cell lysis or macromolecular denaturation from very high temperature or high-dose irradiation. Thus, the existence of a broadly applicable and even potentially universal death pathway that we propose does not exclude the existence of other lethal mechanisms.
The stable, ptsI-cyaA-ROS–mediated tolerance we describe is unique, as these tolerance mutations do not affect bacterial growth. That provides a clear separation of the tolerance we describe from reduced killing due to a growth defect, a caveat that affects the “high-survival” phenotype for most, if not all, growth defect-based tolerance: in such cases, reduced killing can derive from either slow growth or tolerance unrelated to growth. In growth defect-associated tolerance, a key role is played by the stringent response (49–51). Genes involved in growth-defective tolerance include amino acyl tRNA synthetases (metG), ribosephosphate diphosphokinase (prs), ATP-dependent proteases (clpX, clpP), and putative toxin-antitoxin modules (vapBC) (19, 52, 53). Whether ROS is involved is not known. Another type of tolerance derives from increases in endogenous hydrogen sulfide or nitric oxide. These compounds affect intracellular ROS accumulation (54, 55).
Since the ptsI mutant in our study displayed no growth defect, we considered the possibility that the Luria-Bertani (LB) growth medium contributed to tolerance. This medium is poor in sugar but rich in small peptides and amino acids that do not rely on the PTS for carbon-source transport. The mutant did not grow in M9 medium containing glucose as the sole carbon source, consistent with the PTS being the major transporter of glucose. The mutant did grow in M9 plus pyruvate, albeit more slowly than with LB; no growth rate difference was observed between wild-type and ptsI mutant cultures (SI Appendix, Fig. S9 A and B). When we measured ciprofloxacin-mediated killing, which was slower than with LB medium, ptsI-mediated tolerance was observed (SI Appendix, Fig. S9C). Thus, tolerance was not medium-specific, nor did it derive from a growth defect.
Tolerance has two important consequences. First, it likely favors the emergence of resistance by preventing an antimicrobial from rapidly reducing the bacterial burden during infection. Resistant mutants can then spontaneously emerge from large surviving bacterial populations. Those mutant subpopulations are then enriched, as they survive the selective conditions (56). Indeed, recent work associates the emergence of clinical resistance with tolerance during infection by Staphylococcus aureus, even during combination therapy (56). We note that the tolerant mutants we reported exhibited small increases in MIC for some antimicrobials (SI Appendix, Tables S1 and S3); such increases would have made them appear even more tolerant to killing than when we normalized drug concentrations to MIC for killing assays in the present work (we used the same fold of MIC rather than an absolute drug concentration, which is generally not the case in clinical practice). A second consequence, even without the emergence of resistance, is the loss of lethal activity with patients having compromised immune systems (57–59). Tolerance may increase treatment failure and the frequency of relapse (60, 61). Thus, understanding bacterial death well enough to limit the emergence of tolerance is likely to be clinically important.
Limiting tolerance may be difficult, since involvement of metabolism in the death process implies that many different genes can mutate to confer the pan-tolerant phenotype, which makes the emergence of pan-tolerance a high-probability event. At present, the extent of pan-tolerance in pathogen populations is unknown, because susceptibility assays used for surveys measure MIC rather than tolerance. Although agar-plate assays for tolerance have been reported (62), measuring tolerance with large numbers of clinical isolates remains labor intensive. Regardless of the current prevalence of tolerance, we expect that unrestricted, massive disinfectant consumption will increase the prevalence of tolerance and eventually contribute to the emergence of antimicrobial resistance.
Materials and Methods
Bacteria and Reagents.
E. coli K-12 strains (SI Appendix, Table S1) were grown aerobically at 37 °C in LB medium (63) with shaking at 200 rpm. Colony formation was on LB agar at 37 °C. Bacteriophage P1-mediated transduction (64) or CRISPR-based allelic exchange (24) was used for strain construction. Flow cytometry reagents were purchased from Becton Dickinson. Tryptone, yeast extract, powder for LB broth and agar, and carboxy-H2DCFDA were obtained from Thermo Fisher Scientific. Other reagents, including antimicrobials (ciprofloxacin, oxolinic acid, kanamycin, ampicillin, tetracycline, moxifloxacin, and chloramphenicol), phenol, dimethyl 2-oxoglutarate, cyclic AMP, sodium pyruvate, and DMSO were purchased from Sigma-Aldrich. Gentamicin, amikacin, hydrogen peroxide, chlorhexidine, ethanol, isopropanol, 1-butanol, potassium dichromate, sodium hypochlorite solution (5.2%), hydrogen chloride, and sodium hydroxide were purchased from Sangon Biotech. Meropenem (Shenghuaxi Pharmaceutical) and ceftriaxone (Roche) were from Zhongshan Hospital (Xiamen, China) pharmacy.
Susceptibility Determinations.
MIC was determined using a staggered, twofold broth dilution method for each antibiotic and disinfectant. Midlog-phase cultures were diluted 1:5,000 in LB broth to ∼105 cells/mL, mixed with various amounts of each stressor, and incubated for 24 h. MIC was determined as the lowest drug concentration that inhibited culture growth, measured by turbidity, by at least 90% relative to an untreated control.
Bacterial Killing Assays.
Bacterial cultures grown overnight were diluted by 100-fold into fresh medium and then grown to exponential phase (OD600∼0.25) before cultures were aliquoted into glass culture tubes and incubated with various antimicrobials either at a fixed concentration (e.g., a particular fold of MIC) for various times or at various drug concentrations (normalized to MIC) for a fixed time. For UV light-mediated killing, 10 μL of serially diluted bacterial samples were spotted in triplicate on LB agar before exposing the agar plates to UV light at a fixed intensity (0.056 mW/cm2) for various times. Survival following lethal treatment was determined by plating culture aliquots on agar incubated at 37 °C for 24 h. At the end of incubation, colony-forming units were determined visually. Percent survival was determined relative to an untreated control sampled at the time lethal treatment was initiated. When dimethyl 2-oxoglutarate, cyclic AMP, or DMSO were used to inhibit or potentiate killing by disinfectants or antimicrobials, they were added at noninhibitory/subinhibitory concentrations 40, 15, or 0 min before initiation of lethal treatments.
Enrichment of Tolerant Mutants.
To obtain phenol-tolerant mutants, cultures of the wild-type (strain 1) were grown to exponential phase (OD600 ∼0.25) in 30 mL LB medium at 37 °C with shaking at 200 rpm, and they were then treated with 3.5 mg/mL phenol for 20 min, which killed roughly 99% of the cells. Cells were concentrated by centrifugation (4,600 × g, 4 °C, 5 min) and resuspended in 20 mL LB medium. Centrifugation and culture resuspension were performed two more times to remove residual phenol. Cultures were then diluted 100-fold, grown overnight, and subjected to an additional round of enrichment. After 10 rounds of enrichment, treatment with 3.5 mg/mL phenol for 20 min killed less than 70% of cells. These cultures were washed, plated on phenol-free agar and on agar containing 1× MIC of phenol. No colony grew on phenol-containing agar while hundreds grew on phenol-free agar, indicating that few phenol-resistant mutants were enriched. When colonies recovered on phenol-free agar were tested for killing and growth inhibition by phenol, their MIC for phenol was similar to that of the parental, wild-type strain, but survival was much higher (>100-fold). Thus, they were considered tolerant to phenol. One representative tolerant mutant and the parental wild-type strain were subjected to comparative whole-genome sequencing to identify mutations involved in tolerance.
Mutants that were tolerant to antibiotics were obtained in a similar way, using three different agents sequentially at 5× MIC for 3 h (ampicillin), 2.5 h (ciprofloxacin), and 0.5 h (kanamycin) to minimize the selection of resistant mutants from masking enrichment of tolerant mutants. Cultures were treated with each agent once in a round for two rounds, and then they were plated on drug-free agar to obtain single colonies for subsequent confirmation of the tolerance phenotype (little change in MIC but greatly reduced killing). The enrichment was stopped after two rounds, because two rounds allowed sufficient enrichment of tolerant mutants while minimizing the presence of resistant mutants that would overwhelm the tolerant population. Representative tolerant mutants and the wild-type parental strain were subjected to comparative whole-genome sequencing to identify mutations involved in tolerance.
Whole-Genome Sequencing.
Chromosomal DNA was isolated from overnight cultures of wild-type and mutant strains using the Bacterial Chromosome DNA Isolation Kit (Tiangen Biotech) according to the vendor’s technical manual. DNA samples were sent to Novogene for whole-genome sequencing and comparative sequence analysis against the E. coli BW25113 whole-genome sequence (GenBank accession no.: CP009273.1). Nonsynonymous mutations found in mutants but not in the wild-type strain were reconstructed by CRISPR-based allelic exchange (24) in the wild-type parental strain for verification of the tolerance phenotype.
RNA-Seq Analysis.
Exponentially growing cultures (OD600 = 0.3) of the wild-type strain and the ΔptsI mutant were treated with 5× MIC ciprofloxacin for 0 or 90 min. Samples were collected by centrifugation (4,600 × g, 4 °C, 10 min), frozen using liquid nitrogen, and stored at −80 °C before being shipped to Novogene for RNA extraction, library construction, and RNA-seq. Briefly, total RNAs were prepared using the RNA Extraction Kit (Tiangen Biotech). Sequence libraries were generated using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s recommendations; index codes were added to attribute sequences to each sample. The rRNAs were removed by the Ribo-zero Kit (Illumina) to enrich mRNA, followed by treatment with fragmentation buffer (NEBNext RNA First Strand Synthesis Reaction Buffer, New England Biolabs) to break the mRNA into short fragments. One strand of cDNA was synthesized using a six-base random primer and mRNAs as templates followed by synthesis of the corresponding double-stranded cDNAs. The purified double-stranded cDNAs were first end-repaired, A-tailed, and ligated to the sequencing linker; fragments (150∼200 bp) were selected using the AMPure XP system (Beckman Coulter) before PCR amplification was performed and the products were purified using the AMPure XP system to obtain the final library (65). All samples were sequenced on an Illumina HiSeq 2500/Miseq instrument. HTSeq v0.6.1 was used to count the read numbers mapped to each gene (66, 67). RNA-seq data were analyzed using the DEGSeq R package (v1.20.0) (68–71), and the results were deposited into the Gene Expression Omnibus (GEO) database (accession no: GSE133439; access token: qfuleowglfulfah).
Plasmid Construction, Complementation, and Allelic Exchange.
Plasmids used for complementation and CRISPER-based genetic knockout or allelic exchange (24) were constructed by PCR amplification of the targeted wild-type or mutant gene fragments using primers listed in SI Appendix, Table S4, followed by cloning the fragments into pACYC184 or pTargetF after digestion of both plasmids and fragments with the appropriate restriction enzymes (enzyme cutting sites for each cloning are shown as bold, underlined bases in the primer sequences in SI Appendix, Table S4). After ligation with T4 DNA ligase, the fragment–plasmid ligation mixtures were introduced into E. coli DH5α by bacterial transformation, and transformants were selected on agar containing the appropriate antibiotic. Recombinant plasmids were isolated and confirmed by DNA sequencing before they were used for complementation or mutant construction. The N20 sequences for Cas9 recognition were designed to target the kanamycin-resistance cassette of the Keio mutant (72) of interest when the wild-type allele or mutant alleles containing single-base mutations for the targeted gene were constructed. The back-cross of a wild-type allele into a strain containing a point mutation in the same gene was carried out by targeting the mutant allele with an N20-mediated Cas9 cleavage followed by providing a wild-type template refractory to Cas9 cleavage due either to the absence of mutant-specific N20 or to introduction of a synonymous mutation that abolishes the NGG PAM site needed for Cas9 cleavage. A frame-shift mutation in cyaA was found in the Keio collection ptsI mutant during the present work; this cyaA mutation was restored to wild-type by CRISPR-based allelic exchange for all strains that were prepared from the Keio-collection ptsI mutant. Double-mutant construction was performed either by bacteriophage P1-mediated transduction (64, 73) or by CRISPER-mediated introduction of the second, marker-less mutation after using pCP20-based kanamycin marker removal from the first mutation locus (24, 72).
Measurement of Cellular ATP Levels.
Exponentially growing cultures of wild-type, the ptsI mutant (strain 2), the cyaA mutant (strain 17), and the crp mutant (strain 34) were treated with 3.5 mg/mL phenol or 5 MIC of ciprofloxacin for various times before cells were concentrated by centrifugation (12,000 × g, 2 min). Cell pellets were washed two times with cold saline (0.9% NaCl) before they were resuspended in lysis buffer provided in the assay kit. Cells were lysed by repeated (10-s treatment followed by 10-s rest) ultrasonic treatment with a VC-750 ultrasonic processor (Sonics & Materials) at 40% power setting on ice for a total of 2 min. After lysis, cell debris was removed by centrifugation (12,000 × g, 2 min) and the supernatant was used for determination of ATP levels using a firefly luciferase-luciferin–based Enhanced ATP Assay Kit (Beyotime Biotechnology) and a luminometer (CLARIOstar, BMG Labtech) according to the vendor’s technical manual. The amount of ATP was calculated and normalized to cell numbers.
Measurement of Intracellular cAMP.
Exponentially growing cultures of wild-type and the ptsI mutant (strain 2) were treated with 3.5 mg/mL phenol or 5 MIC of ciprofloxacin for various times before cells were concentrated by centrifugation (12,000 × g, 2 min). Cell pellets were washed two times with cold saline (0.9% NaCl) before they were resuspended in lysis buffer provided in the assay kit. Cells were lysed by repeated (10-s treatment followed by 10-s rest) ultrasonic treatment with a VC-750 ultrasonic processor (Sonics & Materials) at 40% power setting on ice for a total of 2 min. After lysis, cell debris was removed by centrifugation (12,000 × g, 2 min), and the supernatant was used for determination of cAMP levels using a monoclonal anti-cAMP antibody-based direct cAMP ELISA Kit (NewEast Biosciences) and a luminometer (CLARIOstar, BMG Labtech) according to the vendor’s technical manual. The amount of cAMP was calculated and normalized to cell numbers.
Measurement of ROS by Flow Cytometry.
Intracellular ROS accumulation was measured using an ROS-sensitive fluorescence dye as described previously (7). Carboxy-H2DCFDA (74, 75) (final concentration 10 μM) was added to cultures for detection of total intracellular ROS. A culture lacking the fluorescent dye was included as a control for autofluorescence (little was detected). At various times after phenol or ciprofloxacin treatment, the fluorescence intensity of bacterial cells was measured using fluorescence-based flow cytometry with a CytoFLEX A00-1-1102 flow cytometer (Beckman Coulter; distributor: Suzhou Xitogen Biotechnologies Co., Ltd.). All tubes with cultures were wrapped with aluminum foil to avoid light. Samples (200 µL), taken at various times, were chilled on ice and subsequently analyzed by flow cytometry. A total of 100,000 cells was analyzed at a speed of 35 µL/min for each sample to determine fluorescence values. The detection parameters were 20-mV laser power, 533/30-nm band pass filter (FL1-channel). Data were analyzed using FlowJo software. Each data point represents the average of at least three independent measurements.
Statistical Considerations.
At least three biological replicates were obtained for all experiments except for RNA-seq analysis. Each data point represents the mean of replicate experiments; error bars show SEMs, unless otherwise stated.
Supplementary Material
Acknowledgments
We thank Marila Gennaro, Matthew Neiditch, Bo Shopsin, Katsunori Sugimoto, and Jason Yang for critical comments; and Yang Liu, Liang Yang, and Yingdan Zhang for assistance in RNA-sequencing data analysis. The work was supported in part by grants from National Natural Science Foundation of China (82172316, 81473251, 81661138005) and by an intramural bridging fund from the Public Health Research Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118566119/-/DCSupplemental.
Data Availability
Data supporting the findings of this study are all shown in the main text and SI Appendix. The RNA-seq data are available from the GEO database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE133439). Bacterial mutant strains and plasmids constructed in the work are listed in SI Appendix, Tables S1 and S4; they are available upon request.
References
- 1.Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J., A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Belenky P., et al. , Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 13, 968–980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dwyer D. J., et al. , Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A. 111, E2100–E2109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohanski M. A., Dwyer D. J., Wierzbowski J., Cottarel G., Collins J. J., Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 135, 679–690 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobritz M. A., et al. , Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A. 112, 8173–8180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong Y., Li L., Luan G., Drlica K., Zhao X., Contribution of reactive oxygen species to thymineless death in Escherichia coli. Nat. Microbiol. 2, 1667–1675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong Y., Zeng J., Wang X., Drlica K., Zhao X., Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. U.S.A. 116, 10064–10071 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foti J. J., Devadoss B., Winkler J. A., Collins J. J., Walker G. C., Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science 336, 315–319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keren I., Wu Y., Inocencio J., Mulcahy L. R., Lewis K., Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339, 1213–1216 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Imlay J. A., Cell death from antibiotics without the involvement of reactive oxygen species. Science 339, 1210–1213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drlica K., Zhao X., Bacterial death from treatment with fluoroquinolones and other lethal stressors. Expert Rev. Anti Infect. Ther. 19, 601–618 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Dwyer D. J., Collins J. J., Walker G. C., Unraveling the physiological complexities of antibiotic lethality. Annu. Rev. Pharmacol. Toxicol. 55, 313–332 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Imlay J. A., Diagnosing oxidative stress in bacteria: Not as easy as you might think. Curr. Opin. Microbiol. 24, 124–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X., Drlica K., Reactive oxygen species and the bacterial response to lethal stress. Curr. Opin. Microbiol. 21, 1–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brauner A., Fridman O., Gefen O., Balaban N. Q., Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Handwerger S., Tomasz A., Antibiotic tolerance among clinical isolates of bacteria. Annu. Rev. Pharmacol. Toxicol. 25, 349–380 (1985). [DOI] [PubMed] [Google Scholar]
- 17.Balaban N. Q., et al. , Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuomanen E., Durack D. T., Tomasz A., Antibiotic tolerance among clinical isolates of bacteria. Antimicrob. Agents Chemother. 30, 521–527 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuo M., et al. , Genetic and transcriptomic analyses of ciprofloxacin-tolerant Staphylococcus aureus isolated by the replica plating tolerance isolation system (REPTIS). Antimicrob. Agents Chemother. 63, e02019-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutscher J., et al. , The bacterial phosphoenolpyruvate:carbohydrate phosphotransferase system: Regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol. Mol. Biol. Rev. 78, 231–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krin E., Sismeiro O., Danchin A., Bertin P. N., The regulation of enzyme IIA(Glc) expression controls adenylate cyclase activity in Escherichia coli. Microbiology (Reading) 148, 1553–1559 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Lister J., On the antiseptic principle in the practice of surgery. BMJ 2, 246–248 (1867). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenberg S., Klauck G., Pesavento C., Klauck E., Hengge R., The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signalling cascade in E. coli biofilm control. EMBO J. 32, 2001–2014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y., et al. , Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin-Reisman I., Brauner A., Ronin I., Balaban N. Q., Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl. Acad. Sci. U.S.A. 116, 14734–14739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craig W. A., Choosing an antibiotic on the basis of pharmacodynamics. Ear Nose Throat J. 77 (6, suppl.), 7–11, discussion 11–12 (1998). [PubMed] [Google Scholar]
- 27.Doucette C. D., Schwab D. J., Wingreen N. S., Rabinowitz J. D., α-Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 7, 894–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tchieu J. H., Norris V., Edwards J. S., Saier M. H. Jr., The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3, 329–346 (2001). [PubMed] [Google Scholar]
- 29.Park Y. H., Lee B. R., Seok Y. J., Peterkofsky A., In vitro reconstitution of catabolite repression in Escherichia coli. J. Biol. Chem. 281, 6448–6454 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Deutscher J., Francke C., Postma P. W., How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harman J. G., McKenney K., Peterkofsky A., Structure-function analysis of three cAMP-independent forms of the cAMP receptor protein. J. Biol. Chem. 261, 16332–16339 (1986). [PubMed] [Google Scholar]
- 32.Karimova G., Ladant D., Ullmann A., Relief of catabolite repression in a cAMP-independent catabolite gene activator mutant of Escherichia coli. Res. Microbiol. 155, 76–79 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Barth E., et al. , Interplay of cellular cAMP levels, sigmaS activity and oxidative stress resistance in Escherichia coli. Microbiology (Reading) 155, 1680–1689 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donovan G. T., Norton J. P., Bower J. M., Mulvey M. A., Adenylate cyclase and the cyclic AMP receptor protein modulate stress resistance and virulence capacity of uropathogenic Escherichia coli. Infect. Immun. 81, 249–258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molina-Quiroz R. C., et al. , Cyclic AMP regulates bacterial persistence through repression of the oxidative stress response and SOS-dependent DNA repair in uropathogenic Escherichia coli. MBio 9, e02144-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwan B. W., Osbourne D. O., Hu Y., Benedik M. J., Wood T. K., Phosphodiesterase DosP increases persistence by reducing cAMP which reduces the signal indole. Biotechnol. Bioeng. 112, 588–600 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Kolb A., Busby S., Buc H., Garges S., Adhya S., Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62, 749–795 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Fridovich I., Superoxide radical: An endogenous toxicant. Annu. Rev. Pharmacol. Toxicol. 23, 239–257 (1983). [DOI] [PubMed] [Google Scholar]
- 39.Hassan H. M., Fridovich I., Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch. Biochem. Biophys. 196, 385–395 (1979). [DOI] [PubMed] [Google Scholar]
- 40.Dabrowski A., Gabryelewicz A., Dabrowska M., Chyczewski L., Effect of dimethylsulfoxide-hydroxyl radical scavenger on cerulein-induced acute pancreatitis in rats. Tokai J. Exp. Clin. Med. 16, 43–50 (1991). [PubMed] [Google Scholar]
- 41.Repine J. E., Pfenninger O. W., Talmage D. W., Berger E. M., Pettijohn D. E., Dimethyl sulfoxide prevents DNA nicking mediated by ionizing radiation or iron/hydrogen peroxide-generated hydroxyl radical. Proc. Natl. Acad. Sci. U.S.A. 78, 1001–1003 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., et al. , Inhibitors of reactive oxygen species accumulation delay and/or reduce the lethality of several antistaphylococcal agents. Antimicrob. Agents Chemother. 56, 6048–6050 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Zhao X., Contribution of oxidative damage to antimicrobial lethality. Antimicrob. Agents Chemother. 53, 1395–1402 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stincone A., et al. , The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 90, 927–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin P. P., et al. , Construction and evolution of an Escherichia coli strain relying on nonoxidative glycolysis for sugar catabolism. Proc. Natl. Acad. Sci. U.S.A. 115, 3538–3546 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aquilano K., Baldelli S., Ciriolo M. R., Glutathione: New roles in redox signaling for an old antioxidant. Front. Pharmacol. 5, 196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dahan-Grobgeld E., et al. , Reversible induction of ATP synthesis by DNA damage and repair in Escherichia coli. In vivo NMR studies. J. Biol. Chem. 273, 30232–30238 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Yang J. H., et al. , A white-box machine learning approach for revealing antibiotic mechanisms of action. Cell 177, 1649–1661.e9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hobbs J. K., Boraston A. B., (p)ppGpp and the stringent response: An emerging threat to antibiotic therapy. ACS Infect. Dis. 5, 1505–1517 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Bryson D., Hettle A. G., Boraston A. B., Hobbs J. K., Clinical mutations that partially activate the stringent response confer multidrug tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 64, e02103-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dutta N. K., et al. , Inhibiting the stringent response blocks Mycobacterium tuberculosis entry into quiescence and reduces persistence. Sci. Adv. 5, eaav2104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fridman O., Goldberg A., Ronin I., Shoresh N., Balaban N. Q., Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Roy S., et al. , Role of ClpX and ClpP in Streptococcus suis serotype 2 stress tolerance and virulence. Microbiol. Res. 223-225, 99–109 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Gusarov I., Shatalin K., Starodubtseva M., Nudler E., Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science 325, 1380–1384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shatalin K., Shatalina E., Mironov A., Nudler E., H2S: A universal defense against antibiotics in bacteria. Science 334, 986–990 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Liu J., Gefen O., Ronin I., Bar-Meir M., Balaban N. Q., Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Kłodzińska S. N., Priemel P. A., Rades T., Nielsen H. M., Combining diagnostic methods for antimicrobial susceptibility testing—A comparative approach. J. Microbiol. Methods 144, 177–185 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Potel G., et al. , Identification of factors affecting in vivo aminoglycoside activity in an experimental model of gram-negative endocarditis. Antimicrob. Agents Chemother. 36, 744–750 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson L. R., Shanholtzer C. J., Tests for bactericidal effects of antimicrobial agents: Technical performance and clinical relevance. Clin. Microbiol. Rev. 5, 420–432 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Britt N. S., et al. , Relationship between vancomycin tolerance and clinical outcomes in Staphylococcus aureus bacteraemia. J. Antimicrob. Chemother. 72, 535–542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gold B., Nathan C., Targeting phenotypically tolerant Mycobacterium tuberculosis. Microbiol. Spectr. 5, 10.1128/microbiolspec.TBTB2-0031-2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gefen O., Chekol B., Strahilevitz J., Balaban N. Q., TDtest: Easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci. Rep. 7, 41284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertani G., Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300 (1951). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomason L. C., Costantino N., Court D. L., E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 79, 1.17.1–1.17.8 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Wang Z., Gerstein M., Snyder M., RNA-seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders S., Huber W., Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love M. I., Anders S., Kim V., Huber W., RNA-seq workflow: Gene-level exploratory analysis and differential expression. F1000 Res. 4, 1070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L., Feng Z., Wang X., Wang X., Zhang X., DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Baba T., et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wall J. D., Harriman P. D., Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology 59, 532–544 (1974). [DOI] [PubMed] [Google Scholar]
- 74.Bass D. A., et al. , Flow cytometric studies of oxidative product formation by neutrophils: A graded response to membrane stimulation. J. Immunol. 130, 1910–1917 (1983). [PubMed] [Google Scholar]
- 75.Brandt R., Keston A. S., Synthesis of diacetyldichlorofluorescin: A stable reagent for fluorometric analysis. Anal. Biochem. 11, 6–9 (1965). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are all shown in the main text and SI Appendix. The RNA-seq data are available from the GEO database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE133439). Bacterial mutant strains and plasmids constructed in the work are listed in SI Appendix, Tables S1 and S4; they are available upon request.