Abstract
The defining trait of obligate anaerobes is that oxygen blocks their growth, yet the underlying mechanisms are unclear. A popular hypothesis was that these microorganisms failed to evolve defences to protect themselves from reactive oxygen species (ROS) such as superoxide and hydrogen peroxide, and that this failure is what prevents their expansion to oxic habitats. However, studies reveal that anaerobes actually wield most of the same defences that aerobes possess, and many of them have the capacity to tolerate substantial levels of oxygen. Therefore, to understand the structures and real-world dynamics of microbial communities, investigators have examined how anaerobes such as Bacteroides, Desulfovibrio, Pyrococcus and Clostridium spp. struggle and cope with oxygen. The hypoxic environments in which these organisms dwell — including the mammalian gut, sulfur vents and deep sediments — experience episodic oxygenation. In this Review, we explore the molecular mechanisms by which oxygen impairs anaerobes and the degree to which bacteria protect their metabolic pathways from it. The emergent view of anaerobiosis is that optimal strategies of anaerobic metabolism depend upon radical chemistry and low-potential metal centres. Such catalytic sites are intrinsically vulnerable to direct poisoning by molecular oxygen and ROS. Observations suggest that anaerobes have evolved tactics that either minimize the extent to which oxygen disrupts their metabolism or restore function shortly after the stress has dissipated.
The primordial atmosphere of Earth was virtually anoxic, containing less than 10−5 of the present level of molecular oxygen (O2)1,2. It was in this circumstance that life appeared 3.8 billion years ago (Ga) and in which the basic biochemical mechanisms and metabolic pathways of organisms were established3,4. Photosystem II appeared approximately 3 Ga; it enabled ancient bacteria to acquire reducing equivalents from water, and thereby liberated photosynthetic microorganisms from a dependence upon electron sources such as hydrogen and hydrogen sulfide5–7. It also produced molecular O2 as a by-product. Because O2 crosses cell membranes at high rates, the early users of this photosystem did not suffer from elevated levels of intracellular O2. Furthermore, the O2 that diffused out of these cells was chemically scavenged by abiotic reactions with environmental reductants, notably dissolved ferrous iron and sulfur species, and for the next one thousand million years the seas and atmosphere remained hypoxic6,7. However, by ~2.5 Ga these reductants had largely been titrated, and molecular O2 levels gradually rose. Atmospheric O2 stabilized at ~10% of contemporary levels, before it finally rose again starting 0.8 Ga8.
Reducing equivalents.
Chemical species that are capable of transferring the equivalent of one electron in redox reactions.
In response to this opportunity, anaerobic respiratory chains of some bacteria evolved such that they could exploit O2 as a novel electron acceptor. Another population of microorganisms maintained their anaerobic styles of metabolism, and their contemporary descendants dwell in sediment and gut habitats that are hypoxic and anoxic zones. Such environments are effectively shielded from O2 by the vigorous respiratory actions of adjacent layers of aerobic and facultative anaerobic bacteria9–11. These anaerobes are notable for their inability to sustain their core metabolism when they are exposed to O2. It was tempting for microbiologists to conclude that these anaerobes failed to evolve in response to oxygenation of the planet, and that this failure is what constrains them to anoxic microenvironments. One of the aims of this Review is to correct that view: anaerobic microorganisms actually share most of the oxidative defences of their aerobic brethren. Instead, their persistent O2 sensitivity is a by-product of the types of chemistry that optimize anaerobic energy production.
In this Review, we present evidence that anaerobes have evolved to tolerate — or even exploit — the occasional incursion of low levels of O2. However, excess O2 arrests their growth, and we review in vitro and in vivo studies showing that O2 does so by disabling a small group of specialized enzymes that have key roles in anaerobic metabolism. Both molecular O2 itself and superoxide (O2−) that is derived from it are implicated. Oxic–anoxic interfaces are also likely sites of environmental hydrogen peroxide (H2O2) generation, and studies reveal that anaerobes respond robustly to protect themselves from it. Finally, we propose that anaerobes are likely to have well-developed strategies to resume growth after a period of O2 exposure, suggesting that this is a prime topic for further exploration.
Anaerobes experience variable O2
Dissolved O2 levels plummet in habitats that contain substantial nutritional opportunities but lack convective systems to replenish the O2 that is consumed by local microorganisms (FIG. 1). O2 penetrates poorly into mud and sludge and sub-seafloor layers. Zones near hydrothermal vents are hypoxic both due to the insolubility of O2 in the super-heated waters that feed them as well as chemical reactions between O2 and the sulfur species that the vents discharge12,13. O2 gradients have been especially well characterized in the mammalian gut. Although the diverse anatomical geography of the gut creates a range of O2 microenvironments10,14, in general O2 levels rapidly decline between the gut epithelium, which receives an O2 supply from the vasculature, and the lumen, where O2 is completely consumed by facultative anaerobic bacteria15. Whereas the solubility of O2 at 37 °C in laboratory culture media is ~210 μM, the vascularized submucosa and colonic muscle wall contain 60–120 μM O2, the apical mucosal surface adjacent to it is limited to 1–10 μM and the bulk of the lumen is essentially anoxic16,17 (FIG. 1). Accordingly, ~90% of enteric bacteria are categorized as obligate anaerobes. This label is functionally defined as denoting organisms that do not require O2 to grow well, and whose growth is blocked by the levels of O2 to which they were exposed in laboratory tests. However, this black-and-white definition turns out to obscure a substantial range of O2 tolerance that is pertinent to the structures of microbial communities. For example, in the healthy gut Bacteroides fragilis is found in high numbers at the mucosal surface, where the O2 concentration is elevated18,19. Although this bacterium cannot grow in fully air-saturated laboratory environments, it possesses both a classic anaerobic-type NrdD ribonucleotide reductase, which is poisoned by O2, and an aerobic-type NrdAB enzyme that actually requires molecular O2 for its activation20. Thus, the presence of NrdAB suggests some capacity to tolerate aeration. Indeed, after traumatic injury B. fragilis is frequently isolated from abscesses in the peritoneum, which is substantially oxygenated (6% O2)18. By contrast, its relative Bacteroides thetaiotaomicron, another abundant gut resident, resides deeper in the lumen, lacks the NrdAB isozyme and is less likely to opportunistically infect aerated tissues. Other Bacteroides spp. — Bacteroides ovatus, Bacteroides uniformis, Bacteroides vulgatus, and so on — exhibit a range of O2 sensitivities (TABLE 1).
Fig. 1. Lifestyles of anaerobes.
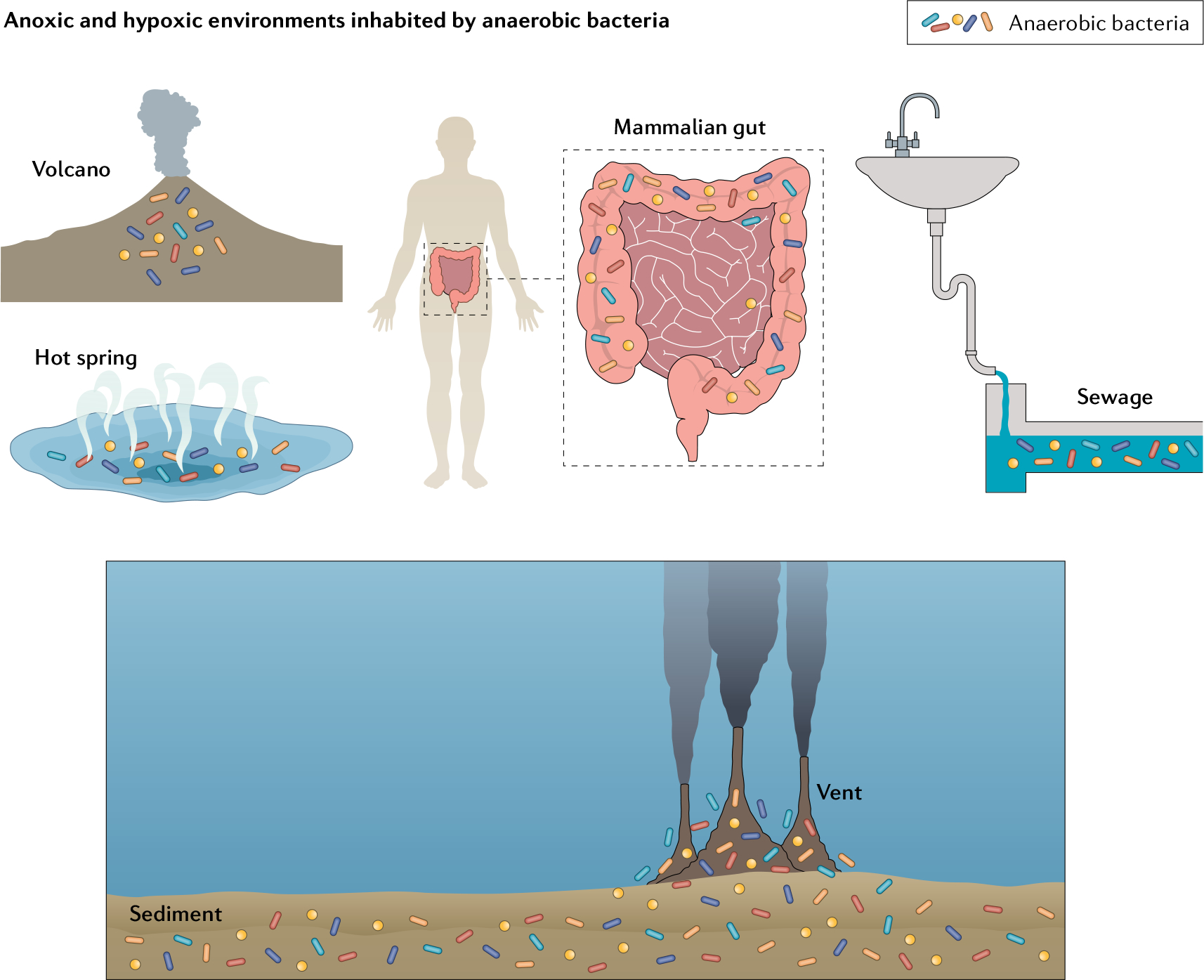
Obligate anaerobes are principal occupants of diverse environments that have limited rates of convective oxygenation. Natural systems such as the intestinal tract, rumen, buried sediments and hot springs have slow oxygen (O2) entry and rapid consumption by peripheral microbes, thereby establishing hypoxic or anoxic conditions.
Table 1 |.
O2 tolerance of obligate anaerobes
| Genus | Species | O2 level (v/v) | Growth | Refs |
|---|---|---|---|---|
| Desulfovibrio | D. vulgaris | 0.04% | Normal | 29 |
| 0.08% | Arrested | |||
| Pyrococcus | P. furiosus | 8% | Grew well | 31 |
| Geobacter | G. sulfurreducens | 10% or less in the headspace | Grew with O2 as a terminal electron acceptor | 128 |
| Air | Tolerated at least 24 h of exposure | |||
| Bacteroides | B. caccae, B. distasonis, B. ovatus, B. thetaiotaomicron, B. uniformis, B. vulgatus | 0.03% (0.3 μM dissolved O2)a | This level of O2 had no inhibiting effect on growth | 56 |
| B. fragilis | 0.1% (1 μM dissolved O2)a | Slow growth in the first 24 h; after that, growth at the anaerobic rate | 56 | |
| 0.2% (2 μM dissolved O2)a | Slow decrease in culture viability | |||
| B. oralis | 0.4% or less | Grew | 129 | |
| Air | Tolerated 24 h | |||
| B. melaninogenicus | 2.5% or less | Grew | 129 | |
| Air | Tolerated exposure (48–72 h) | |||
| Faecalibacterium | F. prausnitzii | Sterile air | Formed a growth rim on agar media | 130 |
| Clostridium | C. sordellii, C. putrificum, C. perfringens | 10% or less | Grew | 129,131 |
| Air | Tolerated up to 72 h | |||
| Peptostreptococcus, Fusobacteria | P. elsdenii, F. nucleatum | Air | Tolerated (48–72 h) | 129 |
v/v, volume/volume.
Calculations were based on the concentration of saturated dissolved oxygen (O2) in water at 37 °C (210 μM).
Similarly, many Clostridium spp. can thrive in liquid media under conditions that are more hypoxic than anoxic. Members of this genus reside in low-O2 soil, marine sediments and the human gut, where they ferment a broad array of organic compounds, including simple and complex carbohydrates and amino acids. However, although they were once regarded as stringent anaerobes, Clostridium butylicum, Clostridium acetobutylicum and Clostridium aminocalericum have been found to grow under continuous microoxic conditions; the acetogenic bacteria Clostridium magnum and Clostridium glycolicum can grow with 1–6% headspace O2; and Clostridium sordellii and Clostridium putrificum tolerate up to 10% O2 (REFS21–25). Desulfovibrio spp. thrive in a similarly wide range of habitats, primarily through the dissimilatory reduction of sulfur and nitrogen species; yet sulfate-reducing Desulfovibrio vulgaris Hildenborough appears to prefer 0.02–0.04% O2 for growth26–29. Thermotoga spp. are bacterial thermophiles that grow well in anoxic cultures but can tolerate up to 0.5% O2 in the gas phase30 — which still pales relative to the anaerobic hyperthermophilic archaeon Pyrococcus furiosus, which grows well in the presence of 8% O2 (REFS31,32). Implicitly, anaerobes experience — and even welcome — some degree of oxygenation.
Anaerobes have evolved to confront O2
Recent studies confirm that anaerobes are highly adapted to O2 exposure. Early surveys reported that superoxide dismutase activity is minimal or absent from many anaerobes33,34. However, subsequent work revealed that these microorganisms instead use superoxide reductases to degrade this O2-derived species31,35,36. Intracellul O2− is generated as a by-product of metabolic activity in oxic environments, so the implication is that in many anaerobes some level of metabolism continues even when O2 levels rise.
Even more strikingly, the advent of genomic sequencing revealed that a substantial number of supposedly anaerobic bacteria possess O2-directed respiratory systems37–39. Two general types of respiratory systems have been found: soluble systems that use a cytoplasmic rubredoxin:oxygen oxidoreductase, and membrane-bound terminal oxidases that receive electrons from a classic quinone-based respiratory chain40–43 (FIG. 2). Many anaerobes — including Bacteroides spp., Desulfovibrio spp. and the acetogen Moorella thermoacetica (originally Clostridium thermoaceticum) — have both42–46.
Fig. 2. O2-dependent respiration in anaerobes.
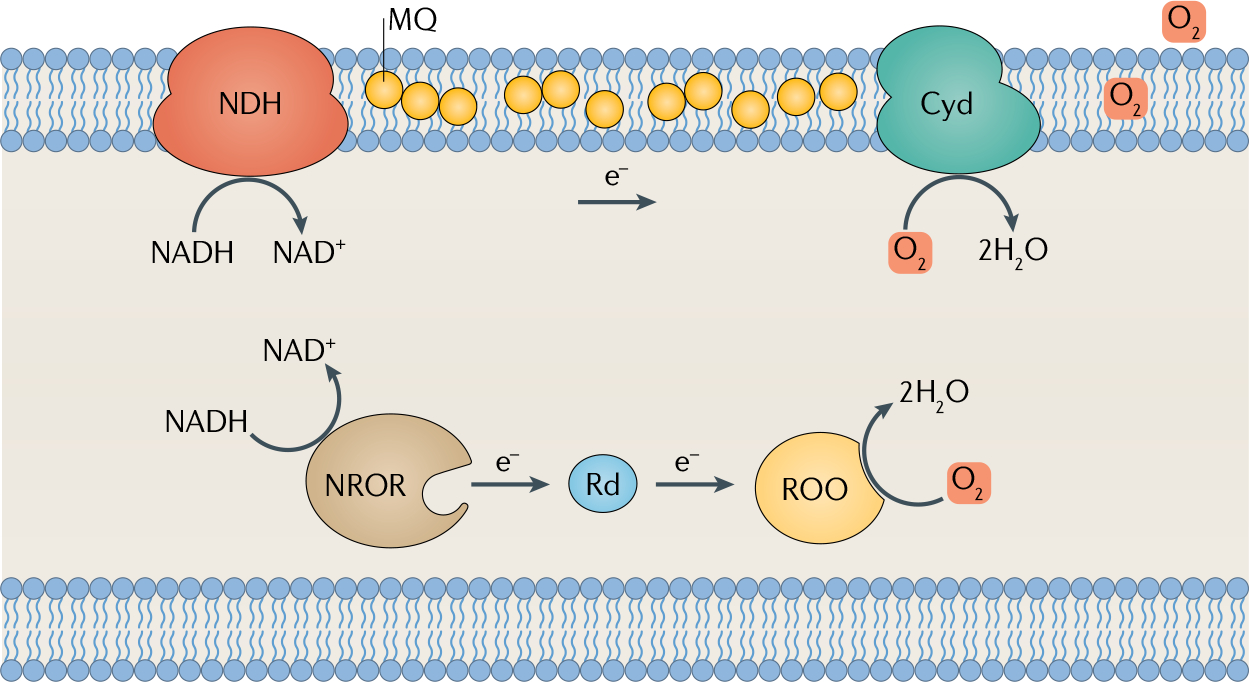
Two types of oxygen (O2)-directed respiration are found in anaerobes. The membrane-bound cytochrome bd oxidase (Cyd) receives electrons from menaquinone (MQ) and upstream dehydrogenases; and the soluble rubredoxin:oxygen oxidoreductase (ROO) obtains electrons from reduced rubredoxin. NDH, NADH dehydrogenase; NROR, NADH:rubredoxin oxidoreductase; Rd, rubredoxin.
This discovery prompted two explanations as to why putative anaerobes would contain such enzymes. One idea is that these systems are defensive, scavenging O2 in order to protect sensitive features of their metabolism from it. The other is that these bacteria consume O2 for exactly the same reason as do their aerobic peers: to enhance energy conservation.
The O2-depletion hypothesis seems obvious but, in fact, is plausible only in limited circumstances. Because O2 equilibrates almost instantly across membranes, respiratory chains cannot lower the O2 concentration inside a bacterium below that of its immediate extracellular environment47. Therefore, the only way that respiration would shield cells from O2 is if a community collectively cleared O2 from its habitat. This idea has been proposed to explain the ability of Bacteroides spp. to create co-infections in erstwhile oxic tissues alongside more fastidiously anaerobic partners48. Similarly, because the viscosity of biofilms limits mixing with the macroenvironment, respiration in biofilm-forming organisms can establish local anoxia and support anaerobic processes such as nitrogen fixation49,50. However, when a bacterium is a minor member of a diverse consortium, as in the gut, one would not expect it to contribute resources for O2 clearance.
Proton motive force.
The transmembrane electrochemical gradient, established by metabolic proton translocation, that powers the membrane proteins that synthesize ATP and import substrates.
Antibonding orbitals.
High-energy molecular orbitals; they are typically incompletely filled and are the orbitals involved in electron-transfer reactions.
The other possibility is that the benefit is energetic. The soluble rubredoxin system enhances ATP production by allowing more substrate to flow to ATP synthesis rather than into redox-balancing pathways51–53. This strategy resembles that of lactic acid bacteria that use O2 as a direct acceptor for soluble lactate and pyruvate oxidases54,55. By contrast, the membrane-bound chain generates proton motive force by conventional proton translocation. In an important demonstration of this idea, a study showed that the cytochrome bd oxidase of B. fragilis can enhance its growth when at low but finite O2 levels56. Accordingly, the bacterium was classified as a nanoaerophile. The presence of O2 - directed respiratory chains confirms that many ‘anaerobes’ can remain metabolically active in O2-containing habitats — and it muddles the definition of anaerobe.
In short, both laboratory-culture experiments and genomic surveys show that O2 sensitivity is a matter of degree. Microorganisms that cannot tolerate full aeration nevertheless have evolved strategies that protect themselves from a lower level of O2, and many have enzymatic machinery that even allow them to exploit it. The evolution of these systems is an appropriate response to the complexity of natural environments.
O2 toxicity and growth arrest
The question as to why air-level O2 arrests the growth of many bacteria and archaea is long-standing. Microorganisms occupy their particular habitats because their central metabolism is well-suited to degrading nutrients that are found there; a reciprocal hypothesis is that anaerobes cannot occupy fully aerobic habitats because O2 is somehow incompatible with their core metabolic strategies.
Those strategies, of course, are diverse. Anaerobic respiration is a catch-all phrase that denotes electron transfer from a range of reductants (for example, molecular hydrogen, lactic acid and hydrogen sulfide) to a similarly broad range of acceptors (for example, carbon dioxide, sulfate and ferric iron). The underlying molecular mechanisms are equally dissimilar. Fermenting organisms may specialize in catabolizing substrates such as short-chain fatty acids, or in degrading carbohydrates or peptides acquired from deceased primary producers. Yet despite the diversity of their metabolic machineries, biochemical studies support the view that the metabolic efficiency of these organisms hinges upon a few key enzymes that are O2-sensitive. The energy-producing pathways of methanogens and Clostridia, for example, rely upon enzymes that are quickly poisoned by O2 in aerobic buffers57,58. The study of anaerobic enzymology has thrived in recent years, and it has become clear that a few functional moieties are particular targets of O2.
O2 poisons some radical-dependent enzymes whereas others are protected.
Anaerobic microorganisms derive most of their energy by splitting their growth substrates into smaller products. Carbohydrates and some amino acids are metabolized through glycolysis to pyruvate — which is subsequently decarboxylated to yield acetyl-CoA, an intermediate in the short pathway that produces acetate and ATP. In non-respiring organisms, the challenge is to split pyruvate without producing NADH: further NADH formation would require reduction of some acetyl-CoA in order to achieve redox balance, thereby cutting into the ATP yield (Supplementary Figure 1). In this regard, the anaerobic enzyme pyruvate formate-lyase (PFL) out performs better-known pyruvate dehydro genase, by converting pyruvate to acetyl-CoA and formic acid (FIG. 3). Splitting pyruvate so that the bonding electrons remain with the C1 carboxylate is contrary to the natural tendency of divalent chemistry, and so PFL circumvents this problem by engaging a univalent mechanism59,60. PFL is primed for this function by an activase that abstracts a single electron from a glycine residue, creating a resting-state glycyl radical species that initiates a radical-based cleavage59,61. This special enzyme improves ATP production during carbohydrate fermentations by 50% or more and enables growth on some substrates that would otherwise be unable to support growth.
Fig. 3. O2 inactivates pyruvate formate-lyase.
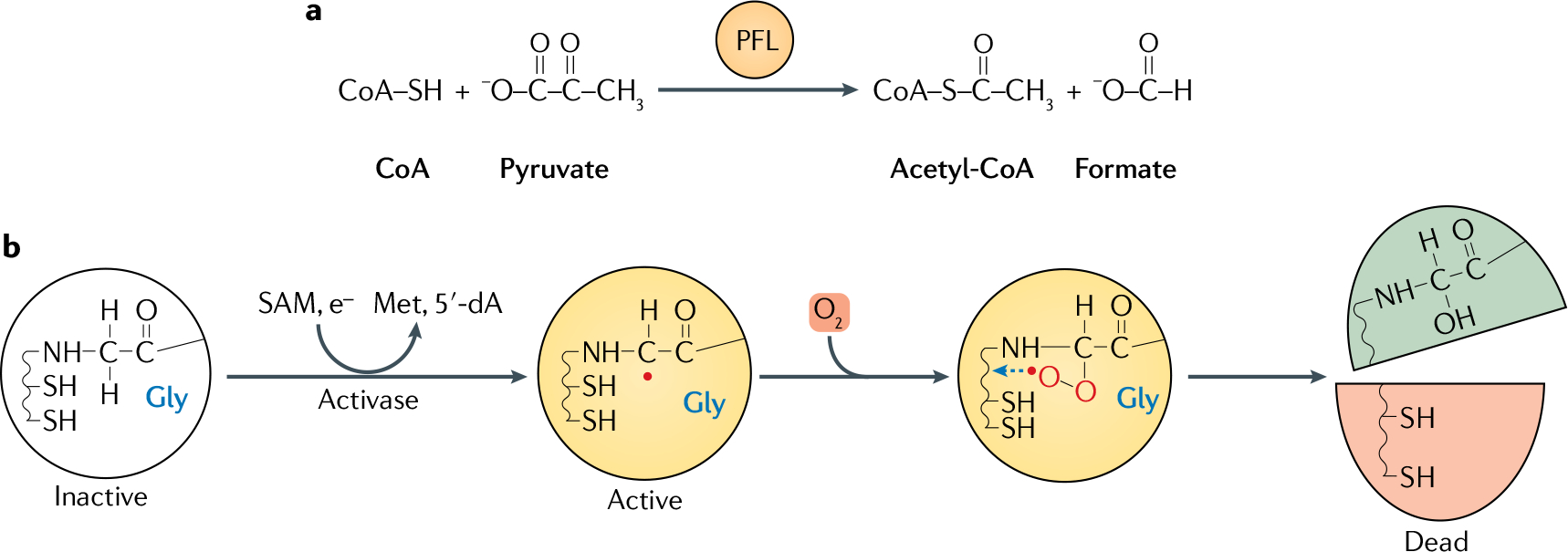
a | Pyruvate formate-lyase (PFL) catalyses pyruvate breakdown without producing NADH; this strategy is energetically economical because it avoids consuming acetyl-CoA to reoxidize NADH. Acetyl-CoA is then available for ATP synthesis (not shown). b | PFL is a prototype of the family of glycyl-radical enzymes. The glycyl radical is produced as a post-translational modification by PFL activating enzyme (activase), which activates PFL through a S-denosylmethionine (SAM)-dependent reaction, in which the monoelectronic reduction of SAM generates a 5-deoxyadenosyl radical (5′-dA) and methionine as by-products. Oxygen (O2) inactivates PFL by adducting the glycyl radical, forming a hydroperoxyl radical species that finally cleaves the polypeptide.
The drawback of radical-based chemistry is that radicals are incompatible with O2. Molecular O2 has a singular electronic structure: it possesses an even number of electrons, but its two outer-most electrons are spin-paired in separate antibonding orbitals62. That is, the molecule is a diradical (Supplementary Figure 2). Radicals react rapidly with other radicals, and so O2 reacts directly with PFL (FIG. 3). A peroxyl radical species is formed on the erstwhile glycyl radical, and then attacks the polypeptide chain and cleaves the protein backbone63. Not only is the catalytic radical lost, but the protein domain that carries the radical dissociates from the protein. Radical–radical reactions can occur at diffusion-limited rates, so even low levels of O2 inactivate PFL in seconds64, both in vitro and in vivo. In short, the fermentation strategy that optimizes ATP yield requires an enzyme mechanism that is intrinsically sensitive to molecular O2.
The anaerobic NrdD ribonucleotide reductase is another glycyl-radical enzyme that conducts a difficult reaction, and it is similarly inactivated by O2. However, unlike PFL activity, ribonucleotide reduction is also an essential function in aerated cells; therefore, alternative forms of the ribonucleotide reductase have evolved. The di-iron enzyme operative in mammals has an active site similar to that of NrdD — but instead of an exposed glycyl radical, it carries a resting-state tyrosyl radical that is buried deep within the polypeptide to ensure that it is inaccessible to O2. Other aerobes rely upon a cobalamin cofactor whose initiating adenosyl radical appears and disappears only within the course of the reaction. The evolution of the latter isozymes represents evolutionary adjustments to the aeration of Earth.
O2 disrupts low-potential metal centres.
That O2 has the capacity to oxidize biomolecules seems self-evident — but its diradical electronic structure prevents its reaction with most biomolecules. O2 cannot receive a spin-paired electron pair, and most organic molecules are incapable of transferring a single electron. For this reason, O2 cannot directly oxidize amino acids, carbohydrates, lipids or nucleic acids — the structural molecules from which cells are built. This oxidation barrier is why molecular O2 was able to accumulate in a world full of reduced carbon.
However, the half-occupied orbitals of O2 are capable of accepting single electrons. Its moderate univalent reduction potential (−160 mV) allows O2 to oxidize a subset of biomolecules that are competent at low-potential univalent electron transfers: flavins, quinones and metal centres. These adventitious reactions65 are not inherently destructive, as the oxidized forms of these moieties are part of their natural catalytic cycles. However, molecular O2 can also oxidize the metal centres of certain enzymes to redox states that lie outside their normal catalytic valences, and such events can destroy the enzyme activity.
This effect was first documented for simple dehydratases of the aconitase family, which employ a [4Fe–4S]2+ cluster to bind and withdraw a hydroxide anion from the substrate. Although the cluster has a non-redox role in this reaction, it can nevertheless be chemically oxidized by molecular O2, converting the cluster to a [4Fe–4S]3+ state that spontaneously degrades to an inactive [3Fe–4S]+ form66–68. This reaction occurs quickl enough (the half-time is ~30 min) to frustrate study of these enzymes in oxic buffers, but is slow enough that the enzymes are functional in vivo — with further assistance from the cluster-rebuilding activities inside the cell.
Many anaerobes, however, use low-potential metals in enzymes that are crucial to their core metabolism — and over-oxidation of those metal centres can be catastrophic. Most anaerobes transfer electrons to low-potential acceptors: methanogens reduce CO2 to methane, Desulfovibrio spp. reduce elemental sulfur to hydrogen sulfide and Bacteroides spp. reduce protons to H2. To ensure that the terminal electron-transfer reaction is not energetically uphill, the electron movement from substrate to oxidant has to occur through metal centres with potentials that are similarly low — with the unfortunate consequence that their accidental electron leakage to O2 will be strongly favoured and rapid. Pyruvate:ferredoxin oxidoreductase (PFOR) is an example that has been closely examined. PFOR is an alternative enzyme to PFL, dissimilating pyruvate to generate acetyl-CoA without NADH formation (Supplementary Figure 1); however, instead of generating formate, PFOR transfers the C1 bonding electrons to ferredoxin, a small iron–sulfur protein that is a general univalent electron carrier. Ferredoxin subsequently delivers the electrons to a hydrogenase. PFOR, like pyruvate dehydrogenase, uses thiamine to decarboxylate pyruvate, but the electrons then move through a series of iron–sulfur clusters to the enzyme surface, where ferredoxin binds69,70. When PFOR is mixed with oxic buffer, its activity quickly disappears. Its clusters are simultaneously bleached, suggesting that cluster over-oxidation by O2 is responsible for the activity loss70.
Similar events occur in vitro at the low-potential iron centres of hydrogenases, at both the molybdenum cofactor and iron enzyme of nitrogenase and at the Ni(I) centre of methyl-CoM reductase (FIG. 4): molecular O2 inactivates them all, and spectroscopic signatures imply that metal oxidations are responsible71. The full details of the oxidation processes, and the disposition of the final enzymes, have not yet been demonstrated72–74. However it seems a safe assumption that similar events transpire in vivo and are responsible for the cessation of hydrogen evolution, nitrogen fixation and methanogenesis, respectively, when these microorganisms are aerated.
Fig. 4. Structures of O2-sensitive metalloenzymes.
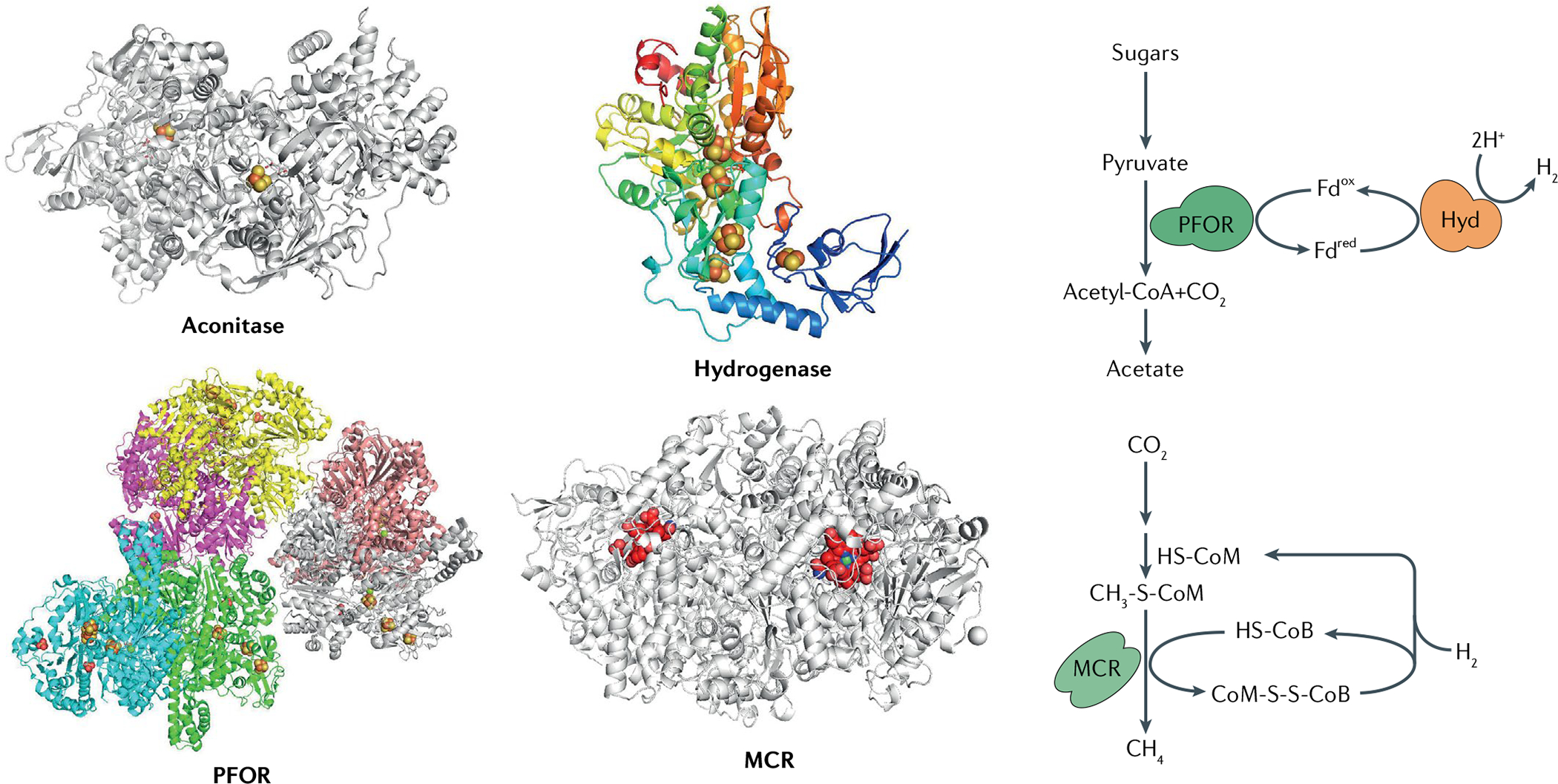
Molecular oxygen (O2) directly inactivates diverse metalloenzymes featuring low-potential metal centres at or near the protein surface. The structures of representative O2-sensitive metalloenzymes are shown (left), alongside their pivotal positions in anaerobic metabolism (right): aconitase B from Escherichia coli (Protein Data Bank (PDB) ID 1L5J); pyruvate:ferredoxin oxidoreductase (PFOR) from Moorella thermoacetica (PDB ID 6CIN); iron-only hydrogenase (Hyd) from Clostridium pasteurianum (PDB ID 1FEH); and methyl-coenzyme M reductase (MCR) from Methanothermobacter marburgensis Marburg (PDB ID 1MRO). Iron–sulfur clusters are represented by yellow and red balls, nickel-containing F430 of MCR is shown in red with nickel in green. CoM-SH, coenzyme M; CoB-SH, coenzyme B; Fdox, oxidized ferredoxin; Fdred, reduced ferredoxin.
Those enzymes all catalyse low-potential redox reactions. However, metal-centre oxidation is apparently also responsible for the O2 sensitivity of a family of non-redox enzymes that are exclusively found in anaerobes. Clostridial consumption of amino acids occurs by Stickland fermentations75, in which the exergonic oxidation of one amino acid must be redox-balanced by reduction of another to an aliphatic product, such as propionate. The latter process requires the dehydration of non-activated substrates. Dehydrations in aerobes invariably rely upon a carbonyl group adjacent to the proton that is to be removed; resonance provides enough anion stabilization that deprotonation is possible. However, anaerobes must operate upon more-reduced substrates, and some lack a stabilizing carbonyl or carboxylate moiety. To solve this problem, Bacteroides spp. employ a long carbon-shuffling pathway to activate the substrate for deprotonation48; the pathway is expensive, requiring not only ample synthesis of protein but also of biotin and cobalamin to activate key enzymes (Supplementary Figure 3). By contrast, Clostridia spp. employ a single-enzyme radical mechanism in which an iron–sulfur cluster injects a low-potential electron into the substrate and then withdraws it at the conclusion of the catalytic cycle76. The enzyme is initially activated by a separate [4Fe–4S]-containing activase, which receives an electron from ferredoxin and then uses ATP hydrolysis to drive the electron into the dehydratase cluster. Both clusters must operate at low potentials in order to push the electron onto the hydroxyacyl-CoA substrate — and therefore both centres are vulnerable to destructive oxidation by molecular O2. Thus, low-potential metal centres may be requisite even for non-redox processes that operate upon the aliphatic, non-activated substrates that anaerobes must process.
Most enzymes in anaerobes are O2-tolerant, as are the equivalent enzymes in aerobes. However, in studying the remarkable enzymatic functions of anaerobes — for instance, cleaving pyruvate without disturbing the redox balance or transferring electrons to low-potential oxidants — researchers found that the key enzymes retained activity only when maintained in anoxic glove boxes. Those in vitro studies provide compelling explanations for the O2 sensitivity of anaerobes — but in vivo studies are needed to confirm that these same enzyme problems recur in vivo.
Anoxic glove boxes.
Boxes (chambers) that provide a strict anaerobic atmosphere of 0–5 ppm molecular oxygen (O2). A palladium catalyst eliminates O2 by using hydrogen gas as a co-reactant; a vacuum airlock reduces O2 levels prior to transfer of items into the glove box.
Studies of O2 poisoning in vivo: confirmation and expansion of in vitro results.
Some enzymes, such as radical S-denosylmethionine (SAM) and aconitase class enzymes, lose activity in oxic buffers but work perfectly well inside oxic cells. This observation warns against a simple extrapolation of in vitro events into the living cell. If the rate of enzyme oxidation is moderate, countervailing repair processes77 may sustain high steady-state activity even when cells are O2-replete. Furthermore, substrates generally stabilize the metal centres to which they bind77, thereby likely lowering the rate of enzyme damage in vivo. These quantitative considerations influence the amount of O2 exposure that an enzyme can tolerate in vivo. To date, there have been only limited physiological studies of how O2 impinges upon the ongoing metabolism of various anaerobes.
The linkage between O2 and metabolic dysfunction has been explored in a top-down way in the case of strains of Bacteroides and Desulfovibrio spp. Under anoxic conditions, the gut bacterium B. thetaiotaomicron efficiently ferments glucose to a mixture of succinate, propionate and acetate, but when it is aerated, glucose consumption stops78. Growth resumes after anoxia is restored. Its normal fermentation pathways are depicted in FIG. 5. Carbohydrate catabolism initially proceeds through glycolysis, with concomitant reduction of NAD+. The NADH thus formed is reoxidized by reduction of phosphoenolpyruvate (PEP) to succinate. This process entails electron flow through an energy-conserving anaerobic respiratory chain. Residual PEP is converted to pyruvate and then decarboxylated, either by PFOR or by PFL70,79 (FIG. 5).
Fig. 5. Metabolism in Bacteroides spp. is blocked upon aeration and resumes in anoxia.
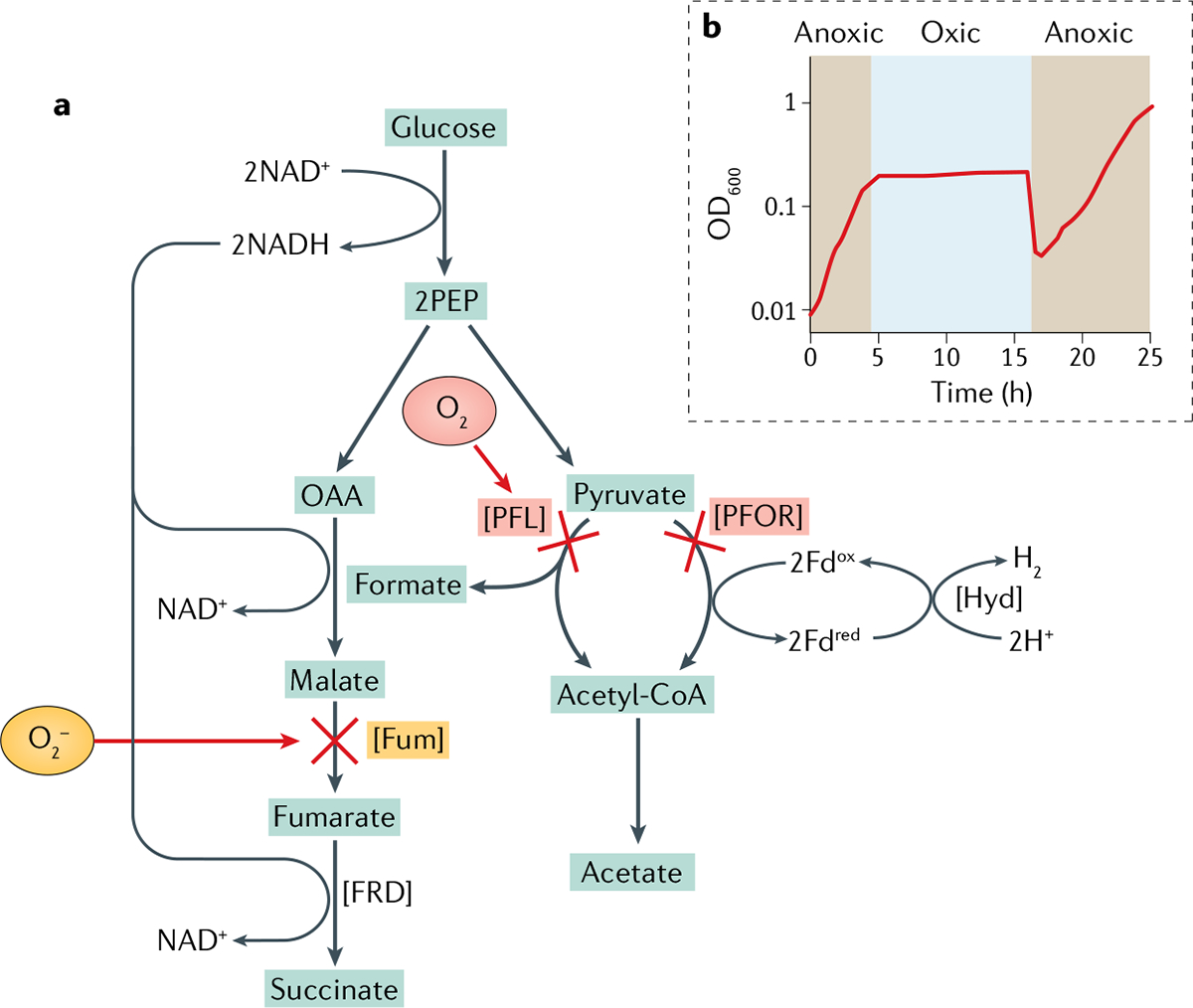
Oxygen (O2) poisoning results from inactivation of several enzymes. a | In aerated cells, endogenous O2− inactivates fumarase in the redox-balancing succinate branch of central metabolism. At the same time, molecular O2 directly inactivates both pyruvate formate-lyase (PFL) and pyruvate:ferredoxin oxidoreductase (PFOR), creating a bottleneck in the pathway leading to acetate. Either block is sufficient to prohibit growth. Enzymes indicated in square brackets. b | Cell viability is not lost during the period of O2 exposure. When anoxia is restored, growth soon resumes (drop in OD600 reflects dilution into anoxic medium). Fdox, oxidized ferredoxin; Fdred, reduced ferredoxin; FRD, fumarate reductase; Fum, fumarase; Hyd, hydrogenase; OAA, oxaloacetic acid; PEP, phosphoenolpyruvate. Part b reprinted from REF.44, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Metabolic analysis of O2-poisoned cells revealed two points of inhibition: at the decarboxylation of pyruvate by PFOR and PFL, and at the dehydration of malate to fumarate by fumarase78,79. Either block would be sufficient to prohibit glucose fermentation. A common model of obligate anaerobiosis posits that toxicity arises from the reactive oxygen species (ROS) O2− and/or H2O2 (REF.33); however, as predicted by in vitro studies, the inactivation of both PFOR and PFL was shown to be mediated directly by molecular O2 itself70,79,80. In fact, PFOR cannot be oxidized by O2− or H2O2, because these species must bind directly to the metals that they oxidize — and those in PFOR are fully coordinated by polypeptide. By contrast, the electronic structure of O2 allows electrons to hop to it by outer-sphere processes, and so can oxidize the metal centres of PFOR if it merely gets close to them.
PFL is hypersensitive to O2, but even when cells were fully aerated it required tens of minutes for PFOR to lose activity79. The pace of inactivation depended upon O2 concentration, with little activity loss at 5% O2 and lower, indicating that PFOR may continue to support the metabolism of B. thetaiotaomicron in low-O2 regions of the intestine.
Interestingly, the sediment-dwelling sulfate-reducing bacterium Desulfovibrio africanus also dissimilates pyruvate with PFOR — but its enzyme features a polypeptide extension that substantially augments its O2 resistance even beyond that of the B. thetaiotaomicron PFOR. O2 exposure triggers the formation of a disulfide bond that locks the extension over the PFOR clusters in a way that may shield them from the close approach of O2 (REFS81,82). The enzyme remains active, albeit with a lower turnover number. Many Desulfovibrio spp. live in habitats with O2 levels that are less controlled than those of the mammalian intestine, and so this structural alteration may enable the bacterium to tolerate oxidative conditions. No analogous adjustment is known that can improve the stability of PFL.
Endogenous ROS impose a second layer of oxidative stress.
The other metabolic block in aerated B. thetaiotaomicron results from the deactivation of fumarase68,78 (FIG. 5). However, in contrast to PFOR and PFL, the pathway block at fumarase cannot be ascribed to direct damage by molecular O2, and this observation suggests a second mechanism of oxidative poisoning. Although eukarotes and some bacteria employ metal-independent fumarases, most anaerobes — including Bacteroides spp. — have a class I fumarase of the aconitase class with a [4Fe–4S] cluster as its catalytic cofactor. The cluster of these enzymes protrudes into the active site so that an iron atom can directly bind the substrate, and this solvent exposure makes it vulnerable to the approach of oxidants83–87 (Supplementary Figure 4a). Molecular O2 can slowly oxidize the fumarase cluster, but it does so on a long timescale that has little biological impact68. However, studies with the enzymes of Escherichia coli established that O2− and H2O2 can avidly bind and oxidize the cluster. The oxidized cluster is unstable and loses its catalytic iron atom, and the enzyme is inactivated. Other hydratases and dehydratases of the same catalytic class are also vulnerable to these oxidants83,88,89. The failure of fumarase in aerated B. thetaiotaomicron suggested that O2− and H2O2 rise to toxic levels. Indeed, it was shown that endogenous O2− is the culprit, and that the [4Fe–4S] enzymes aconitase and isopropylmalate isomerase are similarly poisoned68.
According to sequence analyses, O2-sensitive [4Fe–4S] dehydratases are common to other anaerobes too, including the genera Desulfovibrio, Clostridia and Thermoanaerobacter90. These enzymes are indispensable for the biosyntheses of amino acids belonging to branched-chain (Leu, Ile and Val) and α-ketoglutarate (Glu, Gln, Pro and Arg) families, suggesting that the metabolic failures observed in Bacteroides spp. may occur generally whenever enough O2 encroaches upon anaerobic communities.
It is notable that whereas some microorganisms — and higher organisms — have acquired oxidant-resistant versions of fumarase, others continue to rely upon more vulnerable isozymes. Why is this? The apparent answer is that the cluster-free fumarases are less catalytically efficient, analogous to the D. africanus PFOR91,92. On balance, high-turnover — albeit oxidant-sensitive — isozymes may be the better choice for microorganisms that dwell in niches that are rarely oxygenated.
ROS studies in E. coli have determined that excess O2− can also inactivate a family of enzymes that use a single Fe(II) cofactor to catalyse non-redox reactions93,94 (Supplementary Figure 4b). In fact, two representatives of this family — ribulose-5-phosphate 3-epimerase and peptide deformylase — also lost activity when B. thetaiotaomicron was fully aerated68.
Why is O2 - toxic in anaerobes?
So, although molecular O2 directly mediates some cell injuries to anaerobes, its derivative O2− can also disrupt metabolism. Yet the enzymes that O2− inactivates in aerated B. thetaiotaomicron remain fully functional in aerated E. coli. Why the difference?
The superoxide-sensitive enzymes of Bacteroides spp. are no more intrinsically reactive with O2− than are their homologues from facultative and aerobic bacteria. Indeed, when E. coli and B. thetaiotaomicron enzymes were swapped between the two species, both enzymes remained active in aerated E. coli, but both quickly lost activity in B. thetaiotaomicron68. This result led to the recognition that ROS are simply generated in higher volume in the anaerobe.
The rate of ROS formation in a bacterium can be discerned by tracking the release of H2O2 from a mutant whose scavenging systems are knocked out95. Because O2− and H2O2 are made by similar processes65, these measurements provide predictions about O2− production as well. In one study, a B. fragilis mutant was constructed that has little ability to scavenge H2O2, and the authors found that when the strain was aerated, H2O2 effluxed from these cells several-fold faster than it effluxes from non-scavenging E. coli mutants43. A similar experiment showed that aerated B. thetaiotaomicron produced H2O2 ten times more quickly than E. coli strains did96. Thus, it is not surprising that the aeration of Bacteroides spp. causes superoxide-sensitive pathways to fail and growth to stop.
In general, when cells grow in oxic environments, molecular O2 adventitiously abstracts electrons from the reduced flavins or metal centres of some redox enzymes, thereby forming O2− and H2O2 (REFS44,97,98). Because these events occur in proportion to collision frequency, the rate of ROS production is proportionate to the O2 concentration. Indeed, the rate of superoxide-mediated enzyme damage in B. thetaiotaomicron is modest at low O2 concentrations but is crippling upon full aeration68.
The source of all these ROS is not certain. B. thetaiotaomicron has an anaerobic respiratory chain that carries large fluxes of electrons through its flavins and metal centres, but follow-up work indicated that this system is unlikely to be the primary producer of ROS34,98,99. The deletion of the rubredoxin-dependent electron chain (FIG. 2) caused only a modest drop in H2O2 production44. One attractive candidate is the ferredoxin-mediated flow of electrons from pyruvate to hydrogenase (FIG. 5). The PFOR–ferredoxin–hydrogenase pathway involves a series of low-potential ferredoxin-type metal centres. The reduction potentials of ferredoxins typically range from −350 to −455 mV, which makes electron transfer to molecular O2 (−160 mV) favourable100,101. Indeed, biochemical reports have shown that O2 oxidizes the ferredoxins of Clostridium pasteurianum and spinach, generating O2− (REF.102). For now, this point remains unsettled. The primary ROS source remains of interest, as its identity might shed light on the possibility that endogenous ROS-mediated enzyme damage is a common phenomenon when anaerobes encounter O2.
Defences against environmental H2O2
Damage to enzymes from molecular O2 or endogenous O2− can block the growth of anaerobes, but this damage is not lethal: once anoxia is restored, enzymes can be repaired or replaced, and growth can resume. However, DNA damage is potentially deadly. DNA oxidation is precipitated by the Fenton reaction, in which cellular ferrous iron transfers an electron to H2O2. The resultant hydroxyl radical can oxidize both nucleotide bases and sugars, creating impassible lesions that block DNA replication.
Facultative anaerobic bacteria activate H2O2 stress responses only when H2O2 flows into the cell from the environment. By contrast, full aeration is sufficient to activate those stress responses in many anaerobes, presumably due to the high rate of internal H2O2 formation from the autoxidation of their low-potential electron carriers. One study showed that B. fragilis responds to aeration by inducing catalase, the alkyl hydroperoxidase (AhpC), thioredoxin peroxidase (Tpx) and rubrerythrin peroxidases103. A similar response was observed for B. thetaiotaomicron96. C. acetobutylicum represses H2O2 - scavenging enzymes using PerR, a transcriptional repressor that is deactivated when H2O2 oxidizes its Fe(II) cofactor; this system, too, is activated by simple aeration104. If this response is not triggered quickly enough, internal Fenton chemistry is lethal.
Surprisingly, various evidence suggests that bacteria may need to deal with H2O2 even when bacteria are still in the anaerobic gut. For example, the enteric bacterium E. coli induces a cytochrome c peroxidase (Ccp) upon H2O2 exposure, but only when O2 is absent105. This arrangement refutes the idea that H2O2 stress only occurs in highly oxygenated habitats. The apparent purpose of the membrane-bound enzyme is to enable the bacterium to use H2O2 as an alternative respiratory electron acceptor106. Bacteroides spp. are among the many other bacteria that carry Ccp homologues103.
How does H2O2 arise in hypoxic habitats (BOX 1)? Reduced sulfur species are released by sulfate-reducing bacteria in the intestinal lumen, and they may converge with O2 that seeps in from the epithelium, triggering abiotic ROS formation at the oxic–anoxic interface. Furthermore, some lactic acid bacteria that thrive at such zones generate H2O2 as a stoichiometric product of their central metabolism55,107. Similar events are likely at analogous interfaces in soil and elsewhere. The importance of all this H2O2 is underscored by the ubiquity and high titres of bacterial defences against it. Experimental studies and genomic sequence data have revealed that anaerobes protect themselves from environmental H2O2 by deploying most of the same defensive tactics that were originally identified in aerobes. Bacteroides, Desulfovibrio, Pyrococcus and Clostridium spp. synthesize both NADH peroxidase (Ahp) and rubrerythrins to scavenge H2O2; Bacteroides and Desulfovibrio spp. have catalase in addition21,25,26,31,32,96,103,108,109.
Box 1 |. Oxidative stress in hypoxic environments.
Natural anoxic environments may be intermittently oxygenated, leading to the formation of reactive oxygen species (ROS). At the periphery of deep-sea vents, cool oxygenated seawater mixes with hydrothermal vent fluid; oxygen (O2) oxidizes hydrogen sulfide (H2S) and forms extracellular superoxide (O2−) and hydrogen peroxide (H2O2). This ROS-forming reaction also occurs at the interface of sediment and water in marine environments. In the mammalian gastrointestinal tract, oxygenation can be enhanced by disruptive stresses. For example, inflammatory bowel diseases, including Crohn’s disease and ulcerative colitis, are associated with dysbiosis of the colon microbiota that may be mediated by enhanced oxygenation. Oxygenation can also increase upon changes in the host physiology that accompany antibiotic treatment15,120,132,133 or by the presence of pathogens. This O2 plausibly triggers several sources of colonic H2O2. Sulfide that is generated by luminal sulfur-reducing bacteria may react abiotically with O2 that it encounters at the oxic–anoxic interface, producing H2O2 (REFS134–136). Gut inflammation releases H2O2 that is produced by host macrophages121,122,137. Some intestinal commensal bacteria even actively induce ROS production, mainly in the form of H2O2 within enterocytes. For example, members of the genus Lactobacillus produce and shed small formylated peptides, which are perceived via formyl peptide receptors localized on the apical surface of gut epithelia. Those receptors then activate NADPH oxidases (NOX) that generate ROS. To deal with perturbations such as these, anaerobes possess inducible defences against exogenous oxidants.
Anaerobes sense H2O2 and launch these responses using the same ROS-sensing transcription factors that were originally discovered in aerobes. OxyR is the paradigmatic H2O2-sensing transcription factor110. It was originally identified and characterized in E. coli, but is broadly distributed among anaerobic bacteria103,111,112. The greatest threat of H2O2 is that it will react with intracellular Fe(II) to generate lethal DNA damage; therefore, although the full regulon can vary from one bacterium to another, it generally includes enzymes that scavenge H2O2 (catalase and peroxidases) and that sequester intracellular Fe(II) (the mini-ferritin Dps)113,114.
Other bacteria employ the iron-based sensing system of PerR115,116 to regulate their H2O2 stress responses. This regulon is distributed among some anaerobes and has been characterized in C. acetobutylicum and D. vulgaris Hildenborough104,117,118. It includes many of the same genes that OxyR controls in other bacteria, including Ahp, catalase and Dps. Therefore, our view today is almost fully reversed from that of earlier days: anaerobes are obviously subjected to H2O2 stress in their lifestyle, and they have evolved a full suite of strategies to defend themselves against it.
It is worth noting that early work may have led researchers to overestimate the lethal effects of aeration upon anaerobes. Laboratory experiments that tested this phenomenon often abruptly introduced O2 directly into erstwhile anoxic media; such media often contained high levels of reduced sulfur species, including cysteine, sulfide or even dithionite. Consequent redox reactions would have generated unnaturally high concentrations of H2O2, which could diffuse into cells and produce lethal DNA damage119.
Reduction potential and growth
It is sometimes asserted that even in the absence of O2 species some anaerobes cannot grow in an environment with a high redox potential. This formulation obscures more than illuminates: no single reduction potential accurately describes an environment. The redox partners that comprise growth substrates — for example, H2 and CO2 for methanogens, or lactate and nitrate for denitrifiers — are not in thermodynamic equilibrium with one another, and in fact the organism derives energy from catalysing their approach towards equilibrium. Instead, the statement that a high redox potential interferes with the growth of an anaerobe typically means that a key reduced growth substrate is insufficiently available. In natural mixed populations, the addition of sulfate, for example, can suppress the growth of methanogens because it favours sulfate-reducing bacteria in their mutual competition for H2 (REF.110). In pure cultures, some microorganisms require hydrogen sulfide, rather than sulfate, as their source of essential sulfur atoms; others may be able to import only ferrous iron rather than its ferric form. It is preferable to identify the specific problem rather than to ascribe it to a putative overarching reduction potential.
How do anaerobes rebound from aeration?
Recovery from oxygenation is likely to be a key event in the lifestyle of anaerobes — whether for Desulfovibrio spp. that dwell near oxic interfaces or for the methanogens that cope with cycles of desiccation and rainfall that bring oxygenated water into soil crevasses. The issue is highly pertinent to the establishment and stability of the gut microbiome: not only do these microorganisms encounter stress when transiting through the aerobic world from one host to another, but disruptive antibiotic treatments and inflammation events increase the penetration of O2 into the intestinal lumen120–122. Opportunist pathogens such as B. fragilis and Clostridium perfringens must survive aeration when they disseminate into oxic tissues. However, our understanding of how cells recover from oxidative stress is more advanced in aerobes and facultative anaerobic bacteria. A take-home lesson from that work is that those bacteria employ various strategies to repair and reactivate enzymes that were damaged by O2− or H2O2. Iron–sulfur clusters are repaired with the help of inducible cluster-assembly systems78,87,123,124, and mononuclear Fe(II) enzymes are re-metallated, either with iron itself or with oxidant-resistant manganese93,125 The same reactivations occur when stressed anaerobes return to anoxic habitats.
So, it seems logical that anaerobes must also have evolved systems that allow them to cope with O2− mediated injuries to glycyl-radical enzymes and metalloproteins. The speed with which growth resumes after periods of aeration (FIG. 5) tends to support this conjecture. Ironically, one elegant tactic to restore the activity of O2-cleaved PFL was actually discovered in E. coli, a facultative bacterium that uses PFL in anoxic environments. After a period of O2 exposure, the resumption of anoxia stimulates the synthesis of GrcA, a small protein that physically resembles the glycyl-radical domain of PFL that is lost upon aeration126. GrcA binds to the remaining PFL fragment and, in collaboration with the PFL-activating enzyme, regenerates an active radical-containing complex. However, GrcA seems to be restricted to facultative bacteria – organisms whose movement between anoxic and oxic habitats is likely to be frequent. GrcA is absent from anaerobes that do not tolerate full aeration. So, do they, too, have mechanisms that allow them to recover after O2 exposure?
It has been pointed out that an individual bacterium that colonizes a new habitat likely spends as much time in transit as it does in its preferred niche127. Therefore, it is almost certain that evolution has equipped anaerobes not only to survive O2 stress but also to recover swiftly from it. The tactics by which they do so are largely unstudied, and we regard this as the next challenge in this field.
Conclusions
As obligate anaerobes have become genetically accessible, and as researchers have become much better at investigating their physiologies and the biochemical mechanisms of O2-sensitive enzymes, the field has finally acquired enough knowledge to put together the puzzle of why O2 is so disruptive to them. The goal is both to illuminate the molecular bases of their incapacity to grow in oxic environments and to reveal the adaptations that provide them with some level of O2 tolerance.
The notion that anaerobes simply neglected to evolve antioxidant defences has been refuted: these bacteria possess the majority of the same antioxidant defences and regulatory systems that are found in facultative anaerobic bacteria and aerobes. Instead, their intractable problem with O2 derives from the chemical mechanisms by which they optimize their anaerobic success. Low-potential electron flow is necessary to achieve redox-balanced fermentations and to effect anaerobic respiration, but it relies upon univalent carriers that are naturally vulnerable to adventitious oxidation by molecular O2. The over-oxidation of metal centres to unstable valences is destructive to the key enzymes that underlie the redox processes that are fundamental to much of anaerobic physiology. Those oxidation reactions are also likely responsible for the excessive ROS that are formed when anaerobes are aerated; such ROS cause another layer of damage. Meanwhile, many anaerobes also employ free-radical mechanisms to enable difficult chemistry — and the free-radical nature of O2 causes it to disrupt those catalysts by reacting directly with them. Thus, it seems unavoidable that the most effective anaerobes — those that predominate in anoxic habitats — are incapable of thriving when exposed to O2.
We have moved a long way from the early hypothesis that anaerobes shelter in environments from which O2 and ROS are completely excluded. What remains to be determined are details of the injuries, measurements of damage rates and elucidation of as yet unknown defensive tactics. Ultimately, we wish to identify the circumstances and dynamics by which O2 disrupts and reshapes important microbial communities.
Supplementary Material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31970101), the National Institutes of Health (NIH) (GM049640), the Natural Science Foundation of Guangdong Province (No. 2019A1515011685) and the Research Start-up Project of Shantou University (NTF18018).
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Microbiology thanks F. Barras; T. Schmidt, who co-reviewed with M. Hoosta; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41579-021-00583-y.
References
- 1.Slesak I, Kula M, Slesak H, Miszalski Z & Strzalka K How to define obligatory anaerobiosis? An evolutionary view on the antioxidant response system and the early stages of the evolution of life on Earth. Free. Radic. Biol. Med 140, 61–73 (2019). [DOI] [PubMed] [Google Scholar]
- 2. Lyons TW, Reinhard CT & Planavsky NJ The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315 (2014). This paper describes current thinking on the history of O2 on Earth.
- 3.Gould SB et al. Adaptation to life on land at high O2 via transition from ferredoxin- to NADH-dependent redox balance. Proc. R. Soc. Lond. B Biol. Sci 286, 20191491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemmey H & Badham N Oxygen in the Precambrian atmosphere: an evaluation of the geological evidence. Geology 10, 141–146 (1982). [Google Scholar]
- 5.Fischer WW, Hemp J & Johnson JE Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet. Sci 44, 647–683 (2016). [Google Scholar]
- 6.Schad M, Konhauser KO, Sanchez-Baracaldo P, Kappler A & Bryce C How did the evolution of oxygenic photosynthesis influence the temporal and spatial development of the microbial iron cycle on ancient Earth? Free. Radic. Biol. Med 140, 154–166 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Hamilton TL, Bryant DA & Macalady JL The role of biology in planetary evolution: cyanobacterial primary production in low-oxygen Proterozoic oceans. Environ. Microbiol 18, 325–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bekker A et al. Dating the rise of atmospheric oxygen. Nature 427, 117–120 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Muller M et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev 76, 444–495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman ES et al. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc. Natl Acad. Sci. USA 115, 4170–4175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng L, Kelly CJ & Colgan SP Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am. J. Physiol. Cell Physiol 309, C350–C360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marie B, Genard B, Rees JF & Zal F Effect of ambient oxygen concentration on activities of enzymatic antioxidant defences and aerobic metabolism in the hydrothermal vent worm, Paralvinella grasslei. Mar. Biol 150, 273–284 (2006). [Google Scholar]
- 13.Lesser MP Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol 68, 253–278 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Rivera–Chavez F, Lopez CA & Baumler AJ Oxygen as a driver of gut dysbiosis. Free. Radic. Biol. Med 105, 93–101 (2017). [DOI] [PubMed] [Google Scholar]
- 15. Espey MG Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free. Radic. Biol. Med 55, 130–140 (2013). This study details that intestinal anaerobes may encounter a range of O2 concentrations, depending upon their proximity to the epithelium.
- 16.Kelly CJ & Colgan SP Breathless in the gut: Implications of luminal O2 for microbial pathogenicity. Cell Host Microbe 19, 427–428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwerdtfeger LA, Nealon NJ, Ryan EP & Tobet SA Human colon function ex vivo: dependence on oxygen and sensitivity to antibiotic. PLoS ONE 14, e0217170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawyer RG, Spengler MD, Adams RB & Pruett TL The peritoneal environment during infection. The effect of monomicrobial and polymicrobial bacteria on pO2 and pH. Ann. Surg 213, 253–260 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renvall S & Niinikoski J Intraperitoneal oxygen and carbon dioxide tensions in experimental adhesion disease and peritonitis. Am. J. Surg 130, 286–292 (1975). [DOI] [PubMed] [Google Scholar]
- 20.Smalley D, Rocha ER & Smith CJ Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J. Bacteriol 184, 895–903 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitham JM, Tirado-Acevedo O, Chinn MS, Pawlak JJ & Grunden AM Metabolic response of Clostridium ljungdahlii to oxygen exposure. Appl. Environ. Microbiol 81, 8379–8391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talukdar PK, Olguin-Araneda V, Alnoman M, Paredes-Sabja D & Sarker MR Updates on the sporulation process in Clostridium species. Res. Microbiol 166, 225–235 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Tracy BP, Jones SW, Fast AG, Indurthi DC & Papoutsakis ET Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr. Opin. Biotechnol 23, 364–381 (2012). [DOI] [PubMed] [Google Scholar]
- 24. Kint N et al. How the anaerobic enteropathogen Clostridioides difficile tolerates low O2 tensions. mBio 11, e01559–20 (2020). This study shows that Clostridioides difficile responds to O2 exposure by inducing enzymes that scavenge O2 and H2O2.
- 25.Kawasaki S et al. Adaptive responses to oxygen stress in obligatory anaerobes Clostridium acetobutylicum and Clostridium aminovalericum. Appl. Environ. Microbiol 71, 8442–8450 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fournier M et al. Function of oxygen resistance proteins in the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol 185, 71–79 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silaghi-Dumitrescu R, Ng KY, Viswanathan R & Kurtz DM Jr. A flavo-diiron protein from Desulfovibrio vulgaris with oxidase and nitric oxide reductase activities. Evidence for an in vivo nitric oxide scavenging function. Biochem. 44, 3572–3579 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Rowan F et al. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis. Colon. Rectum 53, 1530–1536 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Johnson MS, Zhulin IB, Gapuzan ME & Taylor BL Oxygen-dependent growth of the obligate anaerobe Desulfovibrio vulgaris Hildenborough. J. Bacteriol 179, 5598–5601 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Fourn C et al. An oxygen reduction chain in the hyperthermophilic anaerobe Thermotoga maritima highlights horizontal gene transfer between Thermococcales and Thermotogales. Environ. Microbiol 13, 2132–2145 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Thorgersen MP, Stirrett K, Scott RA & Adams MWW Mechanism of oxygen detoxification by the surprisingly oxygen-tolerant hyperthermophilic archaeon, Pyrococcus furiosus. Proc. Natl Acad. Sci. USA 109, 18547–18552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strand KR et al. Oxidative stress protection and the repair response to hydrogen peroxide in the hyperthermophilic archaeon Pyrococcus furiosus and in related species. Arch. Microbiol 192, 447–459 (2010). [DOI] [PubMed] [Google Scholar]
- 33.McCord JM, Keele BB Jr. & Fridovich I An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc. Natl Acad. Sci. USA 68, 1024–1027 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chance B, Sies H & Boveris A Hydroperoxide metabolism in mammalian organs. Physiol. Rev 59, 527–605 (1979). [DOI] [PubMed] [Google Scholar]
- 35.Jenney FE Jr., Verhagen MF, Cui X & Adams MW Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286, 306–309 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Niviere V & Fontecave M Discovery of superoxide reductase: an historical perspective. J. Biol. Inorg. Chem 9, 119–123 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Degli Esposti M, Mentel M, Martin W & Sousa FL Oxygen reductases in alphaproteobacterial genomes: physiological evolution from low to high oxygen environments. Front. Microbiol 10, 499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiss MC et al. The physiology and habitat of the last universal common ancestor. Nat. Microbiol 1, 16116 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Morris RL & Schmidt TM Shallow breathing: bacterial life at low O2. Nat. Rev. Microbiol 11, 205–212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wildschut JD, Lang RM, Voordouw JK & Voordouw G Rubredoxin:oxygen oxidoreductase enhances survival of Desulfovibrio vulgaris Hildenborough under microaerophilic conditions. J. Bacteriol 188, 6253–6260 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Victor BL, Baptista AM & Soares CM Dioxygen and nitric oxide pathways and affinity to the catalytic site of rubredoxin:oxygen oxidoreductase from Desulfovibrio gigas. J. Biol. Inorg. Chem 14, 853–862 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Sund CJ et al. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol. Microbiol 67, 129–142 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Meehan BM, Baughn AD, Gallegos R & Malamy MH Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc. Natl Acad. Sci. USA 109, 12153–12158 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Z & Imlay JA The fumarate reductase of Bacteroides thetaiotaomicron, unlike that of Escherichia coli, is configured so that it does not generate reactive oxygen species. mBio 8, e01873–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borisov VB, Gennis RB, Hemp J & Verkhovsky MI The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 1807, 1398–1413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das A, Silaghi-Dumitrescu R, Ljungdahl LG & Kurtz DM Jr. Cytochrome bd oxidase, oxidative stress, and dioxygen tolerance of the strictly anaerobic bacterium Moorella thermoacetica. J. Bacteriol 187, 2020–2029 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ligeza A, Tikhonov AN, Hyde JS & Subczynski WK Oxygen permeability of thylakoid membranes: electron paramagnetic resonance spin labeling study. Biochim. Biophys. Acta 1365, 453–463 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Wexler HM Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev 20, 593–621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poole RK & Hill S Respiratory protection of nitrogenase activity in Azotobacter vinelandii — roles of the terminal oxidases. Biosci. Rep 17, 303–317 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Rakoff-Nahoum S, Foster KR & Comstock LE The evolution of cooperation within the gut microbiota. Nature 533, 255–259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L et al. Rubredoxin oxidase, a new flavo-hemoprotein, is the site of oxygen reduction to water by the “strict anaerobe” Desulfovibrio gigas. Biochem. Biophys. Res. Commun 193, 100–105 (1993). [DOI] [PubMed] [Google Scholar]
- 52.Silaghi-Dumitrescu R et al. A flavodiiron protein and high molecular weight rubredoxin from Moorella thermoacetica with nitric oxide reductase activity. Biochemistry 42, 2806–2815 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Silva G, Oliveira S, LeGall J, Xavier AV & Rodrigues-Pousada C Analysis of the Desulfovibrio gigas transcriptional unit containing rubredoxin (rd) and rubredoxin–oxygen oxidoreductase (roo) genes and upstream ORFs. Biochem. Biophys. Res. Commun 280, 491–502 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Spellerberg B et al. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol 19, 803–813 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Seki M, Iida K, Saito M, Nakayama H & Yoshida S Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol 186, 2046–2051 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baughn AD & Malamy MH The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427, 441–444 (2004). This paper demonstrates that the cytochrome bd oxidase of B. fragilis enhances its growth in the presence of nanomolar concentrations of O2.
- 57. Kim J, Hetzel M, Boiangiu CD & Buckel W Dehydration of (R)-2-hydroxyacyl-CoA to enoyl-CoA in the fermentation of α-amino acids by anaerobic bacteria. FEMS Microbiol. Rev 28, 455–468 (2004). This paper explains how ‘archerases’ manage the dehydration of non-activated substrates, yet are O2-sensitive because of the mechanism that is involved.
- 58.Thauer RK Methyl (alkyl)-coenzyme M reductases: nickel F-430-containing enzymes involved in anaerobic methane formation and in anaerobic oxidation of methane or of short chain alkanes. Biochemistry 58, 5198–5220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner AF, Frey M, Neugebauer FA, Schäfer W & Knappe J The free radical in pyruvate formate-lyase is located on glycine-734. Proc. Natl Acad. Sci. USA 89, 996–1000 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shibata N & Toraya T Molecular architectures and functions of radical enzymes and their (re)activating proteins. J. Biochem 158, 271–292 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Sawers G & Watson G A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase. Mol. Microbiol 29, 945–954 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Naqui A, Chance B & Cadenas E Reactive oxygen intermediates in biochemistry. Annu. Rev. Biochem 55, 137–166 (1986). [DOI] [PubMed] [Google Scholar]
- 63.Knappe J, Elbert S, Frey M & Wagner AF Pyruvate formate-lyase mechanism involving the protein-based glycyl radical. Biochem. Soc. Trans 21, 731–734 (1993). [DOI] [PubMed] [Google Scholar]
- 64. Zhang W, Wong KK, Magliozzo RS & Kozarich JW Inactivation of pyruvate formate-lyase by dioxygen: defining the mechanistic interplay of glycine 734 and cysteine 419 by rapid freeze-quench EPR. Biochemistry 40, 4123–4130 (2001). This paper details how O2 cleaves the PFL 2 polypeptide.
- 65.Imlay JA The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol 11, 443–454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gardner PR & Fridovich I Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem 266, 19328–19333 (1991). [PubMed] [Google Scholar]
- 67.Hausladen A & Fridovich I Superoxide and peroxynitrite inactivate aconitases, but nitric oxide does not. J. Biol. Chem 269, 29405–29408 (1994). [PubMed] [Google Scholar]
- 68.Lu Z, Sethu R & Imlay JA Endogenous superoxide is a key effector of the oxygen sensitivity of a model obligate anaerobe. Proc. Natl Acad. Sci. USA 115, E3266–E3275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frey M Hydrogenases: hydrogen-activating enzymes. Chembiochem 3, 153–160 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Ragsdale SW Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem. Rev 103, 2333–2346 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Pandelia ME, Lubitz W & Nitschke W Evolution and diversification of Group 1 [NiFe] hydrogenases. Is there a phylogenetic marker for O2-tolerance? Biochim. Biophys. Acta 1817, 1565–1575 (2012). [DOI] [PubMed] [Google Scholar]
- 72. Kubas A et al. Mechanism of O2 diffusion and reduction in FeFe hydrogenases. Nat. Chem 9, 88–95 (2017). This paper explores how molecular O2 attacks iron-only hydrogenases.
- 73.Swanson KD et al. [FeFe]-hydrogenase oxygen inactivation is initiated at the H cluster 2Fe subcluster. J. Am. Chem. Soc 137, 1809–1816 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Stripp ST et al. How oxygen attacks [FeFe] hydrogenases from photosynthetic organisms. Proc. Natl Acad. Sci. USA 106, 17331–17336 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hilton MG The metabolism of pyrimidines by proteolytic Clostridia. Arch. Microbiol 102, 145–149 (1975). [DOI] [PubMed] [Google Scholar]
- 76.Buckel W et al. Enzyme catalyzed radical dehydrations of hydroxy acids. Biochim. Biophys. Acta 1824, 1278–1290 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Gardner PR & Fridovich I Inactivation-reactivation of aconitase in Escherichia coli. A sensitive measure of superoxide radical. J. Biol. Chem 267, 8757–8763 (1992). [PubMed] [Google Scholar]
- 78.Pan N & Imlay JA How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol. Microbiol 39, 1562–1571 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Khademian M & Imlay JA Do reactive oxygen species or does oxygen itself confer obligate anaerobiosis? The case of Bacteroides thetaiotaomicron. Mol. Microbiol 114, 333–347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reddy SG et al. Dioxygen inactivation of pyruvate formate-lyase: EPR evidence for the formation of protein-based sulfinyl and peroxyl radicals. Biochemistry 37, 558–563 (1998). [DOI] [PubMed] [Google Scholar]
- 81.Vita N, Hatchikian EC, Nouailler M, Dolla A & Pieulle L Disulfide bond-dependent mechanism of protection against oxidative stress in pyruvateferredoxin oxidoreductase of anaerobic Desulfovibrio bacteria. Biochemistry 47, 957–964 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Pieulle L et al. Study of the thiol/disulfide redox systems of the anaerobe Desulfovibrio vulgaris points out pyruvate:ferredoxin oxidoreductase as a new target for thioredoxin 1. J. Biol. Chem 286, 7812–7821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Flint DH, Tuminello JF & Emptage MH The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem 268, 22369–22376 (1993). This article presents the first analysis of the mechanism by which oxidants degrade enzymic iron–sulfur clusters.
- 84.Flint DH & Allen RM Iron–sulfur proteins with nonredox functions. Chem. Rev 96, 2315–2334 (1996). [DOI] [PubMed] [Google Scholar]
- 85.Jang S & Imlay JA Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron–sulfur enzymes. J. Biol. Chem 282, 929–937 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liochev SI & Fridovich I Modulation of the fumarases of Escherichia coli in response to oxidative stress. Arch. Biochem. Biophys 301, 379–384 (1993). [DOI] [PubMed] [Google Scholar]
- 87.Lu Z & Imlay JA A conserved motif liganding the [4Fe–4S] cluster in [4Fe–4S] fumarases prevents irreversible inactivation of the enzyme during hydrogen peroxide stress. Redox Biol 26, 101296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuo CF, Mashino T & Fridovich I α,β-Dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J. Biol. Chem 262, 4724–4727 (1987). [PubMed] [Google Scholar]
- 89.Gardner PR & Fridovich I Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J. Biol. Chem 266, 1478–1483 (1991). [PubMed] [Google Scholar]
- 90.Shimoyama T, Rajashekhara E, Ohmori D, Kosaka T & Watanabe K MmcBC in Pelotomaculum thermopropionicum represents a novel group of prokaryotic fumarases. FEMS Microbiol. Lett 270, 207–213 (2007). [DOI] [PubMed] [Google Scholar]
- 91.Flint DH Initial kinetic and mechanistic characterization of Escherichia coli fumarase A. Arch. Biochem. Biophys 311, 509–516 (1994). [DOI] [PubMed] [Google Scholar]
- 92.Woods SA, Schwartzbach SD & Guest JR Two biochemically distinct classes of fumarase in Escherichia coli. Biochim. Biophys. Acta 954, 14–26 (1988). [DOI] [PubMed] [Google Scholar]
- 93.Sobota JM, Gu M & Imlay JA Intracellular hydrogen peroxide and superoxide poison 3-deoxy-D-arabinoheptulosonate 7-phosphate synthase, the first committed enzyme in the aromatic biosynthetic pathway of Escherichia coli. J. Bacteriol 196, 1980–1991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu MZ & Imlay JA Superoxide poisons mononuclear iron enzymes by causing mismetallation. Mol. Microbiol 89, 123–134 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li X & Imlay JA Improved measurements of scant hydrogen peroxide enable experiments that define its threshold of toxicity for Escherichia coli. Free. Radic. Biol. Med 120, 217–227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mishra S & Imlay JA An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol. Microbiol 90, 1356–1371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Korshunov S & Imlay JA Two sources of endogenous hydrogen peroxide in Escherichia coli. Mol. Microbiol 75, 1389–1401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Imlay JA & Fridovich I Assay of metabolic superoxide production in Escherichia coli. J. Biol. Chem 266, 6957–6965 (1991). [PubMed] [Google Scholar]
- 99.Dan Dunn J, Alvarez LA, Zhang X & Soldati T Reactive oxygen species and mitochondria: a nexus of cellular homeostasis. Redox Biol 6, 472–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meyer J Ferredoxins of the third kind. FEBS Lett. 509, 1–5 (2001). [DOI] [PubMed] [Google Scholar]
- 101.Valentine RC Bacterial ferredoxin. Bacteriol. Rev 28, 497–517 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Misra HP & Fridovich I The generation of superoxide radical during the autoxidation of ferredoxins. J. Biol. Chem 246, 6886–6890 (1971). [PubMed] [Google Scholar]
- 103.Rocha ER, Herren CD, Smalley DJ & Smith CJ The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe 9, 165–173 (2003). [DOI] [PubMed] [Google Scholar]
- 104.Hillmann F et al. The role of PerR in O2-affected gene expression of Clostridium acetobutylicum. J. Bacteriol 191, 6082–6093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Partridge JD, Poole RK & Green J The Escherichia coli yhjA gene, encoding a predicted cytochrome c peroxidase, is regulated by FNR and OxyR. Microbiology 153, 1499–1509 (2007). [DOI] [PubMed] [Google Scholar]
- 106.Khademian M & Imlay JA Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc. Natl Acad. Sci. USA 114, E6922–E6931 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pericone CD, Park S, Imlay JA & Weiser JN Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol 185, 6815–6825 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Figueiredo MC, Lobo SA, Carita JN, Nobre LS & Saraiva LM Bacterioferritin protects the anaerobe Desulfovibrio vulgaris Hildenborough against oxygen. Anaerobe 18, 454–458 (2012). [DOI] [PubMed] [Google Scholar]
- 109.Rocha ER, Owens G Jr. & Smith CJ The redoxsensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol 182, 5059–5069 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raskin L, Rittmann BE & Stahl DA Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl. Environ. Microbiol 62, 3847–3857 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Rocha ER & Smith CJ Ferritin-like family proteins in the anaerobe Bacteroides fragilis: when an oxygen storm is coming, take your iron to the shelter. Biometals 26, 577–591 (2013). This paper shows that when O2 is sensed, the anaerobe B. fragilis sequesters its iron to avoid lethal Fenton chemistry.
- 112.Li X et al. Transcriptomic analysis reveals hub genes and subnetworks related to ROS metabolism in Hylocereus undatus through novel superoxide scavenger trypsin treatment during storage. BMC Genomics 21, 437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andrews SC, Robinson AK & Rodriguez-Quinones F Bacterial iron homeostasis. FEMS Microbiol. Rev 27, 215–237 (2003). [DOI] [PubMed] [Google Scholar]
- 114.Zeth K Dps biomineralizing proteins: multifunctional architects of nature. Biochem. J 445, 297–311 (2012). [DOI] [PubMed] [Google Scholar]
- 115. Lee JW & Helmann JD The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367 (2006). This study reports how the PerR transcription factor uses Fenton chemistry to sense the presence of H2O2.
- 116.Marinho HS, Real C, Cyrne L, Soares H & Antunes F Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol 2, 535–562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mukhopadhyay A et al. Cell-wide responses to low–oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol 189, 5996–6010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hillmann F, Fischer RJ, Saint-Prix F, Girbal L & Bahl H PerR acts as a switch for oxygen tolerance in the strict anaerobe Clostridium acetobutylicum. Mol. Microbiol 68, 848–860 (2008). This study demonstrates that the activation of a peroxide defence system enables some growth of an obligate anaerobe in the presence of O2.
- 119.Liochev SI & Fridovich I The role of O2.− in the production of HO•: in vitro and in vivo. Free. Radic. Biol. Med 16, 29–33 (1994). [DOI] [PubMed] [Google Scholar]
- 120.Tulstrup MV et al. Antibiotic treatment affects intestinal permeability and gut microbial composition in wistar rats dependent on antibiotic class. PLoS ONE 10, e0144854 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rigottier-Gois L Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. ISME J. 7, 1256–1261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Winter SE et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Keyer K & Imlay JA Inactivation of dehydratase [4Fe–4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J. Biol. Chem 272, 27652–27659 (1997). [DOI] [PubMed] [Google Scholar]
- 124.Jang S & Imlay JA Hydrogen peroxide inactivates the Escherichia coli Isc iron–sulphur assembly system, and OxyR induces the Suf system to compensate. Mol. Microbiol 78, 1448–1467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Imlay JA, Sethu R & Rohaun SK Evolutionary adaptations that enable enzymes to tolerate oxidative stress. Free. Radic. Biol. Med 140, 4–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bowman SEJ et al. Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme. J. Biol. Inorg. Chem 24, 817–829 (2019). This structural analysis shows how a complementary protein can reactivate O2-damaged PFL.
- 127.Savageau MA Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat 122, 732–744 (1983). [Google Scholar]
- 128.Lin WC, Coppi MV & Lovley DR Geobacter sulfurreducens can grow with oxygen as a terminal electron acceptor. Appl. Environ. Microbiol 70, 2525–2528 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tally FP, Stewart PR, Sutter VL & Rosenblatt JE Oxygen tolerance of fresh clinical anaerobic bacteria. J. Clin. Microbiol 1, 161–164 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khan MT et al. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic–anoxic interphases. ISME J. 6, 1578–1585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rolfe RD, Hentges DJ, Barrett JT & Campbell BJ Oxygen tolerance of human intestinal anaerobes. Am. J. Clin. Nutr 30, 1762–1769 (1977). [DOI] [PubMed] [Google Scholar]
- 132.Jernberg C, Lofmark S, Edlund C & Jansson JK Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66 (2007). [DOI] [PubMed] [Google Scholar]
- 133.Kelly CJ et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Imlay JA Where in the world do bacteria experience oxidative stress? Environ. Microbiol 21, 521–530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Macfarlane GT, Gibson GR & Cummings JH Comparison of fermentation reactions in different regions of the human colon. J. Appl. Bacteriol 72, 57–64 (1992). [DOI] [PubMed] [Google Scholar]
- 136.Pitcher MC, Beatty ER & Cummings JH The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 46, 64–72 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhu H & Li YR Oxidative stress and redox signaling mechanisms of inflammatory bowel disease: updated experimental and clinical evidence. Exp. Biol. Med 237, 474–480 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


