Significance
Eosinophils contribute to type 2 immunity against helminths and allergens. The small intestine harbors eosinophils with incompletely understood pathophysiological roles. Here, we show that intestinal eosinophils include two subsets. One expresses the inhibitory receptor Clec4a4 and the inhibitory ligand PD-L1 and is unique to the small intestine; the other manifests a proinflammatory phenotype. Both subsets are blood derived. Remarkably, Clec4a4+ eosinophils were instructed by the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor that imprints many gut immune cells. Selective AHR deletion in eosinophils depleted Clec4a4+ eosinophils, augmented innate lymphocytes producing type 2 cytokines, and enhanced helminth clearance. We conclude that Clec4a4+ eosinophils have immunomodulatory functions, which could be harnessed for the therapy of food allergies and eosinophilic gastrointestinal disorders.
Keywords: eosinophil, intestine, helminth, aryl hydrocarbon receptor, allergy
Abstract
C-type lectin domain family 4, member a4 (Clec4a4) is a C-type lectin inhibitory receptor specific for glycans thought to be exclusively expressed on murine CD8α− conventional dendritic cells. Using newly generated Clec4a4-mCherry knock-in mice, we identify a subset of Clec4a4-expressing eosinophils uniquely localized in the small intestine lamina propria. Clec4a4+ eosinophils evinced an immunomodulatory signature, whereas Clec4a4− eosinophils manifested a proinflammatory profile. Clec4a4+ eosinophils expressed high levels of aryl hydrocarbon receptor (Ahr), which drove the expression of Clec4a4 as well as other immunomodulatory features, such as PD-L1. The abundance of Clec4a4+ eosinophils was dependent on dietary AHR ligands, increased with aging, and declined in inflammatory conditions. Mice lacking AHR in eosinophils expanded innate lymphoid cells of type 2 and cleared Nippostrongylus brasiliensis infection more effectively than did wild-type mice. These results highlight the heterogeneity of eosinophils in response to tissue cues and identify a unique AHR-dependent subset of eosinophils in the small intestine with an immunomodulatory profile.
Eosinophils are a small subset of granulocytes with acidophilic cytoplasmic granules that are generated in the bone marrow (BM) from myeloid progenitors; circulate in the bloodstream; and reside at low numbers in hematopoietic organs, the lung, adipose tissue, and the gastrointestinal tract (1–3). Eosinophil-specific granules contain cationic proteins, such as major basic protein, eosinophil cationic protein, eosinophil-derived neurotoxin, and eosinophil peroxidase (EPX), that have cytotoxic, ribonuclease, and oxidative activities (3–5). Eosinophils also produce lipid mediators, such as cysteinyl leukotriene C4, which activate cognate cysteinyl leukotriene receptors (CysLTRs) to promote vascular permeability, mucus secretion, and smooth muscle contraction (6). Moreover, eosinophils contain intracellular granules that store preformed cytokines and chemokines, which enable these leukocytes to respond to rapid stimuli and release soluble factors through classical exocytosis, piecemeal degranulation, and eosinophil cytolysis (7). Through this armamentarium of soluble mediators, eosinophils primarily contribute to host defense against helminth infections. Inappropriate recruitment, accumulation, and activation of eosinophils are associated with allergic inflammatory diseases, such as asthma, atopic dermatitis, and food allergy, as well as primary eosinophilic gastrointestinal disorders. Signals that recruit and activate eosinophils during inflammation, such as eotaxins (CCL11, CCL24, CCL26), interleukins (IL-5, IL-13, and IL-33), have been extensively investigated (8–11); however, little is known about the signals that maintain tissue eosinophils quiescent in the steady state.
Clec4a4 (C-type lectin domain family 4, member a4; also known as DCIR2) is a mouse cell surface receptor that belongs to the C-type lectin receptor (CLR) family (12, 13). The carbohydrate recognition domain of Clec4a4 specifically binds to bisecting N-acetylglucosamine glycans expressed on normal cells (14). The cytoplasmic domain of Clec4a4 contains an immunoreceptor tyrosine-based inhibition motif (15) that recruits SHP protein tyrosine phosphatases, which mediate a downstream inhibitory pathway (16). Clec4a4 is the cell surface molecule recognized by the monoclonal antibody 33D1 that defines CD8α− conventional dendritic cells (cDCs) (17–21), which specialize in antigen presentation by major histocompatibility complex (MHC) class II to CD4+ T cells (17, 22). Clec4a4 deficiency was shown to enhance the ability of CD8α− cDCs to produce cytokines in response to toll-like receptor ligands and activate T and B cell immune responses against specific antigens (19).
Here, we generated a Clec4a4 reporter mouse and found that Clec4a4 is expressed not only by CD8α− cDCs but also by a subset of eosinophils that are uniquely present in the small intestine lamina propria (siLP), where they represent the largest eosinophil population. We demonstrated that these Clec4a4+ eosinophils display a transcriptional profile that cumulatively denotes an immunomodulatory profile, as it is dominated by genes encoding inhibitory receptors, ligands for inhibitory receptors expressed on other cells, leukotriene degrading enzymes, and cholesterol biosynthetic enzymes. Conversely, the relatively minor population of Clec4a4– eosinophils selectively displayed a phenotype prone to inflammation. Parabiosis demonstrated that both subsets are blood borne. The gene expression program of Clec4a4+ eosinophils was in part driven by the aryl hydrocarbon receptor (AHR), which induced expression of Clec4a4 and other immunomodulatory molecules while suppressing proinflammatory programs. The abundance of Clec4a4+ eosinophils increased progressively with age and depended on dietary AHR ligands, but it diminished in various models of helminth infection and inflammation in parallel with an increase of Clec4a4– eosinophils. Clec4a4−/− mice, in which both eosinophils and CD8α− cDCs were freed from Clec4a4 inhibition, cleared Heligmosomoides polygyrus more efficiently than did wild-type (WT) mice in early stages of the infection. Moreover, EoCre × Ahrfl/fl mice, which selectively lack AHR and AHR-induced genes (including Clec4a4) in eosinophils, cleared Nippostrongylus brasiliensis infection more effectively than control mice, perhaps due to increased expansion of unfettered type 2 innate lymphoid cells (ILC2s), which mediate innate antihelminth responses (23, 24). We envision that AHR and its dietary ligands imprint a subset of eosinophils residing in the siLP with an immunomodulatory program. During parasitic infections and other pathogenic insults, changes in the tissue environment reverse the ratio between AHR-imprinted eosinophils and eosinophils with an inflammatory program to elicit an effective host response. This finding may open therapeutic avenues for reprogramming intestinal eosinophils in food allergies as well as primary eosinophilic gastrointestinal disorders of unknown etiology (25).
Results
The Small Intestine Contains Clec4a4+ and Clec4a4− Eosinophil Subsets.
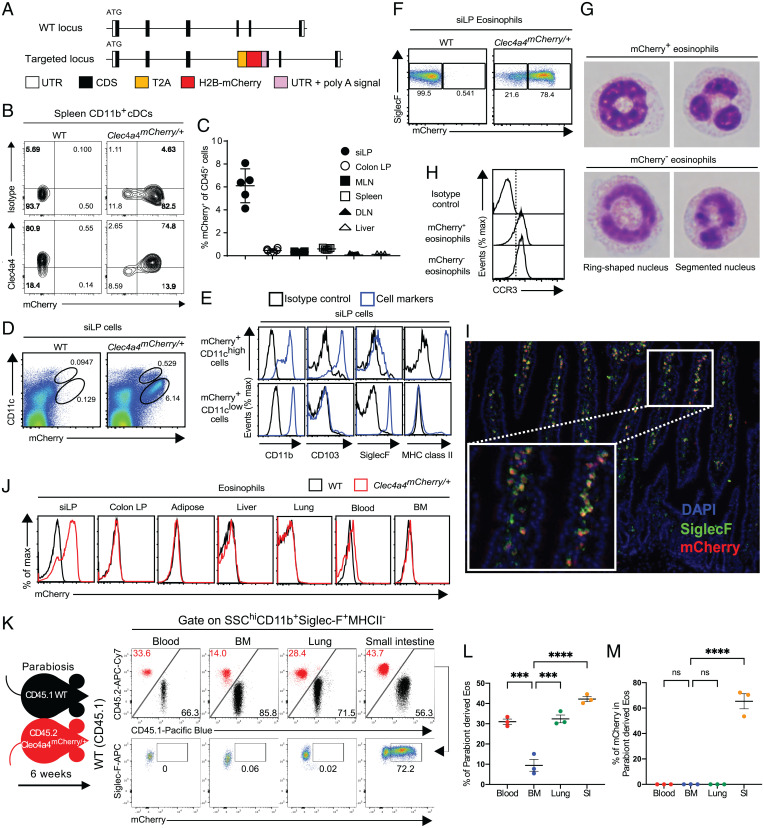
To identify Clec4a4-expressing cells, we generated Clec4a4-mCherry knock-in (Clec4a4mCherry) mice (Fig. 1A). In spleens of Clec4a4mCherry/+ mice, mCherry was selectively expressed in CD11b+ dendritic cells (DCs) stained by the 33D1 monoclonal antibody, which recognizes Clec4a4 (17) (Fig. 1B). This result verified that mCherry messenger RNA was selectively transcribed under the control of the Clec4a4 promoter and recapitulated the expression pattern of Clec4a4 protein. We next surveyed mCherry expression in CD45+ hematopoietic cells from various tissues of Clec4a4mCherry/+ mice. A large number of mCherry+ CD45+ cells were observed in the siLP (Fig. 1C). These cells encompassed two subsets with distinct levels of CD11c expression (Fig. 1D). CD11chigh mCherry+ cells expressed CD11b, CD103, and MHC class II (Fig. 1E), thereby corresponding to the extensively characterized intestinal CD103+ CD11b+ cDCs (26). However, the CD11clow mCherry+ subset expressed CD11b and SiglecF, a cell surface marker of eosinophils, suggesting expression of Clec4a4 on eosinophils. To corroborate this result, we examined mCherry expression in siLP eosinophils that were gated as side scatter (SSC)high, MHC class II–, CD11b+, and SiglecF+ cells (27). Indeed, mCherry was expressed in ∼70 to 80% of siLP eosinophils, whereas the mCherry– subset represented a minor fraction (Fig. 1F and SI Appendix, Fig. S1A). Both mCherry+ and mCherry– eosinophils displayed ring-shaped and segmented nuclei indicative of mature eosinophils (Fig. 1G). Both subsets expressed the CC chemokine receptor 3 (CCR3), a common eosinophil marker and a receptor for eotaxin (Fig. 1H) (1–3). Analysis of intestinal sections of Clec4a4mCherry/+ mice by fluorescence microscopy at steady state showed that most of the SiglecF+ Clec4a4+ eosinophils were present in the lamina propria of the intestinal villi (Fig. 1I). Examination of mCherry expression in eosinophils of colon, adipose tissue, liver, lung, blood, and BM revealed that Clec4a4 expression was restricted to siLP eosinophils (Fig. 1J).
Fig. 1.
A heterogeneous population of Clec4a4+ and Clec4a4− eosinophils resides in the small intestine. (A) Targeting strategy to generate Clec4a4mCherry/+ mice. CDS, coding sequence; H2B, human histone H2B; T2A, T2A self-cleaving peptide; UTR, untranslated region. (B) Expression of Clec4a4 and mCherry in splenic CD11b+ cDCs. (C) Frequency of mCherry+ cells among CD45+ cells (gated on live cells) in the indicated tissues (n = 3 to 7). DLN, skin-draining lymph node; LP, lamina propria; MLN, mesenteric lymph node. (D) mCherry expression on CD11chigh and CD11clow CD45+ cells derived from siLP (gated on live cells). (E) Expression of CD11b, CD103, Siglec-F, and MHC class II on mCherry+ CD11chigh CD45+ cells and mCherry+ CD11clow CD45+ cells derived from siLP (gated on live cells). (F) mCherry expression in Siglec-F+ eosinophils derived from siLP (gated on SSChigh MHC class II− CD11b+ cells). (G) Representative pictures of FACS-sorted mCherry+ and mCherry− eosinophils from siLP. (H) CCR3 expression in mCherry+ and mCherry− eosinophils in siLP. (I) Representative image showing localization of Siglec-F+mCherry+ eosinophils in siLP villi of Clec4a4mCherry/+ mice. Inset is an enlargement of I. DAPI, 4′,6-diamidino-2-phenylindole. (J) mCherry expression in eosinophils in the indicated tissues. (K) Parabiosis of CD45.1 WT and CD45.2 Clec4a4mCherry/+ mice indicates that eosinophils recirculate in siLP, blood, and lung but much less so in BM; Clec4A4 mCherry is only expressed by eosinophils in the siLP. (L) Frequency of parabiont-derived eosinophils in the indicated tissues. (M) Frequency of parabiont-derived mCherry+ eosinophils in the indicated tissues. Results are shown as mean ± SEM. P values were calculated using one-way ANOVA and the Tukey's multiple comparisons test or the two-tailed Student’s t test. ns, not significant. ***P < 0.001; ****P < 0.0001.
Since many immune cell types found in the small intestinal mucosa consist of long-term tissue-resident cells adapted to environmental cues, we sought to examine whether this is the case for Clec4a4+ eosinophils. We performed parabiosis, joining WT (CD45.1) and Clec4a4mCherry/+ (CD45.2) mice for 6 wk. Analysis of CD45.1 WT mice revealed a substantial proportion of CD45.2+ eosinophils in the small intestine (Fig. 1 K and L), ∼60% of which were mCherry+ (Fig. 1M), suggesting that Clec4a4+ eosinophils are mainly blood borne rather than derived from a local resident population. Interestingly, although the frequency of parabiont-derived eosinophils in the small intestine was similar to that observed in blood and lung, only cells in the small intestine expressed mCherry, indicating that Clec4a4 is induced by the unique environment of the small intestine. Accordingly, mCherry was not expressed in activated lung eosinophils purified from a model of house dust mite–induced inflammation (SI Appendix, Fig. S1B). Within the small intestine, the representation of Clec4a4+ eosinophils varied in different segments, decreasing from the proximal to most distal regions (SI Appendix, Fig. S1C). Taken together, these results demonstrate that the small intestine contains Clec4a4+ and Clec4a4− eosinophil subsets in the steady state and that the Clec4a4+ subset is unique to the small intestine due to the impact of the microenvironment on blood-derived eosinophils.
Clec4a4+ and Clec4a4− Eosinophils Display Distinct Receptor Repertoires.
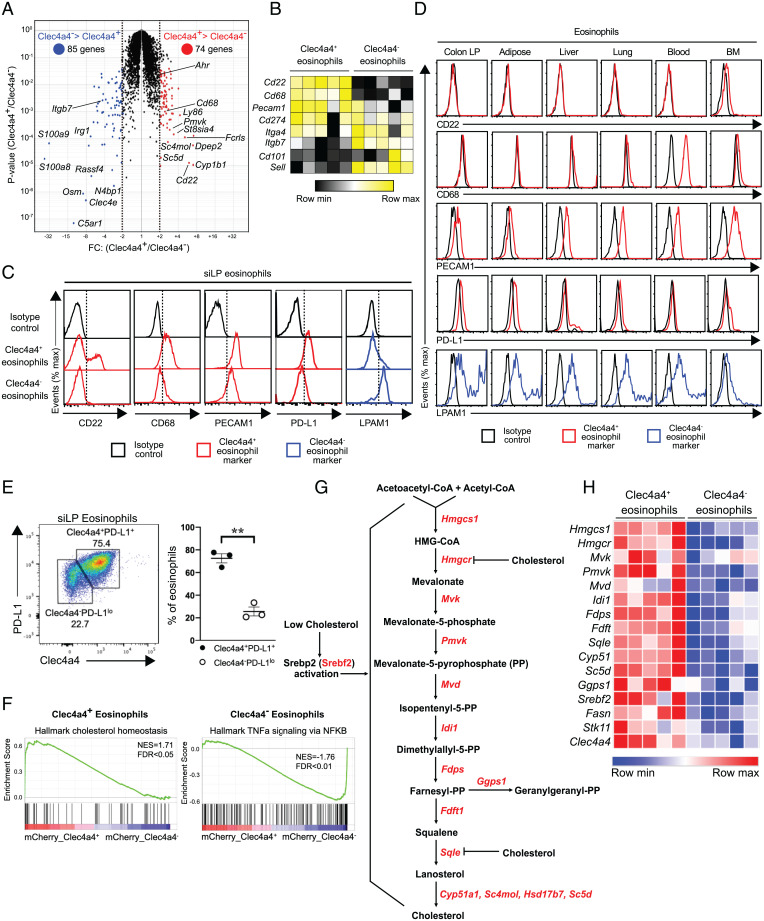
To gain more insight into the characteristics of Clec4a4+ eosinophils, we examined genes differentially expressed between fluorescence-activated cell sorting (FACS)-sorted Clec4a4+ and Clec4a4− eosinophils by microarray analysis (the sorting strategy is shown in SI Appendix, Fig. S1A). Seventy-four transcripts were expressed at least twofold higher in Clec4a4+ eosinophils than in the Clec4a4− population, whereas 85 transcripts had the opposite pattern (Fig. 2 A and B and SI Appendix, Fig. S2 A and B). Clec4a4+ eosinophils abundantly expressed Cd22, Cd68, Pecam1, and Cd274 (encoding PD-L1), whereas Itgb7 (encoding the β7-integrin) was more highly expressed by Clec4a4− eosinophils. We confirmed by flow cytometry that a fraction of siLP Clec4a4+ eosinophils expressed CD22, a receptor of the sialoadhesin family that transmits inhibitory signals; all Clec4a4+ eosinophils expressed the lysosomal marker CD68, the adhesion molecule PECAM1, and CD274 (PD-L1), which is the ligand for the inhibitory receptor PD-1 (Fig. 2C). Conversely, Clec4a4− eosinophils expressed the lymphocyte Peyer’s patch adhesion molecule 1 (LPAM1), which consists of the α4β7-integrin heterodimer. A survey of these cell surface markers in the eosinophils of colon lamina propria, adipose tissue, liver, lung, blood, and BM showed that CD22 was exclusively expressed in siLP eosinophils, corroborating a recent study (28) (Fig. 2D); CD68 was expressed by blood eosinophils, and PECAM1 was expressed by liver, lung, blood, and BM eosinophils. LPAM1 was expressed in all Clec4a4− eosinophils of various tissues. Overall, these data validate the identification of two phenotypically distinguishable subsets of siLP eosinophils. The lung was recently reported to include a subset of Siglec-FintCD62L+CD101low–resident eosinophils and a subset of Siglec-FhiCD62L−CD101hi–recruited inflammatory eosinophils (29). However, these subsets were phenotypically distinct from Clec4a4+ and Clec4a4− siLP subsets, as both CD62L and CD101 were weakly expressed in siLP eosinophils (SI Appendix, Fig. S2C).
Fig. 2.
Transcriptomic profiling and characterization of Clec4a4+ and Clec4a4− eosinophil subsets. (A) Volcano plot of differentially expressed genes in mCherry+ and mCherry− eosinophils (n = 5 replicates in each group). Transcripts with FC (fold change) > 2 and P < 0.05 in mCherry+ and mCherry− eosinophils are depicted in red and blue, respectively. (B) Heat map of selected genes specific to Clec4a4+ and Clec4a4− eosinophils. (C) Differential expression of cell surface (CD22, PECAM1, PD-L1, and LPAM1) and intracellular markers (CD68) by Clec4a4+ and Clec4a4− siLP eosinophils. (D) Expression of the markers indicated in C by eosinophils from the indicated tissues. LP, lamina propria. (E, Left) Expression of Clec4a4 and PD-L1 in eosinophils derived from siLP. (E, Right) Frequency of Clec4a4+PD-L1+ and Clec4a4−PD-L1lo eosinophils in the siLP. (F) Enrichment plots demonstrating up-regulated cholesterol homeostasis and TNFα signaling via NF-κB in mCherry+ and mCherry− eosinophils, respectively. NES, normalized enrichment score; FDR, false discovery rate. (G) Biochemical pathways for de novo cholesterol synthesis. CoA, coenzyme A; HMG, hydroxymethylglutaryl. (H) Heat map of genes associated with cholesterol synthesis pathways in mCherry+ and mCherry− eosinophils. Results are shown as mean ± SEM. P values were calculated using the two-tailed Student’s t test. **P < 0.01.
We noted that Clec4a4+ eosinophils expressed higher amounts of signal-regulatory protein alpha (SIRPα) (SI Appendix, Fig. S2D), a receptor that delivers intracellular inhibitory signals upon recognition of its ubiquitously expressed ligand CD47 (30, 31). Given that CD22 and Clec4a4 have also been shown to be potent inhibitory receptors specific for glycans expressed by B cells (32) and DCs (19), respectively, the concerted expression of these inhibitory receptors suggests that Clec4a4+ eosinophils are fairly refractory to activating stimuli. Notably, Clec4a4+ eosinophils expressed PD-L1 (Fig. 2 C–E), which inhibits ILC2, T cells, and myeloid cells expressing PD-1 (33, 34). Thus, Clec4a4+ eosinophils may further maintain a noninflammatory environment by inhibiting immune responses by ILC2s, T cells, and myeloid cells. Conversely, the Clec4a4– subset expressed LPAM1, which interacts with mucosal cell adhesion molecule-1 expressed in vascular endothelium and is required for the homing of eosinophils into the gastrointestinal tract (2), suggesting that Clec4a4– eosinophils may be recently recruited from the blood.
Clec4a4+ and Clec4a4− Eosinophils Express Alternative Immunomodulatory and Inflammatory Programs.
We next explored transcriptome profiles of Clec4a4+ and Clec4a4− eosinophils for different functional properties. Clec4a4+ eosinophils selectively expressed Dipeptidase 2 (Dpep2), a membrane-bound dipeptidase that hydrolyses the leukotriene D4 into leukotriene E4 (Fig. 2A and SI Appendix, Fig. S2 A and B). LTE4 is the weakest activator of CysLTR1 (6), and DPEP2 has been reported to modulate macrophage inflammation and nuclear factor kappa B (NF-κB) signaling independent of its enzymatic activity (35). Another striking feature of the Clec4a4+ eosinophil transcriptome was the concerted expression of many genes controlling the de novo cholesterol biosynthesis pathway (e.g., Hmgcs1, Pvmk, Idi1, Fdps, Fdft1, Sqle, Sc5d, Cyp51, Sc4mol) (36) (Fig. 2 F–H). Cholesterol synthesis may be necessary to generate membranes for efficient intracellular storage of eosinophil granules and for secretory vesicles (7).
Clec4a4− eosinophils evinced a clear proinflammatory and tissue repair profile exemplified by the preferential expression of genes involved in inflammation (S100a8, S100a9, Tnf, Il6) and fibrosis (Oncostatin M, Osm) (Fig. 2A and SI Appendix, Fig. S2 A and B). The expression of Irak3, components of the NF-κB pathway (e.g., Nfkb1, Nfkb2, Nfkbia), Fas, and components of the inflammasome (e.g., Nlrp3, Casp4) corroborated the proinflammatory predisposition of Clec4a4− eosinophils (Fig. 2F and SI Appendix, Fig. S2E). A similar gene expression pattern was observed in BM eosinophils stimulated by IL-33, which activates the NF-κB pathway (37). Overall, these results demonstrate that Clec4a4+ and Clec4a4– siLP eosinophilic subsets have distinct immunomodulatory and proinflammatory programs, respectively.
AHR Drives the Profile of Clec4a4+ Eosinophils.
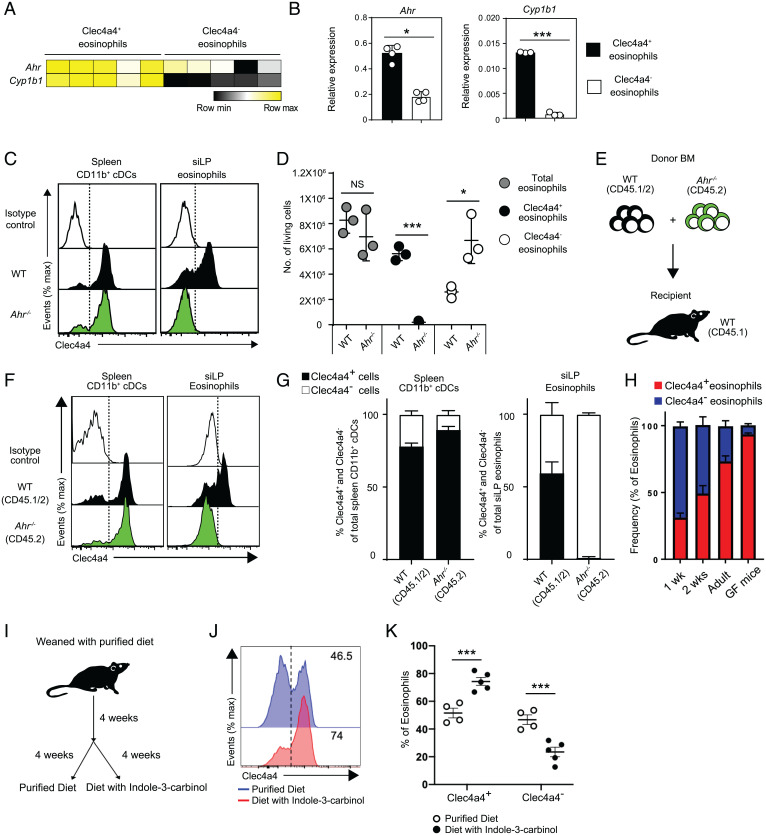
We asked which transcription factors could drive the different programs of siLP eosinophils. We noted that Clec4a4+ eosinophils preferentially expressed the Ahr gene and its target Cyp1b1, which we confirmed by qRT-PCR analysis (Fig. 3 A and B). AHR is a member of the Per–Arnt–Sim superfamily of transcription factors that sense environmental signals. In the gut, AHR senses indole compounds that are generated by degradation of nutritional tryptophan by the microbiota or are directly introduced with certain foods, such as cruciferous vegetables (38–42). AHR signaling sustains development and function of many cell types in the intestine, including ILC3s, Th17, DCs, macrophages, intraepithelial lymphocytes (IELs), and enteric neurons (41–45). To validate the impact of AHR on Clec4a4+ eosinophils, we examined Ahr−/− mice. Ahr−/− siLP eosinophils and DCs did not express Clec4a4, while Ahr−/− spleen CD11b+ cDCs maintained Clec4a4 (Fig. 3C and SI Appendix, Fig. S3A). We also noted that the absence of Clec4a4+ eosinophils in Ahr−/− mice was paralleled by a compensatory increase in Clec4a4– eosinophils, such that the total number of eosinophils was comparable in Ahr−/− and WT mice (Fig. 3D). Thus, AHR is required for Clec4a4 expression, while the overall pool of siLP eosinophils is AHR independent in the steady state. To see whether the requirement of AHR was cell intrinsic or cell extrinsic, we performed a mixed BM chimera experiment. After lethal irradiation, CD45.1+ WT mice were transplanted with a 50:50 mixture of CD45.1/2+ WT BM and CD45.2+ Ahr−/− BM (Fig. 3E). Both WT BM and Ahr−/− BM successfully reconstituted splenocytes (SI Appendix, Fig. S3B) and spleen CD11b+ Clec4a4+ cDCs (Fig. 3 F and G) in about a 50:50 WT/Ahr−/− ratio (Fig. 3 F and G). Small intestinal eosinophils were reconstituted at a 30:70 WT/Ahr−/− ratio (SI Appendix, Fig. S3B), indicating that Ahr−/− eosinophils have a competitive reconstitution advantage over WT eosinophils for reasons that remain to be determined. Notwithstanding, only WT BM reconstituted both Clec4a4+ and Clec4a4– eosinophils, whereas Ahr−/− BM could only generate Clec4a4– eosinophils (Fig. 3 F and G). We conclude that the requirement of AHR for Clec4a4 expression in eosinophils is cell intrinsic.
Fig. 3.
Development of the Clec4a4+ eosinophil subset is dependent on aging and on the AHR pathway. (A) Heat map of Ahr and Cyp1b1 in mCherry+ and mCherry− eosinophils. (B) qRT-PCR of Ahr and Cyp1b1 transcripts in Clec4a4+ and Clec4a4− eosinophils (n = 4 replicates). (C) Clec4a4 expression in splenic CD11b+ cDCs and siLP eosinophils from WT and Ahr−/− mice. (D) Absolute numbers of Clec4a4+ and Clec4a4− eosinophils in WT and Ahr−/− mice (n = 3 in each group). (E) Experimental strategy for mixed BM chimeras. (F) Clec4a4 expression in splenic CD11b+ cDCs and siLP eosinophils derived from WT and Ahr−/− BM cells after reconstitution. (G) Frequency of Clec4a4+ and Clec4a4− splenic CD11b+ cDCs and siLP eosinophils in BM chimeras. (H) Frequency of Clec4a4+ and Clec4a4− siLP eosinophils in WT mice at 1, 2, and 10 to 12 wk of age (n = 3, 6, and 7 replicates, respectively) and in GF mice (n = 5). (I) Experimental time line for feeding mice with a purified diet free of AHR ligands or a purified diet containing the AHR ligand I3C. (J) Representative Clec4a4 expression in siLP eosinophils derived from mice fed with a purified diet free of AHR ligands or a purified diet containing AHR ligands. (K) Frequency of Clec4a4+ and Clec4a4− siLP eosinophils in mice fed with a purified diet free of AHR ligands (n = 4) or a purified diet containing AHR ligands (n = 5). Results are shown as mean ± SEM. P values were calculated using the two-tailed Student’s t test. NS, not significant. *P < 0.05; ***P < 0.001.
We next asked whether AHR is just required for Clec4a4 expression or for the differentiation of the entire Clec4a4+ eosinophil program. Since all eosinophils were Clec4a4– in Ahr−/− mice, we selected genes differentially expressed between Clec4a4+ and Clec4a4– eosinophils of WT mice and ascertained their expression in Clec4a4– eosinophils of Ahr−/− mice. Among the genes more abundantly expressed in WT Clec4a4+ than in WT Clec4a4– eosinophils, 20 genes, including Cd22, were more highly expressed in Clec4a4− eosinophils of Ahr−/− mice than in Clec4a4− eosinophils of WT mice, indicating AHR independency (SI Appendix, Fig. S3C). Conversely, 21 of the genes highly expressed in WT Clec4a4+ but not in WT Clec4a4– eosinophils were expressed at low levels in Clec4a4− eosinophils of Ahr−/− mice, indicating AHR dependency; these genes included Dpep2 as well as genes involved in cholesterol metabolism, including Idi1, Sc5d, Hmgcs1, and Hsd17b7 (SI Appendix, Fig. S3C).
Among the genes expressed less in WT Clec4a4+ than in WT Clec4a4– eosinophils, 17 genes were expressed at higher levels in Ahr−/− eosinophils, indicating suppression by AHR (SI Appendix, Fig. S3D). These genes included many inflammatory molecules, such as Il6, S100a8, and S100a9. In comparison with their relatively high expression in Clec4a4− eosinophils of WT mice, 10 genes were equally suppressed in Clec4a4+ eosinophils of WT mice and Clec4a4− eosinophils of Ahr−/− mice, including Tnf, Nlrp3, and Fas (SI Appendix, Fig. S3D). Taken together, these results demonstrate that the differentiation of Clec4a4+ eosinophils is both AHR dependent and independent. Genes either induced or suppressed in an AHR-independent fashion in Clec4a4+ eosinophils may be controlled by other transcription factors, such as NR4A2 (also called Nurr1), which is more abundantly expressed in Clec4a4+ than in Clec4a4– eosinophils (SI Appendix, Fig. S3C) and has been shown to control inflammation and lipid metabolism (46).
Differentiation of Clec4a4+ Eosinophils Is Partly Dependent on Dietary AHR Ligands.
Since AHR senses indole compounds generated by intestinal microbiota or introduced with certain nutrients, we hypothesized that Clec4a4+ eosinophils may develop upon colonization of the intestinal tract by a mature microbiota and/or introduction of a normal diet after weaning. Consistent with this hypothesis, Clec4a4+ cells within siLP eosinophils were more abundant in young adult mice than in 1- or 2-wk-old mice (Fig. 3H). To directly test the impact of microbiota on siLP eosinophils, we examined germ-free (GF) mice but found that Clec4a4+ eosinophils were equally represented in GF and control specific pathogen–free (SPF) mice (Fig. 3H). We also treated SPF mice with antibiotics and then, colonized them with Lactobacilluls reuteri 100-23, which produces large amounts of indole ligands when fed a tryptophan-rich diet (44, 47). However, enrichment of microbiota with indole-producing L. reuteri did not expand Clec4a4+ eosinophils further than that seen in mice colonized with an L. reuteri mutant unable to produce AHR ligands (SI Appendix, Fig. S3E). We hypothesized that the lack of bacterial-derived AHR ligands may be compensated by dietary ligands. To test the impact of dietary AHR ligands, weaned mice were fed a purified diet lacking AHR ligands for 4 wk and then, kept for an additional 4 wk on this diet with or without additional indole-3-carbinol (I3C), a plant-derived precursor of AHR ligands (Fig. 3I). The population of Clec4a4+ eosinophils declined in mice fed the purified diet but was restored by the I3C supplement (Fig. 3 J and K), corroborating a dependency of Clec4a4+ eosinophils on nutritional AHR ligands.
Clec4a4+ Eosinophils Wane during Intestinal Inflammation.
We sought to assess the presence of Clec4a4+ and Clec4a4– eosinophils in response to infection with H. polygyrus, a natural intestinal parasite of mice (48). In this experimental model, BALB/c mice are infected with H. polygyrus by oral gavage with infective larvae, which penetrate into the submucosa of the small intestine and develop into adult worms that return into the intestinal lumen, mate, and produce eggs that are excreted in the feces. We noted that after infection with H. polygyrus, Clec4a4+ eosinophils declined by day 8 postinfection, whereas Clec4a4− eosinophils expanded in compensation (SI Appendix, Fig. S4A). A similar decrease of Clec4a4+ eosinophils paralleled by a compensatory increase of Clec4a4− eosinophils was observed in other intestinal inflammatory conditions, such as those induced in a model of high-fat diet (SI Appendix, Fig. S4B) and in a model of food allergy caused by gastrointestinal intolerance to ovalbumin (SI Appendix, Fig. S4 C–F). These findings raise the possibility that Clec4a4+ eosinophils become Clec4a4− in pathological settings, acquiring a proinflammatory program more suitable for host responses. In addition, newly recruited Clec4a4− eosinophils may fail to acquire the AHR-induced profile.
Clec4a4 and AHR Imprinting Attenuate Host Defense against Parasites.
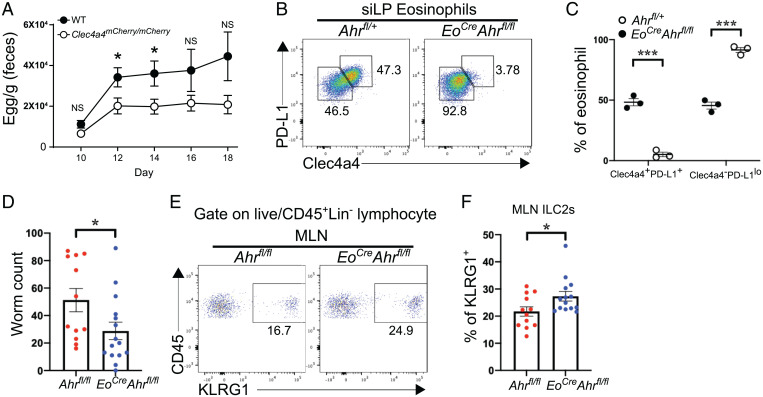
Given that Clec4a4 is an inhibitory receptor, we hypothesized that a deletion of Clec4a4 may augment the early responses against parasites, such as H. polygyrus. To test this, we infected homozygous Clec4a4mCherry/mCherry mice, in which both endogenous Clec4a4 genes are disrupted by mCherry. Mice were infected with H. polygyrus and examined for the numbers of eggs in the feces on days 10, 12, 14, 16, and 18 postinfection. Clec4a4mCherry/mCherry mice had lower egg counts than did WT mice on days 12 and 14 postinfection (Fig. 4A), suggesting that the release of quiescent eosinophils from Clec4a4-mediated inhibition enhances early host responses against helminth infection.
Fig. 4.
Clec4a4+ eosinophils modulate type 2 immunity during helminth infection. (A) Egg counts in feces of WT and Clec4a4-deficient (Clec4a4mCherry/mCherry) mice (n = 13 in each group) at the indicated time points during H. polygyrus infection. (B) Expression of Clec4a4 and PD-L1 in siLP eosinophils derived from Ahrfl/+ and EoCreAhrfl/fl mice. (C) Frequency of Clec4a4+PD-L1+ and Clec4a4−PD-L1lo siLP eosinophils in Ahrfl/+ and EoCreAhrfl/fl mice (n = 3 each group). (D) Worm counts in Ahrfl/fl (n = 12) and EoCreAhrfl/fl (n = 15) mice at day 6 postinfection with N. brasiliensis. (E) Representative percentages of KLRG1+ ILC2s among CD45+ cells in mesenteric lymph nodes (MLNs) of Ahrfl/fl and EoCreAhrfl/fl mice. A gate was applied on lineage-negative cells. Lineage markers included CD3, CD4, CD8, CD19, and NK1.1. (F) Frequency of ILC2s in MLNs from Ahrfl/fl (n = 12) and EoCreAhrfl/fl (n = 14) mice at day 6 postinfection with N. brasiliensis. Results are shown as mean ± SEM. P values were calculated using the two-tailed Student’s t test or one-way ANOVA and the Tukey's multiple comparisons test. NS, not significant. *P < 0.05; ***P < 0.001.
Since Clec4a4 is expressed in both eosinophils and CD11b+ DCs, both of which may contribute to host response, we sought to more selectively establish the impact of Clec4a4 and possibly, other AHR-induced genes on the clearance of helminth infection. For this purpose, we crossed mice with floxed alleles of Ahr (Ahrfl/fl) with an EoCre deleter mice, in which Cre recombinase is driven by the endogenous promoter of Epx (49). A marked reduction of Clec4a4+ eosinophils was evident in the small intestine of EoCre × Ahrfl/fl mice (Fig. 4B). This finding was consistent with the cell-intrinsic requirement of AHR for Clec4a4 expression observed in the mixed BM chimera experiment (Fig. 3F). The expression of PD-L1 on small intestine eosinophils was also markedly reduced in EoCre × Ahrfl/fl mice (Fig. 4 B and C). We next challenged EoCre × Ahrfl/fl mice with H. polygyrus. Fecal content of H. polygyrus eggs and granuloma counts were similar in EoCre × Ahrfl/fl mice and control mice (SI Appendix, Fig. S5). We further infected mice with N. brasiliensis, which elicits a more rapid gastrointestinal expulsion than H. polygyrus. While clearance of H. polygyrus infection in C57BL/6 mice requires up to 20 wk and a sustained T cell response (50), adult N. brasiliensis worms are normally expelled from the intestines of C57BL/6 mice by day 7 postinfection. N. brasiliensis counts were lower in EoCre × Ahrfl/fl mice than in control mice on day 6 postinfection (Fig. 4D). We noted that enhanced worm expulsion was paralleled by an increase in ILC2s in mesenteric lymph nodes (Fig. 4 E and F). ILC2s mediate antiparasitic responses through secretion of IL-5 and IL-13 (23, 24). Since ILC2s express the inhibitory receptor PD-1 (34) and Clec4a4+ eosinophils express PD-L1, the lack of this inhibitory interaction in EoCre × Ahrfl/fl mice may facilitate ILC2 expansion. We conclude that AHR-imprinted Clec4a4+ eosinophils attenuate intestinal parasite clearance mechanisms.
Discussion
Eosinophils contribute to host defense against helminth infections through the release of granule proteins, leukotrienes, and cytokines but can also cause allergic inflammation (1–3). Using a newly generated reporter mouse for the inhibitory receptor Clec4a4, we have identified a unique population of eosinophils exclusively found in the siLP. In addition to Clec4a4, the receptor repertoire of these cells included other inhibitory receptors, such as CD22 (28) and SIRPα (30, 31). The concomitant expression of PD-L1, which inhibits ILC2s, T cells, and myeloid cells expressing PD-1, and DPEP2, which degrades leukotrienes, indicates that Clec4a4+ eosinophils may be both refractory to activation and prone to restrain intestinal immune responses. This large population of Clec4a4+ eosinophils was accompanied by a smaller population of Clec4a4– eosinophils with a proinflammatory and tissue repair program specifying tumor necrosis factor (TNF), IL-6, and OSM among other soluble mediators. These cells were poorly represented in the steady state but expanded in inflammatory conditions, while Clec4a4+ eosinophils declined. Gut Clec4a4+ and Clec4a4– eosinophils were phenotypically distinct from other eosinophil subsets recently reported in the lung, such as lung-resident eosinophils (29) and airway-recruited inflammatory eosinophils (29). We conclude that eosinophils, like other immune cell types, exhibit a tissue diversity that reflects the influence of microenvironmental factors.
Remarkably, the phenotype of Clec4a4+ eosinophils was in part instructed by AHR, which also controls the development and function of many other gut immune cell populations, including ILC3s, Th17, DCs, macrophages, and IELs, as well as enteric neurons (41–45). AHR drove expression of Clec4a4 as well as other immunomodulatory molecules, such as DPEP2 and PD-L1. A very recent study independently corroborated that eosinophils of the gastrointestinal tract display an AHR-dependent profile (51). In the gut, AHR is activated by indoles derived from vegetables that contain tryptophan catabolic products (38–42) and microbial degradation of nutritional tryptophan (47). Accordingly, Clec4a4+ eosinophils were depleted in mice fed an AHR ligand–deprived diet but restored by a supplement of nutritional I3C; although not formally confirmed by us, I3C is known to generate the biologically active AHR ligand indolo[3,2-b]carbazole in the acidic pH of the stomach. Clec4a4+ eosinophils were similarly represented in GF mice and SPF mice; moreover, this population did not preferentially expand in mice with a microbiota enriched with AHR ligand–producing bacteria, suggesting redundancy between dietary and microbiota-generated AHR ligands. Future studies will be necessary to test the relative impact of dietary and microbiota-derived AHR ligands in the generation of Clec4a4+ eosinophils by comparing the abundance of these cells in GF and SPF mice fed a purified diet and reconstituted with microbes either competent or unable to produce AHR ligands.
The instruction of Clec4a4+ eosinophils by AHR ligands is reminiscent of the tissue imprinting previously reported in gut-resident immune cells, such as ILCs and macrophages. However, our parabiosis experiment demonstrated that Clec4a4+ eosinophils are blood derived, while tissue-resident ILCs and macrophages are long-lived tissue cells that renew from local progenitors (52–54). We conclude that blood eosinophils adapt to the environment and acquire Clec4a4+ features soon after reaching the small intestine. Notably, during intestinal H. polygyrus infection or models of inflammation, the representation of Clec4a4+ eosinophils rapidly declined, paralleled by an increase of Clec4a4– eosinophils. We envision that Clec4a4+ eosinophils derive from blood Clec4a4– eosinophils instructed by AHR signaling. In inflammatory conditions, this imprinting may weaken or vanish because of decreased AHR expression or less accessible AHR ligands in the altered tissue environment. Further studies are needed to test this hypothesis.
We noted that the functional impact of AHR-imprinted eosinophils in antihelminth defense was different in the two models of helminth infection in EoCre × Ahrfl/fl mice that we tested. Clearance of H. polygyrus infection was similar in EoCre × Ahrfl/fl and control mice, a result that was also recently reported (51). Since H. polygyrus infection lasts up to 20 wk in B6 mice and requires a robust Th2 response for worm clearance, this result probably reflects the limited contribution of eosinophils to this infection model (55, 56). Conversely, N. brasiliensis was more effectively cleared in EoCre × Ahrfl/fl mice. This phenotype correlated with an increased frequency of ILC2s in the mesenteric lymph nodes, which may reflect release of PD-1+ ILC2s from the inhibitory constraints imposed by PD-L1 expressed on eosinophils (34). Whether Th2 responses are also altered in this scenario remains to be established. We envision that the immunomodulatory effect of AHR-dependent eosinophils may preferentially emerge in this model because N. brasiliensis infection in C57BL/6 mice elicits a more rapid and robust gastrointestinal immune response in which eosinophils may play a more prominent role (57).
To date, human counterparts of gut Clec4a4+ and Clec4a4– eosinophils have not been identified. In the mouse, Clec4a4 is one of four closely linked CLR genes, Clec4a1 to -4 (also known as Dcir1 to -4); in humans, only one DCIR molecule exists, which is expressed by CD14+ monocytes, CD15+ granulocytes, DC subsets, and B cells in the peripheral blood (15, 58–60). Thus, more eosinophil-specific markers should be investigated. The identification of an AHR-imprinted eosinophil subset in human intestine may open therapeutic avenues aimed at reprogramming eosinophils in food allergies and primary eosinophilic gastrointestinal disorders that are caused by abnormal eosinophil activation (25).
Materials and Methods
Experimental details on animals, cell extraction from tissues, antibody staining for flow cytometry and sorting, microarray analyses, parasitic infections, and statistical analyses for this study are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Motohiro Kobayashi for advice and helpful discussions, Hong-Erh Liang and Richard Locksley for the gift of N. brasiliensis larvae, Ken Murphy and Arifumi Iwata for providing lung eosinophils from house dust mite–treated mice, the Hope Center Animal Surgery Core for performing parabiosis experiments, and the Genome Technology Access Center (GTAC) in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. GTAC is partially supported by National Cancer Institute Cancer Center Support Grant P30 CA91842 (to the Siteman Cancer Center) and National Center for Research Resources Institute of Clinical and Translational Sciences/Clinical and Translational Science Awards Grant UL1 TR000448. J.K. is supported by The Naito Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and Daiichi Sankyo Foundation of Life Science grants for studying overseas. S.J. is supported by the Promotion Project of Education, Research, and Medical Care from Shinshu University Hospital. The generation of Clec4a4 reporter mice was supported by Mucosal Immunology Studies Team NIH Grant AI095542.
Footnotes
Reviewers: B.E., Leiden University Medical Centre; and G.L., Malaghan Institute.
Competing interest statement: M. Colonna receives research support from Pfizer, Aclaris, Ono, NGM Biopharmaceutical, Oncorus, and VigilNeuro; serves on the scientific advisory boards of Vigil Neuro and NGM Biopharmaceutical; and is consultant for Cell Signaling Technologies. All other authors have nothing to disclose.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204557119/-/DCSupplemental.
Data Availability
Microarray data reported in this paper have been deposited in Gene Expression Omnibus (accession no. GSE199370). All study data are included in the article and/or SI Appendix.
References
- 1.Weller P. F., Spencer L. A., Functions of tissue-resident eosinophils. Nat. Rev. Immunol. 17, 746–760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothenberg M. E., Hogan S. P., The eosinophil. Annu. Rev. Immunol. 24, 147–174 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Gleich G. J., Adolphson C. R., Leiferman K. M., The biology of the eosinophilic leukocyte. Annu. Rev. Med. 44, 85–101 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Acharya K. R., Ackerman S. J., Eosinophil granule proteins: Form and function. J. Biol. Chem. 289, 17406–17415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamann K. J., et al. , In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J. Immunol. 144, 3166–3173 (1990). [PubMed] [Google Scholar]
- 6.Kanaoka Y., Austen K. F., Roles of cysteinyl leukotrienes and their receptors in immune cell-related functions. Adv. Immunol. 142, 65–84 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Spencer L. A., Bonjour K., Melo R. C., Weller P. F., Eosinophil secretion of granule-derived cytokines. Front. Immunol. 5, 496 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponath P. D., et al. , Cloning of the human eosinophil chemoattractant, eotaxin. Expression, receptor binding, and functional properties suggest a mechanism for the selective recruitment of eosinophils. J. Clin. Invest. 97, 604–612 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell H. D., et al. , Molecular cloning, nucleotide sequence, and expression of the gene encoding human eosinophil differentiation factor (interleukin 5). Proc. Natl. Acad. Sci. U.S.A. 84, 6629–6633 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lantero S., Alessandri G., Spallarossa D., Scarso L., Rossi G. A., Stimulation of eosinophil IgE low-affinity receptor leads to increased adhesion molecule expression and cell migration. Eur. Respir. J. 16, 940–946 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Cherry W. B., Yoon J., Bartemes K. R., Iijima K., Kita H., A novel IL-1 family cytokine, IL-33, potently activates human eosinophils. J. Allergy Clin. Immunol. 121, 1484–1490 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerscher B., Willment J. A., Brown G. D., The dectin-2 family of C-type lectin-like receptors: An update. Int. Immunol. 25, 271–277 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geijtenbeek T. B., Gringhuis S. I., Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 9, 465–479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagae M., et al. , Recognition of bisecting N-acetylglucosamine: Structural basis for asymmetric interaction with the mouse lectin dendritic cell inhibitory receptor 2. J. Biol. Chem. 288, 33598–33610 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer-Wentrup F., et al. , DCIR is endocytosed into human dendritic cells and inhibits TLR8-mediated cytokine production. J. Leukoc. Biol. 85, 518–525 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Flornes L. M., et al. , Identification of lectin-like receptors expressed by antigen presenting cells and neutrophils and their mapping to a novel gene complex. Immunogenetics 56, 506–517 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Dudziak D., et al. , Differential antigen processing by dendritic cell subsets in vivo. Science 315, 107–111 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Kanazawa N., et al. , DCIR acts as an inhibitory receptor depending on its immunoreceptor tyrosine-based inhibitory motif. J. Invest. Dermatol. 118, 261–266 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Uto T., et al. , Clec4A4 is a regulatory receptor for dendritic cells that impairs inflammation and T-cell immunity. Nat. Commun. 7, 11273 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussenzweig M. C., Steinman R. M., The cell surface of mouse lymphoid dendritic cells. Immunol. Today 3, 65–68 (1982). [DOI] [PubMed] [Google Scholar]
- 21.Neubert K., et al. , Antigen delivery to CD11c+CD8- dendritic cells induces protective immune responses against experimental melanoma in mice in vivo. J. Immunol. 192, 5830–5838 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki S., et al. , CD8+ CD205+ splenic dendritic cells are specialized to induce Foxp3+ regulatory T cells. J. Immunol. 181, 6923–6933 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price A. E., et al. , Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. U.S.A. 107, 11489–11494 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ricardo-Gonzalez R. R., Molofsky A. B., Locksley R. M., ILC2s—development, divergence, dispersal. Curr. Opin. Immunol. 75, 102168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothenberg M. E., Eosinophilic gastrointestinal disorders (EGID). J. Allergy Clin. Immunol. 113, 11–28 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Muzaki A. R., et al. , Intestinal CD103(+)CD11b(-) dendritic cells restrain colitis via IFN-γ-induced anti-inflammatory response in epithelial cells. Mucosal Immunol. 9, 336–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlens J., et al. , Common gamma-chain-dependent signals confer selective survival of eosinophils in the murine small intestine. J. Immunol. 183, 5600–5607 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Wen T., et al. , The pan-B cell marker CD22 is expressed on gastrointestinal eosinophils and negatively regulates tissue eosinophilia. J. Immunol. 188, 1075–1082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesnil C., et al. , Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Invest. 126, 3279–3295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiffert M., et al. , Human signal-regulatory protein is expressed on normal, but not on subsets of leukemic myeloid cells and mediates cellular adhesion involving its counterreceptor CD47. Blood 94, 3633–3643 (1999). [PubMed] [Google Scholar]
- 31.Verjan Garcia N., et al. , SIRPα/CD172a regulates eosinophil homeostasis. J. Immunol. 187, 2268–2277 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Nitschke L., Carsetti R., Ocker B., Köhler G., Lamers M. C., CD22 is a negative regulator of B-cell receptor signalling. Curr. Biol. 7, 133–143 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Freeman G. J., et al. , Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192, 1027–1034 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor S., et al. , PD-1 regulates KLRG1+ group 2 innate lymphoid cells. J. Exp. Med. 214, 1663–1678 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X., Yue Y., Xiong S., Dpep2 emerging as a modulator of macrophage inflammation confers protection against CVB3-induced viral myocarditis. Front. Cell. Infect. Microbiol. 9, 57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein J. L., Brown M. S., Regulation of the mevalonate pathway. Nature 343, 425–430 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Tsuzuki H., et al. , Functional interleukin-33 receptors are expressed in early progenitor stages of allergy-related granulocytes. Immunology 150, 64–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockinger B., Di Meglio P., Gialitakis M., Duarte J. H., The aryl hydrocarbon receptor: Multitasking in the immune system. Annu. Rev. Immunol. 32, 403–432 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Lamas B., Natividad J. M., Sokol H., Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol. 11, 1024–1038 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Hubbard T. D., et al. , Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci. Rep. 5, 12689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cella M., Colonna M., Aryl hydrocarbon receptor: Linking environment to immunity. Semin. Immunol. 27, 310–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cervantes-Barragan L., Colonna M., AHR signaling in the development and function of intestinal immune cells and beyond. Semin. Immunopathol. 40, 371–377 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Obata Y., et al. , Neuronal programming by microbiota regulates intestinal physiology. Nature 578, 284–289 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Cervantes-Barragan L., et al. , Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 357, 806–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura A., et al. , Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J. Exp. Med. 206, 2027–2035 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodríguez-Calvo R., Tajes M., Vázquez-Carrera M., The NR4A subfamily of nuclear receptors: Potential new therapeutic targets for the treatment of inflammatory diseases. Expert Opin. Ther. Targets 21, 291–304 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Zelante T., et al. , Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Maizels R. M., et al. , Immune modulation and modulators in Heligmosomoides polygyrus infection. Exp. Parasitol. 132, 76–89 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle A. D., et al. , Homologous recombination into the eosinophil peroxidase locus generates a strain of mice expressing Cre recombinase exclusively in eosinophils. J. Leukoc. Biol. 94, 17–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reynolds L. A., Filbey K. J., Maizels R. M., Immunity to the model intestinal helminth parasite Heligmosomoides polygyrus. Semin. Immunopathol. 34, 829–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diny N. L., et al. , The aryl hydrocarbon receptor contributes to tissue adaptation of intestinal eosinophils in mice. J. Exp. Med. 219, e20210970 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortez V. S., et al. , Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity 44, 1127–1139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gasteiger G., Fan X., Dikiy S., Lee S. Y., Rudensky A. Y., Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ginhoux F., Guilliams M., Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Knott M. L., Matthaei K. I., Foster P. S., Dent L. A., The roles of eotaxin and the STAT6 signalling pathway in eosinophil recruitment and host resistance to the nematodes Nippostrongylus brasiliensis and Heligmosomoides bakeri. Mol. Immunol. 46, 2714–2722 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Strandmark J., et al. , Eosinophils are required to suppress Th2 responses in Peyer’s patches during intestinal infection by nematodes. Mucosal Immunol. 10, 661–672 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Knott M. L., et al. , Impaired resistance in early secondary Nippostrongylus brasiliensis infections in mice with defective eosinophilopoeisis. Int. J. Parasitol. 37, 1367–1378 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Bates E. E., et al. , APCs express DCIR, a novel C-type lectin surface receptor containing an immunoreceptor tyrosine-based inhibitory motif. J. Immunol. 163, 1973–1983 (1999). [PubMed] [Google Scholar]
- 59.Meyer-Wentrup F., et al. , Targeting DCIR on human plasmacytoid dendritic cells results in antigen presentation and inhibits IFN-alpha production. Blood 111, 4245–4253 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Klechevsky E., et al. , Cross-priming CD8+ T cells by targeting antigens to human dendritic cells through DCIR. Blood 116, 1685–1697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data reported in this paper have been deposited in Gene Expression Omnibus (accession no. GSE199370). All study data are included in the article and/or SI Appendix.