Abstract
The thermal stability and catalytic activity of phospholipase A1 from Serratia sp. strain MK1 were improved by evolutionary molecular engineering. Two thermostable mutants were isolated after sequential rounds of error-prone PCR performed to introduce random mutations and filter-based screening of the resultant mutant library; we determined that these mutants had six (mutant TA3) and seven (mutant TA13) amino acid substitutions. Different types of substitutions were found in the two mutants, and these substitutions resulted in an increase in nonploar residues (mutant TA3) or in differences between side chains for polar or charged residues (mutant TA13). The wild-type and mutant enzymes were purified, and the effect of temperature on the stability and catalytic activity of the enzymes was investigated. The melting temperatures of the TA3 and TA13 enzymes were increased by 7 and 11°C, respectively, compared with the melting temperature of the wild-type enzyme. Thus, we found that evolutionary molecular engineering was an effective and efficient approach for increasing thermostability without compromising enzyme activity.
Enzymes can be tailored for optimal performance in industrial applications by evolutionary molecular engineering, which is also called directed evolution or in vitro evolution (2, 26). Many successfully engineered proteins have been described, and enzymes with enhanced properties, such as sufficient stability (12, 38), a high level of activity (34), altered substrate specificity (36), and the ability to interact correctly with surfaces (9), have been developed for industrial applications (3). Judging from the recent successes of irrational design approaches, such as mutagenic PCR (6) and DNA shuffling (33) followed by screening for improved properties, directed evolution may be more efficient than rational design involving both iterative computer design and site-directed mutagenesis.
Thermostability is often a primary goal when workers try to improve the properties of an industrial enzyme, since high temperatures in industrial processes provide such benefits as increased substrate solubility, decreased viscosity of the medium, and a lower risk of microbial contamination. In other words, thermostable enzymes are of considerable biotechnological interest since their enhanced stability could greatly reduce enzyme replacement costs or permit processes to be carried out at high temperatures. Recently, many attempts have been made to understand the principles underlying the stability of proteins; introducing disulfide bonds (25), chemical cross-links (20), salt bridges (11), and metal binding sites (22) and increasing intramolecular hydrophobic packing (37) have all been proposed. However, the use of these techniques is limited to the enzymes whose three-dimensional structures have been determined. In the case of enzymes for which extensive information concerning structure and function is not available, directed evolution is a powerful tool for studying or engineering thermostability and catalytic activity of the enzymes.
Phospholipase A1 hydrolyzes the 1-acyl group of a phospholipid to lysophospholipid and fatty acid. Recently, it has been reported that phospholipase A1 and the well-characterized enzyme phospholipase A2 play important biological roles in both phospholipidosis, a pathological condition in which phospholipids accumulate in lysosomes (24), and virulence factors for bacterial and fungal pathogenesis (15, 29). In addition to its physiological roles, phospholipase A1 is of particular interest for industrial applications as it yields 2-acyl-lysophospholipids. Lysophospholipids are excellent emulsifiers and are particularly suitable for use in many industrial applications, such as food technology and the cosmetics and pharmaceutical industries. Lysophospholipids are able to enhance emulsification in oil-water emulsions due to increased solubility in water, form emulsions which are more stable to changing pH and temperature conditions, and keep emulsions stable in the presence of magnesium or calcium ions. Lysophospholipids also have several physiological functions, including a role in platelet aggregation and a role as a signaling molecule (8). They also affect ripening and storage characteristics of fruits, leaves, and green plant tissue (21).
Phospholipase A1 gene plaA of Serratia sp. strain MK1, encodes a 321-amino-acid monomer and has been cloned as described previously (32). Even though phospholipase A1 has been found in various organisms (5, 7, 15, 23, 29), the use of phospholipase A1 for phospholipid modification has been limited due to its low stability and poor availability. Moreover, crystallographic and structural data for phospholipase A1 have not been obtained yet. Since the evolutionary protein engineering technique was a more useful approach for modifying enzymes in the absence of such information, we attempted to use evolutionary molecular engineering to improve phospholipase A1 for practical purposes. The evolutionary engineering technique used in this study involved preparing protein variants by using mutagenic PCR, expressing the protein, and then screening for mutants with improved thermostability. In this study, we enhanced the catalytic activity and the thermostability of phospholipase A1 from Serratia sp. strain MK1 by the evolutionary technique. To do this, we developed a filter-based screening system with which we could identify both properties on a single processed filter; with this system the in which catalytic activity of a heat-treated mutant library could be assayed on a phosphatidylcholine-containing gel at a normal temperature. Below we describe two thermostable phospholipase A1 mutants which exhibited higher levels of activity at the temperatures examined than the wild type exhibited.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Escherichia coli XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 [F′ proAB lacIq ZΔ15 Tn10 (Tetr)]} (Stratagene, San Diego, Calif.) was used as the host strain. Plasmid pMJ1 encoding phospholipase A1 gene plaA of Serratia sp. strain MK1 (32) was used as a template in PCR mutagenesis for evolutionary molecular engineering. Plasmid pSTV28 (Takara Shuzo, Shiga, Japan) was used to construct a mutant enzyme library. Recombinant strains were grown in Luria-Bertani (LB) medium or TYSPN medium (20 g of Bacto Tryptone [Difco Laboratories] per liter, 10 g of yeast extract per liter, 5 g of Na2HPO4 per liter 10 g of KNO3 per liter, 5 g of NaCl per liter). When necessary, antibiotics, such as ampicillin (100 μg/ml) or chloramphenicol (50 μg/ml), were added to the media.
DNA manipulation and sequencing.
Standard recombinant DNA manipulation techniques were used for isolation of plasmid DNA, restriction digestion, ligation, and transformation into E. coli (28). All restriction enzymes, DNA-modifying enzymes, and related reagents used for DNA manipulation were purchased from Sigma, New England Biolabs Inc., or Boehringer Mannheim. DNA fragments required for subcloning experiments were gel purified with a QIAGEN kit (QIAGEN, Hilden, Germany). DNA was sequenced by cycle sequencing by using an ABI PRISM BigDye Primer cycle sequencing kit and AmpliTaq DNA polymerase (Perkin-Elmer, Foster City, Calif.).
Plasmid construction.
Plasmids pTA3 and pTA13, which were isolated from the second random mutant library, were derivatives of plasmid pSTV28 (Takara Shuzo) containing the evolved phospholipase A1 genes. Plasmids containing six consecutive histidine-tagged (6× His-tagged) and evolved phospholipase A1 genes were generated from plasmids pTA3 and pTA13, respectively, by PCR amplification by using cloned Pfu polymerase (Stratagene) and a primer that included 6× His tag-encoding sequences. Construction of a plasmid harboring the 6× His-tagged and native phospholipase A1 genes was identical to construction of thermostable variants, except that plasmid pMJ1 was used as a template. The derivatives were used for purification of wild-type phospholipase A1 and two thermostable phospholipase A1 variants.
Random mutagenesis and mutant library construction.
Random mutagenesis of the plaA gene was carried out by performing mutagenic PCR. Two oligonucleotides flanked by EcoRI or BamHI restriction sites, 5′-CGGAATTCTTTGAGTTTTACCTCTGCGATCG-3′ and 5′-GCGGATCCCATCAGGCATTGGCCATCGCCTCC-3′, were used as forward and reverse primers, respectively. In order to obtain both the desired level of mutation (three or four amino acid substitutions) and base substitutions without mutational bias, the conditions used for PCR random mutagenesis were optimized; a 100-μl reaction mixture contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 7 mM MgCl2, 0.1 mM MnCl2, 0.2 mM dATP, 0.2 mM dGTP, 1 mM dCTP, 1 mM dTTP, 25 pmol of each oligonucleotide primer, 5 ng of template DNA, and 5 U of Taq polymerase (Takara Shuzo). Equal amounts of the mutant plasmids isolated from the thermostable mutants that gave positive signals during the first screening were pooled and used as templates for the second cycle of PCR random mutagenesis. The second PCR was performed with an automatic thermal cycler (Ericomp, San Diego, Calif.) for 25 cycles consisting of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min. The mutagenic PCR products were gel purified by using a QIAGEN kit and were ligated with EcoRI- and BamHI-digested pSTV28. E. coli XL1-Blue was transformed with the resulting ligated DNA and plated onto LB agar plates containing 50 μg of chloramphenicol per ml.
Thermostability screening.
E. coli cells harboring the randomly mutated phospholipase A1 genes were grown on LB agar plates containing chloramphenicol and then transferred to nylon membranes (Hybond-N; Amersham-Pharmacia). The nylon membranes containing the transferred cells were laid on fresh LB agar plates containing chloramphenicol and incubated at 37°C for 5 h and then at 4°C for 2 h. The membranes were laid on a series of filter papers that had been wetted as follows: (i) with a cell wall-weakening solution containing 25 mM Tris-HCl (pH 8.0), 1 mg of lysozyme per ml, and 1 mM EDTA (pH 8.0) for 15 min; (ii) with a cell-lysing solution containing 10 mM Tris-HCl (pH 8.0) and 0.1% (wt/vol) Triton X-100 for 30 min; and (iii) with 50 mM Tris-HCl (pH 8.0) for 30 min. Then the processed membranes were incubated at 70°C for 2 h during the first screening or at 80°C for 3 h during the second screening. Positive colonies that produced transparent halos were selected by incubating the heat-treated filters at 42°C for 2 h on activity indicator plates (PCY plates) containing phosphatidylcholine. Each PCY plate contained a 1% agarose gel containing 0.8% (wt/vol) phosphatidylcholine, 20 mM CaCl2, and 0.4% taurocholic acid emulsified in 50 mM Tris-HCl (32). Under these conditions, wild-type phospholipase A1 did not produce a detectable halo on an indicator plate.
Enzyme purification.
For purification of native phospholipase A1 and evolved thermostable phospholipase A1, a 6× His affinity tag was fused to the C terminus of each enzyme as described above. Each E. coli strain harboring recombinant wild-type or thermostable mutant phospholipase A1 was grown at 30°C in TYSPN medium. Crude cell extract in a solution containing 50 mM sodium phosphate (pH 8.0), 300 mM NaCl, 10 mM imidazole, 0.05% Triton X-100, and 10% glycerol was purified with nickel-nitrilotriacetic acid agarose resin (QIAGEN) and imidazole-containing buffer. After extensive washing, the bound protein was eluted with 250 mM imidazole-containing phosphate buffer. The active eluates were applied to a Sephacryl S-200 column (Amersham-Pharmacia). The buffer used was 75 mM CaCl2–300 mM NaCl in 50 mM sodium phosphate buffer. Fractions containing enzymatic activity were pooled and then concentrated by applying them to a centrifugal filtration device (Centricon Concentrators, Amicon Millipore). The protein was dialyzed against 50 mM phosphate (pH 8.0) containing 0.1 mM phenylmethylsulfonyl fluoride protease inhibitor. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to determine the purity of the protein.
Enzyme activity.
Free fatty acids released by catalysis of phospholipase A1 were measured by the pH stat titration method. The activity was measured at an assay temperature of 37°C with an autotitrator and an autoburette (Radiometer Copenhagen, Lyon, France). An aqueous emulsion containing 3.4 mM phosphatidylcholine, 10 mM CaCl2, and 2.6 mM deoxycholic acid was used as the substrate. Phospholipase A1 activity was expressed in micromoles of phosphatidylcholine hydrolyzed per minute. The protein concentration of each purified enzyme was determined by using the Bio-Rad protein assay reagent (Bio-Rad Life Science Group, Hercules, Calif.).
Thermal stability and kinetic parameters.
Heat treatment of the purified enzymes was performed with a programmable temperature controller. An enzyme in 50 mM sodium phosphate (pH 8.0) was incubated at different temperatures and cooled immediately in an ice bath. The phospholipase A1 activity remaining after the heat treatment was measured by using the pH stat titration method described above.
The Km and kcat values were estimated from the intercepts of Lineweaver-Burk plots. The enzyme activity was measured as described above, except that the concentrations of the phosphatidylcholine substrate used were 200, 100, 67, 50, and 40 μM.
RESULTS
Isolation of thermostable mutant phospholipase A1.
The thermal stability of phospholipase A1 was improved without reducing the enzyme activity by constructing a mutant library by mutagenic PCR and by using filter-based visual screening that was sufficiently sensitive and rapid. The library of phospholipase A1 variants was expressed in E. coli XL1-Blue, immobilized spatially on nylon membranes, and then treated with three types of cell lysis solutions. Simple visual screening was based on the formation of a clear zone as a result of enzymatic hydrolysis of phosphatidylcholine. Thermostable variants of phospholipase A1 produced clear halos when they were blotted onto filters, subjected to heat treatment at 70°C (in the first screening) or at 80°C (in the second screening), and then incubated at 42°C on an indicator plate (PCY plate) containing phosphatidylcholine. This screening system was very useful for simultaneously identifying two properties, such as thermostability and catalytic activity.
Plasmid pMJ1 containing the 960-bp plaA gene (GenBank accession no. U37262) was used as a template in the first mutagenic PCR in order to introduce random point mutations into the phospholipase A1-encoding region. After the first cycle of PCR mutagenesis and screening, eight positive clones containing thermostable phospholipase A1 variants were isolated from approximately 10,000 colonies. The phospholipase A1 activities of these mutants were detected on indicator plates (PCY plates) after heat treatment at 70°C for 2 h, although no halo was detected under the same conditions in the case of the wild-type enzyme (Fig. 1). Plasmids encoding each mutant phospholipase A1 were isolated from eight clones and pooled (equal amounts of all plasmids). A possible limitation of sequential engineering approaches is that a particular mutational pathway or its branches may be brought about by selecting only one highly enhanced mutant to parent subsequent generations. Therefore, we used this plasmid mixture as template DNA in the second mutagenesis step in order to obtain as many available evolutionary pathways to thermostability as possible. The resultant mutagenic PCR products were subcloned in order to prepare a mutant library for the second screening. After a heat treatment which involved incubating the second mutant library at 80°C for 3 h, we finally isolated two clones from which two plasmids (pTA3 and pTA13) were prepared. These two plasmids were sequenced to identify the locations of the substituted amino acid residues.
FIG. 1.
Thermostable phospholipase A1 variants on an activity assay gel containing phosphatidylcholine. An enzyme-blotted membrane was heat treated at 70°C for 2 h, placed on an activity assay gel, and then incubated at 42°C for 2 h. Thermostable mutants isolated from the first round of mutagenesis and screening (S1 through S8) produced transparent halos on the assay gel. C1 was the control consisting of E. coli harboring plasmid pSTV28, and C2 was a mutant that did not exhibit thermostability in the first round. Wild-type phospholipiase A1 (C3) did not produce a halo under the same conditions.
DNA sequences and thermal stabilities of wild-type phospholipase A1 and mutant phospholipase A1 variants.
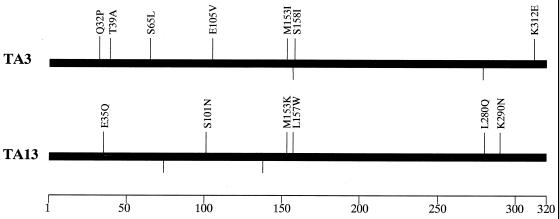
DNA sequences of the two thermostable phospholipase A1 genes were determined. Two phospholipase A1 mutants, TA3 and TA13, had seven and six amino acid substitutions, respectively (Fig. 2). Mutant TA3 had substitutions that resulted in amino acids which had uncharged and nonpolar side chains (Q32P, T39A, E105V, M153I, and S158I). On the other hand, substitutions in amino acids that had polar or charged side chains were found in mutant TA13 (E35Q, S101N, M153K, L157W, L280Q, and K290N).
FIG. 2.
Amino acid substitutions in the evolved phospholipase A1. The deduced amino acid changes are indicated; the numbers are the positions in the wild-type phospholipase A1 sequence (GenBank accession no. AAD10476). The bar represents the 320 amino acids of the phospholipase A1 coding region, and the line at the bottom indicates the relative positions of mutations. Nucleotide changes that did not result in amino acid changes are indicated below the proteins.
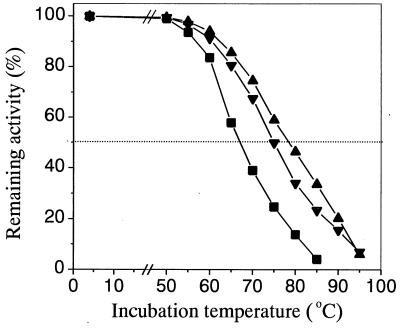
We placed the 6× His tag at the C terminus of each protein in order to purify the wild type and the thermostable mutants (TA3 and TA13). Each recombinant strain which produced the 6× His-tagged proteins was cultured in TYSPN medium, and the wild-type phospholipase A1 and mutant phospholipase A1 variants were purified as described above. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis revealed that the wild-type phospholipase A1 and evolved phospholipase A1 variants were purified to homogeneity (data not shown). The thermal stabilities of the purified enzymes were estimated by measuring the remaining activities at 40°C after heat treatment at various temperatures (Fig. 3). Figure 3 shows that the melting temperatures (Tm) of thermostable mutant enzymes TA3 and TA13 (Tm is the temperature at which 50% inactivation occurs after heat treatment for 20 min) were higher by 7 and 11°C, respectively, than the Tm of the wild-type enzyme.
FIG. 3.
Effect of temperature on the stability of wild-type phospholipase A1 and mutant phospholipase A1 variants. Each purified enzyme was diluted to the same concentration with 50 mM sodium phosphate buffer (pH 8.0) and treated at different temperatures for 20 min. The remaining activities were expressed as percentages of the original activities. Three independent experiments were carried out, and values whose error ranges were within 5% were averaged. Symbols: ■, wild-type enzyme; ▾, mutant TA3; ▴, mutant TA13.
Activities of thermostable mutant phospholipase A1 variants.
The kinetic parameters of the wild-type and mutant enzymes are shown in Table 1. The wild-type phospholipase A1 and mutant phospholipase A1 variants exhibited typical Michaelis-Menten saturation kinetics when the initial velocity was plotted against the concentration of the substrate. The Michaelis constant (Km) of the mutant TA3 enzyme was slightly (8.2%) less than the Km of the wild-type enzyme. In contrast, the Km of the mutant TA13 enzyme was slightly (5.2%) more than the Km of the wild-type enzyme. The catalytic constant (kcat) of the mutant TA3 enzyme was 2.8% lower than the kcat of the wild type enzyme, whereas the kcat of the mutant TA13 enzyme was 21.8% higher. As a result of the considerable increase in kcat, the catalytic efficiency of mutant TA13 was 16% higher than the catalytic efficiency of the wild type. These results imply that acquisition of thermostability by evolutionary engineering is possible without a significant loss of catalytic activity. Moreover, if a screening strategy which reflects catalytic activity at the normal temperature as well as stability is devised, a variant having both properties, such as mutant TA13 in this study, can be obtained.
TABLE 1.
Kinetic parameters of wild-type phospholipase A1 and mutant phospholipase A1 variants with phosphatidylcholine at pH 8.0 and 37°Ca
| Enzyme | Km (μM) | kcat (s−1) | kcat/Km (s−1 · μM) |
|---|---|---|---|
| Wild type | 114.5 | 11.23 | 0.098 |
| TA3 | 105.1 | 10.92 | 0.104 |
| TA13 | 120.4 | 13.67 | 0.114 |
All values are averages based on values from at least duplicate assays.
DISCUSSION
We created a thermostable phospholipase A1 by evolutionary molecular engineering by using a combination of mutagenic PCR and filter-based screening. The method which we used to tailor enzymes is evolutionary random mutagenesis followed by a filter-based assay, not site-specific mutagenesis guided by a detailed knowledge of the three-dimensional structure of the enzyme. The filter-based assay was useful for identifying improved thermostability because each variant produced in a large number of colonies can be blotted directly onto filters and handled easily in a heat treatment procedure. Moreover, if the filter-based assay is combined with appropriate visual screening that depends on enzymatic activity, several properties, such as increased thermal stability and increased resistance to organic solvents, are likely to be screened consecutively with one processed filter, while catalytic activity is maintained. Using this screening method, we obtained an evolved phospholipase A1 that exhibited increased catalytic activity and increased thermostability compared to the wild-type enzyme.
Two thermostable mutants were obtained after two rounds of random mutagenesis and screening, and then they were sequenced in order to identify the amino acid substitutions. Amino acid residues having nonpolar side chains were substituted for amino acid residues having charged or polar side chains in all of the mutations of TA3 except the K312E mutation. It has been noted that isoleucine and alanine allow tighter packing in hydrophobic cores (4, 19, 31) and that proline can give extra stability to loops (35). Therefore, one possible explanation for the thermostability of the TA3 enzyme is that the amino acid changes generated by directed evolution might have caused the structure to change so that the hydrophobic-core packing was tighter. In contrast to TA3, however, most of the mutations in TA13 resulted in changes in side chains in polar or charged residues. It has been proposed that creating paired charges and hydrogen bonding is important in the stabilization of proteins (1, 14, 16). Although the estimate obtained without a precise three-dimensional structure of phospholipase A1 is somewhat imperfect, stabilization in TA13 is believed to be achieved by the formation of additional hydrogen bonds or charged pairs. In recent years, workers have proposed a variety of theories to explain the enhanced thermostability of proteins in terms of amino acid composition (13, 27). Judging from a variety of the proposed mechanisms (1, 4, 13, 14, 16, 27, 31, 35), mutants TA3 and TA13 could have obtained enhanced thermal stability through different evolutionary paths. In other words, it is thought that the pathway which results in greater thermostability in mutant TA3 is the pathway involved in the increase in internal hydrophobicity; on the other hand, the pathway in mutant TA13 is the pathway involved in creation of additional hydrogen bonds or charged pairs. We, therefore, suggest that diverse enzymes that evolve from the different evolutionary pathways can be obtained, unless the available evolutionary pathways are restricted to thermostability pathways.
Although sometimes stabilization of an enzyme is achieved with a significant loss of catalytic activity (30), we obtained phospholipase A1 mutants that exhibited both increased activity and increased thermostability. Molecular flexibility of an enzyme is essential for both binding to substrates and catalysis. It has been proposed that thermophilic and mesophilic proteins exhibit similar degrees of flexibility at their respective optimum temperatures (17), although thermophilic proteins are thought to be less flexible than mesophilic proteins at lower temperatures. Moreover, the specific activity of a thermophilic enzyme at its optimum temperature is often comparable to the specific activity of a homologous enzyme from a mesophile at its optimum temperature (10). It has been reported frequently that thermostability and catalytic activity at low temperatures are not mutually exclusive, suggesting that activity and thermostability are at least partially independent properties and that these two properties are not incompatible in an enzyme (12, 38). Thus, if an evolutionary engineering experiment is not limited to one property (i.e., thermostability) but allows workers to determine both properties (i.e., thermostability and catalytic activity), as shown in our screening study, thermostability and catalytic activity can be enhanced simultaneously in an enzyme. When the catalytic activities were measured at a number of temperatures in order to verify this, our evolved thermostable phospholipase A1 variants not only were stable at the temperatures examined but also exhibited high levels of catalytic activity (data not shown). Thus, our results also indicate that evolutionary molecular engineering is a powerful technique for enhancing the properties (i.e., thermostability and catalytic activity) of an enzyme that is to be used at a high temperature.
Now we are investigating the applied aspects of the engineered thermostable phospholipase A1. It is frequently noted that a higher temperature is used in enzyme reactors to produce useful phospholipid derivatives due to the insolubility of phospholipids at room temperature. Therefore, we are attempting to produce lysophospholipids by using the novel phospholipase A1 that is more thermostable and more active than the wild-type enzyme.
ACKNOWLEDGMENTS
This work was supported by a grant from Doosan Serdary Research Laboratories and the Ministry of Science and Technology of Korea and by grant 95-0502-01-02-3 from the Korea Science and Engineering Foundation.
REFERENCES
- 1.Aguilar C F, Sanderson I, Moracci M, Ciaramella M, Nucci R, Rossi M, Pearl L H. Crystal structure of the beta-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus: resilience as a key factor in thermostability. J Mol Biol. 1997;271:789–802. doi: 10.1006/jmbi.1997.1215. [DOI] [PubMed] [Google Scholar]
- 2.Arnold F H, Moore J C. Optimizing industrial enzymes by directed evolution. Adv Biochem Eng Biotechnol. 1997;58:1–14. doi: 10.1007/BFb0103300. [DOI] [PubMed] [Google Scholar]
- 3.Arnold F H, Volkov A A. Directed evolution of biocatalysts. Curr Opin Chem Biol. 1999;3:54–59. doi: 10.1016/s1367-5931(99)80010-6. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin E, Xu J, Hajiseyedjavadi O, Baase W A, Matthews B W. Thermodynamic and structural compensation in “size-switch” core repacking variants of bacteriophage T4 lysozyme. J Mol Biol. 1996;259:542–559. doi: 10.1006/jmbi.1996.0338. [DOI] [PubMed] [Google Scholar]
- 5.Brok R G, Brinkman E, van Boxtel R, Bekkers A C, Verheij H M, Tommassen J. Molecular characterization of enterobacterial pldA genes encoding outer membrane phospholipase A. J Bacteriol. 1994;176:861–870. doi: 10.1128/jb.176.3.861-870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadwell R C, Joyce G F. Randomization of genes by PCR mutagenesis. PCR Methods Applic. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 7.Dawson R M, Irvine R F, Hemington N, Hirasawa K. The alkaline phospholipase A1 of rat liver cytosol. Biochem J. 1983;209:865–872. doi: 10.1042/bj2090865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durieux M E, Lynch K R. Signalling properties of lysophosphatidic acid. Trends Pharmacol Sci. 1993;14:249–254. doi: 10.1016/0165-6147(93)90021-b. [DOI] [PubMed] [Google Scholar]
- 9.Egmond M R, Antheunisse W P, Ravestein P, Mooren A T, de Vlieg J. Engineering surface charges in a subtilisin. Adv Exp Med Biol. 1996;379:219–228. doi: 10.1007/978-1-4613-0319-0_23. [DOI] [PubMed] [Google Scholar]
- 10.Fagain C O. Understanding and increasing protein stability. Biochim Biophys Acta. 1995;1252:1–14. doi: 10.1016/0167-4838(95)00133-f. [DOI] [PubMed] [Google Scholar]
- 11.Fairman R, Chao H G, Lavoie T B, Villafranca J J, Matsueda G R, Novotny J. Design of heterotetrameric coiled coils: evidence for increased stabilization by Glu(−)-Lys(+) ion pair interactions. Biochemistry. 1996;35:2824–2829. doi: 10.1021/bi952784c. [DOI] [PubMed] [Google Scholar]
- 12.Giver L, Gershenson A, Freskgard P O, Arnold F H. Directed evolution of a thermostable esterase. Proc Natl Acad Sci USA. 1998;95:12809–12813. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haney P J, Badger J H, Buldak G L, Reich C I, Woese C R, Olsen G J. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc Natl Acad Sci USA. 1999;96:3578–3583. doi: 10.1073/pnas.96.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendsch Z S, Jonsson T, Sauer R T, Tidor B. Protein stabilization by removal of unsatisfied polar groups: computational approaches and experimental tests. Biochemistry. 1996;35:7621–7625. doi: 10.1021/bi9605191. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman D R. Allergens in hymenoptera venom. XXVI. The complete amino acid sequences of two vespid venom phospholipases. Int Arch Allergy Immunol. 1994;104:184–190. doi: 10.1159/000236728. [DOI] [PubMed] [Google Scholar]
- 16.Huyghues-Despointes B M, Baldwin R L. Ion-pair and charged hydrogen-bond interactions between histidine and aspartate in a peptide helix. Biochemistry. 1997;36:1965–1970. doi: 10.1021/bi962546x. [DOI] [PubMed] [Google Scholar]
- 17.Jaenicke R. Stability and folding of ultrastable proteins: eye lens crystallins and enzymes from thermophiles. FASEB J. 1996;10:84–92. doi: 10.1096/fasebj.10.1.8566552. [DOI] [PubMed] [Google Scholar]
- 18.Makriyannis, A., R. L. Duclos, and D. J. Fournier. April 1998. Phospholipid compounds and use therefor. U.S. patent 5,744,459.
- 19.Matsumura M, Becktel W J, Matthews B W. Hydrophobic stabilization in T4 lysozyme determined directly by multiple substitutions of Ile 3. Nature. 1988;334:406–410. doi: 10.1038/334406a0. [DOI] [PubMed] [Google Scholar]
- 20.Noda Y, Fukuda Y, Segawa S. A two-dimensional NMR study of exchange behavior of amide hydrogens in a lysozyme derivative with an extra cross-link between Glu35 and Trp108—quenching of cooperative fluctuations and effects on the protein stability. Biopolymers. 1997;41:131–143. doi: 10.1002/(SICI)1097-0282(199702)41:2<131::AID-BIP2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 21.Palta, J. P., and K. M. Farag. June 1992. Plant and fruit treatment with lysophosphatidylethanolamine. U.S. patent 5,126,155.
- 22.Pantoliano M W, Whitlow M, Wood J F, Rollence M L, Finzel B C, Gilliland G L, Poulos T L, Bryan P N. The engineering of binding affinity at metal ion binding sites for the stabilization of proteins: subtilisin as a test case. Biochemistry. 1988;27:8311–8317. doi: 10.1021/bi00422a004. [DOI] [PubMed] [Google Scholar]
- 23.Pete M J, Ross A H, Exton J H. Purification and properties of phospholipase A1 from bovine brain. J Biol Chem. 1994;269:19494–19500. [PubMed] [Google Scholar]
- 24.Reasor M J, Kacew S. An evaluation of possible mechanisms underlying amiodarone-induced pulmonary toxicity. Proc Soc Exp Biol Med. 1996;212:297–304. doi: 10.3181/00379727-212-44019. [DOI] [PubMed] [Google Scholar]
- 25.Reiter Y, Brinkmann U, Jung S H, Pastan I, Lee B. Disulfide stabilization of antibody Fv: computer predictions and experimental evaluation. Protein Eng. 1995;8:1323–1331. doi: 10.1093/protein/8.12.1323. [DOI] [PubMed] [Google Scholar]
- 26.Rubingh D N. Protein engineering from a bioindustrial point of view. Curr Opin Biotechnol. 1997;8:417–422. doi: 10.1016/s0958-1669(97)80062-6. [DOI] [PubMed] [Google Scholar]
- 27.Russell R J, Taylor G L. Engineering thermostability: lessons from thermophilic proteins. Curr Opin Biotechnol. 1995;6:370–374. doi: 10.1016/0958-1669(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoichet B K, Baase W A, Kuroki R, Matthews B W. A relationship between protein stability and protein function. Proc Natl Acad Sci USA. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shortle D, Stites W E, Meeker A K. Contributions of the large hydrophobic amino acids to the stability of staphylococcal nuclease. Biochemistry. 1990;29:8033–8041. doi: 10.1021/bi00487a007. [DOI] [PubMed] [Google Scholar]
- 32.Song J K, Kim M K, Rhee J S. Cloning and expression of the gene encoding phospholipase A1 from Serratia sp. MK1 in Escherichia coli. J Biotechnol. 1999;72:103–114. doi: 10.1016/s0168-1656(99)00096-6. [DOI] [PubMed] [Google Scholar]
- 33.Stemmer W P. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taguchi S, Ozaki A, Momose H. Engineering of a cold-adapted protease by sequential random mutagenesis and a screening system. Appl Environ Microbiol. 1998;64:492–495. doi: 10.1128/aem.64.2.492-495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe K, Kitamura K, Suzuki Y. Analysis of the critical sites for protein thermostabilization by proline substitution in oligo-1,6-glucosidase from Bacillus coagulans ATCC 7050 and the evolutionary consideration of proline residues. Appl Environ Microbiol. 1996;62:2066–2073. doi: 10.1128/aem.62.6.2066-2073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yano T, Oue S, Kagamiyama H. Directed evolution of an aspartate aminotransferase with new substrate specificities. Proc Natl Acad Sci USA. 1998;95:5511–5515. doi: 10.1073/pnas.95.10.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yutani K, Ogasahara K, Tsujita T, Sugino Y. Dependence of conformational stability on hydrophobicity of the amino acid residue in a series of variant proteins substituted at a unique position of tryptophan synthase alpha subunit. Proc Natl Acad Sci USA. 1987;84:4441–4444. doi: 10.1073/pnas.84.13.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Arnold F H. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 1999;12:47–53. doi: 10.1093/protein/12.1.47. [DOI] [PubMed] [Google Scholar]