Abstract
Porcine deltacoronavirus (PDCoV) is an emerging enteropathogenic coronavirus that has the potential for cross-species infection. Many viruses have been reported to induce endoplasmic reticulum stress (ERS) and activate the unfolded protein response (UPR). To date, little is known about whether and, if so, how the UPR is activated by PDCoV infection. Here, we investigated the activation state of UPR pathways and their effects on viral replication during PDCoV infection. We found that PDCoV infection induced ERS and activated all three known UPR pathways (inositol-requiring enzyme 1 [IRE1], activating transcription factor 6 [ATF6], and PKR-like ER kinase [PERK]), as demonstrated by IRE1-mediated XBP1 mRNA cleavage and increased mRNA expression of XBP1s, ATF4, CHOP, GADD34, GRP78, and GRP94, as well as phosphorylated eIF2α expression. Through pharmacologic treatment, RNA interference, and overexpression experiments, we confirmed the negative role of the PERK–eIF2α pathway and the positive regulatory role of the ATF6 pathway, but found no obvious effect of IRE1 pathway, on PDCoV replication. Taken together, our results characterize, for the first time, the state of the ERS response during PDCoV infection and identify the PERK and ATF6 pathways as potential antiviral targets.
Keywords: Porcine deltacoronavirus (PDCoV), ER stress, Unfolded protein response (UPR), Viral replication
1. Introduction
Porcine deltacoronavirus (PDCoV) is a single-stranded, positive-sense RNA virus that belongs to the genus Deltacoronavirus in the family Coronaviridae (Woo et al., 2012). PDCoV infection causes acute diarrhea and often dehydration or even death in nursing piglets (Ma et al., 2015, Xu et al., 2018). To date, PDCoV infection has been reported in many countries, including the United States, South Korea, Canada, China, Thailand, Lao People’s Democratic Republic, Vietnam, Japan, Peru, and Mexico, resulting in significant economic losses (Ma et al., 2015, Tang et al., 2021). Furthermore, recent studies reported that chickens, calves, turkeys, and even humans are also susceptible to PDCoV infection (Boley et al., 2020, Jung et al., 2017, Lednicky et al., 2021, Liang et al., 2019). The significant threat PDCoV poses to human and animal health has sparked increasing interest in studying this emerging virus.
During virus infection, the endoplasmic reticulum (ER) is often stressed and overwhelmed with excessive production of viral proteins, resulting in ER stress (ERS) (Fung and Liu, 2014, Xue and Feng, 2021). To deal with ERS and maintain protein homeostasis, infected cells always initiate the unfolded protein response (UPR). The UPR comprises three known signaling branches: PKR-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme 1 (IRE1). When misfolded proteins accumulate in the ER, the ER chaperone immunoglobulin heavy-chai-binding protein (GRP78/BIP) preferentially binds these proteins and is sequestered away from PERK, ATF6, and IRE1. Subsequently, PERK and IRE1 homodimerize, causing autophosphorylation and activation, while released ATF6 relocalizes to the Golgi complex, where it is cleaved and activated. Once activated, IRE1 activates the endonuclease domain and mediates the cleavage of unspliced X-box-binding protein 1 (XBP1u) mRNA by removing an intron (containing approximately 26 nucleotides), generating the spliced X-box-binding protein 1 (XBP1s), which promotes the expression of chaperones participating in ERS-associated protein degradation (ERAD) and lipid biosynthesis, such as EDEM and ERdj4 (Lee et al., 2003). The active form of ATF6 translocates to the nucleus and promotes the expression of GRP78, GRP94, calnexin, calreticulin, and ER protein 57 kDa (ERp57) (Higa and Chevet, 2012). In the PERK pathway, activated PERK phosphorylates the α subunit of eIF2 (eIF2α) at serine residue 51, which is followed by global translation inhibition; this phosphorylation of eIF2α also promotes ATF4 expression, which in turn stimulates GADD34 and CHOP (Harding et al., 2000). GADD34 can interact with protein phosphatase 1 (PP1) to dephosphorylate eIF2α, and this process serves as a negative feedback loop to recover host protein translation (Novoa et al., 2001).
Previous studies have shown that many viruses, including coronaviruses, can induce ERS and modulate the UPR with varying degrees of selectivity. For example, severe acute respiratory syndrome coronavirus (SARS-CoV) infection activates the IRE1 pathway (DeDiego et al., 2011); African swine fever virus (AFSV) infection activates the ATF6 pathway (Galindo et al., 2012); both the IRE1 and ATF6 arms of the UPR are triggered in cells infected with rotavirus, Zika virus (ZIKA), or West Nile virus (WNV) (Ambrose and Mackenzie, 2011, Tan et al., 2018, Trujillo-Alonso et al., 2011); and all three arms of the UPR are activated after infection by transmissible gastroenteritis virus (TGEV) (Xue et al., 2018), porcine reproductive and respiratory syndrome (PRRSV) (Catanzaro and Meng, 2020), mouse hepatitis virus (MHV) (Bechill et al., 2008), or SARS-CoV-2 (Echavarria-Consuegra et al., 2021). However, there currently are no reports on whether and, if so, how the UPR is activated during PDCoV infection.
In the present study, we showed that PDCoV infection activates all three UPR arms (IRE1, ATF6 and PERK), and we identified the negative role of the PERK pathway and the positive role of the ATF6 pathway in PDCoV replication, providing new insight into PDCoV-induced activation of the UPR.
2. Materials and methods
2.1. Cells, viruses, and reagents
PDCoV strain CHN-HN-2014 (GenBank accession number: KT336560) was isolated in China in 2014 from a piglet with severe diarrhea (Dong et al., 2016). IPI-2I cells (porcine intestinal epithelial cells) and LLC-PK1 cells (porcine kidney epithelial cells) were cultured and maintained at 37 °C and 5 % CO2 in Dulbecco’s Modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % heat-inactivated fetal bovine serum (FBS) (PAN-biotech, Bavaria, Germany). Antibodies against GRP78, GRP94, phospho-eIF2α, and eIF2α were purchased from Cell Signaling Technology (Danvers, MA, USA) or Abcam (Cambridge, MA, USA). Mouse anti-hemagglutinin (HA) antibodies were purchased from Medical and Biological Laboratories (Nagoya, Japan). Anti-β-actin antibody was purchased from ABclonal (Wuhan, China). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Beyotime (Shanghai, China). Inhibitor 4µ8c (B1874) was obtained from APExBIO (Houston, TX, USA). Inhibitors ISRIB (SML0843) and thapsigargin (TG, T9033) was purchased from Sigma. Inhibitors salubrinal (HY-15486), AEBSF (HY-12821), and STF-083010 (HY-15845) were purchased from MedChemExpress (Monmouth Junction, NJ, USA).
2.2. Cloning and construction of plasmids
The full-length cDNA of porcine eIF2α was amplified from LLC-PK1 cells by using primers peIF2α-F/R ( Table 1) and was then cloned into pCAGGS-HA-N with an N-terminal HA tag to yield pCAGGS-HA-eIF2α. The plasmid pCAGGS-HA-eIF2αS51A containing an unphosphorylatable mutant form of eIF2α was generated using polymerase chain reaction (PCR)-mediated site-directed mutagenesis to introduce the Ser51Ala substitution as described previously (Xue et al., 2018).
Table 1.
Primers used for quantitative real-time RT-PCR and plasmid construction.
| Primer | Nucleotide sequence (5’- 3’) |
|---|---|
| qGRP78-F | ATGGCCGTGTGGAGATCATC |
| qGRP78-R | GAGCTGGTTCTTGGCTGCAT |
| qGRP94-F | TACCAGACGGGCAAGGACAT |
| qGRP94-R | AAGAGATACCCTGACCGCAG |
| qCalreticulin-F | ATCTCTGGCAGGTCAAGTCT |
| qCalreticulin-R | TGTCTTTCATTTGCTTTTCTG |
| qATF6-F | TTGGGATTCTACCCTGTTTGC |
| qATF6-R | TTTCATAAGTTTCCTTTGCTGC |
| qXBP1s-F | GAGTCCGCAGCAGGTG |
| qXBP1s-R | CCGTCAGAATCCATGGGG |
| qEDEM1-F | TTGACTCTTGTTGATGCATTGGA |
| qEDEM1-R | GCTTTCTGGAACTCGGATGAAT |
| qERdj4-F | CAGAGAGATTGCAGAAGCATATGA |
| qERdj4-R | GCTTCTTGGATCGAGTGTTTTG |
| qATF4-F | CCC TTTACGTTCTTGCAAACTC |
| qATF4-R | GCTTCCTATCTCCTTCCGAGA |
| qCHOP-F | CTCAGGAGGAAGAGGAGGAAG |
| qCHOP-R | GCTAGCTGTGCCACTTTCCTT |
| qGADD34-F | AAG AGC CTG GAG AGA GGA GAG |
| qGADD34-R | GTCCCCAGGTTTCCAAAAGCA |
| qPDCoV-nsp16-F | GCCCTCGGTGGTTCTATCTT |
| qPDCoV-nsp16-R | TCCTTAGCTTGCCCCAAATA |
| qGAPDH-F | CCTTCCGTGTCCCTACTGCCAAC |
| qGAPDH-R | GACGCCTGCTTCACCACCTTCT |
| peIF2α-F | ACGGGTACCATGGCGCCGTCCGCGCCGCTCCTGA |
| peIF2α-R | CCGCTCGAGTTAAATACCCAGTTCCAAAT |
2.3. RNA extraction and quantitative real-time PCR
LLC-PK1 or IPI-2I cells were mock-infected or infected with PDCoV at a multiplicity of infection (MOI) of 5 for different lengths of time (3, 6, 9, or 12 h) or were treated with TG for 6 h as a positive control. Cellular total RNA was extracted using TRIzol reagent (Invitrogen) and reverse transcribed into cDNA by reverse transcriptase (Roche, Mannheim, Germany). Each quantitative real-time PCR experiment was performed at least three times via the SYBR green PCR assay using the cDNA described above as a template. Target gene mRNA levels were normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers used in these experiments are listed in Table 1.
2.4. Reverse transcription (RT)-PCR analysis of XBP1 mRNA
ERS-induced processing of XBP1 mRNA was evaluated as previously described (Chen et al., 2020). Briefly, total RNA was isolated from PDCoV-infected cells at various post-infection time points. The cDNA was reverse transcribed from 1 µg of total RNA using reverse transcriptase and then amplified with DNA polymerase using the primers XBP1-F (5′-AAACAGAGTAGCAGCTCAGACTGC-3′ and XBP1-R 5′-TCCTTCTGGGTAGACCTCTGGGAG-3′). The amplified fragments of 448 bp and 474 bp in length, respectively representing the spliced (XBP1s) and unspliced (XBP1u) forms of XBP1, were separated in 3% polyacrylamide–Tris-borate-EDTA and visualized by goldview staining.
2.5. RNA interference experiments
The small interfering RNA (siRNA) sequences against IRE1, ATF6, and GRP78 and a non-target control siRNA were designed as described previously (Xue et al., 2018, Zhou et al., 2016). These siRNAs were synthesized by GenePharma Co. Ltd (Shanghai, China) and transfected into LLC-PK1 cells using Lipofectamine 3000 (Invitrogen) in accordance with the manufacturer’s instructions. At 12 h post-transfection, these cells were infected with PDCoV (MOI = 5) for 12 h and then harvested for the measurement of viral replication using quantitative real-time RT-PCR and median tissue culture infectious dose (TCID50) assays.
2.6. Western blot analysis
LLC-PK1 or IPI-2I cells were mock-infected or infected with PDCoV (MOI = 5). At 3, 6, 9, and 12 h post-infection (hpi), cells were collected and subjected to sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Darmstadt, Germany). After being treated with 5 % nonfat milk in phosphate buffer solution with 0.1 % polysorbate-20, the membrane was incubated first with primary antibodies and then with HRP-conjugated secondary antibodies (Beyotime). Finally, the membrane was washed three times and then visualized via enhanced chemiluminescence reagents (Bio-Rad, CA, USA).
2.7. Treatment of cells with pharmaceutic inhibitors
LLC-PK1 cells were pretreated with different pharmaceutic inhibitors for 1 h at 37 °C, then mock-infected or infected with PDCoV (MOI = 5) for 1 h at 37 °C. After the unabsorbed viruses were removed, these cells continued being cultured in the presence of various inhibitors or the corresponding concentration of solvent dimethylsulfoxide (DMSO) or H2O. At 12 hpi, the cells were harvested and analyzed by quantitative real-time RT-PCR, western blot, and TCID50 assays.
2.8. Cell viability assay
The cell viability was measured using the cell counting kit-8 (Beyotime) in accordance with the manufacturer’s protocol.
2.9. Statistical analysis
Statistical differences were determined by one-way ANOVAs or Student’s t-test performed using GraphPad Prism 5.0 software (GraphPad Software, CA, USA). For all experiments, differences were considered to be statistically significant when the corresponding p-values were < 0.05.
3. Results
3.1. PDCoV infection activates the IRE1/XBP-1 pathway
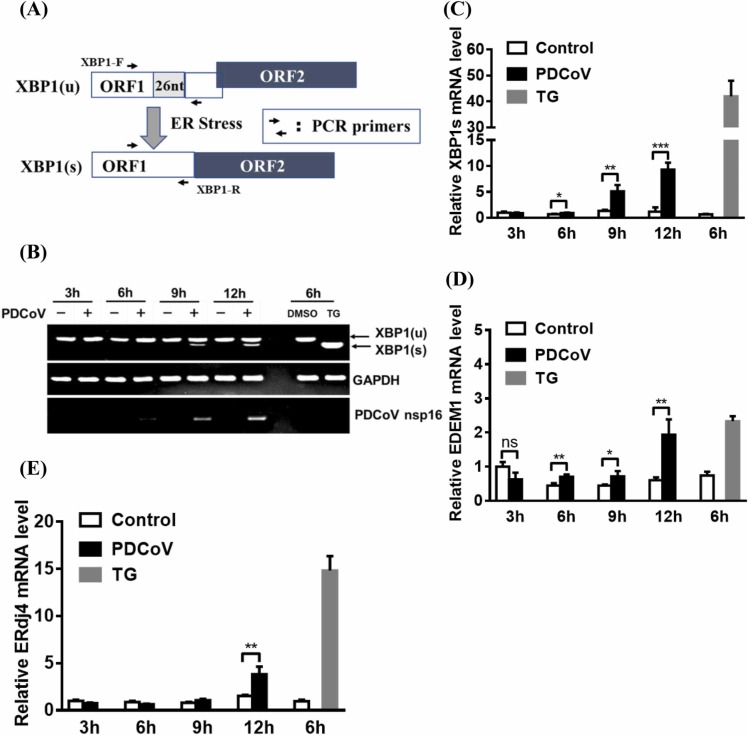
To determine whether the IRE1 pathway is activated in response to PDCoV infection, LLC-PK1 cells were infected with PDCoV for 3, 6, 9, or 12 h. Cellular total mRNA was exacted and subjected to RT-PCR analysis for amplifying both XBP1u and XBP1s to evaluate the splicing of XBP1 mRNA, which is a representation of IRE1 activity ( Fig. 1A). Cells treated with a pharmacological inducer of ERS, TG that had no effect on LLC-PK1 cell viability by concentration (1 µM; Fig. S2A), were used as a positive control. As shown in Fig. 1B, the 474-nt RT-PCR product representing the XBP1u mRNA was the predominant product detected from the mock-infected cells, whereas a 448-nt RT-PCR product representing the XBP1s mRNA was the predominant product observed from the TG-treated cells. As the PDCoV infection progressed, there was a gradual increase in the amount of XBP1s mRNA that could be observed (Fig. 1B). The results of a quantitative real time RT-PCR assay also demonstrate this gradual increase in the amount of XBP1s mRNA (Fig. 1C). These results suggest that the IRE1/XBP1 pathway is activated during PDCoV infection.
Fig. 1.
PDCoV infection induces the splicing of XBP1 mRNA and increases the transcription of XBP1s target genes. (A) Schematic diagram of the XBP1u to XBP1s conversion process mediated by ERS. (B) RT-PCR analysis of XBP1s and XBP1u mRNA. LLC-PK1 cells were mock-infected or infected with PDCoV (MOI = 5) for 3, 6, 9, or 12 h. Cellular total RNA was extracted and subjected to RT-PCR analysis to detect the unspliced and spliced forms of XBP1 mRNA, viral genomic RNA, and GAPDH mRNA. Mock-infected and TG-treated LLC-PK1 cells were used as negative and positive controls, respectively. (C–E) PDCoV-infected LLC-PK1 cells were harvested at 3, 6, 9, or 12 h. Cellular total RNAs were extracted and subjected to quantitative real-time RT-PCR analysis for XBP1s (C), EDEM1 (D), and ERdj4 (E) mRNA. The presented results are the means and standard deviations of data from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, nonsignificant difference.
XBP1s encodes a transcription factor, so we further investigated whether XBP1s-responsive genes are transcriptionally activated in PDCoV-infected cells. The results show that TG-treated cells had approximately 2-fold and 12-fold higher mRNA levels of EDEM1 and ERdj4, respectively, compared with uninfected-cells; additionally, PDCoV-infected cells displayed an obvious increase in both EDEM1 and ERdj4 mRNA levels at 12 hpi (Fig. 1D and 1E).
Because PDCoV is an enteropathogenic coronavirus, the IRE1/XBP-1 pathway was further assessed in PDCoV-infected IPI-2I cells by quantitative real-time RT-PCR. As expected, the XBP1s mRNA level increased gradually over the course of PDCoV infection, which is consistent with the results shown above for similarly treated LLC-PK1 cells (Fig. S1A). Together, these results indicate that PDCoV infection activates the IRE1/XBP-1 pathway.
3.2. PDCoV infection activates the ATF6 pathway
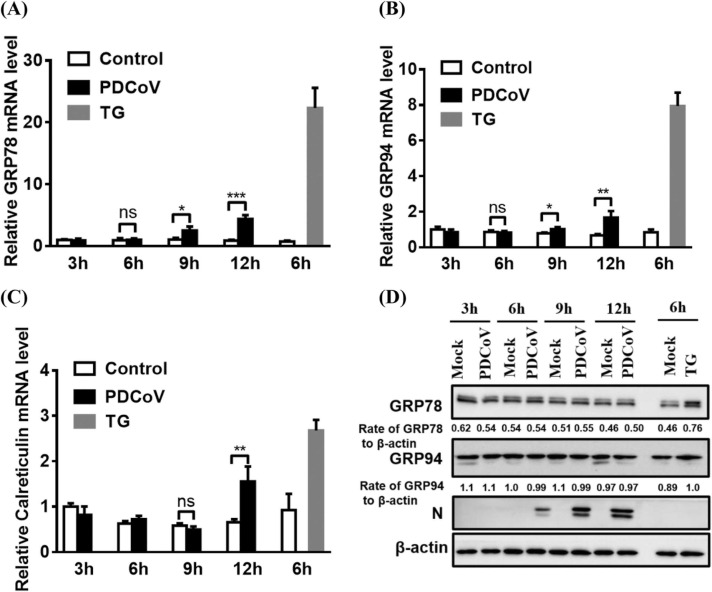
To determine whether the ATF6 pathway is activated in response to PDCoV infection, we first investigated ATF6 cleavage by western blot and ATF6 nuclear translocation by immunofluorescence assay. However, no cleavage and no nuclear translocation of ATF6 were observed in either PDCoV-infected or TG-treated cells (data not shown). It is likely that the antibodies used in our experiments did not efficiently recognize the amino-terminus of porcine ATF6. Similar cases have been described in previous reports (Echavarria-Consuegra et al., 2021, Shi et al., 2022).
As an alternative approach, we investigated whether the ATF6-responsive genes (GRP78, GRP94, and calreticulin) are transcriptionally activated during PDCoV infection. In contrast with mock-infected cells, PDCoV-infected cells displayed a significant increase in the levels of these gene transcripts at 12 hpi ( Fig. 2A–C). We further detected the protein expression of GRP78 and GRP94 in PDCoV-infected LLC-PK1 or IPI-2I cells. The results show that, compared with mock-infected cells, PDCoV-infected cells displayed no significant increase in the expression of these two chaperones over a 12h time course, suggesting that the expression of ATF6 target genes is subtly modulated (Fig. 2D and Fig. S1B). Together, these results show that PDCoV infection activates the ATF6 pathway.
Fig. 2.
PDCoV infection activates the ATF6 pathway. (A–C) LLC-PK1 cells were mock-infected or infected with PDCoV (MOI = 5) for 3, 6, 9, or 12 h. Cellular total RNA was extracted and subjected to quantitative real-time RT-PCR analysis for the detection of GRP78, GRP94, and calreticulin mRNA. Mock-infected and TG-treated LLCPK1 cells were used as negative and positive controls, respectively. (D) PDCoV-infected LLC-PK1 cells were harvested at 3, 6, 9, or 12 h and subjected to western blot analysis for the detection of GRP78, GRP94, and PDCoV N protein. Target protein expression was quantitatively estimated by ImageJ software and presented as the density value relative to that of the β-actin. The presented results are the means and standard deviations of data from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, nonsignificant difference.
3.3. PDCoV infection activates the PERK–eIF2α pathway and induces host translation attenuation
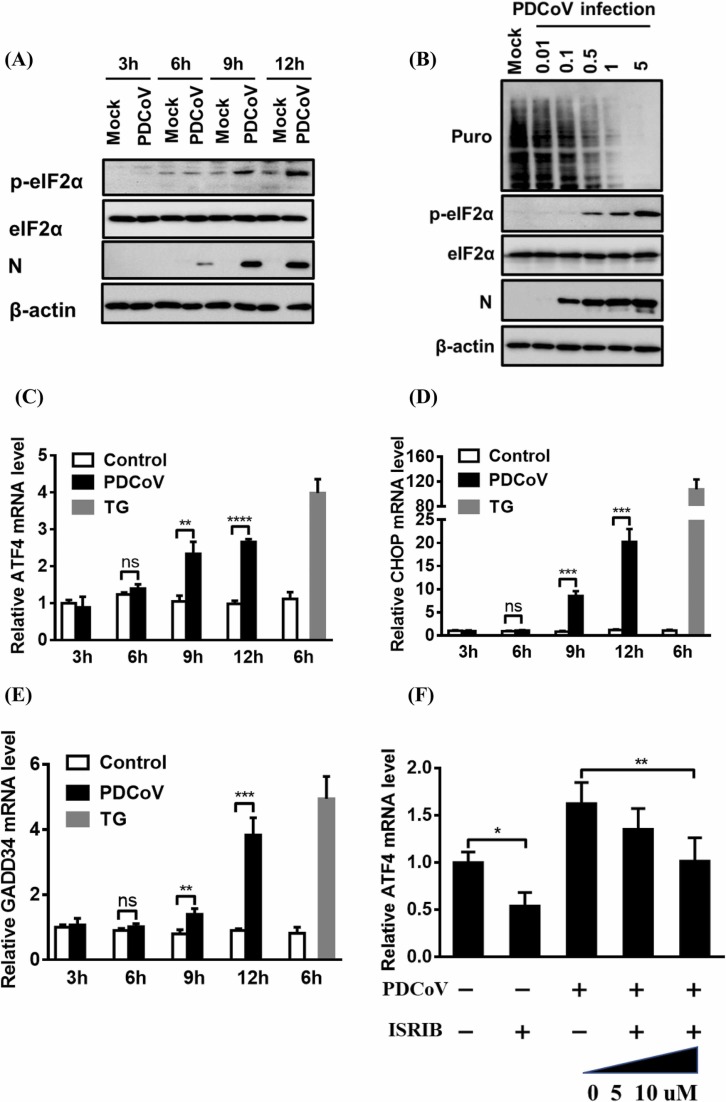
To determine the phosphorylation status of eIF2α during PDCoV infection, LLC-PK1 and IPI-2I cells were each infected with PDCoV for 3, 6, 9, or 12 h and then used in a western blot assay. As shown in Fig. 3A and Fig. S1C, compared with that in the mock-infected cells, the level of phosphorylated eIF2α in PDCoV-infected cells significantly increased over the course of PDCoV infection, while no obvious change in the steady-state level of eIF2α was observed in either mock- or PDCoV-infected cells.
Fig. 3.
PDCoV infection induces activation of the PERK–eIF2α pathway and host translation attenuation. (A) Western blot analysis of eIF2α phosphorylation. LLC-PK1 cells were mock-infected or infected with PDCoV (MOI = 5) for 3, 6, 9, or 12 h. Cells were collected and subjected to western blot analysis for the detection of phosphorylated eIF2α, total eIF2α, and PDCoV N protein. β-actin was used as a protein-loading control. (B) Analysis of protein synthesis in PDCoV-infected cells. LLC-PK1 cells were infected with PDCoV at various doses (MOI = 0.01, 0.1, 0.5, 1, or 5) for 12 h and then treated with puromycin for 25 min, before being used in a western blot analysis with an antibody against puromycin. (C–E) Determination of the transcription of ATF4 and its downstream genes. LLC-PK1 cells were mock-infected or infected with PDCoV (MOI = 5) for 3, 6, 9, or 12 h. The cells were then collected and subjected to quantitative real-time RT-PCR analysis of ATF4 (C), CHOP (D), and GADD34 (E) mRNA. (F) Inhibition of the PERK–eIF2α pathway in LLC-PK1 cells via treatment with the PERK-specific inhibitor ISRIB. LLC-PK1 cells were treated with ISRIB and then infected with PDCoV as described in the Materials and methods, before being using in a quantitative real-time RT-PCR analysis of ATF4 mRNA. The presented results are the means and standard deviations of data from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, nonsignificant difference.
To test whether PDCoV infection induces host translation attenuation, we monitored translation using a short pulse of puromycin, which can be incorporated into nascent polypeptides and can be detected by western blot with an antibody against puromycin (Li et al., 2018). As shown in Fig. 3B, PDCoV infection inhibited cellular protein translation in a dose-dependent manner, while PDCoV mRNA resisted translation attenuation, as demonstrated by the increasing viral N protein expression over the course of infection. We further tested the mRNA expression levels of several factors, such as ATF4, CHOP, and GADD34, that can be activated under high levels of phosphorylated eIF2α. As expected, the mRNA levels of ATF4 and of its two crucial target genes CHOP and GADD34 significantly increased as the infection progressed in both LLC-PK1 (Fig. 3C–E) and IPI-2I cells (Fig. S1D–F).
To further examine whether the PERK–eIF2α–ATF4 branch is activated, the PERK-specific inhibitor ISRIB was used to treat LLC-PK1 cells prior to their infection with PDCoV. The results show that, compared with the control DMSO treatment, ISRIB treatment dose-dependently reduced the PDCoV-induced ATF4 mRNA induction (Fig. 3F). LLC-PK1 cell viability was confirmed to be unchanged by ISRIB concentration (5 or 10 µM; Fig. S2B). These results demonstrate that PDCoV infection activates the PERK–eIF2α–ATF4 axis and induces host translation attenuation.
3.4. The IRE1 pathway is not responsible for PDCoV replication
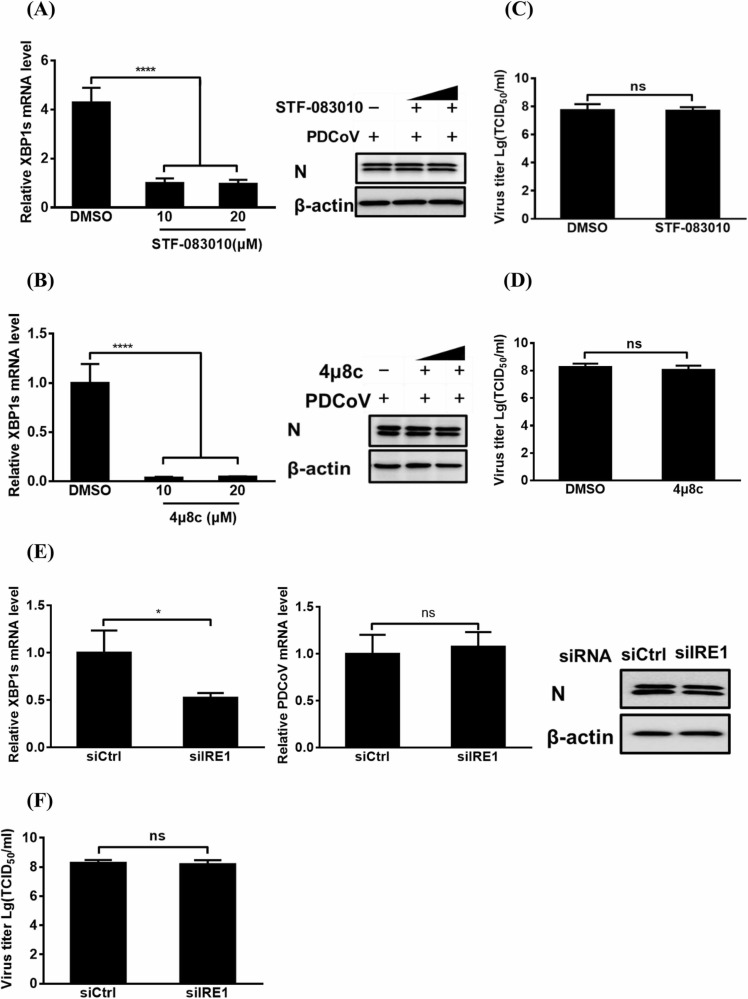
To explore whether IRE1 is required for PDCoV replication, we first investigated the effects of STF-083010 and 4µ8c, two specific IRE1 inhibitors, on PDCoV replication. As shown in Fig. 4A and 4B, compared with the control DMSO treatment, different concentrations (10 or 20 µM) of STF-083010 or 4µ8c significantly inhibited the XBP1s mRNA expression but had no obvious effect on PDCoV proliferation as evidenced by a lack of significant reduction N protein expression. Additionally, the virus titers in the supernatants from cells treated with DMSO were not significantly different from those of cells treated with the highest concentration (20 µM) of STF-083010 or 4µ8c (Fig. 4C and 4D). Importantly, no cytotoxicity was observed in the cells treated with either STF-083010 or 4µ8c at the concentrations used in our experiments (Fig. S2C and S2D). Furthermore, we silenced IRE1 with specific siRNAs for 12 h before infecting the cells with PDCoV. The IRE1 knockdown resulted in a notable downregulation of XBP1s mRNA expression but had no significant negative effect on viral replication, as evidenced by the lack of an obvious difference in the levels of viral genomic RNA and N protein expression (Fig. 4E) and in the virus titer (Fig. 4F). These results demonstrate that IRE1 activation is not required for PDCoV replication.
Fig. 4.
The effect of IRE1 pathway activation on PDCoV replication. (A, B) The effect of RE1 inhibitors STF-083010 (A) and 4µ8c (B) on PDCoV replication. LLC-PK1 cells were treated with various concentrations (10 or 20 µM) of inhibitors STF-083010 or 4µ8c and then infected with PDCoV (MOI = 5) as described in the Materials and methods, before being used in quantitative real-time RT-PCR or western blot analysis for relative XBP1s mRNA and N protein expression. (C, D) The cellular supernatants from cells treated with DMSO or the highest concentration (20 µM) of STF-083010 (C) or 4µ8c (D) were subjected to virus titer detection. (E, F) The effect of IRE1 pathway inhibition on viral replication via genetic approach. LLC-PK1 cells were transfected with siRNA against IRE1 or negative control siRNA for 12 h and then infected with PDCoV (MOI = 5) for 12 h, before being used in quantitative real-time RT-PCR, western blot (E), or TCID50 (F) assay for viral replication. The presented results are the means and standard deviations of data from three independent experiments. *p < 0.05; ****p < 0.0001; ns, nonsignificant difference.
3.5. ATF6 pathway activation contributes to PDCoV replication
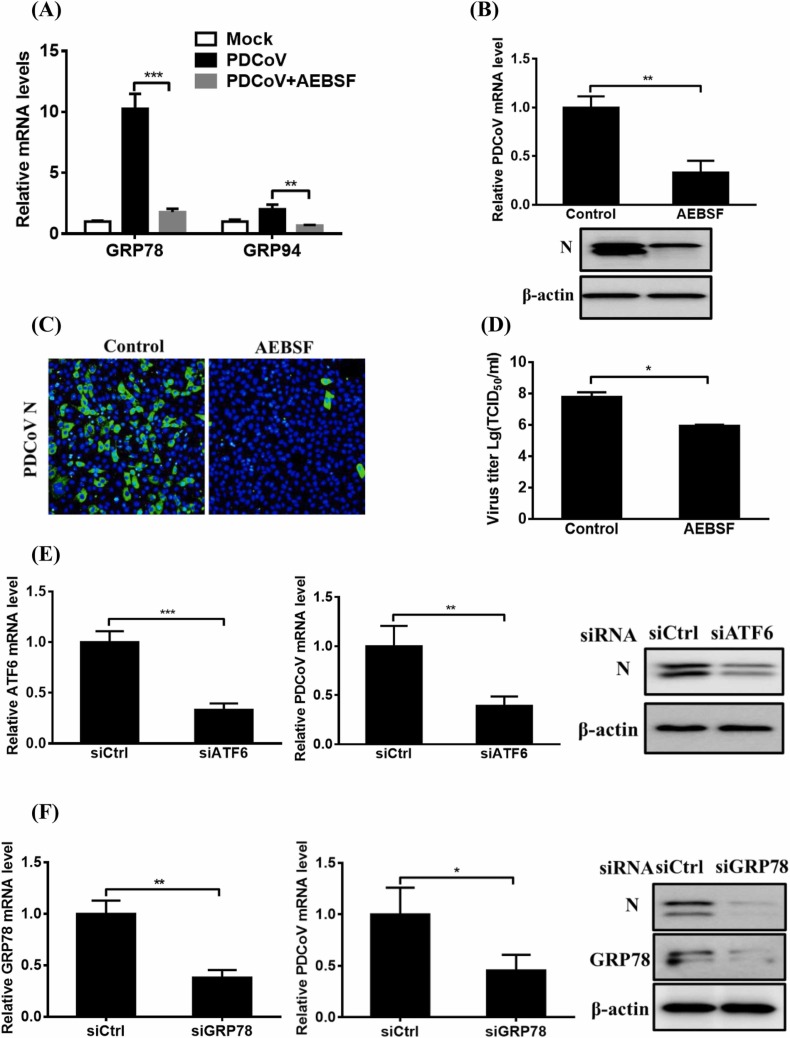
To clarify the role of the ATF6 pathway in PDCoV replication, we assessed PDCoV infection in the presence of AEBSF, an ATF6-specific inhibitor. As shown in Fig. 5A–D, compared with the control treatment, AEBSF treatment significantly inhibited the mRNA expression of GRP78 and GRP94, the target genes of ATF6, and also suppressed PDCoV replication, as evidenced by significant inhibitions of viral genomic RNA, N protein expression, and virus titer. LLC-PK1 cell viability was unaffected by AEBSF treatment at the concentration used (50 µM; Fig. S2E).
Fig. 5.
The effect of ATF6 pathway activation on PDCoV replication. (A–D) The effect of ATF6 pathway inhibitor AEBSF on PDCoV replication. LLC-PK1 cells were treated with the indicated concentration of AEBSF (50 µM) and then infected with PDCoV for 12 h as described in the Materials and methods, before being used in quantitative real-time RT-PCR, western blot, immunofluorescence assay, or TCID50 analysis for determining the relative levels of GRP78 and GRP94 mRNA (A), viral genomic RNA (B), N protein expression (B, C), and virus titer (D). (E, F) The effect of ATF6 or GRP78 knockdown on PDCoV replication. LLC-PK1 cells were transfected with siRNA against ATF6 (E) or GRP78 (F) or with negative control siRNA for 12 h, and then infected with PDCoV for 12 h. Cell samples were harvested and subjected to quantitative real-time RT-PCR and western blot assay for assessing viral replication. The presented results are the means and standard deviations of data from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, nonsignificant difference.
We also examined PDCoV infection in cells with reduced ATF6 expression resulting from siRNA treatment. Compared with the control siRNA, ATF6-specific siRNA notably reduced ATF6 mRNA expression, viral genomic RNA levels, and N protein expression (Fig. 5E). To further measure the effect of the ATF6 pathway on PDCoV replication, we used siRNA to knockdown its effector molecule GRP78. The results show that GRP78-specific siRNA notably reduced GRP78 expression, viral genomic RNA levels, and N protein expression (Fig. 5F). These findings suggest that ATF6 pathway activation is required for PDCoV replication.
3.6. Activation of the PERK–eIF2α pathway inhibits PDCoV replication
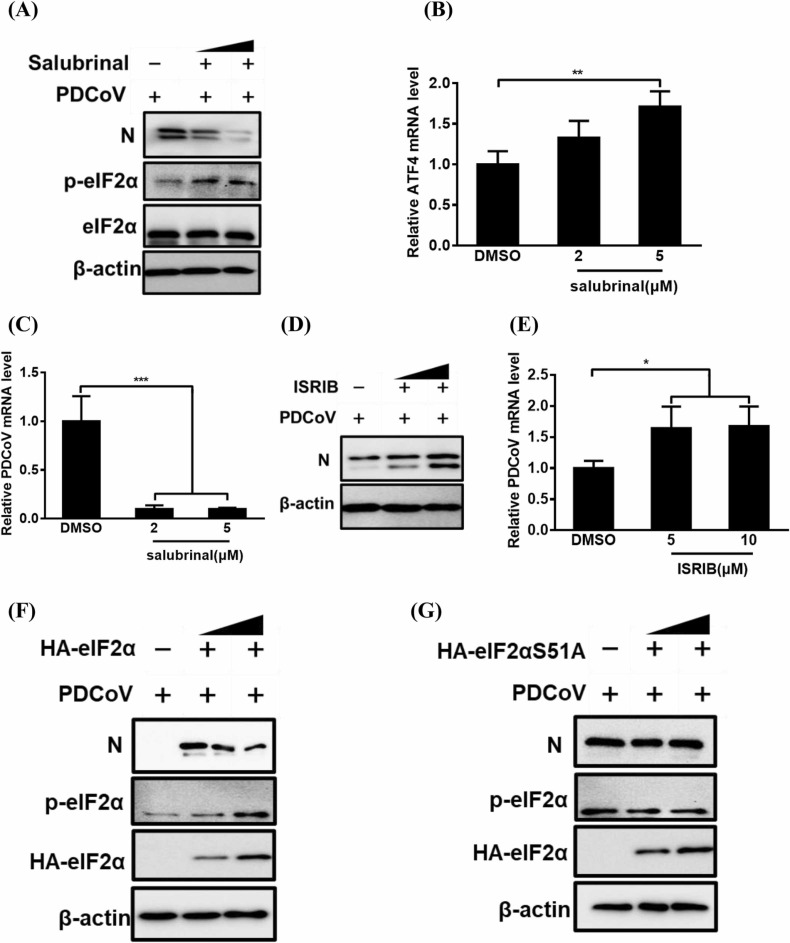
To determine the effect of the PERK–eIF2α pathway on PDCoV replication, we first tested the effect of salubrinal, an eIF2α dephosphorylation inhibitor, on PDCoV replication. As shown in Fig. 6A-C, compared with DMSO treatment, salubrinal treatment notably promoted eIF2α phosphorylation and ATF4 mRNA expression, and it inhibited PDCoV replication, as evidenced by the obvious dose-dependent reduction in N protein expression and viral genomic RNA levels. LLC-PK1 cell viability was confirmed to be unchanged by the concentrations of salubrinal used in our experiments (2 and 5 µM; Fig. S2F).
Fig. 6.
The effect of PERK-eIF2α pathway activation on PDCoV replication. (A–E) The effect of salubrinal (A-C) or ISRIB (D, E) on PDCoV replication. LLC-PK cells were treated with salubrinal or ISRIB and then used in assays as described in panel B of Fig. 5. Collected samples were subjected to western blot (A, D) or quantitative real-time RT-PCR analysis (B, C and E) for determining the expression of N protein, relative ATF4 mRNA and viral genomic RNA. (F, G) LLC-PK1 cells were transfected with pCAGGS-HA-eIF2α (F) or pCAGGS-HA-eIF2αS51A (G) for 12 h and then infected with PDCoV for 12 h, before being used in a western blot analysis for detecting the p-eIF2α, HA-eIF2α, and PDCoV N protein expression as a means of evaluating viral replication. The presented results are the means and standard deviations of data from three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
We next disrupted the PERK–eIF2α pathway with the PERK-specific inhibitor ISRIB. Compared with the control DMSO treatment, ISRIB treatment significantly promoted PDCoV replication in a dose-dependent manner, as evidenced by the increased N protein expression and viral genomic RNA levels (Fig. 6D and 6E). To further elucidate the role of eIF2α Ser-51 phosphorylation in PDCoV replication, we investigated PDCoV infection in LLC-PK1 cells that expressed wildtype eIF2α (HA-eIF2α) or its unphosphorylatable mutant form (HA-eIF2αS51A). As expected, PDCoV infection was inhibited in the cells expressing wildtype eIF2α, whereas no significant decrease in PDCoV infection was found in cells expressing eIF2αS51A (Fig. 6F and 6G). Together, these results show that PERK–eIF2α pathway activation suppresses PDCoV replication.
4. Discussion
This study is the first to report the activation and modulation of all three UPR pathways (PERK, IRE1, and ATF6) during PDCoV infection. We further confirmed that activation of the PERK–eIF2α pathway suppresses PDCoV replication, while ATF6 pathway activation is required for PDCoV replication. Thus, both the PERK and ATF6 pathways can serve as potential antiviral targets.
Our work shows that both the IRE1 and ATF6 pathways are activated during PDCoV infection. However, the activation of IRE1 is not essential for PDCoV replication. Previous studies reported that IRE1 activation promotes the replication of influenza A (IAV) and classical swine fever virus (CSFV) (Hassan et al., 2012, He et al., 2017), whereas it inhibits the replication of respiratory syncytial virus (RSV) (Hassan et al., 2014) and is not associated with the replication of dengue virus (DNV-2) (Yu et al., 2006). Taken together, the IRE1 pathway displays different effects during infection with different viruses.
Given that the IRE1–XBP1 pathway has been confirmed to play vital roles in a wide range of biological process, including immunity, inflammation, and metabolism (Kaufman and Cao, 2010), we speculated that its function may also focus on other aspects, such as the modulation of inflammatory and innate immunity responses, rather than only on relieving ERS. For example, TLR2 and TLR4 specifically activate the IRE1-dependent expression of XBP1s to induce the production of inflammatory cytokines IL-6 and TNF-α (Martinon et al., 2010). Indeed, the expression of inflammatory cytokines (IL-6, IL-8, and TNF-α) is significantly increased after PDCoV infection, both in vitro and in vivo (Jung et al., 2018). However, the relationship between IRE1 and inflammation awaits confirmation by further experiments.
Generally, activation of the ATF6 pathway is more beneficial for viral replication, as the pro-survival downstream effectors of these signals promote the folding of misfolded proteins. Here, we confirmed that ATF6 activation facilitated PDCoV replication. GRP78 mRNA or protein levels are extensively used as a proxy for ATF6 pathway activation. Our results show that GRP78 mRNA levels increased in PDCoV-infected cells, as compared with those in mock-infected cells, in a time-dependent manner, while no corresponding significant increase in GRP78 protein expression was observed. The reason for this apparent discrepancy is not clear, although both upregulations and downregulations of GRP78 at the protein level have been reported during infection with other viruses. For example, Porcine epidemic diarrhea virus (PEDV) and TGEV infection induce ERS and promote GRP78 protein expression (Sun et al., 2021, Xue et al., 2018). Porcine hemagglutinating encephalomyelitis virus (PHEV) infection activates all three UPR arms and increases GRP78 expression at both the mRNA and protein levels (Shi et al., 2022). PRRSV targets the UPR master regulator GRP78 for degradation despite inducing an increase in its mRNA levels in a time-dependent manner (Gao et al., 2019). Additionally, despite MHV-infected cells displaying a transcriptional upregulation of GRP78, its protein was not detectable by western blot in these cells (Echavarria-Consuegra et al., 2021). Thus, UPR pathways are intricate and there is extensive crosstalk among them as well as with other cell signaling pathways, all of which are subtly modulated during virus and host interactions (Fung and Liu, 2014). We also speculate that ATF6 pathway activation may contribute to a stabilization of GRP78 protein abundance throughout PDCoV infection and facilitate viral replication because we found that an siRNA-induced knockdown of GRP78 reduced its protein expression and significantly inhibited PDCoV replication. We could not rule out the possibility that the expressions of other ATF6 target genes were selectively upregulated. Indeed, ASFV infection activates the ATF6 pathway of the UPR, leading to an induction of ER chaperones calreticulin and calnexin but not ERp57 and GRP78 (Galindo et al., 2012). Taken together, ATF6 pathway activation is required for PDCoV replication but is subtly modulated during PDCoV infection.
Unlike MHV infection, PDCoV infection promotes the transcriptions of ATF4 and its target genes CHOP and GADD34 (Bechill et al., 2008), and it notably induces ongoing host cellular translation attenuation. We speculate that this translation attenuation may be responsible for the minimal expression of UPR target genes. Rotavirus infection activates the IRE1 and ATF6 pathways, but these pathways are impaired at the translation level by viral nsp3 protein (Trujillo-Alonso et al., 2011). Whether PDCoV also encodes a protein with a similar function should be investigated. Translation attenuation has also been reported in infections with other viruses, such as SARS-CoV, HCoV-OC43, MHV, feline infectious peritonitis virus (FIPV), TGEV, PEDV, and herpes simplex virus type 1 (Bechill et al., 2008, Esclatine et al., 2004, Favreau et al., 2009, Xue et al., 2018). Generally, global translation inhibition induced by phosphorylated eIF2α represents a vital host cellular antiviral response, which is detrimental to viral replication. For example, the induction of PERK suppresses PEDV and TGEV replication (Wang et al., 2014, Xue et al., 2018). Inhibition of protein synthesis does not appear to be beneficial for MHV replication in vitro (Raaben et al., 2007). However, many viruses, such as foot-and-mouth disease virus (FMDV), DNV (Lee et al., 2018, Ranjitha et al., 2020), and human cytomegalovirus (HCMV) (Yu et al., 2013), can also utilize the PERK pathway to support viral replication. Our work demonstrates that activation of the PERK–eIF2α pathway suppressed PDCoV infection, implying that the inhibition of host protein synthesis induced by PDCoV might be detrimental for viral replication via downregulating cellular factors that are necessary for viral infection. Taken together, the PERK–eIF2α pathway displays variable effects during infection with different viruses.
The ERS and UPR are associated with signaling pathways involved in many different processes, such as apoptosis, autophagy, inflammation, and innate immunity. For example, PEDV induces autophagy in Vero cells via ROS-dependent ERS, mediated through the PERK and IRE1 pathways (Sun et al., 2021). Infectious bronchitis virus (IBV) activates ERS sensor IRE1 and MAP kinase ERK to modulate autophagy (Fung and Liu, 2019). SARS-CoV envelope protein regulates the cell stress response and apoptosis (DeDiego et al., 2011). SARS-CoV ORF3a protein induces ERS and antagonizes the interferon (IFN) signaling pathway (Minakshi et al., 2009). Previous studies have demonstrated that PDCoV infection induces apoptosis, autophagy, and inflammatory responses and inhibits IFN production and signaling transduction (Duan et al., 2021, Jung et al., 2018, Luo et al., 2016, Qin et al., 2019). Here, we demonstrated that PDCoV infection activates ERS and induces UPR activation. Whether PDCoV-induced ERS participates in the regulation of apoptosis, autophagy, inflammation, and IFN responses is an interesting question that deserves further exploration.
5. Conclusion
In summary, our data confirm that PDCoV infection activates the IRE1, ATF6, and PERK pathways of the UPR. We also demonstrate the negative role of the PERK pathway and the positive role of the ATF6 pathway on PDCoV replication. These findings lay the foundation for further dissecting the precise role of ERS and the UPR in PDCoV infection and will help identify molecular targets for the development of new antiviral strategies.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work is supported by the National Key Research and Development Program of China (2021YFD1801104), the National Natural Science Foundation of China (31902247, 32072846, 31730095) and China Postdoctoral Science Foundation (2019T120670, 2018M640717).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.vetmic.2022.109494.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- Ambrose R.L., Mackenzie J.M. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 2011;85:2723–2732. doi: 10.1128/JVI.02050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechill J., Chen Z., Brewer J.W., Baker S.C. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 2008;82:4492–4501. doi: 10.1128/JVI.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boley P.A., Alhamo M.A., Lossie G., Yadav K.K., Vasquez-Lee M., Saif L.J., Kenney S.P. Porcine deltacoronavirus infection and transmission in poultry, United States. Emerg. Infect. Dis. 2020;26:255–265. doi: 10.3201/eid2602.190346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzaro N., Meng X.J. Induction of the unfolded protein response (UPR) suppresses porcine reproductive and respiratory syndrome virus (PRRSV) replication. Virus Res. 2020;276 doi: 10.1016/j.virusres.2019.197820. [DOI] [PubMed] [Google Scholar]
- Chen Q., Men Y., Wang D., Xu D., Liu S., Xiao S., Fang L. Porcine reproductive and respiratory syndrome virus infection induces endoplasmic reticulum stress, facilitates virus replication, and contributes to autophagy and apoptosis. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-69959-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Alvarez E., Oliveros J.C., Zhao J., Fett C., Perlman S., Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLOS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Fang L., Yang H., Liu H., Du T., Fang P., Wang D., Chen H., Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan C., Liu Y., Hao Z., Wang J. Ergosterol peroxide suppresses porcine deltacoronavirus (PDCoV)-induced autophagy to inhibit virus replication via p38 signaling pathway. Vet. Microbiol. 2021;257 doi: 10.1016/j.vetmic.2021.109068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarria-Consuegra L., Cook G.M., Busnadiego I., Lefevre C., Keep S., Brown K., Doyle N., Dowgier G., Franaszek K., Moore N.A., Siddell S.G., Bickerton E., Hale B.G., Firth A.E., Brierley I., Irigoyen N. Manipulation of the unfolded protein response: a pharmacological strategy against coronavirus infection. PLOS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esclatine A., Taddeo B., Roizman B. Herpes simplex virus 1 induces cytoplasmic accumulation of TIA-1/TIAR and both synthesis and cytoplasmic accumulation of tristetraprolin, two cellular proteins that bind and destabilize AU-rich RNAs. J. Virol. 2004;78:8582–8592. doi: 10.1128/JVI.78.16.8582-8592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau D.J., Desforges M., St-Jean J.R., Talbot P.J. A human coronavirus OC43 variant harboring persistence-associated mutations in the S glycoprotein differentially induces the unfolded protein response in human neurons as compared to wild-type virus. Virology. 2009;395:255–267. doi: 10.1016/j.virol.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. The ER stress sensor IRE1 and MAP kinase ERK modulate autophagy induction in cells infected with coronavirus infectious bronchitis virus. Virology. 2019;533:34–44. doi: 10.1016/j.virol.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo I., Hernaez B., Munoz-Moreno R., Cuesta-Geijo M.A., Dalmau-Mena I., Alonso C. The ATF6 branch of unfolded protein response and apoptosis are activated to promote African swine fever virus infection. Cell Death Dis. 2012;3 doi: 10.1038/cddis.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Chai Y., Song J., Liu T., Chen P., Zhou L., Ge X., Guo X., Han J., Yang H. Reprogramming the unfolded protein response for replication by porcine reproductive and respiratory syndrome virus. PLOS Pathog. 2019;15 doi: 10.1371/journal.ppat.1008169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hassan I., Gaines K.S., Hottel W.J., Wishy R.M., Miller S.E., Powers L.S., Rutkowski D.T., Monick M.M. Inositol-requiring enzyme 1 inhibits respiratory syncytial virus replication. J. Biol. Chem. 2014;289:7537–7546. doi: 10.1074/jbc.M113.510594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan I.H., Zhang M.S., Powers L.S., Shao J.Q., Baltrusaitis J., Rutkowski D.T., Legge K., Monick M.M. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J. Biol. Chem. 2012;287:4679–4689. doi: 10.1074/jbc.M111.284695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Xu H., Gou H., Yuan J., Liao J., Chen Y., Fan S., Xie B., Deng S., Zhang Y., Chen J., Zhao M. CSFV infection up-regulates the unfolded protein response to promote its replication. Front. Microbiol. 2017;8:2129. doi: 10.3389/fmicb.2017.02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa A., Chevet E. Redox signaling loops in the unfolded protein response. Cell Signal. 2012;24:1548–1555. doi: 10.1016/j.cellsig.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Jung K., Hu H., Saif L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017;162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Miyazaki A., Hu H., Saif L.J. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet. Microbiol. 2018;221:49–58. doi: 10.1016/j.vetmic.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman R.J., Cao S. Inositol-requiring 1/X-box-binding protein 1 is a regulatory hub that links endoplasmic reticulum homeostasis with innate immunity and metabolism. EMBO Mol. Med. 2010;2:189–192. doi: 10.1002/emmm.201000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J.A., Tagliamonte M.S., White S.K., Elbadry M.A., Alam M.M., Stephenson C.J., Bonny T.S., Loeb J.C., Telisma T., Chavannes S., Ostrov D.A., Mavian C., Beau De Rochars V.M., Salemi M., Morris J.G., Jr. Independent infections of porcine deltacoronavirus among Haitian children. Nature. 2021;600:133–137. doi: 10.1038/s41586-021-04111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., Glimcher L.H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.R., Kuo S.H., Lin C.Y., Fu P.J., Lin Y.S., Yeh T.M., Liu H.S. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci. Rep. 2018;8:489. doi: 10.1038/s41598-017-18909-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Fang L., Zhou Y., Tao R., Wang D., Xiao S. Porcine reproductive and respiratory syndrome virus infection induces both eIF2alpha phosphorylation-dependent and -independent host translation shutoff. J. Virol. 2018;92:e00600–e00618. doi: 10.1128/JVI.00600-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Zhang H., Li B., Ding Q., Wang Y., Gao W., Guo D., Wei Z., Hu H. Susceptibility of chickens to porcine deltacoronavirus infection. Viruses. 2019;11:573. doi: 10.3390/v11060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Fang L., Dong N., Fang P., Ding Z., Wang D., Chen H., Xiao S. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology. 2016;495:10–17. doi: 10.1016/j.virol.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang Y., Liang X., Lou F., Oglesbee M., Krakowka S., Li J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 2015;6 doi: 10.1128/mBio.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F., Chen X., Lee A.H., Glimcher L.H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 2010;11:411–418. doi: 10.1038/ni.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi R., Padhan K., Rani M., Khan N., Ahmad F., Jameel S. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLOS One. 2009;4 doi: 10.1371/journal.pone.0008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I., Zeng H., Harding H.P., Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Du E.Z., Luo W.T., Yang Y.L., Zhang Y.Q., Wang B., Huang Y.W. Characteristics of the life cycle of porcine deltacoronavirus (PDCoV) in vitro: replication kinetics, cellular ultrastructure and virion morphology, and evidence of inducing autophagy. Viruses. 2019;11:455. doi: 10.3390/v11050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Groot Koerkamp M.J., Rottier P.J., de Haan C.A. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 2007;9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjitha H.B., Ammanathan V., Guleria N., Hosamani M., Sreenivasa B.P., Dhanesh V.V., Santhoshkumar R., Sagar B.K.C., Mishra B.P., Singh R.K., Sanyal A., Manjithaya R., Basagoudanavar S.H. Foot-and-mouth disease virus induces PERK-mediated autophagy to suppress the antiviral interferon response. J. Cell Sci. 2020;134 doi: 10.1242/jcs.240622. [DOI] [PubMed] [Google Scholar]
- Shi J., Li Z., Xu R., Zhang J., Zhou Q., Gao R., Lu H., Lan Y., Zhao K., He H., Gao F., He W. The PERK/PKR-eIF2alpha pathway negatively regulates porcine hemagglutinating encephalomyelitis virus replication by attenuating global protein translation and facilitating stress granule formation. J. Virol. 2022;96 doi: 10.1128/JVI.01695-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Jin J., Wang L., Wang J., Zhou H., Zhang Q., Xu X. Porcine epidemic diarrhea virus infections induce autophagy in Vero cells via ROS-dependent endoplasmic reticulum stress through PERK and IRE1 pathways. Vet. Microbiol. 2021;253 doi: 10.1016/j.vetmic.2020.108959. [DOI] [PubMed] [Google Scholar]
- Tan Z., Zhang W., Sun J., Fu Z., Ke X., Zheng C., Zhang Y., Li P., Liu Y., Hu Q., Wang H., Zheng Z. ZIKV infection activates the IRE1-XBP1 and ATF6 pathways of unfolded protein response in neural cells. J. Neuroinflamm. 2018;15:275. doi: 10.1186/s12974-018-1311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P., Cui E., Song Y., Yan R., Wang J. Porcine deltacoronavirus and its prevalence in China: a review of epidemiology, evolution, and vaccine development. Arch. Virol. 2021;166:2975–2988. doi: 10.1007/s00705-021-05226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-Alonso V., Maruri-Avidal L., Arias C.F., Lopez S. Rotavirus infection induces the unfolded protein response of the cell and controls it through the nonstructural protein NSP3. J. Virol. 2011;85:12594–12604. doi: 10.1128/JVI.05620-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J.R., Sun M.X., Ni B., Huan C., Huang L., Li C., Fan H.J., Ren X.F., Mao X. Triggering unfolded protein response by 2-Deoxy-D-glucose inhibits porcine epidemic diarrhea virus propagation. Antivir. Res. 2014;106:33–41. doi: 10.1016/j.antiviral.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Zhong H., Zhou Q., Du Y., Chen L., Zhang Y., Xue C., Cao Y. A highly pathogenic strain of porcine deltacoronavirus caused watery diarrhea in newborn piglets. Virol. Sin. 2018;33:131–141. doi: 10.1007/s12250-018-0003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Feng L. The role of unfolded protein response in coronavirus infection and its implications for drug design. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.808593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Fu F., Ma Y., Zhang X., Li L., Feng L., Liu P. The PERK arm of the unfolded protein response negatively regulates transmissible gastroenteritis virus replication by suppressing protein translation and promoting type I interferon production. J. Virol. 2018;92:e00431–18. doi: 10.1128/JVI.00431-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.Y., Hsu Y.W., Liao C.L., Lin Y.L. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 2006;80:11868–11880. doi: 10.1128/JVI.00879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Pierciey F.J., Jr., Maguire T.G., Alwine J.C. PKR-like endoplasmic reticulum kinase is necessary for lipogenic activation during HCMV infection. PLOS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Qi B., Gu Y., Xu F., Du H., Li X., Fang W. Porcine circovirus 2 deploys PERK pathway and GRP78 for its enhanced replication in PK-15 cells. Viruses. 2016;8:56. doi: 10.3390/v8020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material