Abstract
Acinetobacter baumannii is a bacterium found in most places, especially in clinics and hospitals, and an important agent of nosocomial infections. The presence of class D enzymes such as OXA-type carbapenemases in A. baumannii is proven to have a key function in resistance to carbapenem. The aim of the current study is to determine the blaOXA -type carbapenemase genes and antimicrobial resistance among clinically isolated samples of A. baumannii. We assessed 100 clinically isolated specimens of A. baumannii from patients in intensive care units of educational hospitals of Hamadan, West of Iran. The A. baumannii isolates' susceptibility to antibiotics was performed employing disk diffusion method. Multiplex polymerase chain reaction was used to identify the bla OXA-24-like , bla OXA-23-like , bla OXA-58-like , and bla OXA-51-like genes. The bla OXA-23-like , bla OXA-24-like , and bla OXA-58-like genes' prevalence were found to be 84, 58, and 3%, respectively. The highest coexistence of the genes was for bla OXA-51/23 (84%) followed by bla OXA-51/24-like (58%). The bla OXA-51/23- like pattern of genes is a sort of dominant gene in resistance in A. baumannii from Hamadan hospitals. The highest resistance to piperacillin (83%) and ciprofloxacin (81%) has been observed in positive isolates of bla OXA-23-like . The A. baumannii isolates with bla OXA-58-like genes did not show much resistance to antibiotics. Based on the results of the phylogenetic tree analysis, all isolates have shown a high degree of similarity. This study showed the high frequency of OXA -type carbapenemase genes among A. baumannii isolates from Hamadan hospitals, Iran. Thus, applying an appropriate strategy to limit the spreading of these strains and also performing new treatment regimens are necessary.
Keywords: Acinetobacter baumannii, nosocomial infection, OXA -type carbapenemase
Introduction
Acinetobacter baumannii is an oxidase-negative, gram-negative, nonmotile, nonfermentative coccobacilli and it is found inside most places, especially in hospitals and other health care facilities. 1 2 3 The bacterium is mostly originating from the intensive care units (ICUs). It can cause septicemia, infections of the skin, meningitis, endocarditis, urinary tract infections, soft tissues infections, and ventilator-associated pneumonia in patients in ICU wards. 4 5 6 7 Therefore, infections caused by the bacterium need early and effective antimicrobial therapy. The carbapenems are the most effective antibacterial agents in the case of clinical isolates of A. baumannii . 8 Nowadays, due to excessive and inappropriate use of these drugs, the resistance to carbapenems has increased. 9 A. baumannii can become carbapenem resistant by the acquisition of plasmid-mediated carbapenem hydrolyzing classes of A, B, and D metallo-β-lactamase enzymes. 10 Class D enzymes such as OXA-type carbapenemases, especially the intrinsic presence of bla OXA-58-like , bla OXA-24-like , bla OXA-23-like , bla OXA-51 , and β-lactamases in A. baumannii , are proven to have a vital role in resistance to carbapenem. 11 12 13 The OXA -carbapenemase genes can be encoded by chromosome or plasmid and are mostly associated with insertion elements, particularly ISAbal . 8 With the rise of pan-drug-resistant (PDR) and multidrug-resistant (MDR) strains of A. baumannii all around the world, the bacterium has become a very serious threat for health care organizations. 10 14 Thus, the current study focused to discover the blaOXA -type carbapenemase genes, which, according to previous studies, contribute to more antibiotic resistance among clinical isolates of A. baumannii.
Materials and Methods
Bacterial Isolation and Identification
The cross-sectional research has been performed on 100 A. baumannii isolates from clinical specimens in ICU wards in educational hospitals of Hamadan, West of Iran, from December 2016 to December 2017. A. baumannii isolates were identified using the biochemical tests and polymerase chain reaction (PCR) targeting the carbapenemase gene of blaOXA-51-like. 15 16
Susceptibility Testing
The CLSI—Clinical and Laboratory Standards Institute—(CLSI, 2018) method of disk-agar diffusion 17 and interpretation by CLSI breakpoint were used to measure the susceptibility of A. baumannii isolates to Meropenem (10 µg), Imipenem (10 µg), Amikacin (30 µg), Gentamicin (10 µg), Ciprofloxacin (5 µg), Doxycycline (30 µg), Piperacillin (100 µg), Tobramycin (10 µg), Cefepime (30 µg), and Ampicillin/sulbactam (20 µg) antimicrobial agents. ATCC 27853- Pseudomonas aeruginosa was deployed in susceptibility testing as the control strain.
Multiplex-PCR Assay
Alkaline lysis method was used for extraction of deoxyribonucleic acid (DNA). 15 The primers for PCR amplification of bla OXA-58-like , bla OXA-23-like , bla OXA-51-like , and bla OXA-24-like were used as described before ( Table 1 ). 18
Table 1. Primer sequences used for multiplex polymerase chain reaction amplification of bla OXA genes variants .
| Primer name | 5′ to 3′ sequence prime | Product size (bp) | Annealing temperature (°C) | Reference | |
|---|---|---|---|---|---|
| OXA-51-like | F | TAATGCTTTGATCGGCCTTG | 353 | 53 | Woodford (2006) 18 |
| R | TGGATTGCACTTCATCTTGG | ||||
| OXA-23-like | F | GATCGGATTGGAGAACCAGA | 501 | 53 | Woodford (2006) 18 |
| R | ATTTCTGACCGCATTTCCAT | ||||
| OXA-24-like | F | GGTTAGTTGGCCCCCTTAAA | 246 | 53 | Woodford (2006) 18 |
| R | AGTTGAGCGAAAAGGGGATT | ||||
| OXA-58-like | F | AAGTATTGGGGCTTGTGCTG | 599 | 53 | Woodford (2006) 18 |
| R | CCCCTCTGCGCTCTACATAC | ||||
Multiplex-PCR was performed in a 50-µL reaction volume containing 5 µL PCR buffer 10X with MgCl 2 , 5 µL deoxynucleotide triphosphates (2 mm), 2 µL from each reverse and forward primers (10 pmol), 0.4 µL Taq Polymerase (5U/µL), 6 µL DNA template, and 29.6 µL nuclease-free deionized water. The amplification conditions were as follows: initial denaturation at 94°C for 5 minutes followed by 30 cycles at 94°C for 30 seconds, 53°C for 40 seconds, and 72°C for 50 seconds and a final extension at 72°C for 6 minutes. Gel electrophoresis of the amplicons was performed on 1.5% agarose gel. The findings were analyzed employing a transilluminator device. Bands with 353 bp, 501 bp, 246 bp, and 599 bp were the representatives of bla OXA-51-like , bla OXA-23-like , bla OXA-24-like , and bla OXA-58-like , respectively.
Sequencing
Three PCR products of each gene, which was separately performed, were purified and sequenced directly by Sanger dideoxy-sequencing technology in two directions using a capillary DNA analyzer (Applied Biosystems, Waltham, Massachusetts, United States).
Phylogenetic Analysis of OXA-type Variant
The phylogenetic analysis was conducted independently for the OXA -type variant. The tree is drawn to scale, with branch lengths measuring the number of substitutions per site. The sequences were aligned and manually edited in consensus positions and compared with that of the sequences from GenBank. A phylogenetic tree was built by the maximum likelihood method utilizing the Tamura-Nei model with Mega software (v.7). 19
Statistical Analysis
Statistical analysis was done using Statistical Package for the Social Sciences 23.0 (SPSS, Chicago, Illinois, United States). The Pearson chi-squared test and Fisher's exact test were used to analyze the qualitative data. All p -values < 0.05 were considered statistically significant.
Results
The highest resistance to piperacillin (83%) and ciprofloxacin (81%) has been observed in blaOXA23-like, and the highest resistance and sensitivity were observed for ciprofloxacin (57%) and ampicillin/sulbactam (28%) in blaOXA-24-like positive A. baumannii isolates, respectively. The A. baumannii isolates with blaOXA-58-like genes did not show much resistance to antibiotics. Considering the presence of the studied genes, 3, 58, and 84% of the 100 samples were carrying blaOXA-58-like, blaOXA-24-like, and blaOXA-23-like genes, respectively ( Fig. 1 and Table 2 ). Among the 100 A. baumannii isolates, 97.9% of the imipenem-resistant isolates were positive for at least one OXA-type gene. Moreover, 95.9% and 1% of the meropenem and colistin sulfate resistant isolates were carrying OXA-type genes.
Fig. 1.
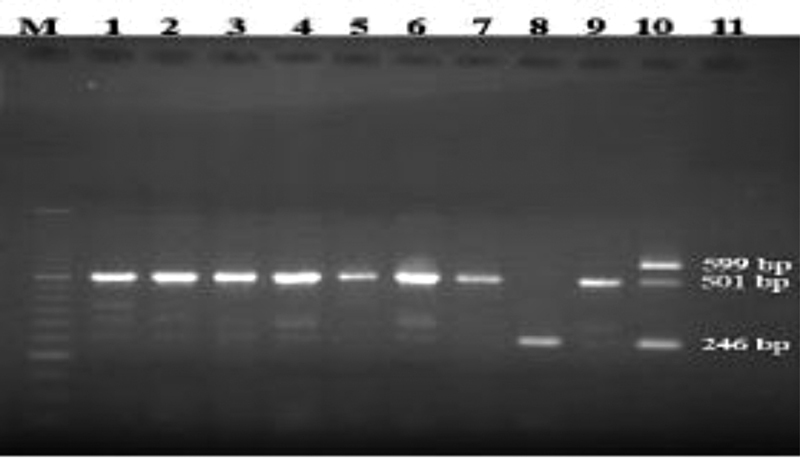
Gel electrophoresis of the blaOXA gene among the Acinetobacter baumannii isolates. M: 50 bp marker; 1–10: the isolates; 11: negative control. Bands with 501 bp, 246 bp, and 599 bp are the representatives of blaOXA-23-like, blaOXA-24-like, and blaOXA-58-like, respectively.
Table 2. OXA-type patterns among carbapenem-resistant Acinetobacter baumannii isolates .
| Gene pattern | No (%) |
|---|---|
| OXA-51 | 100 (100) |
| OXA-23 | 84 (84) |
| OXA-58 | 3 (3) |
| OXA-24 | 58 (58) |
| OXA-51/OXA-23 | 84 (84) |
| OXA-51/OXA-24 | 58 (58) |
| OXA-51/OXA-58 | 3 (3) |
| OXA-51/OXA-23/OXA-24 | 5 (5) |
| OXA-51/OXA-24/OXA-58 | 2 (2) |
| OXA-51/OXA-23/OXA-24/OXA-58 | 1 (1) |
Results of Phylogenetic Tree Analysis of OXA-type Variant
The sequences received from the isolates were confirmed as the OXA -type variants and were deposited in the GenBank under the accession numbers as follows: bla OXA-51-like : KJ451411, KJ451412, and KJ451413; bla OXA-23-like : KJ451414 and KJ451415; and bla OXA-24-like : KJ451416, KJ451417, and KJ451418 ( Fig. 2 ). Analysis of the phylogenic tree demonstrates that all isolates have shown a high degree of similarity.
Fig. 2.
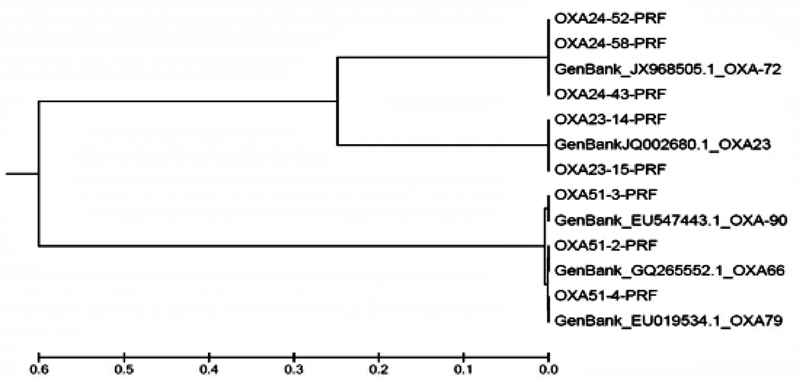
Phylogenetic tree built based on the sequence isolates by maximum likelihood method using the Tamura-Nei model. Based on the results, all isolates have shown a high degree (95–100%) of similarity.
Correlation between Contributions of bla OXA Genes with Antibiotic Resistance
The relationship between blaOXA-types and resistance to different antibiotics was determined by statistical analysis. The prevalence of oxacillinases enzymes encoding genes in antibiotic-resistant isolates was higher than the susceptible strains, but there was no statistically significant relationship ( Table 3 ).
Table 3. Statistical analysis of the correlation between the presence of bla OXA -type genes and antibiotic resistance in Acinetobacter baumannii isolates .
| Gene | OXA23 | Fisher exact test | Exact sig.(two-sided) | OXA24 | Fisher exact test | Exact sig.(two-sided) | OXA58 | Fisher exact test | Exact sig.(two-sided) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antibiotic | Positive | Negative | Positive | Negative | Positive | Negative | |||||||
| Meropenem | R | 79% | 14% | 0.671 | 0.692 | 56% | 37% | 1.855 | 0.374 | 3% | 90% | 1.608 | 1 |
| I | 2% | 0 | 2% | 0% | 0% | 2% | |||||||
| S | 4% | 1% | 2% | 3% | 0% | 5% | |||||||
| Imipenem | R | 73% | 12% | 2.37 | 0.429 | 52% | 33% | 2.066 | 0.351 | 2% | 83% | 2.799 | 0.389 |
| I | 1% | 1% | 2% | 0% | 0% | 2% | |||||||
| S | 11% | 2% | 6% | 7% | 1% | 12% | |||||||
| Amikacin | R | 72% | 12% | 1.422 | 0.443 | 51% | 33% | 1.099 | 0.703 | 2% | 82% | 3.212 | 0.217 |
| I | 8% | 1% | 6% | 3% | 0% | 9% | |||||||
| S | 5% | 2% | 3% | 4% | 1% | 6% | |||||||
| Gentamicin | R | 74% | 13% | 2.33 | 0.324 | 54% | 23% | 3.58 | 0.116 | 3% | 84% | 1.29 | 1 |
| I | 1% | 1% | 2% | 0% | 0% | 2% | |||||||
| S | 10% | 1% | 4% | 7% | 1% | 11% | |||||||
| Ciprofloxacin | R | 81% | 14% | 1.37 | 0.549 | 57% | 38% | 1.75 | 0.580 | 3% | 92% | 2.374 | 1 |
| I | 1% | 0% | 0% | 1% | 0% | 1% | |||||||
| S | 4% | 1% | 3% | 1% | 0% | 4% | |||||||
| Doxycycline | R | 24% | 5% | 2.45 | 0.320 | 18% | 11% | 0.287 | 0.902 | 1% | 28% | 2.092 | 0.387 |
| I | 34% | 3% | 21% | 16% | 0% | 37% | |||||||
| S | 27% | 7% | 21% | 13% | 2% | 32% | |||||||
| Piperacillin | R | 83% | 15% | 1.117 | 1 | 59% | 39% | 2.033 | 0.642 | 3% | 95% | 3.541 | 1 |
| I | 1% | 0% | 1% | 0% | 0% | 1% | |||||||
| S | 1% | 0% | 0% | 1% | 0% | 1% | |||||||
| Tobramycin | R | 65% | 11% | 1.386 | 0.575 | 47% | 29% | 0.824 | 0.676 | 3% | 73% | 1.19 | 1 |
| I | 2% | 1% | 2% | 1% | 0% | 3% | |||||||
| S | 18% | 3% | 11% | 10% | 0% | 21% | |||||||
| Cefepime | R | 79% | 13% | 2.089 | 0.443 | 55% | 37% | 0.501 | 1 | 3% | 89% | 1.50 | 1 |
| I | 4% | 2% | 4% | 2% | 0% | 6% | |||||||
| S | 2% | 0% | 1% | 1% | 0% | 2% | |||||||
| Amp/sul | R | 30% | 3% | 2.94 | 0.243 | 21% | 12% | 1.20 | 0.561 | 0% | 33% | 3.098 | 0.155 |
| I | 20% | 2% | 11% | 11% | 2% | 20% | |||||||
| S | 25% | 10% | 28% | 17% | 1% | 44% | |||||||
Abbreviations: Amp/sul, ampicillin/sulbactam; I, intermediate; R, resistant; S, sensitive; sig., significance.
Discussion
Currently, carbapenem resistance in A. baumannii isolates has become a worldwide problem. This study's results demonstrated that significant numbers of clinical isolates of A. baumannii , especially resistant to imipenem and meropenem isolates, were affirmative for the variety of OXA gene expressions. Commonly, the carbapenem antibiotics are highly active against A. baumannii isolates and resistant to β-lactamase enzymes. However, the resistance to these compounds is a problematic issue and is not a result of the presence of a distinct mechanism, while a combination of diverse mechanisms such as enzymatic and nonenzymatic ones are involved. The most significant mechanism of resistance to carbapenems is the enzymatic hydrolysis, arbitrated by the carbapenemases. 11 20 Recently, the number of β-lactamase enzymes in class D has increased significantly. Some of the OXA -type carbapenemases are widely found in A. baumannii isolates. However, many of the OXA -type carbapenemases illustrate simply weak catalytic activity, but resistance to carbapenems may be caused by a combined action of different OXA -type carbapenemase. 21 Feizabadi et al in 2008 studied the susceptibility to antimicrobial and blaOXA genes distribution among Acinetobacter spp. in Tehran hospitals. They reported the coexistence of bla OXA-51/23 and bla OXA-51/24-like in 25% ( n = 32) and 17.9% ( n = 23) of their studied isolates, respectively. Over 70% of their studied carbapenem-resistant isolates had at least two genes encoding OXA-type carbapenemase. 22 In our study, the highest coexistence was for bla OXA-51/23 (84%) followed by bla OXA-51/24-like (58%), which is considerably higher than their report from Tehran and indicates an elevation in the prevalence of A. baumannii isolates carrying these genes over the time. In the current research, comparable to another research performed in Iran, the most commonly found OXA-type carbapenemase was OXA-23 among other carbapenem-resistant isolates, next to OXA-24. 23 24 Zafari et al reported a high prevalence of OXA-type carbapenemases among A. baumannii isolates in Tehran in 2017. They studied bla OXA-51-like , bla OXA - 58-like , bla OXA-23-like , and bla OXA-24-like genes among 100 isolates of carbapenem-resistant A. baumannii . All of their studied carbapenem-resistant A. baumannii isolates were MDR and extensively drug-resistant and even 2% of the isolates were PDR. The bla OXA-24-like and bla OXA-23-like prevalence genes were reported to be 22% and 81%, respectively. They concluded that there are very few therapeutic options for the treatment of carbapenem-resistant A. baumannii infections. 23 In the present study, just similar to the findings of Zafari et al in Tehran, 84, 58, and 3% of the A. baumannii isolates from Hamadan, west of Iran, were carrying bla OXA-23-like , bla OXA-24-like , and bla OXA-58-like genes, respectively. The very high prevalence of drug resistance among A. baumannii isolates was reported in Hamadan in 2013. 24 Bardbari et al investigated the OXA -type gene presence including bla OXA-58 and bla OXA-23 , blaOXA -24 among 75 clinical and 32 environmental strains of the A. baumannii isolates in Hamadan in 2017. Their results showed that 80.4, 30.8, 0, and 30.8% of their studied A. baumannii isolates have bla OXA-23/24 , blaOXA -24 , bla OXA-23 , and bla OXA-58 genes. 25 Their results are similar to the finding of the present study. Given the fact that both studies are conducted in the same area but on different isolates, the similar results can be explained. In a study that was prepared by Zowawi et al, OXA-23 was identified as the highest type (91.5%) of carbapenemases in carbapenem-resistant A. baumannii isolates from hospitals of Bahrain, Saudi Arabia, Oman, Kuwait, United Arab Emirates, and Qatar followed by OXA-24 (4.3%). 26 Also, research works from other countries have reported that OXA-23 as a dominant type in carbapenem-resistant isolates has been accompanying outbreaks. 27 28 Similar to our results, some studies have reported the low prevalence of OXA-58 among carbapenem-resistant isolates from the east (0.5%), and northwest of Iran (3.2%). 26 29 The outcome of our research confirmed the high prevalence of OXA-type carbapenemase between carbapenem antibiotic-resistant A. baumannii bacteria; furthermore, correct prescription of antibiotics and effective infection control monitors for preventing the extent of resistant isolates are required.
Conclusion
This study's results demonstrated that carbapenem resistance is increasing among A. baumannii clinical isolates in our area and is often associated with multidrug resistance. Moreover, the diversity of bla OXA genes was high among these isolates and OXA-23 is the most important carbapenemase mechanism responsible for the resistance phenotype.
Acknowledgment
The authors would like to acknowledge the Vice-Chancellor of Research and Technology, Hamadan University of Medical Sciences, Hamadan, Iran, for the financial support to the present study.
Funding Statement
Funding The present study was supported financially by the Vice-Chancellor of Research and Technology, Hamadan University of Medical Sciences, Hamadan, Iran.
Conflict of Interest None declared.
Consent for Publication
All authors consented for publication of the above paper.
Availability of Data and Material
All data generated or analyzed during this study are included in this published article.
Authors' Contributions
M.Y.A. designed the study, edited the manuscript. M.S. and A.B. were scientific advisor on molecular genetics section. M.K., M.S., and M.S.A. collaborated in collecting samples and doing experiments. S.H.H. collaborated on patient introduction and clinical examination. A.K. wrote the scientific draft and checked the English use and grammar of the manuscript.
References
- 1.Perez F, Hujer A M, Hujer K M, Decker B K, Rather P N, Bonomo R A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2007;51(10):3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeom J, Shin J H, Yang J Y, Kim J, Hwang G S. (1)H NMR-based metabolite profiling of planktonic and biofilm cells in Acinetobacter baumannii 1656-2. PLoS One. 2013;8(03):e57730. doi: 10.1371/journal.pone.0057730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Bonnin R A, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life. 2011;63(12):1061–1067. doi: 10.1002/iub.532. [DOI] [PubMed] [Google Scholar]
- 4.Gulati S, Kapil A, Das B, Dwivedi S N, Mahapatra A K. Nosocomial infections due to Acinetobacter baumannii in a neurosurgery ICU. Neurol India. 2001;49(02):134–137. [PubMed] [Google Scholar]
- 5.Rodríguez Guardado A, Maradona J A, Asensi V. [Postsurgical meningitis caused by Acinetobacter baumannii: study of 22 cases and review of the literature] Rev Clin Esp. 2001;201(09):497–500. doi: 10.1016/s0014-2565(01)70895-8. [DOI] [PubMed] [Google Scholar]
- 6.Wang K W, Chang W N, Huang C R. Post-neurosurgical nosocomial bacterial meningitis in adults: microbiology, clinical features, and outcomes. J Clin Neurosci. 2005;12(06):647–650. doi: 10.1016/j.jocn.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Krol V, Hamid N S, Cunha B A. Neurosurgically related nosocomial Acinetobacter baumannii meningitis: report of two cases and literature review. J Hosp Infect. 2009;71(02):176–180. doi: 10.1016/j.jhin.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 8.Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54(01):24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahcheraghi F, Aslani M M, Mahmoudi H. Molecular study of carbapenemase genes in clinical isolates of Enterobacteriaceae resistant to carbapenems and determining their clonal relationship using pulsed-field gel electrophoresis. J Med Microbiol. 2017;66(05):570–576. doi: 10.1099/jmm.0.000467. [DOI] [PubMed] [Google Scholar]
- 10.Bertini A, Poirel L, Mugnier P D, Villa L, Nordmann P, Carattoli A. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(10):4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opazo A, Domínguez M, Bello H, Amyes S G, González-Rocha G. OXA-type carbapenemases in Acinetobacter baumannii in South America. J Infect Dev Ctries. 2012;6(04):311–316. doi: 10.3855/jidc.2310. [DOI] [PubMed] [Google Scholar]
- 12.Brown S, Young H K, Amyes S G. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin Microbiol Infect. 2005;11(01):15–23. doi: 10.1111/j.1469-0691.2004.01016.x. [DOI] [PubMed] [Google Scholar]
- 13.Higgins P G, Poirel L, Lehmann M, Nordmann P, Seifert H. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(12):5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard A, O'Donoghue M, Feeney A, Sleator R D. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence. 2012;3(03):243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Safari M, Mozaffari Nejad A S, Bahador A, Jafari R, Alikhani M Y. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU) Saudi J Biol Sci. 2015;22(04):424–429. doi: 10.1016/j.sjbs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turton J F, Woodford N, Glover J, Yarde S, Kaufmann M E, Pitt T L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(08):2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CLSI . 28th edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. [Google Scholar]
- 18.Woodford N, Ellington M J, Coelho J M. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27(04):351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(03):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 20.Queenan A M, Bush K.Carbapenemases: the versatile beta-lactamases Clin Microbiol Rev 20072003440–458.table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walther-Rasmussen J, Høiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57(03):373–383. doi: 10.1093/jac/dki482. [DOI] [PubMed] [Google Scholar]
- 22.Feizabadi M M, Fathollahzadeh B, Taherikalani M. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61(04):274–278. [PubMed] [Google Scholar]
- 23.Zafari M, Feizabadi M M, Jafari S, Abdollahi A, Sabokbar A. High prevalence of OXA-type carbapenemases among Acinetobacter baumannii strains in a teaching hospital of Tehran. Acta Microbiol Immunol Hung. 2017;64(04):385–394. doi: 10.1556/030.64.2017.031. [DOI] [PubMed] [Google Scholar]
- 24.Safari M, Saidijam M, Bahador A, Jafari R, Alikhani M Y. High prevalence of multidrug resistance and metallo-beta-lactamase (MβL) producing Acinetobacter baumannii isolated from patients in ICU wards, Hamadan, Iran. J Res Health Sci. 2013;13(02):162–167. [PubMed] [Google Scholar]
- 25.Bardbari A M, Arabestani M R, Karami M. Highly synergistic activity of melittin with imipenem and colistin in biofilm inhibition against multidrug-resistant strong biofilm producer strains of Acinetobacter baumannii. Eur J Clin Microbiol Infect Dis. 2018;37(03):443–454. doi: 10.1007/s10096-018-3189-7. [DOI] [PubMed] [Google Scholar]
- 26.Zowawi H M, Sartor A L, Sidjabat H E, Balkhy H H, Walsh T R, Al Johani S M. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf Cooperation Council States: dominance of OXA-23-type producers. J Clin Microbiol. 2015;53:896–903. doi: 10.1128/JCM.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino M, Poza M, Roca I. Nosocomial outbreak of a multiresistant Acinetobacter baumannii expressing OXA-23 carbapenemase in Spain. Microb Drug Resist. 2014;20(04):259–263. doi: 10.1089/mdr.2013.0127. [DOI] [PubMed] [Google Scholar]
- 28.Dettori M, Piana A, Deriu M G. Outbreak of multidrug-resistant Acinetobacter baumannii in an intensive care unit. New Microbiol. 2014;37(02):185–191. [PubMed] [Google Scholar]
- 29.Kooti S, Motamedifar M, Sarvari J. Antibiotic resistance profile and distribution of oxacillinase genes among clinical isolates of Acinetobacter baumannii in Shiraz teaching hospitals, 2012–2013. Jundishapur J Microbiol. 2015;8(08):e20215. doi: 10.5812/jjm.20215v2. [DOI] [PMC free article] [PubMed] [Google Scholar]


