Abstract
Objective: Hedyotis diffusa-Scutellaria barbata herb pair (HS) has therapeutic effects on a variety of cancers, and this study aims to systematically explore the multiple mechanisms of HS in the treatment of colorectal cancer (CRC). Methods. The active ingredients of HS were obtained from TCMSP, and the potential targets related to these ingredients were screened from the STITCH, SuperPred, and Swiss TargetPrediction databases. Targets associated with CRC were retrieved by Drugbank, TTD, DisGeNET, and GeneCards. We used a Venn diagram to screen the intersection targets and used Cytoscape to construct the herb-ingredient-target-disease network, and the core targets were selected. The Go analysis and KEGG pathway annotation were performed by R language software. We used PyMol and Autodock Vina to achieve molecular docking of core ingredients and targets. Results: A total of 33 active ingredients were obtained from the HS, and 762 CRC-related targets were reserved from the four databases. We got 170 intersection targets to construct the network and found that the four ingredients with the most targets were quercetin, luteolin, baicalein, and dinatin, which were the core ingredients. The PPI analysis showed that the core targets were STAT3, TP53, MAPK3, AKT1, JUN, EGFR, MYC, VEGFA, EGF, and CTNNB1. Molecular docking results showed that these core ingredients had good binding potential with core targets, especially the docking of each component with MAPK obtained the lowest binding energy. HS acts simultaneously on various signaling pathways related to CRC, including the PI3K-Akt signaling pathway, proteoglycans in cancer, and the MAPK signaling pathway. Conclusions: This study systematically analyzed the active ingredients, core targets, and central mechanisms of HS in the treatment of CRC. It reveals the role of HS targeting PI3K-Akt signaling and MAPK signaling pathways in the treatment of CRC. We hope that our research could bring a new perspective to the therapy of CRC and find new anticancer drugs.
1. Introduction
Colorectal cancer (CRC) is a common digestive tract tumor, which has a high incidence rate, a high mortality rate, rapid progress, and easy spread [1]. The incidence rate of CRC is increasing year by year, the age of onset is getting younger and younger, and the death caused by recurrence and metastasis is still a considerable challenge [2]. Modern pharmacological studies have shown that traditional Chinese medicine (TCM) is one of the comprehensive treatments for CRC in addition to surgery, chemotherapy, radiotherapy, immunotherapy, and targeted therapy, which can effectively inhibit the proliferation of cancer cells and improve the quality of life [3–5]. Compared with side effects of chemotherapy, such as myelosuppression, gastrointestinal reaction, liver function damage, and peripheral neuritis, TCM has the characteristics of long-term administration, fewer side effects, and less drug resistance [6, 7].
TCM believes that cancer toxin is the core of tumor pathogenesis. After cancer toxin is produced, it can induce pathological products such as phlegm turbidity, qi stagnation, and blood stasis [8]. Blood stasis is closely related to tumor and runs through the whole process of swelling. Therefore, clearing heat and detoxification, promoting blood circulation, and removing blood stasis play an essential role in tumor treatment [9]. Hedyotis diffusa belongs to the genus Hedyotis of Rubiaceae, and it is bitter, light, and cold in nature [10]. Scutellaria barbata, belonging to Labiatae, is sour in taste and cold in nature [11]. They have the functions of clearing away heat and detoxification, promoting blood circulation and removing blood stasis, anti-inflammatory and analgesic effects, and can be used as adjuvant treatment for colorectal cancer, breast cancer, bladder cancer, lung cancer, liver cancer, gastric cancer, ovarian cancer, and other malignant tumors [12, 13]. Their pharmacological effects include antitumor, anti-inflammatory, antioxidation, antiangiogenic, promoting cell apoptosis, and improving immune capacity [14, 15]. Hedyotis diffusa and Scutellaria barbata herb pair (HS) are widely used to treat cancer, and their combined effects are more than a single use of drugs. Studies have shown that HS can inhibit the proliferation of human breast cancer cells, and the combination of CTX treatment can significantly inhibit breast cancer model mice [16, 17]. In addition, it could also significantly inhibit the growth of H22 hepatoma xenografts in mice [18]. The mixture of ethanol extracts from HS can dramatically inhibit the growth of human colon cancer cell lines compared with the single drug alcohol extract, which indicates that the anticancer effect of the combination of the two drugs will be enhanced. However, these studies focus on a single target or a single pathway, which could not comprehensively and systematically explain the antitumor effect of HS [19].
Network pharmacology emphasizes the combination of bioinformatics, system biology, and pharmacology, which not only explains the interaction between TCM and diseases but also conforms to the systematic and holistic view of TCM [20, 21]. Network pharmacology updates the “one target, one drug” model to the “multicomponent, multitarget” model and clarifies the complex interactions between genes, proteins, and metabolites related to diseases and drugs from the perspective of the network, which provides the possibility for us to systematically study the relationship between TCM and diseases [22, 23]. In this study, network pharmacology was used to analyze the active ingredients, potential targets, and main mechanisms of HS in the treatment of CRC, and to construct the herb-ingredient-target-disease network, so as to provide a reference for the study of the specific mechanism of the drug in the treatment of CRC. The flowchart of our analysis is shown in Figure 1.
Figure 1.
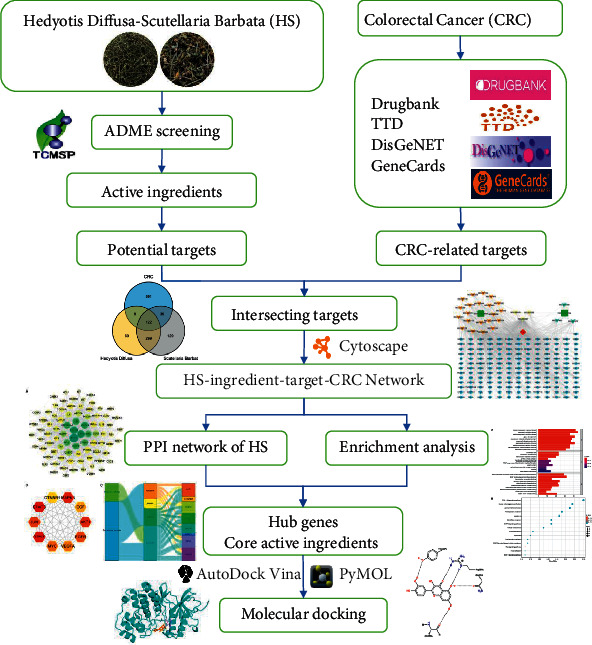
Flowchart of the study.
2. Materials and Methods
2.1. Collection of Active Ingredients
With the common name of a single drug as a keyword, all chemical ingredients of the drug were retrieved by TCMSP [24] (https://tcmspw.com/tcmsp.php). This study used oral bioavailability (OB) and ingredient drug-likeness (DL) as screening conditions for active ingredients.
2.2. Prediction of Potential Targets of HS
We used PubChem [25] (https://pubchem.ncbi.nlm.nih.gov) to search and export the chemical structure data of active ingredients. Since the targets of ingredients without accurate structural information could not be predicted successfully, we decided to remove these ingredients after deleting the duplicate data. The active ingredients were predicted through the STITCH [26] (http://stitch.embl.de/), SuperPred [27] (http://prediction.charite.de/), and Swiss TargetPrediction [28] (http://www.swisstargetprediction.ch/) databases to obtain the corresponding known or predicted targets. The duplicate data had been eliminated, and only the human targets were retained.
The selected active ingredients were imported into the STITCH database for putative target prediction, and the targets with a confidence ≥0.7 were assumed as potential targets. The potential targets of drugs can also be obtained by inputting SMILES into the SuperPred database. The Canonical SMILES of the main active ingredients were uploaded to the Swiss TargetPrediction database, and the probability of each potential target was determined to be greater than or equal to 0.1. The retrieved targets were converted into standardized abbreviations by UniProt [29] (https://www.uniprot.org).
2.3. Collection of CRC-Related Targets
Targets associated with CRC were collected from Drugbank [30] (https://www.drugbank.ca), TTD [31] (http://db.idrblab.net/ttd/), DisGeNET [32] (http://www.disgenet.org/), and GeneCards [33] (https://www.genecards.org). Then the retrieval results of these databases were merged, and only one repeated target was reserved.
2.4. Intersection Targets and Network Construction
The targets of HS and CRC were intersected by R language version 4.1.1 and the common gene was identified as the intersection target of HS and CRC. The herbs, active ingredients, intersection targets, and diseases were introduced into Cytoscape 3.8.0 [34] to construct the herb-ingredient-target-disease network.
2.5. Protein-Protein Interaction (PPI) Analysis
The intersection targets related to HS and CRC were input into the STRING [35] (https://string-db.org/) platform for retrieval. Protein interaction data with high confidence (score >0.7) were selected and saved in a TSV format file. The information of node1, node2, and combined score in the file was imported into the software of Cytoscape to construct a PPI network, and the hub gene was screened by the cytohubba plug-in. The Sankey diagram revealed the relationship between herbs, core ingredients, and hub genes was drawn in R language version 4.1.1.
2.6. Enrichment Analysis
We applied the “clusterProfiler” package to these overlapping genes in order to perform GO enrichment analysis and KEGG analysis [36]. Both the bar plot and dot plot were drawn using the R language version 4.1.1.
2.7. Molecular Docking
We downloaded the SDF format 3D structure file of key ingredients from the PubChem database and converted it to PDB format through Open Babel. We also downloaded the 3D crystal structure of the hub genes through the PDB database (https://www.pdbus.org/), removed ions and water molecules through PyMol 2.4.0 [37], and saved it as a PDB file. Then we repaired the protein structure through the WHAT IF server website (https://swift.cmbi.umcn.nl/servers/html/model.html) and prepared the receptor file in Autodock Vina. Then, the molecular docking simulation was carried out by using Autodock Vina software and visualized by PyMol 2.4.0. The 3D and 2D diagrams of molecular docking models were displayed by PyMol 2.4.0 and PROTEINS PLUS (https://proteins.plus/), respectively.
3. Results
3.1. Screening of Active Ingredients
37 ingredients were found in Hedyotis diffusa and 94 in Scutellaria barbata from TCMSP database. OB ≥ 30% and DL ≥ 0.18 were selected as the screening conditions. There were 7 ingredients from Hedyotis diffusa and 29 from Scutellaria barbata. A total of 33 ingredients were screened. Among them, 3 ingredients were common ingredients of the two herbs, namely quercetin (MOL000098), beta-sitosterol (MOL000358), and stigmasterol (MOL000449). The basic information of active ingredients in HS is shown in Table 1.
Table 1.
Basic information of 33 active ingredients in HS.
| Mol ID | Molecule name | OB (%) | DL | Targets | Herb |
|---|---|---|---|---|---|
| MOL000098 | Quercetin | 46.43 | 0.28 | 369 | H/S |
| MOL000358 | Beta-sitosterol | 36.91 | 0.75 | 49 | H/S |
| MOL000449 | Stigmasterol | 43.83 | 0.76 | 42 | H/S |
| MOL001646 | 2,3-Dimethoxy-6-methyanthraquinone | 34.86 | 0.26 | 0 | H |
| MOL001659 | poriferasterol | 43.83 | 0.76 | 42 | H |
| MOL001663 | (4aS,6aR,6aS,6bR,8aR,10R,12aR,14bS)-10-Hydroxy-2,2,6a,6 b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid | 32.03 | 0.76 | 84 | H |
| MOL001670 | 2-Methoxy-3-methyl-9,10-anthraquinone | 37.83 | 0.21 | 100 | H |
| MOL000006 | Luteolin | 36.16 | 0.25 | 216 | S |
| MOL000173 | Wogonin | 30.68 | 0.23 | 53 | S |
| MOL000351 | Rhamnazin | 47.14 | 0.34 | 100 | S |
| MOL000359 | Sitosterol | 36.91 | 0.75 | 49 | S |
| MOL000953 | CLR | 37.87 | 0.68 | 0 | S |
| MOL001040 | (2R)-5,7-Dihydroxy-2-(4-hydroxyphenyl) chroman-4-one | 42.36 | 0.21 | 83 | S |
| MOL001735 | Dinatin | 30.97 | 0.27 | 120 | S |
| MOL001755 | 24-Ethylcholest-4-en-3-one | 36.08 | 0.76 | 46 | S |
| MOL001973 | Sitosteryl acetate | 40.39 | 0.85 | 23 | S |
| MOL002714 | Baicalein | 33.52 | 0.21 | 180 | S |
| MOL002719 | 6-Hydroxynaringenin | 33.23 | 0.24 | 15 | S |
| MOL002776 | Baicalin | 40.12 | 0.75 | 51 | S |
| MOL002915 | Salvigenin | 49.07 | 0.33 | 100 | S |
| MOL005190 | Eriodictyol | 71.79 | 0.24 | 22 | S |
| MOL005869 | Daucostero_qt | 36.91 | 0.75 | 0 | S |
| MOL008206 | Moslosooflavone | 44.09 | 0.25 | 100 | S |
| MOL012245 | 5,7,4′-Trihydroxy-6-methoxyflavanone | 36.63 | 0.27 | 70 | S |
| MOL012246 | 5,7,4′-Trihydroxy-8-methoxyflavanone | 74.24 | 0.26 | 88 | S |
| MOL012248 | 5-Hydroxy-7,8-dimethoxy-2-(4-methoxyphenyl) chromone | 65.82 | 0.33 | 0 | S |
| MOL012250 | 7-Hydroxy-5,8-dimethoxy-2-phenyl-chromone | 43.72 | 0.25 | 0 | S |
| MOL012251 | Chrysin-5-methylether | 37.27 | 0.2 | 100 | S |
| MOL012252 | 9,19-Cyclolanost-24-en-3-ol | 38.69 | 0.78 | 26 | S |
| MOL012254 | Campesterol | 37.58 | 0.71 | 15 | S |
| MOL012266 | Rivularin | 37.94 | 0.37 | 11 | S |
| MOL012269 | Stigmasta-5,22-dien-3-ol-acetate | 46.44 | 0.86 | 26 | S |
| MOL012270 | Stigmastan-3,5,22-triene | 45.03 | 0.71 | 0 | S |
3.2. Potential Targets of HS
The structures of these active ingredients were obtained in PubChem. We removed six ingredients named MOL001646, MOL000953, MOL005869, MOL012248, MOL012250, and MOL012270, which had no accurate structural information for predicting the target. The structures of the active ingredients were imported into STITCH, SuperPred, and Swiss Target Prediction databases for target prediction. A total of 490 potential targets of Hedyotis diffusa and 589 targets of Scutellaria barbata were obtained.
3.3. Collection of CRC-Related Targets
77, 104, 353, and 390 CRC targets were obtained from four databases, including Drugbank, TTD, DisGeNET, and GeneCards, respectively, shown in the Venn diagram (Figure 2(a)). A total of 762 CRC-related targets were reserved from the four databases.
Figure 2.

(a) Venn diagram showing the CRC-related targets among the four databases. (b) Venn diagram showing the intersecting targets.
3.4. Construction of the Herb-Ingredient-Target-Disease Network
We intersected the potential targets of ingredients and CRC-related targets, and a total of 170 intersecting targets were obtained (Figure 2(b)), among which 131 genes were shared by Hedyotis diffusa and CRC, and 161 genes were shared by Scutellaria barbata and CRC. Then, the two herbs, 27 active ingredients, 170 intersection targets, and CRC disease were imported into Cytoscape 3.8.0 to construct the herb-ingredient-target-disease network (Figure 3). In the network, the more edges a node connects with other nodes, the higher its degree value. The node with a high degree value may be the key node of the network and play a pivotal role in the network. Quercetin had the most significant number of targets, with 222 potential targets, followed by luteolin, baicalein, and dinatin, with 74, 63, and 40 potential targets, respectively. These active ingredients with more targets may be the core ingredients of HS.
Figure 3.

The herb-ingredient-target-disease network of HS against CRC. The green square represents the herb, the inverted triangles of different colors represent the ingredients of different herbs, the blue circle represents the target, the red diamond represents the disease.
3.5. PPI Network Diagram of Intersecting Targets
The 170 intersecting targets obtained above were imported into the STRING 11.0 database for analysis, and the PPI network diagram of intersection targets between HS and CRC was obtained. There were 166 nodes and 1928 edges in the network, and the characteristics of the specific network topology were calculated. The nodes represent the intersection targets, and the edges represent the association between the intersection targets. We used Cytoscape 3.8.0 to show the PPI network diagram and hide the nodes whose degree value is less than 18.5 (median degree value) (Figure 4(a)). The cytohubba plug-in was used to calculate the targets set to screen the core targets further. The top 10 hub genes were STAT3, TP53, MAPK3, AKT1, JUN, EGFR, MYC, VEGFA, EGF, and CTNNB1 (Figure 4(b)). Furthermore, we revealed the relationship between two herbs, four core ingredients, and ten hub genes using Sankey diagram (Figure 4(c)).
Figure 4.

(a) The PPI network of the intersecting targets. (b) The 10 hub genes obtained from the PPI network. (c) The Sankey diagram revealed the relationship between herbs, core ingredients, and hub genes.
3.6. Go and KEGG Analysis of Intersecting Targets
Based on GO enrichment and KEGG analysis, we determined how these intersecting targets function biologically. As a result, the bar plot of the top 10 terms of biological process (BP), cellular component (CC), and molecular function (MF) terms were displayed (Figure 5(a)). In the three categories, changes in the BP of targets were enriched in cellular response to chemical stress and response to oxidative stress; changes in MF were mainly enriched in transcription factor binding, transcription coregulator binding, and ubiquitin-like protein ligase binding, while CC were mainly enriched in membrane raft, membrane microdomain, and transcription regulator complex. Through enrichment and screening of the KEGG pathway, 168 signaling pathways were obtained. Combined with literature research, we screened out the pathways directly related to CRC and showed the top 15 pathways in the dot plot according to the count value. Among them, the PI3K-Akt signaling pathway, proteoglycans in cancer, and MAPK signaling pathway are closely related to CRC, which may be the critical pathways of HS in the treatment of CRC (Figure 5(b)).
Figure 5.

(a) GO enrichment analysis results. (b) KEGG pathway enrichment analysis results.
3.7. Molecular Docking
In this study, we conducted molecular docking of 10 hub genes (STAT3, TP53, MAPK3, AKT1, JUN, EGFR, MYC, VEGFA, EGF, and CTNNB1) with four core ingredients (quercetin, luteolin, baicalein, and dinatin) to evaluate the protein-ligand binding potential. The results showed that the four ingredients had the best docking effect with MAPK, and the binding energies were -8.2 kcal/mol, -8.3 kcal/mol, -8.6 kcal/mol, and -8.0 kcal/mol, respectively (Figure 6).
Figure 6.

Heatmap of the molecular docking efficiency.
Quercetin and luteolin also achieved quite good docking results with TP53, and the docking results were -8.0 kcal/mol and -8.0 kcal/mol, respectively. The 3D diagrams of molecular docking models were displayed by PyMol 2.4.0, which showed the interaction of MAPK with quercetin, luteolin, baicalein, and dinatin (Figure 7). At the same time, the 2D diagrams showed the details of the interaction by using Proteins Plus (Figure 8).
Figure 7.
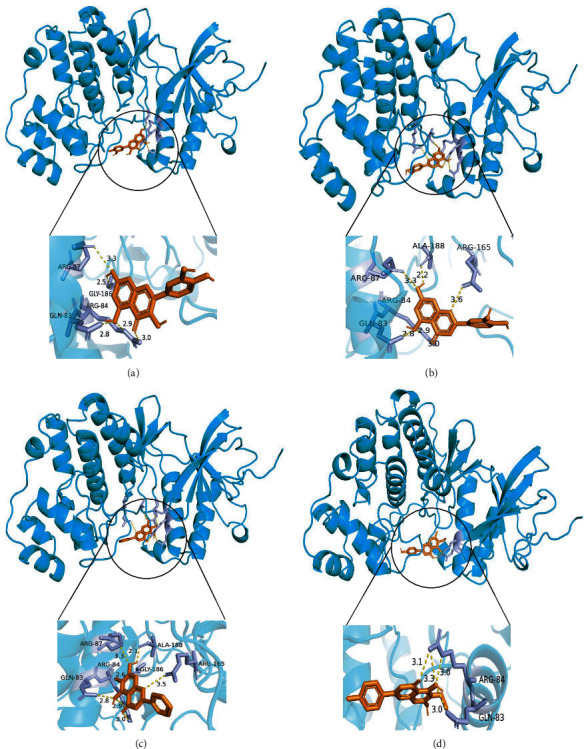
3D diagram of molecular docking models, MAPK3 binds to quercetin (a), luteolin (b), baicalein (c), and dinatin (d).
Figure 8.

2D diagram of molecular docking models shows the details of the interaction, MAPK3 binds to quercetin (a), luteolin (b), baicalein (c), and dinatin (d).
4. Discussion
Both Hedyotis diffusa and Scutellaria barbata are heat-clearing and detoxicating TCMs. Hedyotis diffusa can also activate the blood circulation and relieve pain, while Scutellaria barbata can remove blood stasis and diuresis [38]. They could play a coordinated and synergistic role in treating tumors, and the combined use of the two drugs is not simply superimposed, and their combined application is more effective than a single drug [39]. The antitumor mechanism of Hedyotis diffusa is mainly achieved by enhancing immune function, interfering with the energy metabolism of tumor cells, inducing tumor cell apoptosis, and influencing the mitochondrial pathway [40, 41]. However, the antitumor mechanisms of Scutellaria barbata include inhibiting tumor cell growth, inducing tumor cell apoptosis, inhibiting tumor angiogenesis and metastasis, regulating immunity, and reversing drug resistance of tumor cells [42, 43].
In other words, some of their functions are the same, especially in inducing apoptosis, enhancing cell immunity, and reducing telomerase activity [40]. They also have their own unique functions, which makes them mutually beneficial to play a more significant role. Some studies have shown that in the breast cancer model mice, the HS group can significantly increase the tumor inhibition rate, serum INF–γ and IL-2 levels, and decrease serum TNF-α level [16]. It is reported that when the sample concentration was in the medium concentration range (0.5–1.2 mg/ml), HS had a strong chelating ability to Fe2+, followed by Hedyotis diffusa, and the extract of Scutellaria barbata was relatively weak. It is suggested that the herb pair may inhibit or resist tumor formation by chelating the transition metal ions and blocking the lipid peroxidation chain reaction. In addition, HS can provide the most substantial DNA protection, which may be why it works [44].
In this study, the network pharmacology method was used to analyze the active ingredients, targets, and potential mechanisms of HS in CRC treatment. Through the construction of an herb-ingredient-target-disease network and a PPI network of intersecting targets, the mechanism of action of HS on CRC was systematically analyzed.
In the herb-ingredient-target-disease network, quercetin, luteolin, baicalein, and dinatin have more targets than other ingredients, which may be the core active ingredients of HS. Quercetin is one of the main ingredients of Hedyotis diffusa, and the other three are the main ingredients of Scutellaria barbata. It has been reported that quercetin can not only directly inhibit the proliferation of tumor cells [45, 46], but also plays an antitumor effect by antioxidating [47] and activating antitumor immunity [48] and inhibiting EMT [49, 50]. Luteolin is a natural flavonoid, which can increase ceramide, leading to the apoptosis and death of CRC cells and inhibit the synthesis and metabolism of ceramide complex sphingolipid [51, 52]. In addition, it can inhibit the production of MMP-9 and MMP-2 and up-regulate the expression of TIMP-2, thus playing an antimetastasis role [53–55]. Baicalein is considered to possess antitumor activity, which can effectively disrupt the proliferation, migration, and invasion of CRC cells and decrease the expression of epithelial-mesenchymal transition promoting factors including vimentin, Twist1, and Snail [56, 57]. It also inhibits the proliferation and invasion of human CRC cell lines by reducing the expression of MMP-2 and MMP-9 via regulation of the AKT signaling pathway [58, 59]. Dinatin is also known as hispidulin, and it can induce ROS-mediated apoptosis of human non-small cell lung cancer cells by activating the ER stress pathway and ER stress-induced apoptosis of human liver cancer cells by activating the AMPK/mTOR signaling pathway [60–62]. Moreover, hispidulin treatment significantly inhibited the activity of sphingosine kinase one and consequently promoted ceramide accumulation, thus leading to apoptosis of renal cell carcinoma [63].
In the PPI network,we identified ten hub genes, namely, STAT3, TP53, MAPK3, AKT1, JUN, EGFR, MYC, VEGFA, EGF, and CTNNB1, which may be the targets of HS in the treatment of CRC. The activation of STAT3 could be observed in various tumors, which was closely related to inflammation and immunity and promoted tumor progression as an oncogene [64]. The expression of pSTAT3 increased significantly in tumor-associated fibroblasts and activated angiogenesis-related transcription factors to promote the advancement of CRC [65]. The missense mutation of TP53 (mutp53) was common in CRC, and it was estimated that this mutation existed in more than half of the CRC. Mutp53 limited the binding of SHP and STAT3 and derived cancer growth and invasion by activating STAT3 [66]. MAPK3, also known as ERK1, is a serine/threonine kinase. It was found that the levels of phosphorylated STAT3 and ERK1/2 decreased significantly in 8-gingerol treated CRC cells, resulting in the reduced expression of the downstream target gene c-Myc [67]. The expression of VEGF was regulated by Gab2 and stimulated its downstream genes ERK1/2 and c-Myc in CRC cells [68]. EGFR is a receptor tyrosine kinase, which can act as a regulator of tumor immune monitoring, activate the JAK/STAT3 signaling pathway, and promote the expression of PD-L1 [69]. Overexpression of EGFR and its downstream Ras/Raf/MEK/ERK signaling plays an essential role in regulating cell cycle progression, cell proliferation, and apoptosis [70, 71]. Studies have shown that after EGF treatment, pY291 Fas promoted the nuclear localization of phosphorylated EGFR and phosphorylated STAT3, the expression of cyclin D1, the activation of Akt and MAPK pathways mediated by STAT3 [72]. Transcription factor c-Jun was regulated by phosphatidic acid (a key intermediate of lipid metabolism) and can enhance the transcription of the WEE1 gene, a checkpoint regulator of the cell cycle [73]. The abnormal expression of CTNNB1 was closely related to the progression and metastasis of CRC. Studies have shown that TRAF6 inhibited EMT and CRC metastasis by driving the degradation mechanism of CTNNB1 [74]. The molecular docking results showed that these hub genes and core ingredients have good binding potential, suggesting that HS played a role in the treatment of CRC mainly through these hub genes.
In the pathway enrichment, the PI3K-Akt signaling pathway (hsa04151) has been identified as the critical target of tumor-targeted therapy, which plays a vital role in regulating the proliferation, migration, and apoptosis of tumor cells [75–77]. Phosphoinositide 3-kinase was a member of the intracellular lipid kinases and regulated cell proliferation and differentiation [78]. AKT activation drove both glycolytic metabolism of glucose and mitochondrial metabolism that generated acetyl-CoA, the biosynthetic precursor of fatty acids, cholesterol, and isoprenoid synthesis. Akt signal transduction could activate the mTORC1 complex, and mTORC1 stimulated adipogenesis by regulating SREBP-mediated FASN expression [79]. The PI3K/Akt/mTOR pathway inhibitors provided a promising target for the treatment of CRC. Proteoglycans in the cancer pathway (hsa05205) revealed the role of proteoglycan in the growth, metastasis, and dissemination of cancer cells. For example, biglycan, as a proteoglycans, combined with vascular endothelial growth factor, can promote the progression of CRC by inducing the increase of vascular density [80]. MAPK signaling pathway (hsa04010) could also be associated. MAPK axis was downstream of many membrane receptors, including EGFR, which transmits extracellular signals to the nucleus and regulates various cellular functions [81]. The RAS/MAPK signaling pathway was an essential pathway in the proliferation, differentiation, and invasion of CRC, as activated RAS triggers the activation of RAF and subsequently activated RAF phosphorylates activates MEK, which phosphorylates and activates MAPK/ERK [82]. In addition, we found that colorectal cancer (hsa05210) was directly related to CRC and was an essential regulator of cell proliferation, apoptosis, and genomic stability [15].
5. Conclusion
In this study, we screened four core ingredients (quercetin, luteolin, baicalein, and dinatin) from 33 HS active ingredients and obtained 170 intersection targets related to CRC. The top 10 hub genes were STAT3, TP53, MAPK3, AKT1, JUN, EGFR, MYC, VEGFA, EGF, and CTNNB1, which may be the core targets of HS. Our study shows that HS acts simultaneously on a variety of signaling pathways related to CRC, such as the PI3K-Akt signaling pathway, proteoglycans in cancer, and the MAPK signaling pathway, which provides a reference for the research on the specific mechanism of the drug in the treatment of CRC. However, the limitation of this study is that network pharmacology ignores the content of each ingredient in the drug. In addition, the current research methods ignore the possible production of new compounds in the process of drug decoction. At the same time, another limitation is that network pharmacology has a high false-positive rate. It is necessary to illustrate the molecular level of HS on the treatment of CRC in the future. However, this screening technology, combined with network pharmacology and molecular docking, saves a lot of scientific research resources and helps researchers screen drug ingredients and core targets efficiently. In the follow-up research, on the one hand, we could continue to study the effect of HS on the treatment of CRC at the molecular level. On the other hand, we need to constantly optimize the technology related to drug screening to achieve more accurate drug screening and reduce the false-positive rate. We hope that our research can bring a new perspective to the therapy of CRC and find new anticancer drugs.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 82074061), the National Key Research and Development Program (No. 2017YFC1309204), and the Beijing Municipal Science and Technology Commission (No. Z181100001718150).
Contributor Information
Guofeng Pan, Email: pan.gf1218@163.com.
Benqiang Rao, Email: raobenqiang@bjsjth.cn.
Data Availability
The data used in the study are available upon request to the corresponding author.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
ZY and SL contributed equally to this work. BQR and GFP contributed to conceptualization, supervision, and investigation. ZPY, SL, and HZT performed network pharmacology and analysis. ZPY and SL conducted molecular docking. JXQ, BW, and YYW performed the methodology and software data analysis. ZPY and SL contributed to writing the original draft. BQR and GFP were involved in formal analysis, visualization, reviewing, and editing.
References
- 1.Siegel R. L., Miller K. D., Fedewa S. A., et al. Colorectal cancer statistics. CA A Cancer Journal for Clinicians . 2017;67(3):177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Hong Y., Rao Y. Current status of nanoscale drug delivery systems for colorectal cancer liver metastasis. Biomedicine & Pharmacotherapy . 2019;114 doi: 10.1016/j.biopha.2019.108764.108764 [DOI] [PubMed] [Google Scholar]
- 3.Ooft S. N., Weeber F., Dijkstra K. K., et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Science Translational Medicine . 2019;11(513) doi: 10.1126/scitranslmed.aay2574.eaay2574 [DOI] [PubMed] [Google Scholar]
- 4.Halama N., Zoernig I., Berthel A., et al. Tumoral immune cell exploitation in colorectal cancer metastases can Be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell . 2016;29(4):587–601. doi: 10.1016/j.ccell.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Dekker E., Rex D. K. Advances in CRC prevention: screening and surveillance. Gastroenterology . 2018;154(7):1970–1984. doi: 10.1053/j.gastro.2018.01.069. [DOI] [PubMed] [Google Scholar]
- 6.Fei Z., Lijuan Y., Xi Y., et al. Gut microbiome associated with chemotherapy-induced diarrhea from the CapeOX regimen as adjuvant chemotherapy in resected stage III colorectal cancer. Gut Pathogens . 2019;11(1):p. 18. doi: 10.1186/s13099-019-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmer P., Trebing S., Timmers-Trebing U., et al. Eight-week, multimodal exercise counteracts a progress of chemotherapy-induced peripheral neuropathy and improves balance and strength in metastasized colorectal cancer patients: a randomized controlled trial. Supportive Care in Cancer . 2018;26(2):615–624. doi: 10.1007/s00520-017-3875-5. [DOI] [PubMed] [Google Scholar]
- 8.Wang J. Y., Cheng H. B., Zhou Z. Y. TCM theory of precancerous lesions of colorectal cancer. Journal of Traditional Chinese Medicine . 2018;59(21):24–28. [Google Scholar]
- 9.Yin H., Li T., Jin W. Primary exploration of GU nai-long for the treatment of tumor. Chinese Journal of Basic Medicine in Traditional Chinese Medicine . 2018;024(004):494–496. [Google Scholar]
- 10.Chen R., He J., Tong X., Tang L., Liu M. The Hedyotis diffusa willd. (Rubiaceae): a review on phytochemistry, pharmacology, quality control and pharmacokinetics. Molecules . 2016;21(6):p. 710. doi: 10.3390/molecules21060710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Chen W., Li M., Zhang F., Chen K., Chen W. A review of the ethnopharmacology, phytochemistry, pharmacology, and quality control of Scutellaria barbata D. Don. Journal of Ethnopharmacology . 2020;254 doi: 10.1016/j.jep.2019.112260.112260 [DOI] [PubMed] [Google Scholar]
- 12.Wang Q., Acharya N., Liu Z., et al. Enhanced anticancer effects of Scutellaria barbata D. Don in combination with traditional Chinese medicine components on non-small cell lung cancer cells. Journal of Ethnopharmacology . 2018;217:140–151. doi: 10.1016/j.jep.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z., Qi F., Cui Y., et al. An update on Chinese herbal medicines as adjuvant treatment of anticancer therapeutics. BioScience Trends . 2018;12(3):220–239. doi: 10.5582/bst.2018.01144. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad R., Ali A. M., Israf D. A., Ismail N. H., Shaari K., Lajis N. H. Antioxidant, radical-scavenging, anti-inflammatory, cytotoxic and antibacterial activities of methanolic extracts of some Hedyotis species. Life Sciences . 2005;76(17):1953–1964. doi: 10.1016/j.lfs.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 15.Liu X., Wu J., Zhang D., Wang K., Duan X., Zhang X. A network pharmacology approach to uncover the multiple mechanisms of Hedyotis diffusa willd. On colorectal cancer. Evidence-based Complementary and Alternative Medicine . 2018;2018:1–12. doi: 10.1155/2018/6517034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mo Z. C., Wang M., Luo X. Q. Anti-tumor activity of combination of hedyotic diffusa and Scutellaria barbata. Natural Product Research and Development . 2016;28(02):210–215. [Google Scholar]
- 17.Du J. Y., Xu Y., Wang N. Mechanism of active fractions from hedyotidis herba- scutellariae barbatae herba in inducing apoptosis of MDA- MB- 231 cells. Chinese Journal of Experimental Traditional Medical Formulae . 2018;24(17):99–107. [Google Scholar]
- 18.Li X. J., Wang A. J. Experimental study on antitumor effect of Hedyotis diffusa and Scutellaria barbata powder on H22 hepatoma mice. Jiangsu Journal of Traditional Chinese Medicine . 2013;000(007):66–68. [Google Scholar]
- 19.Jin X. T., Jia Y. X., Li Z. Y. Anti-cancer effect of extracts and combination of extracts of three Chinese medicinal herbs. Journal of Shanxi University (Natural Science Edition) . 2013;036(003):480–483. [Google Scholar]
- 20.Song Y. J., Bao J. M., Zhou L. Y., et al. An analysis of the anti-neuropathic effects of qi she pill based on network pharmacology. Evidence-based Complementary and Alternative Medicine . 2020;2020:1–15. doi: 10.1155/2020/7193832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y., Wang H., Pan Y., Liu T. Investigating the multi-target pharmacological mechanism of Hedyotis diffusa willd acting on prostate cancer: a network pharmacology approach. Biomolecules . 2019;9(10):p. 591. doi: 10.3390/biom9100591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren B., Tan L., Xiong Y., et al. Integrated analysis of the mechanisms of da-chai-hu decoction in type 2 diabetes mellitus by a network pharmacology approach. Evidence-based Complementary and Alternative Medicine . 2020;2020:1–21. doi: 10.1155/2020/9768414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng Z., Liu X., Wu J., et al. Mechanisms of compound kushen injection for the treatment of lung cancer based on network pharmacology. Evidence-based Complementary and Alternative Medicine . 2019;2019:1–15. doi: 10.1155/2019/4637839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics . 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S., Thiessen P. A., Bolton E. E., et al. PubChem substance and compound databases. Nucleic Acids Research . 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D., Santos A., von Mering C., Jensen L. J., Bork P., Kuhn M. Stitch 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research . 2016;44(D1):D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickel J., Gohlke B. O., Erehman J., et al. SuperPred: update on drug classification and target prediction. Nucleic Acids Research . 2014;42(W1):W26–W31. doi: 10.1093/nar/gku477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Research . 2014;42(W1):W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magrane M., Consortium U. P. Database . Vol. 2011. Oxford): 2011. UniProt Knowledgebase: a hub of integrated protein data.bar009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wishart D. S., Feunang Y. D., Guo A. C., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Research . 2018;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y. H., Yu C. Y., Li X. X., et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Research . 2018;46(D1):D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piñero J., Queralt-Rosinach N., Bravo A., et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database . 2015;2015(0) doi: 10.1093/database/bav028.bav028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mok S. R. S., Mohan S., Grewal N., Elfant A. B., Judge T. A. A genetic database can be utilized to identify potential biomarkers for biphenotypic hepatocellular carcinoma-cholangiocarcinoma. Journal of Gastrointestinal Oncology . 2016;7(4):570–579. doi: 10.21037/jgo.2016.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franz M., Lopes C. T., Huck G., Dong Y., Sumer O., Bader G. D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics . 2016;32(2):309–311. doi: 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D., Morris J. H., Cook H., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research . 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G., Wang L.-G., He Q. Y. clusterProfiler: an R Package for comparing biological themes among gene clusters. OMICS A Journal of Integrative Biology . 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F., Yuan C., Wu H. Z., Liu B., Yang Y. F. Bioinformatics, molecular docking and experiments in vitro analyze the prognostic value of CXC chemokines in breast cancer. Frontiers Oncology . 2021;11 doi: 10.3389/fonc.2021.665080.665080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L. W., Li L. N. LIU weisheng’s experience on treating malignant tumor with hedyotic diffusa and Scutellaria barbata. Liaoning Journal of Traditional Chinese Medicine . 2019;10(010):2051–2053. [Google Scholar]
- 39.Luo J. Q., Liu H. B. Anticancer effect of Scutellaria barbata and Hedyotis diffusa willd. Modern oncol . 2014;22(02):0481–0484. [Google Scholar]
- 40.Yan H., Lv Q. T. The progress of the anti - tumor activity of Oldenlandia diffusa, Scutellaria barbaraand their combination. Modern oncol . 2015;23(22):3353–3356. [Google Scholar]
- 41.Yeh Y. C., Chen H. Y., Yang S. H. Hedyotis diffusa Combined with Scutellaria barbata Are the Core Treatment of Chinese Herbal Medicine Used for Breast Cancer Patients: A Population-Based Study. Evid Based Complement Alternat Med. . 2014;2014:p. 202378. doi: 10.1155/2014/202378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao G., Balunas M. J. Current therapeutic role and medicinal potential of Scutellaria barbata in Traditional Chinese Medicine and Western research. Journal of Ethnopharmacology . 2016;182:170–180. doi: 10.1016/j.jep.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 43.Luo J. Q., Liu H. B. Antitumor effect of Scutellaria barbata and Hedyotis diffusa willd. Modern Oncology . 2014;22(002):481–484. [Google Scholar]
- 44.Dong H. H., Cao S. W., Yu Y. Y. Antioxidant and free radical scavenging activities of extracts from Scutellaria barbata, Hedyotis diffusa and their combination. Natural Product Research and Development . 2008;20(005):782–786. [Google Scholar]
- 45.Zhang Z., Li B., Xu P., Yang B. Integrated whole transcriptome profiling and bioinformatics analysis for revealing regulatory pathways associated with quercetin-induced apoptosis in HCT-116 cells. Frontiers in Pharmacology . 2019;10:p. 798. doi: 10.3389/fphar.2019.00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y., Wang T., Chen D., et al. Quercetin preferentially induces apoptosis in KRAS-mutant colorectal cancer cells via JNK signaling pathways. Cell Biology International . 2019;43(2):117–124. doi: 10.1002/cbin.11055. [DOI] [PubMed] [Google Scholar]
- 47.Lin R., Piao M., Song Y., Liu C. Quercetin suppresses AOM/DSS-Induced colon carcinogenesis through its anti-inflammation effects in mice. Journal of Immunology Research . 2020;2020:9242601–10. doi: 10.1155/2020/9242601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., Shen L., Li X., Song W., Liu Y., Huang L. Nanoformulated codelivery of quercetin and alantolactone promotes an antitumor response through synergistic immunogenic cell death for microsatellite-stable colorectal cancer. ACS Nano . 2019;13(11):12511–12524. doi: 10.1021/acsnano.9b02875. [DOI] [PubMed] [Google Scholar]
- 49.Feng J., Song D., Jiang S., et al. Quercetin restrains TGF-β1-induced epithelial-mesenchymal transition by inhibiting Twist1 and regulating E-cadherin expression. Biochemical and Biophysical Research Communications . 2018;498(1):132–138. doi: 10.1016/j.bbrc.2018.02.044. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y., Guo Y., Wang M., Dong H., Zhang J., Zhang L. Quercetrin from Toona sinensis leaves induces cell cycle arrest and apoptosis via enhancement of oxidative stress in human colorectal cancer SW620 cells. Oncology Reports . 2017;38(6):3319–3326. doi: 10.3892/or.2017.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Y., Rao C., Zheng G., Wang S. Luteolin suppresses colorectal cancer cell metastasis via regulation of the miR-384/pleiotrophin axis. Oncology Reports . 2019;42(1):131–141. doi: 10.3892/or.2019.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdel Hadi L., Di Vito C., Marfia G., et al. Sphingosine kinase 2 and ceramide transport as key targets of the natural flavonoid luteolin to induce apoptosis in colon cancer cells. PLoS One . 2015;10(11) doi: 10.1371/journal.pone.0143384.e0143384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandurangan A. K., Kumar S. A. S., Dharmalingam P., Ganapasam S. Luteolin, a bioflavonoid inhibits azoxymethane-induced colon carcinogenesis: involvement of iNOS and COX-2. Pharmacognosy Magazine . 2014;10(Suppl 2):S306–S310. doi: 10.4103/0973-1296.133285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pandurangan A. K., Dharmalingam P., Sadagopan S. K., Ganapasam S. Luteolin inhibits matrix metalloproteinase 9 and 2 in azoxymethane-induced colon carcinogenesis. Human & Experimental Toxicology . 2014;33(11):1176–1185. doi: 10.1177/0960327114522502. [DOI] [PubMed] [Google Scholar]
- 55.Pandurangan A. K., Esa N. M. Luteolin, a bioflavonoid inhibits colorectal cancer through modulation of multiple signaling pathways: a review. Asian Pacific Journal of Cancer Prevention . 2014;15(14):5501–5508. doi: 10.7314/apjcp.2014.15.14.5501. [DOI] [PubMed] [Google Scholar]
- 56.Zeng Q., Zhang Y., Zhang W., Guo Q. Baicalein suppresses the proliferation and invasiveness of colorectal cancer cells by inhibiting Snail-induced epithelial-mesenchymal transition. Molecular Medicine Reports . 2020;21(6):2544–2552. doi: 10.3892/mmr.2020.11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Z., Hou R., Gao S., Song D., Feng Y. Baicalein inhibits proliferation activity of human colorectal cancer cells HCT116 through downregulation of ezrin. Cellular Physiology and Biochemistry . 2018;49(5):2035–2046. doi: 10.1159/000493714. [DOI] [PubMed] [Google Scholar]
- 58.Wang C. Z., Zhang C. F., Luo Y., et al. Baicalein, an enteric microbial metabolite, suppresses gut inflammation and cancer progression in ApcMin/+ mice. Clinical and Translational Oncology . 2020;22(7):1013–1022. doi: 10.1007/s12094-019-02225-5. [DOI] [PubMed] [Google Scholar]
- 59.Rui X., Yan X. I., Zhang K. Baicalein inhibits the migration and invasion of colorectal cancer cells via suppression of the AKT signaling pathway. Oncology Letters . 2016;11(1):685–688. doi: 10.3892/ol.2015.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holzner S., Brenner S., Atanasov A. G., et al. Intravasation of SW620 colon cancer cell spheroids through the blood endothelial barrier is inhibited by clinical drugs and flavonoids in vitro. Food and Chemical Toxicology . 2018;111:114–124. doi: 10.1016/j.fct.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 61.Lv L., Zhang W., Li T., Jiang L., Lu X., Lin J. Hispidulin exhibits potent anticancer activity in vitro and in vivo through activating ER stress in non-small-cell lung cancer cells. Oncology Reports . 2020;43(6):1995–2003. doi: 10.3892/or.2020.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han M., Gao H., Xie J., et al. Retracted article: hispidulin induces ER stress-mediated apoptosis in human hepatocellular carcinoma cells in vitro and in vivo by activating AMPK signaling pathway. Acta Pharmacologica Sinica . 2019;40(5):666–676. doi: 10.1038/s41401-018-0159-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Gao H., Gao M. Q., Peng J. J., Han M., Liu Kl, Han Yt. Retracted article: hispidulin mediates apoptosis in human renal cell carcinoma by inducing ceramide accumulation. Acta Pharmacologica Sinica . 2017;38(12):1618–1631. doi: 10.1038/aps.2017.154. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Jiang H., Liu X., Knolhoff B. L., et al. Development of resistance to FAK inhibition in pancreatic cancer is linked to stromal depletion. Gut . 2020;69(1):122–132. doi: 10.1136/gutjnl-2018-317424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heichler C., Scheibe K., Schmied A., et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut . 2020;69(7):1269–1282. doi: 10.1136/gutjnl-2019-319200. [DOI] [PubMed] [Google Scholar]
- 66.Schulz-Heddergott R., Stark N., Edmunds S. J., et al. Therapeutic ablation of gain-of-function mutant p53 in colorectal cancer inhibits stat3-mediated tumor growth and invasion. Cancer Cell . 2018;34(2):298–314. doi: 10.1016/j.ccell.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu S. M., Yao X. H., Hao Y. H., Pan A. H., Zhou X. W. 8-Gingerol regulates colorectal cancer cell proliferation and migration through the EGFR/STAT/ERK pathway. International Journal of Oncology . 2020;56(1):390–397. doi: 10.3892/ijo.2019.4934. [DOI] [PubMed] [Google Scholar]
- 68.Ding C., Luo J., Fan X., et al. Elevated Gab2 induces tumor growth and angiogenesis in colorectal cancer through upregulating VEGF levels. Journal of Experimental & Clinical Cancer Research . 2017;36(1):p. 56. doi: 10.1186/s13046-017-0524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Y., Zhang C., Liu X., et al. ARIH1 signaling promotes anti-tumor immunity by targeting PD-L1 for proteasomal degradation. Nature Communications . 2021;12(1):p. 2346. doi: 10.1038/s41467-021-22467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Markman B., Javier Ramos F., Capdevila J., Tabernero J. EGFR and KRAS in colorectal cancer. Advances in Clinical Chemistry . 2010;51:71–119. doi: 10.1016/s0065-2423(10)51004-7. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Q., Wang C., Han X., Yang G., Ge Z., Zhang G. Knockdown of ADAM17 inhibits cell proliferation and increases oxaliplatin sensitivity in HCT-8 colorectal cancer through EGFR-PI3K-AKT activation. Biochemical and Biophysical Research Communications . 2018;503(4):2333–2339. doi: 10.1016/j.bbrc.2018.06.158. [DOI] [PubMed] [Google Scholar]
- 72.Ta N. L., Chakrabandhu K., Huault S., Hueber A. O. The tyrosine phosphorylated pro-survival form of Fas intensifies the EGF-induced signal in colorectal cancer cells through the nuclear EGFR/STAT3-mediated pathway. Scientific Reports . 2018;8(1) doi: 10.1038/s41598-018-30804-z.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J., Pan C., Boese A. C., et al. DGKA provides platinum resistance in ovarian cancer through activation of c-JUN-WEE1 signaling. Clinical Cancer Research . 2020;26(14):3843–3855. doi: 10.1158/1078-0432.ccr-19-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu H., Lu X. X., Wang J. R., et al. TRAF6 inhibits colorectal cancer metastasis through regulating selective autophagic CTNNB1/β-catenin degradation and is targeted for GSK3B/GSK3β-mediated phosphorylation and degradation. Autophagy . 2019;15(9):1506–1522. doi: 10.1080/15548627.2019.1586250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng H., Jiang X., Zhang Q., et al. Naringin inhibits colorectal cancer cell growth by repressing the PI3K/AKT/mTOR signaling pathway. Experimental and Therapeutic Medicine . 2020;19(6):3798–3804. doi: 10.3892/etm.2020.8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang S., Dong S. M., Kim B. R., et al. Thioridazine induces apoptosis by targeting the PI3K/Akt/mTOR pathway in cervical and endometrial cancer cells. Apoptosis . 2012;17(9):989–997. doi: 10.1007/s10495-012-0717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scartozzi M., Giampieri R., Maccaroni E., et al. Phosphorylated AKT and MAPK expression in primary tumours and in corresponding metastases and clinical outcome in colorectal cancer patients receiving irinotecan-cetuximab. Journal of Translational Medicine . 2012;10(1):p. 71. doi: 10.1186/1479-5876-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Narayanankutty A. PI3K/Akt/mTOR pathway as a therapeutic target for colorectal cancer: a review of preclinical and clinical evidence. Current Drug Targets . 2019;20(12):1217–1226. doi: 10.2174/1389450120666190618123846. [DOI] [PubMed] [Google Scholar]
- 79.Ventura R., Mordec K., Waszczuk J., et al. Inhibition of de novo Palmitate Synthesis by Fatty Acid Synthase Induces Apoptosis in Tumor Cells by Remodeling Cell Membranes, Inhibiting Signaling Pathways, and Reprogramming Gene Expression. EBioMedicine . 2015;2(8):808–824. doi: 10.1016/j.ebiom.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahrens T. D., Bang-Christensen S. R., Jørgensen A. M., et al. The role of proteoglycans in cancer metastasis and circulating tumor cell analysis. Frontiers in Cell and Developmental Biology . 2020;8:p. 749. doi: 10.3389/fcell.2020.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soleimani A., Rahmani F., Saeedi N., et al. The potential role of regulatory microRNAs of RAS/MAPK signaling pathway in the pathogenesis of colorectal cancer. Journal of Cellular Biochemistry . 2019;120(12):19245–19253. doi: 10.1002/jcb.29268. [DOI] [PubMed] [Google Scholar]
- 82.Cicenas J., Tamosaitis L., Kvederaviciute K., et al. KRAS, NRAS and BRAF mutations in colorectal cancer and melanoma. Medical Oncology . 2017;34(2):p. 26. doi: 10.1007/s12032-016-0879-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in the study are available upon request to the corresponding author.


