Abstract
Pleurotus ostreatus is a white rot basidiomycete that produces several extracellular laccase isoenzymes, including phenol oxidase A1b (POXA1b), POXA2, and POXC. POXC was the most abundant isoenzyme produced under all of the growth conditions examined in this study. Copper was the most efficient inducer of laccase activity among the putative inducers tested. The amounts of all of the previously described laccase isoenzymes increased substantially in copper-supplemented cultures. Under these conditions expression of POX isoenzymes was regulated at the level of gene transcription. It is worth noting that poxa1b mRNA was the most abundant induced transcript at all of the growth times analyzed, and the amount of this mRNA increased until day 7. The discrepancy between the poxa1b transcript and protein amounts can be explained by the presence of a high level of the protein in P. ostreatus cellular extract, which indicated that the POXA1b isoenzyme could be inefficiently secreted and/or that its physiological activity could occur inside the cell or on the cell wall. Moreover, the POXA1b isoenzyme behaved uniquely, as its activity was maximal on the second day of growth and then decreased. An analysis performed with protease inhibitors revealed that the loss of extracellular POXA1b activity could have been due to the presence of specific proteases secreted into the copper-containing culture medium that affected the extracellular POXA1b isoenzyme.
White rot fungi produce various isoforms of extracellular oxidases and peroxidases, which are involved in the degradation of lignin in their natural environments. These enzymes are nonspecific and consequently oxidize a broad spectrum of structurally different substrates, such as highly toxic phenolic compounds and azo dyes (9, 20).
Laccase (p-diphenol:dioxygen oxidoreductase; EC 1.10.3.2) belongs to the group of blue copper oxidases which use oxygen as an electron acceptor to remove hydrogen radicals from phenolic hydroxyl groups (19, 24). The free radicals formed can undergo rearrangements that lead to alkyl-aryl cleavage, oxidation of benzyl alcohols, and cleavage of side chains and aromatic rings (1). In the presence of appropriate redox mediators, laccases can also oxidize nonphenolic substrates (2).
It has been reported that several fungi have more than one laccase-encoding gene. Four different cDNA sequences have been found in Rhizoctonia solani (25), up to five laccase genes have been found in Trametes villosa (26, 27), three genomic sequences have been found in the basidiomycete I-62 (14) and Pleurotus ostreatus (6–8), and two genes have been found in Agaricus bisporus (22). Thus, the biochemical diversity of laccase isoenzymes appears to be due to the multiplicity of laccase genes. Moreover, regulation of the expression of these genes is substantially different in different species.
P. ostreatus is a white rot basidiomycete that produces several laccase isoenzymes, and phenol oxidase C (POXC) is the most abundant isoenzyme produced under all of the growth conditions that have been examined (6, 16). Three other isoenzymes which are secreted by this fungus have also been purified and characterized; these isoenzymes are POXA1w, POXA2, and POXA1b (8, 16). POXA1w exhibits peculiar differences with regard to its metal content. In fact, this enzyme contains two zinc atoms per molecule, one iron atom per molecule, and only one copper atom per molecule (16). Addition of CuSO4 to culture broth results in a substantial increase in the total laccase activity and production of the POXA1b isoenzyme, while POXA1w is almost unaffected when copper is added (8). The main structural characteristics of POXA1b are very similar to those of POXA1w, but POXA1b produces the classical laccase UV-visible light spectrum and contains the four characteristic copper atoms per molecule (8). Both POXA1w and POXA1b are much more stable than the other two P. ostreatus laccases that have been characterized, POXA2 and POXC (8).
Three genes which encode laccase isoenzymes in P. ostreatus have been identified so far; these genes are poxc (previously pox2), pox1 (which codes for a laccase isoenzyme that has not been identified yet), and poxa1b (6–8).
Differential regulation of ligninolytic-enzyme-encoding genes in response to culture conditions has been documented previously (3, 14, 26, 27). In the genus Pleurotus, Leonowicz and Trojanowski (12) studied the effect of ferulic acid as an inducer of a specific laccase isoenzyme in P. ostreatus. Moreover, Muñoz et al. (15) demonstrated that a laccase isoenzyme was induced by wheat straw alkalilignin and vanillic and veratric acids in Pleurotus eryngii. In this paper we describe the effect of copper, the most efficient of the putative inducers tested, on the production of specific laccase isoenzyme patterns. Furthermore, expression of POX isoenzymes is regulated at the level of gene transcription, and the three known laccases appear to be regulated differently. The anomalous behavior of POXA1b induction compared with induction of the other isoenzymes was also investigated in this study.
MATERIALS AND METHODS
Organism and culture conditions.
The white rot fungus P. ostreatus (Jacq.:Fr.) Kummer (type Florida) was maintained by periodic transfer at 4°C on potato dextrose (2.4%) agar plates (Difco) in the presence of 0.5% yeast extract (Difco).
Preparations were incubated as previously described (16). Mycelia were grown in liquid basal medium (24 g of potato dextrose broth per liter, 5 g of yeast extract per liter) or in the same medium supplemented with 150 μM copper sulfate at the time of inoculation.
The effect of protease inhibitors on laccase activity was determined by adding a mixture containing 1 μM antipain, 4.6 μM bestatin, 10 μM chymostatin, 100 μM phenylmethylsulfonyl fluoride, and 2 μM E-64 (all obtained from Boehringer) to the basal medium containing 150 μM CuSO4. These inhibitors were added to the culture broth after 2, 3, and 4 days of incubation. Duplicate cultures were analyzed periodically to determine laccase activity.
All of the experiments were performed at least two times by using two or three replicates for each set of conditions and each time.
Protein purification and enzyme assays.
Extracellular laccases produced by P. ostreatus were purified from liquid cultures as previously described (8, 16, 17).
Laccase activity was determined at 25°C by using the substrate 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) as previously described (16).
Enzyme activities were expressed in international units.
Nondenaturing PAGE.
Polyacrylamide gel electrophoresis (PAGE) was performed at an alkaline pH under nondenaturing conditions. The separating and stacking gels contained 9 and 4% acrylamide, respectively. The buffer solution used for the separating gel contained 50 mM Tris-HCl (pH 9.5), and the buffer solution used for the stacking gel contained 18 mM Tris-HCl (pH 7.5). The electrode reservoir solution contained 25 mM Tris and 190 mM glycine (pH 8.4). The gels were stained to visualize laccase activity by using ABTS as the substrate.
Western blotting.
Proteins were separated on sodium dodecyl sulfate (SDS)-PAGE gels as described by Laemmli (11) and were electroblotted onto ProBlott polyvinylidene difluoride membranes (Applied Biosystems). Electroblotting was performed in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (pH 11)–10% (vol/vol) methanol at 50 V for 180 min at room temperature. The blocking solution contained 5% (wt/vol) dried milk in phosphate-buffered saline supplemented with 0.2% (vol/vol) Triton X-100 (washing buffer). The membrane was washed and incubated with the primary antiserum diluted 1:100 in washing buffer at room temperature for 1 h with continuous shaking. Subsequently, the membrane was washed and incubated as described above with anti-rabbit immunoglobulin G–peroxidase conjugate (Sigma) diluted 1:2,000 in washing buffer. The blots were visualized with a solution containing 100 mM Tris-HCl (pH 7.5), 0.5 mg of 3,3′-diaminobenzidine per ml, 0.03% (wt/vol) NiCl2, and 0.006% (vol/vol) H2O2.
Preparation of mycelium crude extract.
Total extract from P. ostreatus mycelium was prepared as follows. Lyophilized cells were ground in a mortar with a pestle. The ground material was resuspended in cold extraction buffer [200 mM Tris-HCl (pH 8.0), 400 mM (NH4)2SO4, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 7 mM β-mercaptoethanol] and then centrifuged at 4°C for 1 h at 15,000 × g.
Genomic DNA isolation and Southern analysis.
P. ostreatus mycelia were harvested on Mira-cloth, washed twice with distilled water, quickly frozen, and stored at −80°C. High-molecular-weight genomic DNA was isolated from lyophilized mycelia by the method of Raeder and Broda (18). The genomic DNA was digested with several restriction enzymes as specified by the manufacturer (Promega). Denatured DNA fragments were analyzed by electrophoresis on a 0.8% agarose gel in TAE buffer (40 mM Tris acetate, 10 mM EDTA) by using standard protocols and then were transferred to a Hybond-NX nylon membrane (Amersham). The blots were hybridized under high-stringency conditions at 65°C in 0.5 M phosphate buffer–7% SDS–10 mM EDTA (pH 7.2) and washed at the same temperature; the final wash was performed with 1× SSC–0.1% SDS (1× SSC is 15 mM NaCl plus 1.5 mM sodium citrate, pH 7.0).
Radioactive DNA probes were prepared with [α-32P]dATP (Amersham) by random priming with a labeling kit (Amersham).
RNA isolation and Northern analysis.
Mycelia were collected by filtration, washed with sterilized distilled water, quickly frozen, and stored at −80°C. The cells were lyophilized and weighed.
Total RNA was purified by the method of Lucas et al. (13). The RNA concentration was determined spectrophotometrically, and 30-μg portions of total RNA were electrophoresed in a 1% agarose–formaldehyde gel with 20 mM MOPS (morpholinepropanesulfonic acid)–5 mM sodium acetate (pH 7.0)–1 mM EDTA and then blotted onto a Hybond-NX nylon membrane (Amersham) in 10× SSC. Filters were hybridized with appropriately labeled probes as described by Sambrook et al. (21) and washed; the final wash was performed with 0.1× SSC–0.1% SDS at 42°C. The specific activities of the three labeled probes used were comparable, and all analyses were performed with two different RNA preparations. As controls, filters were also hybridized with a 0.65-kb NcoI fragment of the Aspergillus nidulans actin gene, which was kindly provided by Sovan Sarkar, University of Westminster, London, United Kingdom.
Transcripts were quantified by using the actin signal as a loading control and QuantiScan software (Biosoft, Ferguson, Mo.).
RESULTS
Effect of copper on laccase isoenzyme production.
Copper has been reported to be a strong laccase inducer in the fungal species Trametes versicolor and Phanerochaete chrysosporium (3, 4). Previously, we observed a significant increase in laccase activity in copper-supplemented P. ostreatus culture broth and described purification of a new laccase isoenzyme produced under these conditions (POXA1b) (8). It has also been reported (8) that the increase in activity is proportional to the amount of copper added and that the maximal effect (about 30 U/ml) is obtained at a CuSO4 concentration of 150 μM. Under these conditions a 50-fold increase after 6 days of growth was obtained compared to the maximal activity in the basal medium (3 days of growth). The presence of copper did not affect fungal growth since the biomass dry weights at different times were the same in the presence and in the absence of copper. In this study we examined the effect of copper induction in more detail.
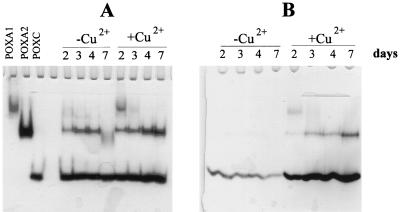
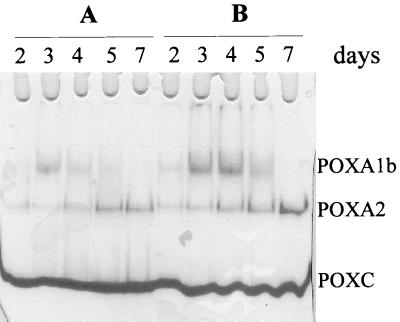
Laccase isoenzymes were separated on native PAGE gels by using purified POXC, POXA2, POXA1b, and POXA1w isoenzymes as standards. Under the conditions used POXA1b and POXA1w had the same electrophoretic mobilities, so these two isoenzymes are indicated as POXA1 in Fig. 1.
FIG. 1.
Zymograms of laccase isoenzymes in the absence and in the presence of 150 μM CuSO4. Samples containing 0.015 U (A) or 0.04 μg (B) of proteins collected at different times (2, 3, 4, and 7 days) were used.
To study production of the different isoenzymes as a function of growth time in the presence or absence of copper, samples containing 0.015 U (Fig. 1A) or 0.04 μg of protein (Fig. 1B) collected at different times were analyzed by native PAGE. A remarkable increase in the amount of POXA2 in the presence of copper after prolonged incubation was observed. The effect of copper on POXA1 activity seemed to be greater, and the presence of copper resulted in an anomalous time course. In fact, as shown in Fig. 1, the POXA1 band was most intense on the second day and then rapidly disappeared. However, the time at which the maximum POXA1 activity occurred varied between the second and third days. It is worth noting that POXA1w production was almost unaffected by the presence of copper and the amount of POXA1w was about 10-fold lower than the amount of POXA1b (8). For this reason the POXA1 band could be essentially attributed to POXA1b.
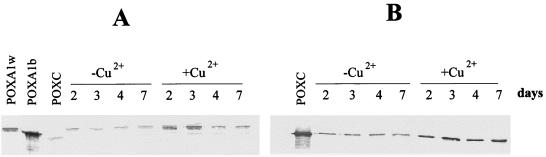
In order to relate the time course of POXA1b activity to protein production, a Western blot analysis was performed by using anti-POXA1b antibodies (Fig. 2A). It has been shown that anti-POXA1b antibodies give weaker recognition signals for POXC, POXA2, and POXA1w. In fact, POXA1b was not the only protein detected in the samples which we analyzed, as shown in Fig. 2A. On the other hand, a more intense band, corresponding to POXA1b, was visible only in the presence of copper after 2 and 3 days. Therefore, the loss of POXA1b activity coincided with the loss of the protein.
FIG. 2.
Western blot analyses of samples containing 0.05 U of protein collected at different times in the absence and in the presence of 150 μM CuSO4. (A) POXA1b antibodies. (B) POXC antibodies.
A time course evaluation of the POXC isoenzyme was unreliable under the native PAGE conditions used to detect all of the other isoenzymes since the POXC results were overloaded. However, the increase in POXC production after copper was added is clearly shown in Fig. 1B; in this experiment samples containing equal amounts of proteins collected at different times were used. It is also evident that there was a decrease in the amount of POXC during fungal growth in the basal medium and that there was an opposite effect in the presence of copper.
The Western blot analysis performed with POXC antibodies (Fig. 2B) revealed that there was an increase in POXC production in the presence of copper, which confirmed the inductive effect of this metal.
Effect of copper on laccase gene transcription.
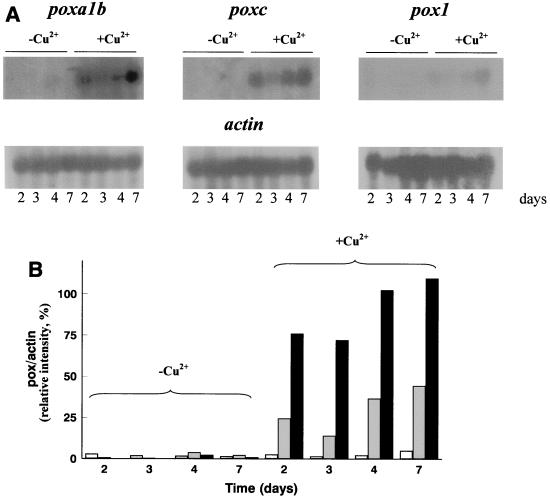
In order to analyze and differentiate the effects of copper on transcription of the different laccase isoenzymes, poxc, pox1, and poxa1b cDNAs were used to detect transcripts of the corresponding genes. Since the level of identity between poxc and pox1 cDNAs is high (83%), Southern blot analyses were performed under very stringent conditions in order to verify the specificity of these probes. Different patterns of bands were obtained for each gene (data not shown), which allowed us to use the cDNAs as selective probes. The results of Northern blot experiments are shown in Fig. 3, which also shows the results of a quantitative analysis of the intensity of each band. Similar results were obtained with RNA preparations from different P. ostreatus cultures. In all cases the actin gene of A. nidulans was used as a loading control. In cultures grown in the absence of copper, detectable amounts of poxc and poxa1b mRNA were present only in cells collected after 3 days of growth. Strong transcriptional induction was observed in the copper-supplemented cultures for poxa1b and poxc genes. poxa1b mRNA was the most abundant transcript at all of the times analyzed, while pox1 mRNA was barely detectable even under inducing conditions. The time courses of the three transcripts seemed to be similar; in all cases the amounts of mRNA decreased after 2 days and there were increases from the third day on (Fig. 3).
FIG. 3.
(A) Northern blot analysis of poxa1b, poxc, and pox1 gene transcripts in the absence and in the presence of copper. (B) Transcript levels expressed as percentages of the actin signal. Solid bars, poxa1b; shaded bars, poxc; open bars, pox1.
Localization of POXA1b activity.
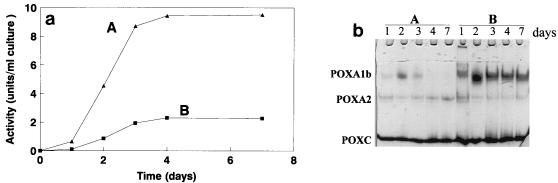
The intracellular laccase activity of mycelia grown in the presence of copper was quantified and compared to the extracellular activity, and native PAGE profiles of both intra- and extracellular isoenzymes were also analyzed (Fig. 4). As Fig. 4 shows, the relative activity of POXA1b compared to the other isoenzymes was higher in the cellular extract than in the culture broth even after prolonged growth. The presence of a larger amount of POXA1b in the cellular extract than in the culture broth was also confirmed by a Western blot analysis (data not shown), in which the time course of extracellular POXA1b activity was similar to that shown in Fig. 2A.
FIG. 4.
(a) Laccase activity per milliliter of culture in the presence of copper in culture broth (line A) and in intracellular protein extract (line B). (b) Zymogram of extracellular (lanes A) and intracellular (lanes B) laccase isoenzymes. Samples that contained 0.015 U of protein and were collected at different times were used.
Effect of protease inhibitors.
In order to verify that the loss of extracellular POXA1b after 2 or 3 days of growth was due to degradation by secreted proteases, copper-amended fungal cultures were grown in the presence and in the absence of protease inhibitors. If the protease inhibitor cocktail was added after 2, 3, or 4 days of growth, a 1.5-fold increase in total laccase activity was observed and the native PAGE band corresponding to POXA1b persisted at least until day 5 (Fig. 5). Moreover, a 7-day filtered broth preparation (100 μl) was incubated with 0.2 U of purified POXA1b in the presence or in the absence of the inhibitor cocktail, and native PAGE was performed after 2 days of incubation (Fig. 6). The results obtained unambiguously demonstrated that the loss of POXA1b was due to the action of extracellular proteases. On the other hand, no decrease in POXA1b activity was observed if a 2-day filtered broth preparation was used under the same incubation conditions. These data also suggest that the inhibitor cocktail had no effect on fungal growth and/or on the secretion mechanism.
FIG. 5.
Zymogram of laccase isoenzymes in copper-supplemented cultures in the absence (lanes A) and in the presence (lanes B) of protease inhibitors. Samples that contained 0.015 U of protein and were collected at different times were used.
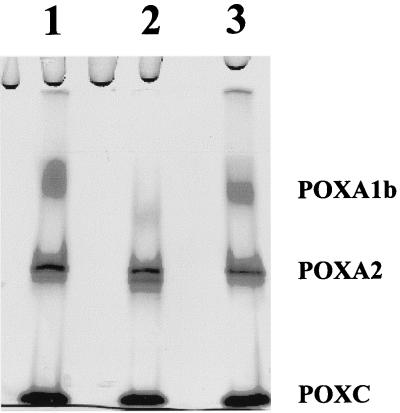
FIG. 6.
Zymogram of purified POXA1b incubated with 7-day filtered culture broth (100 μl). Lane 1, control collected at zero time; lane 2, sample collected after 2 days of incubation without protease inhibitors; lane 3, sample collected after 2 days of incubation in the presence of protease inhibitors. One-microliter aliquots were loaded into the lanes.
DISCUSSION
Laccases are produced as a number of isoenzymes that are encoded by gene families in several fungal species. Many different genes that encode laccase isoenzymes in ligninolytic fungi (14, 22, 25–27), including P. ostreatus (6–8), have been cloned and sequenced, but little work has been done to study the regulation of laccase gene expression. In this study we investigated the effect of copper on laccase production in P. ostreatus and analyzed the effect of copper on transcription and translation of some laccase isoenzymes.
Other potential inducers that were tested (MnSO4, FeCl3, ZnSO4, veratryl alcohol, veratraldehyde, vanillic acid, ferulic acid) did not have a significant effect on total laccase activity but produced different laccase isoenzyme patterns (data not shown). The greatest increase in laccase activity was obtained in the copper-supplemented culture, and production of the three known isoenzymes (POXA1b, POXA2, and POXC) was strongly increased under these conditions.
Northern blot analyses clearly revealed that copper had a marked effect on induction of poxa1b and poxc gene transcription, while the effect on pox1 (which encodes an unidentified laccase) was smaller. Moreover, the POXA1b transcript was the most abundant transcript in the copper-supplemented cultures at all of the times analyzed, although the amount of POXA1b protein produced was significantly less than the amount of POXC protein produced, as shown by the Western blot analysis. The difference between the amounts of POXA1b and POXC revealed by this analysis was less than the difference observed in a zymogram because the specific activity of POXC (about 4,000 U/mg) was greater than the specific activity of POXA1b (about 2,000 U/mg) with the substrate used (ABTS).
Furthermore, the POXA1b isoenzyme exhibited a peculiar time course; in fact, no extracellular POXA1b activity was detected after 3 days. The Western blot analysis also revealed that there was a significant decrease in the concentration of this isoenzyme, which is consistent with the decrease in activity. This behavior is even more unusual considering that the amount of the POXA1b transcript continued to increase after the third day. An analysis of the cellular extract compared to the secreted proteins, performed by native PAGE and Western blotting, revealed that POXA1b was only partially secreted. Therefore, the presence of poxa1b mRNA was justified by the presence of the protein both in the cellular extract and in extracellular broth. Further investigation is needed to determine if the fungal secretion mechanism for POXA1b is inefficient or the physiological activity of this isoenzyme occurs inside the cell or on the cell wall. It is worth recalling that the signal peptide of POXA1b satisfies the criteria for a minimal signal sequence (8).
The loss of POXA1b from the culture broth could have been due to the action of proteases in the copper-containing culture medium that specifically affected this isoenzyme. In fact, addition of protease inhibitors to the culture broth resulted in an increase in the POXA1b activity that was observed until day 5. Furthermore, it has been demonstrated that proteases present in culture broth (after 7 days of growth) are able to degrade purified POXA1b. The specificity and physiological role of these proteases are under investigation.
Several metal responsive element consensus sequences (23) have been identified in promoter regions of the three pox genes (8). These putative metal responsive element sequences are similar to the sequences found in the promoters of metallothionein genes (10), whose expression is induced by a range of heavy metals; a metal-regulatory protein acts both as a metal receptor and as a trans-acting transcription factor.
All three laccase transcripts had similar time courses in the presence of copper. Northern blot analyses revealed that the maximum amounts of transcripts were present on the second day of fungal growth and that there was a subsequent increase from the third day on. This peculiar behavior could have been due to a direct effect of copper induction during the early phase of fungal growth. Free copper ions, as well as the production of a toxic compound, could result in oxidative stress at an advanced stage of fungal growth and could be responsible for late transcriptional induction (5).
ACKNOWLEDGMENTS
We thank Ennio Cocca for performing quantitative analyses of Northern blot data.
This work was supported by grants from the Ministero dell'Università e della Ricerca Scientifica (Progetti di Rilevante Interesse Nazionale grant PRIN 98), the Consiglio Nazionale delle Ricerche (Progetto Finalizzato Biotecnologie), and the Ministero delle Politiche Agricole (Progetto Finalizzato Nazionale sulle Biotecnologie Vegetali).
G. Palmieri and P. Giardina contributed equally to this work.
REFERENCES
- 1.Bourbonnais R, Paice M G. Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267:99–102. doi: 10.1016/0014-5793(90)80298-w. [DOI] [PubMed] [Google Scholar]
- 2.Bourbonnais R, Paice M G. Demethylation and delignification of kraft pulp by Trametes versicolor laccase in the presence of 2,2′-azinobis-(3-ethylbenzthiazoline-6-sulphonate) Appl Microbiol Biotechnol. 1992;36:823–827. [Google Scholar]
- 3.Collins P J, Dobson A D W. Regulation of laccase gene transcription in Trametes versicolor. Appl Environ Microbiol. 1997;63:3444–3450. doi: 10.1128/aem.63.9.3444-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dittmer J K, Patel N J, Dhawale S W, Dhawale S S. Production of multiple laccase isoforms by Phanerochaete chrysosporium grown under nutrient sufficiency. FEMS Microbiol Lett. 1997;149:65–70. [Google Scholar]
- 5.Fernandez-Larrea J, Stahl U. Isolation and characterization of a laccase gene of Podospora anserina. Mol Gen Genet. 1996;252:539–551. doi: 10.1007/BF02172400. [DOI] [PubMed] [Google Scholar]
- 6.Giardina P, Aurilia V, Cannio R, Marzullo L, Amoresano A, Siciliano R, Pucci P, Sannia G. The gene, protein, and glycan structures of laccase from Pleurotus ostreatus. Eur J Biochem. 1996;235:508–515. doi: 10.1111/j.1432-1033.1996.00508.x. [DOI] [PubMed] [Google Scholar]
- 7.Giardina P, Cannio R, Martirani L, Marzullo L, Palmieri G, Sannia G. Cloning and sequencing of a laccase gene from the lignin-degrading basidomycete Pleurotus ostreatus. Appl Environ Microbiol. 1995;61:2408–2413. doi: 10.1128/aem.61.6.2408-2413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giardina P, Palmieri G, Scaloni A, Fontanella B, Faraco V, Cennamo G, Sannia G. Protein and gene structure of a blue laccase from Pleurotus ostreatus. Biochem J. 1999;34:655–663. [PMC free article] [PubMed] [Google Scholar]
- 9.Hammel K. Organopollutant degradation by ligninolytic fungi. In: Young L Y, Cerniglia C E, editors. Microbial transformation and degradation of toxic organic chemicals. New York, N.Y: Wiley-Liss, Inc.; 1995. pp. 331–346. [Google Scholar]
- 10.Imbert J, Culotta V, Furst P, Gedamu L, Hammer D. Regulation of metallothionein gene transcription by metals. In: Eichorn G L, Marzilli L G, editors. Metal-ion induced regulation of gene expression. Vol. 8. New York, N.Y: Elsevier Science Publishing, Inc.; 1990. pp. 139–164. [PubMed] [Google Scholar]
- 11.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 12.Leonowicz A, Trojanowski J. Induction of a new laccase from the fungus Pleurotus ostreatus by ferulic acid. Microbios. 1975;13:167–174. [Google Scholar]
- 13.Lucas M C, Jacobson J W, Giles N H. Characterization and in vitro translation of polyadenylated messenger ribonucleic acid from Neurospora crassa. J Bacteriol. 1977;130:1192–1198. doi: 10.1128/jb.130.3.1192-1198.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansur M, Suarez T, Fernandez-Larrea J, Brizuela M A, Gonzalez A E. Identification of a laccase gene family in the new lignin-degrading basidiomycete CECT 20197. Appl Environ Microbiol. 1997;63:2637–2646. doi: 10.1128/aem.63.7.2637-2646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muñoz C, Guillen F, Martinez A T, Martinez M J. Induction and characterization of laccase in the ligninolytic fungus Pleurotus eryngii. Curr Microbiol. 1997;34:1–5. doi: 10.1007/s002849900134. [DOI] [PubMed] [Google Scholar]
- 16.Palmieri G, Giardina P, Bianco C, Scaloni A, Capasso A, Sannia G. A novel white laccase from Pleurotus ostreatus. J Biol Chem. 1997;272:31301–31307. doi: 10.1074/jbc.272.50.31301. [DOI] [PubMed] [Google Scholar]
- 17.Palmieri G, Giardina P, Marzullo L, Desiderio B, Nitti G, Cannio R, Sannia G. Stability and activity of phenol oxidase from the ligninolytic fungus Pleurotus ostreatus. Appl Microbiol Biotechnol. 1993;39:632–636. doi: 10.1007/BF00205066. [DOI] [PubMed] [Google Scholar]
- 18.Raeder V, Broda P. Preparation and characterization of DNA from lignin degrading fungi. Methods Enzymol. 1988;161B:211–220. [Google Scholar]
- 19.Rheinhammar B. Laccase. In: Lontie R, editor. Copper proteins and copper enzymes. Vol. 3. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 1–35. [Google Scholar]
- 20.Rodriguez E, Pickard M A, Vazquez-Duhalt R. Industrial dye decolorization by laccases from ligninolytic fungi. Curr Microbiol. 1999;38:27–32. doi: 10.1007/pl00006767. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecolar cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Smith N, Shnyreva A, Wood D A, Thurston C S. Tandem organization and highly disparate expression of the two laccase genes lcc1 and lcc2 in the cultivated mushroom Agaricus bisporus. Microbiology. 1998;144:1063–1069. doi: 10.1099/00221287-144-4-1063. [DOI] [PubMed] [Google Scholar]
- 23.Thiele D J. Metal-regulated transcription in eukaryotes. Nucleic Acids Res. 1992;20:1183–1191. doi: 10.1093/nar/20.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurston C F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- 25.Wahleithner J A, Xu F, Brown K M, Brown S H, Golightly E J, Halkier T, Kauppinen S, Pederson A, Schneider P. The identification and characterization of four laccases from the plant pathogenic fungus Rhizoctonia solani. Curr Genet. 1996;29:395–403. doi: 10.1007/BF02208621. [DOI] [PubMed] [Google Scholar]
- 26.Yaver D S, Golightly E J. Cloning and characterization of three laccase genes from the white rot basidiomycete Trametes villosa: genomic organization of the laccase gene family. Gene. 1996;181:95–102. doi: 10.1016/s0378-1119(96)00480-5. [DOI] [PubMed] [Google Scholar]
- 27.Yaver D S, Xu F, Golightly E J, Brown K M, Brown S H, Rey M W, Schneider P, Halkier T, Mondorf K, Dalboge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]