Abstract
The basidiomycete Marasmius quercophilus is commonly found during autumn on the decaying litter of the evergreen oak (Quercus ilex L.), a plant characteristic of Mediterranean forest. This white-rot fungus colonizes the leaf surface with rhizomorphs, causing a total bleaching of the leaf. In synthetic liquid media, this white-rot fungus has strong laccase activity. From a three-step chromatographic procedure, we purified a major isoform to homogeneity. The gene encodes a monomeric glycoprotein of approximately 63 kDa, with a 3.6 isoelectric point, that contains 12% carbohydrate. Spectroscopic analysis of the purified enzyme (UV/visible and electron paramagnetic resonance, atomic absorption) confirmed that it belongs to the “blue copper oxidase” family. With syringaldazine as the substrate, the enzyme's pH optimum was 4.5, the optimal temperature was 75°C, and the Km was 7.1 μM. The structural gene, lac1, was cloned and sequenced. This gene encodes a 517-amino-acid protein 99% identical to a laccase produced by PM1, an unidentified basidiomycete previously isolated from wastewater from a paper factory in Spain. This similarity may be explained by the ecological distribution of the evergreen oak in Mediterranean forest.
Litter mineralization is an important component of biogeochemical cycles in terrestrial environments. Lignin is the most difficult litter polymer to degrade, and the only organisms known to completely mineralize lignin are white-rot fungi (6, 17). In the last two decades, several such organisms have been studied. At present, three main enzymes (i.e., manganese and lignin peroxidases and laccases) (15, 30) are implicated in the biodegradation of lignin. In addition to fundamental studies on lignin mineralization, these enzymes also have potential uses in industrial processes such as the bleaching of paper pulp (1) or the remediation of xenobiotics in effluents (3). There is no clear relationship between the distribution of ligninolytic enzymes and lignin degradation, since white-rot fungi with only one, with a combination of two, or with all three enzymes are known and can degrade lignin (15, 27, 34).
The role of laccases in lignin degradation has only recently become well established (38). Laccases (p-diphenol oxidase, EC 1.10.3.2) are polyphenol oxidases that catalyze the reduction of oxygen to water with a concomitant oxidation of phenolic compounds. They are typically glycoproteins containing 2 to 4 atoms of copper per molecule and are found in plants and fungi (25, 33, 35).
The evergreen oak (Quercus ilex) forms a characteristic forest climax common in the Western Mediterranean area (22, 28). The leaf of Q. ilex, highly lignous and covered by a thick and waxy upper cuticle, is typical of sclerous plants exposed to dryness, particularly during the summer. The white-rot fungus Marasmius quercophilus colonizes dead leaves of Quercus ilex (32). Under favorable temperature and humidity conditions, i.e., in autumn and sometimes in May and June, this fungus becomes predominant among litter fungi and produces many rhizomorphs on leaf surfaces, strongly bleaching the whole leaf. The point where stalks or rhizomorphs come in contact with the leaf cuticle is outlined in black, indicating oxidation of phenolic compounds (16).
We have begun to study the ligninolytic system of M. quercophilus. When it is cultivated on malt medium, the only lignolytic activity produced by this fungus is a strong extracellular laccase (32). Several laccases may be responsible for this activity, since three isoenzymes have previously been detected in the culture supernatant (13, 32). Thus, our objectives in this study were (i) to describe further the extracellular laccase activity of M. quercophilus and (ii) to identify the biochemical properties of the enzyme(s) responsible for this activity. We report here the purification and characterization of the major laccase isoform and the cloning of the structural gene coding for this enzyme. Laccase 1 (LAC1) from M. quercophilus is remarkably stable over a wide range of temperatures. The biochemical properties and the amino acid sequence of this enzyme are very similar to those of a laccase purified from PM1, a previously unidentified basidiomycete, suggesting that M. quercophilus and PM1 could be conspecific.
MATERIALS AND METHODS
Enzyme production.
M. quercophilus was isolated as previously described (32) and propagated as mycelial cultures on malt agar plates (per liter: malt extract, 20 g; agar, 15 g). Precultures were obtained by inoculating mycelial fragments into 1-liter Erlenmeyer flasks containing 200 ml of a medium comprising, per liter, 20 g of malt extract, 0.1% Tween 80, and 0.5 mg of CuSO4; they were then incubated at 28°C on a reciprocal shaker (50 rpm) for 5 days. The preculture was used to inoculate 3-liter Erlenmeyer flasks containing 600 ml of the same medium, which were cultivated under the same conditions. Maximum laccase activity was reached after 5 days.
Laccase assay.
Laccase assay was based on syringaldazine oxidation (1.3 × 10−2 mM) in either 0.1 M phosphate buffer (pH 6.0) or 50 mM acetate buffer (pH 4.5) except where otherwise specified. The increase in A525 (ɛ = 65,000 M−1 cm−1) was monitored spectroscopically at 30°C (21). One unit of laccase oxidizes 1 μmol of syringaldazine per min (21). Syringaldazine was used as a substrate for the determination of the Km through Lineweaver-Burk plots. Potential inhibitors were assayed between 0.05 and 10 mM. Various compounds were tested as possible LAC1 substrates by polarography with a model 781 oxygen meter (Strathkelvin Instruments). The substrates, at a final concentration of 0.1 mM in 0.1 M phosphate buffer (pH 6.0), were the same as those previously tested with the crude extract from the fungus culture (32). Reactions were started by the addition of 0.2 U of laccase.
Enzyme purification.
The liquid culture (2 liters) was harvested, filtered successively through glass microfiber filters GFC and GFD (Whatman Ltd, Maidstone, England), concentrated 40 times by ultrafiltration using YM10 membranes (Amicon; Millipore, Bedford, Mass.), and buffered with 20 mM phosphate (pH 6.0) (buffer A). The subsequent purification steps were carried out at room temperature. The concentrated crude extract was applied to an ion-exchange Q-Sepharose column (2.6 by 40 cm; Amersham Pharmacia Biotech Europe GmbH, Freiburg, Germany) equilibrated with the same buffer. Proteins were eluted with a step gradient of NaCl at 0.17 M for 60 min and at 0.27, 0.35, 0.5, and 1 M concentrations for 40 min each. Fractions containing the major laccase activity were pooled and concentrated, and the buffer was changed to 0.2 M NaCl in 20 mM phosphate (pH 6.0) (buffer B). This solution was subjected to gel filtration using a Sephacryl S 200 column (2.6 by 100 cm; Amersham Pharmacia Biotech Europe GmbH) equilibrated with buffer B and eluted at a flow rate of 0.5 ml/min. The fractions containing laccase activity were again pooled, concentrated, and loaded onto a Chelating Sepharose column (1 by 5 cm; Amersham Pharmacia Biotech Europe GmbH) charged with copper (Cu2+) and equilibrated with buffer B. Proteins were eluted with a linear gradient of NH4Cl (0 to 1 M in 60 min) at a flow rate of 1 ml/min. The fractions containing laccase activity were pooled, and the enzyme purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Enzyme characterization.
Protein concentration was determined by the method of Bradford (5) with bovine serum albumin as a standard. SDS-PAGE analysis of proteins was performed on 7.5 to 12% polyacrylamide gels (19). Proteins were stained with Coomassie brilliant blue G250 (Sigma, St. Louis, Mo.). The molecular mass of laccase was estimated on an SDS-PAGE gel and by gel filtration on a Nucleogel GFC 300-8 column (0.77 by 30 cm; Macherey Nagel GmbH, Düren, Germany). Laccase activity was detected by native electrophoresis by incubating the gel at 25°C in either 0.2 M acetate buffer (pH 3.6) containing 0.2% p-phenylenediamine (modified from the buffer in reference 20) or in 0.1 M phosphate buffer (pH 6.0) containing 1 mM syringaldazine or guaiacol. We estimated the carbohydrate content of the laccase by comparison of the migration of native and N-glycanase (Roche Diagnostics GmbH, Mannheim, Germany)-treated enzyme on an SDS-PAGE gel. The carbohydrate composition was determined by gas chromatography on sillylated hydrolyzed sugars. The copper content was determined from purified laccase (157 μg/ml) in 10 mM phosphate buffer (pH 6.0) by atomic absorption spectroscopy on an Yvon Jobin (Longjumeau, France) JY 38 apparatus. Laccase absorption was determined on a Uvikon 860 spectrophotometer (Kontron Instruments, Milan, Italy). X band electron paramagnetic resonance (EPR) spectra were recorded on a Bruker (Wissembourg, France) ESP 300 spectrophotometer at 9.3 GHz and 16 K in 20 mM phosphate buffer (pH 6.0). The purified protein was subjected to cyanogen bromide treatment as described in reference 24; both N-terminal and internal CNBr peptide sequences were determined by stepwise Edman degradation. The optimum pH for the enzyme was determined using either 0.1 M phosphate buffer (pH 5.5 to 7.5), 0.1 M glycine-HCl solution (pH 3.0 to 5.0), or 50 mM acetate buffer (pH 4.0 to 5.5). The optimum temperature was determined between 30 and 85°C, and thermal stability was assessed between −20 and 90°C using phosphate buffer.
Gene cloning.
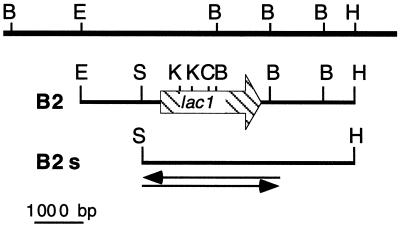
Laccase sequences in databases (GenBank and EMBL) were aligned using the Clustal W tool from the Mac Vector package (Oxford Molecular, Oxford, United Kingdom). Degenerate PCR primers were obtained from Eurogentec (Seraing, Belgium): the forward primer AK2, 5′C(G/I/C)AC(I/G)CAITA(I/C)TGTGA(I/C)GG3′, is based on the consensus peptide STQYCD found in copper-binding region II, and the reverse primer AK3, 5′TG(A/C)CCITCIAT(I/C)(G/C)IIAA(I/C)G3′, is based on the consensus peptide TFSIDGH found in the middle of the protein (nucleotides in parentheses represent minimal variations [degeneracy] for the same position). Genomic DNA was prepared from M. quercophilus as described in reference 29. The 477-bp AK2–AK3 PCR fragment was sequenced and used as a probe in a Southern hybridization experiment. EcoRI/HindIII genomic DNA fragments, ranging between 4,500 and 6,500 bp, purified from the agarose gel, were used to construct a partial genomic library in a pBluescript vector (Stratagene, La Jolla, Calif.). After transformation of XL1-Blue Escherichia coli cells and amplification, the library was screened with the labeled AK2–AK3 amplicon. Five positive clones were analyzed further by restriction mapping, and one, B2, representative of the five, was shortened to obtain a SacI/HindIII subclone (B2s). From this subclone, 3 kb covering the entire lac1 open reading frame was fully sequenced starting from the SacI end (Genomexpress, Grenoble, France).
Nucleotide sequence accession number.
The sequence of the M. quercophilus laccase gene lac1 reported in this paper has been assigned GenBank Data Library accession no. AF162785.
RESULTS
M. quercophilus laccase activity.
When cultured in shake flasks, M. quercophilus synthesizes laccase as the sole ligninolytic activity (32). Maximum laccase activity was obtained on malt medium supplemented with Tween 80 and copper. After 5 days, a syringaldazine oxidation activity of 0.8 U/ml was reached. From these 5-day-old cultures, native electrophoresis of the supernatant revealed an intense band of laccase activity and a few diffuse minor bands (data not shown).
Purification of the major extracellular laccase.
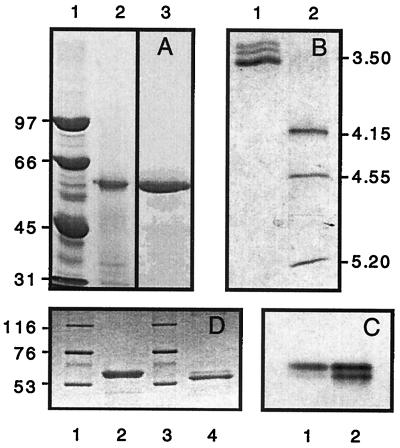
At the end of the purification (Table 1), 19 mg of purified enzyme was obtained with a specific activity of 13 U/mg, which corresponds to a final yield of 31%. The purified LAC1 appears to be a monomeric polypeptide of 62 kDa (by SDS-PAGE [Fig. 1A, lane 3]) or 63 kDa (by gel filtration [data not shown]). On an isoelectric focusing gel, this laccase has an isoelectric point of 3.6 and separates from two minor bands of lower pI (Fig. 1B, lane 1). A similar pattern is seen on native gels where laccase migrates as a doublet, with most of the activity in the upper band (Fig. 1C). This variation is probably due to glycosylation heterogeneity in the preparation (33).
TABLE 1.
Purification of laccase 1 activity from M. quercophilus
| Step | Vol (ml) | Protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Supernatant | 2,000 | 144.0 | 800 | 5.6 | 100 | |
| Ultrafiltration | 50 | 106.2 | 718 | 6.9 | 90 | 1.2 |
| Q-Sepharosea | 26 | 39.0 | 401 | 10.3 | 50 | 1.9 |
| Sephacryl S-200 | 20 | 26.0 | 289 | 11.1 | 36 | 2.0 |
| Chelating Sepharose (Cu)b | 20 | 18.8 | 245 | 13.0 | 31 | 2.4 |
About 60% of the laccase activity eluted as a single peak early in the gradient (within 20 min at 170 mM NaCl), while 10% eluted as two peaks late in the gradient (350 mM NaCl). The fractions corresponding to the first peak were pooled and further purified.
Laccase 1 eluted as a single peak in the fractions corresponding to 0.4 to 0.6 M ammonium sulfate.
FIG. 1.
Electrophoresis of the purified laccase from M. quercophilus. (A) SDS–7.5% PAGE of purified laccase from M. quercophilus. Lane 1, standard proteins; lane 2, 20 μg of crude extract; lane 3, 10 μg of purified laccase. (B) Isoelectric focusing of purified laccase from M. quercophilus. Lane 1, laccase; lane 2, standards. Proteins were stained with Coomassie brilliant blue. (C) Native PAGE of purified laccase from M. quercophilus. Lane 1, 1 μg of purified laccase; lane 2, 3 μg of the same sample. Protein was stained with guaiacol. (D) Lanes 1 and 3, standard proteins; lane 2, untreated laccase; lane 4, laccase treated with N-glycanase. Proteins were stained with Coomassie brilliant blue. Dried gels were scanned with an Agfa Snapscan 1236 piloted with Fotolook 2.09.6 software. Legends were added with Canvas 3.0.6 software.
Glycosylation pattern.
Enzymatic deglycosylation of the purified enzyme with N-glycanase followed by SDS-PAGE analysis allowed us to estimate the sugar content to be around 12% of the molecular mass (Fig. 1D). We used gas chromatography on sillylated hydrolyzed sugars to estimate the composition of the carbohydrate part of the enzyme and found the following: mannose, 58%; glucose, 14.5%; rhamnose, 14.5%; N-acetylglucosamine, 7%; and xylose, 6%.
Spectroscopic characterization.
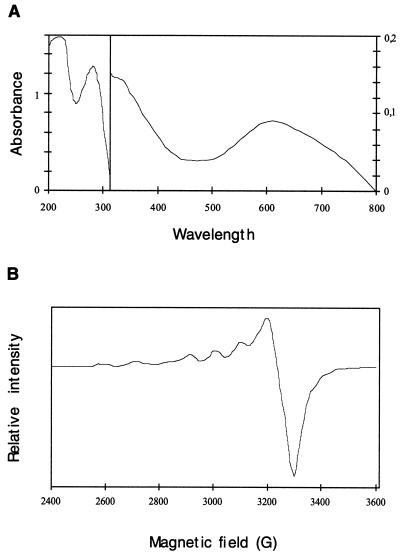
The purified enzyme has the typical color of blue copper oxidases and contains 4.0 atoms of copper per molecule. The presence of a type 1 copper atom was deduced from the UV/visible spectrum (31) of the purified enzyme (Fig. 2A), which shows a broad peak at 611 nm (ɛ = 5,200 M−1 cm−1). In this spectrum, the presence of a shoulder at 333 nm could indicate a binuclear type 2 copper [Cu(II)] complex (12). Finally, the EPR spectrum (Fig. 2B) revealed the presence of two Cu(II) ions in a different coordination environment, which correspond to the type 1 atom copper (/Az/ < 95 10−4 cm−1) and the type 2 copper atom (/Az/ > 140 10−4 cm−1) (12).
FIG. 2.
Spectroscopic characterization of the purified laccase. (A) Absorbance spectrum of purified laccase from M. quercophilus (1.09 mg/ml in 20 mM phosphate buffer, pH 6) at 30°C. (B) EPR spectrum of purified laccase from M. quercophilus (1.09 mg/ml in 20 mM phosphate buffer, pH 6.0) at 9.3 GHz and 16 K.
Physicochemical and kinetic properties.
The optimum pH for the enzyme was 4.5 in 0.1 M glycine-HCl (pH 3.0 to 5.0); the temperature optimum was 75°C, and the enzyme was stable at 60°C for more than 1 h. Laccase activity is 1.6 times higher at 75°C than at 30°C and is stable for 10 min, while the enzyme is totally inactivated within 15 min at higher temperatures (80 to 90°C). After 5 months of storage in phosphate buffer at either −20 or 4°C, the enzyme retains 60% of its initial activity. The Km was 7.1 μM when syringaldazine was used as a substrate. EDTA, sodium thioglycolate, sodium dodecyl dithiocarbamate, and sodium azide were tested as possible inhibitors. EDTA did not inhibit the laccase at the concentration used. Sodium thioglycolate and sodium dodecyl dithiocarbamate inhibited laccase activity at concentrations of >1 mM; sodium azide inhibited the reaction over the entire range of concentrations (0.05 to 10 mM) tested, and the inhibition was total if the concentration was >1 mM.
Substrate spectrum.
Several phenolic compounds were tested as potential laccase substrates both with the crude extract and with the purified enzyme (Table 2). ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] was chosen as a reference because only one oxidation step can occur with this compound. In agreement with the stoichiometry of the reaction, a ratio close to 0.25 mol of O2 consumed per mol of ABTS was found both with the crude extract and with the purified enzyme. Values ranging from 0.4 to 1.3 were found for the other substrates which can undergo multiple oxidation, except for veratrylic alcohol, for which no oxygen consumption was measured. For all but two of the compounds tested, catechol and coumarylic alcohol, the amount of oxygen necessary to oxidize the substrate was higher in the crude extract than in the purified enzyme solution.
TABLE 2.
Comparison of phenolic compound oxidation in the crude extract versus laccase 1 purified solution
| Substrate | O2/substrate (mol/mol)a
|
|
|---|---|---|
| Laccase 1 | Crude extract | |
| ABTS | 0.24 ± 0.01 | 0.26 ± 0.01 |
| Catechol | 0.56 ± 0.02 | 0.53 ± 0.02 |
| 4-Methyl catechol | 0.40 ± 0.01 | 0.55 ± 0.01 |
| Protocatechuic acid | 0.40 ± 0.00 | 0.59 ± 0.02 |
| Guaiacol | 0.47 ± 0.02 | 0.58 ± 0.02 |
| Coniferylic acid | 0.66 ± 0.02 | 0.76 ± 0.03 |
| Coumarylic alcohol | 0.48 ± 0.02 | 0.37 ± 0.02 |
| Sinapylic alcohol | 0.51 ± 0.04 | 0.91 ± 0.04 |
| Ferulic acid | 0.40 ± 0.04 | 0.64 ± 0.03 |
| Gallic acid | 1.00 ± 0.03 | 1.28 ± 0.05 |
| Caffeic acid | 0.40 ± 0.02 | 0.59 ± 0.03 |
| Veratrylic alcohol | 0 | 0 |
Values are means from three experiments.
N-terminal and CNBr peptide sequence analysis.
Twenty micrograms of the purified protein was first reduced, carboxymethylated, and subjected to Edman degradation. The first 20 residues at the amino terminus are SIGPVADLTISNGAVSPDGF. This sequence is identical to that of the amino terminus of the laccase from basidiomycete PM1 (8) (accession number Z12156) and closely related to those of basidiomycete CECT 20197 (23) (accession number U65400). Trametes villosa (36, 37) (accession numbers L49376 and L78077), and Trametes versicolor (4) (accession number Y18012). We also sequenced a 15-kDa internal peptide containing the first consensus copper site (HWHGFFQ) and found that the first 23 residues from this peptide, AFGFAGGRFTINGASFTPPTVPV, were again a perfect match with the PM1 enzyme.
The lac1 structural gene.
The gene encoding LAC1 was cloned from a partial genomic DNA library (Fig. 3). The lac1 gene from M. quercophilus is nearly identical to its counterpart from the unknown basydiomycete PM1 (8). Among the 2,783 nucleotides comparable with the data available for PM1, we found 118 mismatches representing a total of 4.2% divergence. These differences are primarily found in the 3′ noncoding region and in introns (85 differences in the 3′ region, 20 in introns, 12 in exons, and 1 in the 5′ region). At the protein level, there is only one amino acid difference, as codon 199 (exon 6) specifies a serine (AGC) instead of an asparagine (AAC) in the PM1 sequence (8).
FIG. 3.
Restriction map of the cloned genomic DNA. The top line contains restriction sites deduced from a genomic Southern blot (not shown). The hatched arrow on the second line represents the lac1 open reading frame. The bottom line represents clone B2s; arrows represent the portion of the clone which was sequenced on both strands. Restriction enzymes: B, BamHI; C, ClaI; E, EcoRI; H, HindIII; K, KpnI.
DISCUSSION
In this paper, we describe the purification of an extracellular laccase from Marasmius. Members of this genus are saprophytes that colonize decaying litter (11). M. quercophilus can degrade evergreen oak litter. When grown on ground oak leaves, this fungus produces laccase and Mn peroxidase as lignolytic enzymes, but on malt extract, only laccase activity is found (32). The properties of the major extracellular laccase (LAC1) produced by M. quercophilus are typical of laccases associated with white-rot fungi (33). However, unusual characteristics include insensitivity to 10 mM EDTA, heat tolerance (stable for more than 1 h at 60°C), and maximum activity at 75°C. Thus, LAC1 is among the most heat-resistant laccases known (7, 14). The stability of this enzyme over a wide range of temperatures probably reflects structural properties necessary to preserve activity in a Mediterranean ecosystem. As activity at high temperatures is usually an absolute requirement for industrial applications, LAC1 may be a good target for the development of biotechnological tools.
Laccases are nonspecific enzymes that can oxidize numerous compounds. However, variations in substrate utilization may reflect differences in the role played by these enzymes in litter mineralization. Among the phenolic compounds we tested as potential LAC1 substrates, only veratrylic alcohol was not oxidized (Table 2). This substance is a substrate for lignin peroxidase, and so far only laccase from Phlebia radiata has been found to oxidize it (22a). Most of the other substrates were oxidized to a greater extent when tested with the crude extract than when tested with the purified enzyme. This is particularly true for sinapylic alcohol, for which the oxygen consumption per mole of substrate in the crude extract is 1.8 times that found in the purified enzyme fraction. This difference suggests that the crude extract contains at least one other oxidase activity in addition to LAC1. Since neither peroxidase nor tyrosinase activities have been detected in the M. quercophilus malt culture supernatant (32), this oxidase activity may come from the other laccase isozymes secreted by this fungus. Laccases are often encoded by gene families that produce isoforms, allowing a modulation of the degradation potential with subtle variations in activity through differential regulation. To fully understand the involvement and the specific role of M. quercophilus laccases in evergreen oak litter mineralization, it is necessary to study these isoforms.
The lac1 structural gene is very similar (96%) to a laccase gene in PM1 (7) (accession number Z12156). At the protein level, the homology is even more striking because the M. quercophilus and PM1 laccases differ only at residue 199, which has a serine (AGC) in the former and an asparagine (AAC) in the latter. To our knowledge, this is the closest known relationship between two laccases. By comparison, the Coriolus versicolor clv3 gene (26) (accession number D84235) and the T. villosa lcc1 gene (37) (accession number L78077) encode identical proteins except for 6 amino acids. On the other hand, the laccase isolated from Coriolus hirsutus (18) (accession number Q02497) differs at 48 amino acids from its counterpart isolated from C. versicolor (product of the clv3 gene) (37). The PM1 laccase and the M. quercophilus LAC1 share the same optimum pH, the same pI, nearly the same heat resistance, and the same range of specific activity (as measured on guaiacol in acetate buffer [pH 4.5]). The only significant difference is the carbohydrate content: 12 and 6.5% for M. quercophilus and PM1 (7), respectively. These similarities suggest that these fungi may be conspecific, yet they have been isolated in two different states, microscopic for PM1 and macroscopic for M. quercophilus, and from two different media and countries: paper factory wastewater in Spain for PM1 and evergreen oak decaying litter in France for M. quercophilus. This similarity could be explained by the ecological distribution of M. quercophilus, which is closely linked to the distribution of its host (2). This fungus colonizes many types of fallen leaves, primarily from Quercus species but also from Castenea sativa and occasionally from Fagus (2). Moreover, Quercus ilex is widely distributed in Spain (28). Further study of the genomes of these two basidiomycetes should provide more insight into their potential conspecificity.
In conclusion, we have demonstrated that the laccase activity found in the liquid culture of M. quercophilus can be primarily attributed to a single protein present in relatively large amounts. The crude enzyme from malt culture, which may have industrial utility, can reduce the kappa number of a kraft pulp by 21.2% in 5 days (32). To obtain a 24% reduction of the kappa number, Bourbonnais et al. combined a mediator, ABTS, with a laccase purified from Trametes versicolor (4). Therefore, it will be interesting to test the bleaching capability of the purified LAC1 with or without inclusion of a mediator. From an ecological point of view, it is now important to determine the effects of environmental factors on the activity of this enzyme when it is liberated in the litter. For example, minerals present in the litter are probably important, since they can act either as activators or as inhibitors of enzymatic activity. Similarly, the role of humic substances, which are associated with laccases in litter (9, 10), must be established, since they may act as protease inhibitors (10), or contain laccase inhibitors.
ACKNOWLEDGMENTS
We thank the “Société Nationale Elf Aquitaine” for financial support. A. Klonowska is a recipient of an Agence de l'Environement et de la Maîtrise de l'Energie (ADEME) fellowship.
We thank J.-C. Michalski for performing carbohydrate analysis and A. Fournel for the EPR spectroscopy.
REFERENCES
- 1.Akhtar M, Attridge M, Myers G C, Blanchette R A, Kirk T K. Biochemical pulping of loblolly pine with different strains of the white rot fungus Ceriporiopsis subvermispora. TAPPI J. 1992;75:105–109. [Google Scholar]
- 2.Antonin V, Nordeloos M E. A monograph of Marasmius, Collybia and related genera in Europe. Part 1: Marasmius, Setulipes and Marasmiellus Libri botanici. Vol. 8. Eching, Germany: IHW Verlag; 1993. [Google Scholar]
- 3.Bar D P, Aust S D. Mechanisms white rot fungi use to degrade pollutants. Environ Sci Technol. 1994;28:78–87. doi: 10.1021/es00051a724. [DOI] [PubMed] [Google Scholar]
- 4.Bourbonnais R, Paice M G, Reid I D, Lanthier P, Yaguchi M. Lignin oxidation by laccase isoenzymes from Trametes versicolor and role of the mediator 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) in Kraft lignin depolymerization. Appl Environ Microbiol. 1995;61:1876–1880. doi: 10.1128/aem.61.5.1876-1880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Buswell J A, Odier R. Lignin biodegradation. Crit Rev Biotechnol. 1987;6:1–60. [Google Scholar]
- 7.Coll P M, Tabernero C, Santamaria R, Perez P. Characterization and structural analysis of the laccase 1 gene from the newly isolated ligninolytic basidiomycete PM1 (CECT 2971) Appl Environ Microbiol. 1993;59:4129–4135. doi: 10.1128/aem.59.12.4129-4135.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coll P M, Fernandez-Abalos J M, Villanueva J R, Santamaria R, Pérez P. Purification and characterization of a phenoloxidase (laccase) from the lignin-degrading basidiomycete PM1 (CECT 2971) Appl Environ Microbiol. 1993;59:2607–2613. doi: 10.1128/aem.59.8.2607-2613.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Criquet S. Ph.D. thesis. Marseille, France: University of Aix-Marseille III; 1999. [Google Scholar]
- 10.Dilly O, Nannipieri P. Intracellular and extracellular enzyme activity in soil with reference to elemental cycling. Z Pflanzenernähr Bodenkd. 1998;161:243–248. [Google Scholar]
- 11.Durrieu G. Ecologie des champignons. Paris, France: Masson; 1993. [Google Scholar]
- 12.Eggert C, Temp U, Eriksson K-E. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl Environ Microbiol. 1996;62:1151–1158. doi: 10.1128/aem.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnet A-M, Tagger S, Le Petit J. Effects of copper and aromatic inducers on the laccases of the white-rot fungus Marasmius quercophilus. C R Acad Sci Ser III Life Sci. 1999;322:499–503. [Google Scholar]
- 14.Fukushima Y, Kirk T K. Laccase component of the Ceriporiopsis subvermispora lignin-degrading system. Appl Environ Microbiol. 1995;61:872–876. doi: 10.1128/aem.61.3.872-876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatakka A. Lignin-modifying enzymes from selected white-rot fungi: production and role in lignin degradation. FEMS Microbiol Rev. 1994;13:125–135. [Google Scholar]
- 16.Hintikka V. The colonization of litter and wood by basidiomycetes in Finnish forest. In: Frakland J C, Hedger J N, Swift M J, editors. Decomposer basidiomycetes: their biology and ecology. Cambridge, United Kingdom: Cambridge University Press; 1982. pp. 213–226. [Google Scholar]
- 17.Kirk T K, Farell R L. Enzymatic “combustion”: the microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 18.Kojima Y, Tsukuda Y, Kawai Y, Tsukamoto A, Sugiura J, Sakaino M, Kita Y. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Leonowicz A, Trojanowski J. Induction of a new laccase form in the fungus Pleurotus ostreatus by ferulic acid. Microbios. 1975;13:167–174. [Google Scholar]
- 21.Leonowicz A, Grzywnowicz K. Quantitative estimation of laccase form in some white-rot fungi using syringaldazine as a substrate. Enzyme Microb Technol. 1981;3:55–58. [Google Scholar]
- 22.Lossaint P, Rapp M. La forêt Mediterranéenne de chênes verts. In: Lamotte M, Bourlière I, editors. Problèmes d'écologie. Ecosystèmes terrestres. Paris, France: Masson; 1978. pp. 129–185. [Google Scholar]
- 22a.Lundell T, Hatakka A. Participation of Mn(II) in the catalysis of laccase, manganese peroxidase and lignin peroxidase from Phlebia radiata. FEBS Lett. 1994;348:291–296. doi: 10.1016/0014-5793(94)00627-x. [DOI] [PubMed] [Google Scholar]
- 23.Mansur M, Suarez T, Fernandez-Larrea J B, Brizuela M A, Gonzalez A E. Identification of a laccase gene family in the new lignin-degrading basidiomycete CECT 20197. Appl Environ Microbiol. 1997;63:2637–2646. doi: 10.1128/aem.63.7.2637-2646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsudaira P. Limited N-terminal sequence analysis. Methods Enzymol. 1990;182:602–613. doi: 10.1016/0076-6879(90)82047-6. [DOI] [PubMed] [Google Scholar]
- 25.Mayer A M. Polyphenol oxidases in plants. Recent progress. Phytochemistry. 1987;26:11–20. [Google Scholar]
- 26.Mikuni J, Morohoshi M. Cloning and sequencing of a second laccase gene from the white-rot fungus Coriolus versicolor. FEMS Microbiol Lett. 1997;155:79–84. doi: 10.1111/j.1574-6968.1997.tb12689.x. [DOI] [PubMed] [Google Scholar]
- 27.Nerud F, Misurcova Z. Distribution of ligninolytic enzymes in selected white-rot fungi. Folia Microbiol. 1996;41:264–266. [Google Scholar]
- 28.Ozenda P. Les végétaux dans la biosphère. Paris, France: Doin; 1982. [Google Scholar]
- 29.Raeder U, Broda P. Preparation and characterization of DNA from lignin-degrading fungi. Methods Enzymol. 1988;161:211–220. [Google Scholar]
- 30.Reid I D. Biodegradation of lignin. Can J Bot. 1995;73:S1011–S1018. [Google Scholar]
- 31.Rheinhammar B. Laccase. In: Lontie R, editor. Copper proteins and copper enzymes. Boca Raton, Fla: CRC Press, Inc.; 1984. pp. 4–10. [Google Scholar]
- 32.Tagger S, Périssol C, Gil G, Vogt G, Le Petit J. Phenoloxidases of the white-rot fungus Marasmius quercophilus isolated from an evergreen oak litter (Quercus ilex L.) Enzyme Microb Technol. 1998;23:372–379. [Google Scholar]
- 33.Thurston C F. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- 34.Tuor U, Winterhalter K, Fiechter A. Enzymes of white-rot fungi involved in lignin degradation and ecological determinants for wood decay. J Biotechnol. 1995;41:1–17. [Google Scholar]
- 35.Yaropolev A I, Skorobogat'ko O V, Vartanov S S, Varfolomeyev S D. Laccase—properties, catalytic mechanism, and applicability. Appl Biochem Biotechnol. 1994;49:257–280. [Google Scholar]
- 36.Yaver D S, Xu F, Golightly E J, Brown K M, Brown S H, Rey M W, Schneider P, Halkier T, Mondorf K, Dalboge H. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Env Microbiol. 1996;62:834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaver D S, Golightly E J. Cloning and characterization of three laccase genes from the white-rot basidiomycete Trametes villosa: genomic organization of the laccase gene family. Gene. 1996;181:95–102. doi: 10.1016/s0378-1119(96)00480-5. [DOI] [PubMed] [Google Scholar]
- 38.Youn H-D, Hah Y C, Kang S-O. Role of laccase in lignin degradation by white-rot fungi. FEMS Microbiol Lett. 1995;132:183–188. [Google Scholar]