Abstract
A cultivation-independent technique for genetic profiling of PCR-amplified small-subunit rRNA genes (SSU rDNA) was chosen to characterize the diversity and succession of microbial communities during composting of an organic agricultural substrate. PCR amplifications were performed with DNA directly extracted from compost samples and with primers targeting either (i) the V4–V5 region of eubacterial 16S rRNA genes, (ii) the V3 region in the 16S rRNA genes of actinomycetes, or (iii) the V8–V9 region of fungal 18S rRNA genes. Homologous PCR products were converted to single-stranded DNA molecules by exonuclease digestion and were subsequently electrophoretically separated by their single-strand-conformation polymorphism (SSCP). Genetic profiles obtained by this technique showed a succession and increasing diversity of microbial populations with all primers. A total of 19 single products were isolated from the profiles by PCR reamplification and cloning. DNA sequencing of these molecular isolates showed similarities in the range of 92.3 to 100% to known gram-positive bacteria with a low or high G+C DNA content and to the SSU rDNA of γ-Proteobacteria. The amplified 18S rRNA gene sequences were related to the respective gene regions of Candida krusei and Candida tropicalis. Specific molecular isolates could be attributed to different composting stages. The diversity of cultivated bacteria isolated from samples taken at the end of the composting process was low. A total of 290 isolates were related to only 6 different species. Two or three of these species were also detectable in the SSCP community profiles. Our study indicates that community SSCP profiles can be highly useful for the monitoring of bacterial diversity and community successions in a biotechnologically relevant process.
Composting is the biological conversion of solid organic material into usable end products such as fertilizers, substrates for mushroom production, or biogas (methane). Regardless of the product, the active component mediating the biodegradation and conversion processes during composting is the resident microbial community. Therefore, optimization of compost quality is directly linked to the composition and succession of microbial communities in the composting process. This means that tools are required to monitor and characterize microbial communities during the composting processes and to relate microbial communities to compost quality.
Cultivation of microorganisms extracted from compost samples allows one to obtain pure cultures which can be used for further taxonomic or physiological characterizations (2, 4, 11, 13, 14, 16, 47). Rapid molecular PCR-based techniques, such as amplified ribosomal DNA restriction analysis (ARDRA), are useful for comparison of a large number of isolates at the phylogenetic level (48). This technique allowed the characterization of Thermus strains and Bacillus-related bacteria isolated from hot composting material (5, 6). However, since any chosen cultivation approach will inevitably favor the growth of some community members while others are inhibited or not culturable at all, it is unlikely that any cultivation method will allow a full description of the microbial diversity. Therefore, cultivation-independent methods have recently been used to characterize microbial-community successions during composting. These include assessment of the diversity of directly extracted phospholipids (9, 21, 25), measurement of carbon source utilization by substrate-extracted microbial cell consortia (9, 24, 42), and nucleic acid-based techniques (29).
Several protocols have been used to directly extract total DNA from compost material. This approach, in combination with PCR-mediated gene detection, allowed the detection of specific genes of pathogenic microorganisms (37) or monitoring of the fate of recombinant DNA from decaying plant material (26) and the persistence of seeded microorganisms (31). In a recent study, bacterial 16S rRNA gene sequences were amplified from compost DNA and, after cloning in Escherichia coli, were characterized by restriction enzyme analysis (7). The results indicated that some molecular clones were identical to genes of cultivated species, but others could not be matched. Also, the study showed repetitive isolation of identical products. Such repetitive isolations can be avoided if the community DNA-amplified PCR products are electrophoretically separated to generate sequence-specific genetic profiles. Denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) have become popular for this purpose in biodiversity studies of microbial communities from a variety of habitats (8, 12, 22, 32, 43). Kowalchuk and coworkers recently used this technique to characterize ammonia-oxidizing bacteria in composts (29). However, this approach has not yet been used to study microbial-community succession during composting at a broader phylogenetic level.
As an alternative to DGGE and TGGE, we recently developed a protocol which allows the application of single-strand-conformation polymorphism (SSCP) (18, 34) for the cultivation-independent assessment of microbial-community diversity (41). In contrast to DGGE and TGGE, no GC clamp or construction of gradient gels is required, and thus the SSCP method has the potential to be more easily applied (30). We have used the method to compare rhizosphere microbial communities of different plants (41). Here we report on the use of the SSCP approach to characterize microbial-community successions during composting. The method was used to monitor the production of mushroom composts in an 18-day-long, self-heating composting process.
MATERIALS AND METHODS
Composting, sampling, and chemical analyses.
Two separate composting windrows were set up for this investigation. Each windrow consisted of a wooden box (area, 1.6 m2; height, 2.0 m) filled with a mixture of field-grown shredded maize plants (Zea mays) (750 kg), 0.5 m3 of wood chips, and 10% straw-bedded horse manure. This mixture was wetted before composting for the initiation of the self-heating phase. The windrows were turned every 2 to 3 days in order to enhance the composting process and avoid the formation of anaerobic compartments. The temperature was continuously measured at three depths (20, 30, and 50 cm) in the composting pile, using measuring lances to monitor the process. Replicate samples were taken from a 50-cm depth. Chemical properties of the compost samples (pH, organic carbon, total nitrogen, ammonium, and nitrate) were analyzed by standard methods (35). Samples designated for DNA extraction were stored immediately at −20°C.
Isolation and characterization of pure bacterial cultures from compost.
In order to obtain pure cultures from composting material, samples taken at the end of the composting process (18 days) were suspended in sodium polyphosphate solution (0.2%, wt/vol). Dilutions of this suspension were inoculated onto plate count agar (PCA; Oxoid Unipath Ltd., Basingstoke, United Kingdom) supplemented with 50 mg of cycloheximide liter−1 for suppression of fungal growth. The inoculated plates were incubated at 50°C for 16 h before single colonies were transferred to fresh agar and subcultured.
The ARDRA technique was used for the characterization of isolates (48). Colonies of bacterial cells grown were suspended in 50 μl of lysis buffer (0.05 M NaOH–0.25% [wt/vol] sodium dodecyl sulfate) and incubated for 15 min at 95°C. The suspensions were diluted with 450 μl of water and centrifuged in a microcentrifuge at the highest setting for 5 min at room temperature. An aliquot (1 to 5 μl) of the centrifuged solution was used as a template for PCR. A 1,060-bp product from the eubacterial 16S rRNA genes was amplified with primers Ec41(f) and Ec1066(r) as described elsewhere (23). A total of 5 μl of each PCR product was incubated with 5 U of a restriction endonuclease (CfoI and RsaI, separately; both from New England Biolabs, Schwalbach, Germany) and buffer solution supplied by the manufacturer in a final volume of 20 μl for 1 h at 37°C. The restriction fragments were analyzed on 7% (wt/vol) polyacrylamide gels with 1× TBE (Tris-borate-EDTA buffer [40]) and 7 M urea (Roth, Karlsruhe, Germany) using a Multiphor II apparatus (Pharmacia Amersham Biotech, Freiburg, Germany). Products were stained with silver nitrate (3).
Sequencing of the PCR-amplified 16S rRNA genes (1,060 bp) obtained from pure cultures was conducted by IIT Bioservice, Bielefeld, Germany. Alignments and data bank identifications were carried out using BLASTN 2.0.4 (1).
Extraction of DNA from compost material and generation of PCR-SSCP genetic profiles.
Compost samples were ground in liquid nitrogen, and total DNA was extracted from samples of 200 mg (wet weight) using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Compost DNA was eluted from DNeasy columns with 150 μl of manufacturer-supplied AE buffer. A 10−2 dilution of the eluate was used for PCR. The DNA concentration was measured fluorometrically using Pico Green dye (Molecular Probes, Leiden, The Netherlands) and a microtiter plate reader (Fluoroskan II; Labsystems, Helsinki, Finland).
Three different primer systems were used to amplify 16S rRNA or 18S rRNA genes from total community DNA of compost (Table 1). For SSCP, reverse primers were phosphorylated at the 5′ end. Each PCR was performed in a total volume of 100 μl in micro-test tubes (Flat Cap Micro Tubes; MWG Biotech, Ebersberg, Germany). Reaction mixtures contained 1× PCR buffer with 1.5 mM MgCl2, deoxynucleoside triphosphates (200 μM each dATP, dCPT, dGTP, and dTTP), 0.5 μM each primer, 3.75 U of DNA polymerase (Expand-Taq; Roche Diagnostics, Mannheim, Germany), and 2 μl of the diluted DNA extract. To increase amplification efficiencies from compost DNA, T4 gene 32 protein was added at a concentration of 5 μg per reaction (Roche Diagnostics) (46). Cycle conditions for the reactions using Com and NS primers (Table 1) were as follows: initial denaturation at 94°C for 3 min; 35 cycles of 94°C for 60 s, 50°C for 60 s, and 72°C for 70 s; and a final elongation for 5 min at 72°C. For PCR with primers F243 and R531, the annealing temperature was increased to 60°C. PCR products were purified (Qiaquick; Qiagen), and 15 μl of purified product was subsequently digested with 10 U of lambda exonuclease (Pharmacia Amersham Biotech) in a final volume of 50 μl to obtain single-stranded DNA molecules (41). The single-stranded DNA molecules were purified and separated on a 0.6× MDE polyacrylamide gel (FMC Bioproducts, Rockland, Maine) according to our previously described protocol (41). To make the SSCP products visible, gels were silver stained (3).
TABLE 1.
Characterization of primers used for SSCP-based microbial-community analyses
| Primer name | Primer sequence (5′–3′) | Primer target (positions)a | Targeted variable region(s) of the small-subunit rRNA genesb | Primer annealing temp during PCR (°C) | Reference |
|---|---|---|---|---|---|
| Com1 | CAG CAG CCG CGG TAA TAC | Bacteria (519–536) | V4 and V5 | 50 | 41 |
| Com2 | CCG TCA ATT CCT TTG AGT TT | Bacteria (907–926) | |||
| F243 | GGA TGA GCC CGC GGC CTA | Actinomycetes (226–243) | V3 | 60 | 22 |
| R531 | CGG CCG CGG CTG CTG GCA CGT A | Actinomycetes (512–534) | |||
| NS7 | GAG GCA ATA ACA GGT CTG TGA TGC | Fungi (1185–1207) | V8 and V9 | 50 | 49 |
| NS8 | TCC GCA GGT TCA CCT ACG GA | Fungi (1508–1527) |
Isolation and cloning of single products from SSCP community profiles.
Selected products (“bands”) identified in MDE polyacrylamide gels after silver staining were excised with razor blades, and single-stranded DNA was eluted from the gel by a “crush and soak” procedure described by Sambrook et al. (40) and modified by Schwieger and Tebbe (41). The single-strand DNA molecules were reamplified by PCR using the same primers and conditions as for the respective SSCP analysis. The resulting PCR products were analyzed by SSCP for purity and identity by comparing them with the original fragments in the community profiles.
For cloning, the reamplified PCR products were subjected to another PCR. Com primer-amplified products (including V4 and V5; see Table 1) were used in PCR with two phosphorylated primers and Vent polymerase (New England Biolabs). The amplicons generated, which were blunt ended and phosphorylated, were further purified using a purification kit (Qiaquick; Qiagen). These fragments were ligated into the dephosphorylated plasmid vector pUC18 and cut with SmaI (Appligene Oncor, Heidelberg, Germany), and 5 μl of the ligation mix was used for the electroporation of competent E. coli strain JM 109 cells (Promega, Mannheim, Germany). For subcloning of amplicons generated with primer pair NS7–NS8 or F243–R531, PCR-mediated reamplifications were carried out under the same conditions and with the same primers as those used for amplifications from compost DNA, except that the reverse primer was also dephosphorylated. The purified PCR products were ligated into the plasmid vector pGEM-T (Promega) in a reaction volume of 10 μl. Half of the ligation mix was used to transform E. coli JM 109 supercompetent cells by a protocol provided by the manufacturer.
The presence of inserts of the expected sizes was confirmed by PCR with the flanking vector primers M13 forward and M13 reverse (Promega). For this purpose, ampicillin-resistant colonies were treated as described for ARDRA (23), and 2 μl of lysate was used as a template in PCR with the same cycle conditions as those described for the Com primers. The sizes of these PCR products were controlled on 1.2% (wt/vol) agarose gels (40). Amplicons whose sizes corresponded to those of the original fragments were purified (Qiaquick; Qiagen). These purified products were used directly for double-stranded sequence analysis.
DNA sequencing of subcloned SSCP products.
For DNA sequencing, infrared dye 800-labeled M13 sequencing primers (MWG Biotech) were used in combination with the Thermosequenase sequencing kit with 7-deaza GTP (Pharmacia Amersham Biotech). Primer and template concentrations and PCR reagents were selected according to the instructions of the manufacturer (Pharmacia Amersham Biotech). Conditions for the cycle sequencing process, which was conducted in a Primus 96 thermocycler (MWG Biotech), were as follows: initial denaturation at 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 54°C for 20 s, and 72°C for 60 s. The sequences were automatically analyzed on a 6% (wt/vol) polyacrylamide gel (Rapid Gel XL; Pharmacia Amersham Biotech) using a LI-COR 4200 (MWG Biotech). Alignments and database identifications of the consensus sequences were carried out using BLASTN 2.0.4 (1).
Nucleotide sequence accession numbers.
The new DNA sequences described in this study have been deposited in the GenBank sequence database under accession numbers AF213262 to AF213286.
RESULTS
Conditions during the composting process.
The composting process for the preparation of the mushroom compost lasted 18 days. The initial temperature in the composting windrows was 20°C. Within 3 days of incubation the temperature increased to 40°C at a depth of 80 cm, 60°C at 50 cm, and 70°C at 30 cm. During further incubation, interrupted by the turning of the material every 3 to 4 days, the temperatures increased to a maximum of 80°C at all three depths after 10 days. From 10 days to 18 days, during the maturation phase, temperatures decreased slowly but continuously to 65 to 70°C. The pH decreased within the first 2 days from 5.6 to 4.5. Later, the pH increased to 7.0. Nitrate was detectable only during the first 3 days of incubation (threshold of detection, 1 mg kg−1). Ammonia levels increased during the process from 4 to 22 mg kg−1. The total nitrogen concentration decreased from 17 to 14 g kg−1 from day 0 to day 3. Subsequently, from day 3 to day 18, the concentration increased to 20 g kg−1. In summary, the parameters indicated a typical composting process of organic agricultural material as desired for the production of mushroom cultivation substrate.
SSCP genetic profiles of microbial communities during the composting process.
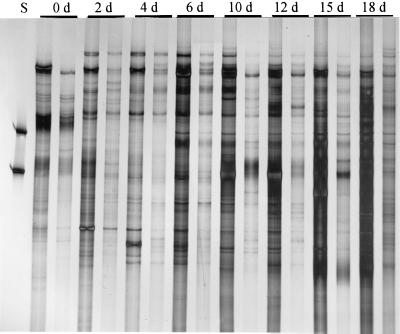
Primers designed to amplify the eubacterial 16S rRNA gene sequences including the variable V4 and V5 regions yielded complex SSCP patterns on polyacrylamide gels. The number of bands increased during the composting process, and at the end of the process (18 days) the profiles consisted of more than 30 different products (bands) (Fig. 1). Specific products (bands) occurred with similar intensities (yields) in profiles obtained from two separate composting windrows. The successions of patterns obtained from the two windrows were similar, which indicated good reproducibility of the DNA extraction method and PCR amplifications.
FIG. 1.
Succession of PCR-amplified products during a composting process as detected by SSCP on a polyacrylamide gel. PCR primers were designed to amplify the hypervariable regions V4 and V5 of eubacterial 16S rRNA genes from directly extracted compost DNA (S, standard DNA). For each day (d), results obtained from two separate composting windrows are shown.
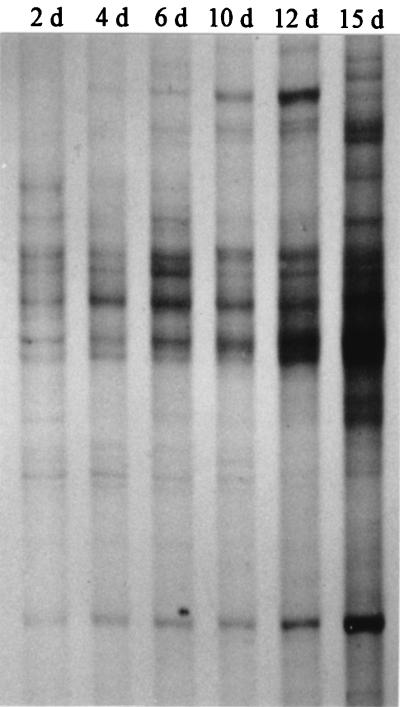
With primers targeting the V3 region of actinomycete 16S rRNA genes, the SSCP patterns consisted of fewer bands except for the last sampling date, when band intensities were too strong to clearly differentiate between products in some gel regions (Fig. 2). The composting-stage-related increase of individual bands in the profiles indicated a succession of community members and suggested that the diversity of actinomycetes increased during compost maturation.
FIG. 2.
SSCP patterns of single-stranded PCR products obtained from amplifications targeting the V3 region of actinomycetes. DNA directly extracted from composting material was used as a template. Days of sampling (d) are indicated for each lane.
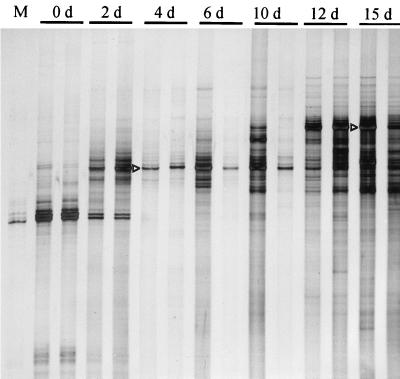
Primers selected to amplify fungus-specific 18S rRNA gene sequences, including the V8 and V9 regions, yielded products from the organic material even before composting (Fig. 3). When these SSCP products were compared with those of other samples, it became evident that one of the dominant products detected at day 0 was identical to the product amplified from 18S rRNA genes of maize, one of the major sources of organic material in this composting process (Fig. 3, lane M). Thus, the selected primers were not exclusively specific for fungal DNA. The disappearance of the maize product after 4 days suggested the degradation of plant DNA during this early composting stage. At this stage (4 days), another dominant product was detected. At the beginning of the composting, this product was the only detectable product. However, during further composting, additional products were detected, first (6 and 10 days) only in one windrow and later (12 and 15 days) in both windrows. Especially for the last two sampling dates shown in Fig. 3, another dominant product with relatively low electrophoretic mobility was detected. Both dominant products (indicated by arrowheads in Fig. 3), as well as other products shown in the gels above (Fig. 1 and 2), were further identified by DNA sequencing.
FIG. 3.
Succession of SSCP patterns obtained from single-stranded DNA products amplified by PCR from directly extracted compost DNA. Primers targeted conserved regions to amplify the V8 and V9 regions of the eukaryotic 18S rRNA gene sequence (M, products obtained from DNA extracted from leaves of maize plants). Days of sampling (d) are given above lanes. For each day, parallel samples were obtained from two separate composting windrows. Arrowheads point to products of the prospective clones CP-1 (4 d) and CP-2 (15 d) (see also Table 2).
Identification of products from community SSCP profiles.
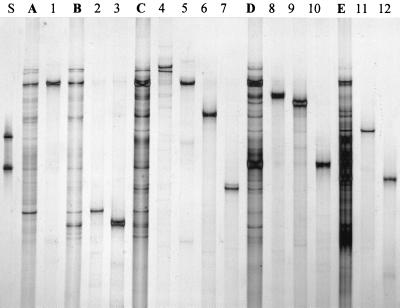
To identify the predominant products by DNA sequencing, a total of 19 different DNA single strands (“bands”) were excised from the three SSCP profiles shown in Fig. 1 to 3. By PCR, the opposite strands were regenerated and the products were reamplified. SSCP gel electrophoresis was used to evaluate the purities and identities of the reamplified products, as shown for products obtained from PCR targeting the hypervariable regions V4 and V5 of eubacterial 16S rRNA genes (Fig. 4). In most cases, reamplification products corresponded to the expected positions in the community patterns and no additional products were observed. These products were then directly used for cloning and DNA sequencing. Table 2 gives the period of detection and the closest relatives found for each molecular isolate.
FIG. 4.
Comparison of PCR-SSCP community patterns with single products isolated from profiles and reamplified by PCR. Patterns obtained with primers amplifying the hypervariable regions V4 and V5 of eubacterial 16S rRNA genes from directly extracted compost DNA are shown in lanes A (sample taken after 2 days of composting), B (4 days), C (6 days), D (10 days), and E (15 days). Amplified products of prospective clones are shown in lanes 1 (clone CB-12), 2 (CB-2), 3 (CB-1), 4 (CB-3), 5 (CB-4), 6 (CB-5), 7 (CB-6), 8 (CB-7), 9 (CB-8), 10 (CB-9), 11 (CB-10), and 12 (CB-11). For identification of clones, see Table 2.
TABLE 2.
Characterization of molecular isolates, cloned from PCR-SSCP genetic profiles of 16S rRNA genes amplified from total DNA extracted from compost
| Primer-targeted gene sequence (region) | Clone | GenBank accession no. | Closest relative based on partial sequence homologya (% similarity) | Age of composting sample used for product isolation (days) | Period of product detection in SSCP community profiles (days) |
|---|---|---|---|---|---|
| 16S rRNA (V4 and V5) | CB-2 | AF213263 | Lactobacillus panis (94.9) | 4 | 2–4 |
| CB-1 | AF213262 | Lactobacillus confusus (99.4) | 2 | 2–6 | |
| CB-3 | AF213264 | Candida krusei (99.6) | 6 | 2–10 | |
| CB-9 | AF213270 | Bacillus thermocatenulatus (92.3) | 10 | 2–18 | |
| CB-12 | AF213273 | Bacillus badius (100.0) | 2 | 2–18 | |
| CB-6 | AF213267 | Azotobacter salinestris (97.1) | 6 | 4–12 | |
| CB-5 | AF213266 | Xanthomonas campestris (96.8) | 6 | 4–12 | |
| CB-7 | AF213268 | Bacillus stearothermophilus (98.1) | 10 | 6–10 | |
| CB-4 | AF213265 | Bacillus caldoxylolyticus (98.6) | 6 | 6–12 | |
| CB-10 | AF213271 | Clostridium thermolacticum (94.1) | 18 | 6–18 | |
| CB-8 | AF213269 | Bacillus fusiformis (96.1) | 10 | 10 | |
| CB-11 | AF213272 | Microbispora bispora (99.2) | 15 | 15–18 | |
| 16S rRNA (V3) | CA-4 | AF213277 | Streptomyces nodosus (98.3) | 10 | 2–15 |
| CA-5 | AF213278 | Streptomyces thermodiastaticus (100.0) | 12 | 2–15 | |
| CA-3 | AF213276 | Streptosporangium vulgare (98.2) | 15 | 2–15 | |
| CA-1 | AF213274 | Bacillus thermoleovorans (92.4) | 12 | 4–12 | |
| CA-2 | AF213275 | Detolaasinbacter sp. (99.0) | 15 | 4–15 | |
| 18S rRNA (V8 and V9) | CP-1 | AF213279 | Candida krusei (99.4) | 4 | 1–18 |
| CP-2 | AF213280 | Candida tropicalis (98.4) | 15 | 10–18 |
Identified by BLASTN 2.0.8 (1).
Most molecular isolates, 12 of 19, were obtained from PCR products generated with primers targeting the eubacterial V4–V5 region (Table 2). Surprisingly, one of the sequenced products was larger (556 bp instead of approximately 400 bp), and this product was, in fact, closely related to a eukaryotic organism, the yeast Candida krusei. Lactobacillus-related sequences were detectable only during the early stage of composting (2 to 6 days). Five sequences were related to Bacillus species, among them one with complete sequence identity for this variable region found in the 16S rRNA gene of Bacillus badius. Sequences related to B. badius and Bacillus thermocatenulatus were detectable throughout the whole composting process (2 to 18 days). One sequence with relatively low similarity was related to the anerobic spore-forming Clostridium thermolacticum. Two sequences were related to γ-Proteobacteria and one sequence to an actinomycete. The latter group was more specifically targeted with primers amplifying the V3 region. In fact, except for one sequence which was related to another Bacillus species, the other four sequences were related to actinomycetes. These four sequences were already detectable at the beginning of the composting, but their intensities, especially those of CA-4, related to Streptomyces nodosus, and CA-5, related to Streptomyces thermodiastaticus, increased continuously during the process (Fig. 2). The two dominant products detected in SSCP profiles of 18S rRNA gene-targeted PCR were related to two yeast sequences, both members of the genus Candida. The same organism which was accidentally detected with primers designed to amplify the V4–V5 16S rRNA gene region after 6 days of incubation (CB-3) was also identified as one of the dominant fungal sequences (CP-1) throughout the composting process.
Characterization of bacteria isolated by cultivation and comparison to molecular isolates.
A collection of 290 strains was isolated under aerobic conditions at 50°C from compost samples taken at the end of the composting process. Incubation of inoculated agar plates under aerobic conditions at 50°C resulted in fast growth of bacteria. In order to avoid contamination of pure cultures, the incubation period before subcultivation of isolates (single colonies) was only 16 h. Using the combined results obtained with two restriction endonucleases (CfoI and RsaI), we selected 22 isolates which were further identified by sequencing of the uncut PCR product. These isolates could be assigned to six different groups (Table 3). The 16S rRNA gene sequences of three groups showed high similarity to those of Bacillus species. Two sequences were related to Pseudomonas species and one to Xanthomonas campestris.
TABLE 3.
Characterization of cultivated thermophilic and heterotrophic bacteria isolated at the end of the hot-composting stage
| ARDRA groupa | Representative strain | Size of sequence (bp) | GenBank accession no. | Closest identified relativeb (% sequence similarity) |
|---|---|---|---|---|
| 1 | Sko08 | 640 | AF213284 | Bacillus coagulans (98.9) |
| 2 | Sko02 | 559 | AF213281 | Bacillus denitrificans (99.8) |
| 3 | Sko06 | 638 | AF213282 | Bacillus smithii (99.8) |
| 4 | Sko14 | 606 | AF213285 | Pseudomonas citronellolis (95.7) |
| 5 | Sko17 | 693 | AF213286 | Pseudomonas stutzeri (95.4) |
| 6 | Sko07 | 637 | AF213283 | Xanthomonas campestris (96.0) |
Characterized by combined results of RsaI and CfoI digests of PCR-amplified 16S rRNA gene sequences.
Identified by BLASTN 2.0.8 (1).
From selected pure cultures of each group, PCR products generated with Com primers (used for eubacterial-community analysis) were loaded onto SSCP gels with community profiles as a reference (data not shown). Similarities in electrophoretic mobility were observed between isolate Sko07 and clone CB-5, both related to X. campestris. Sequence alignments of both showed 295 matches for 299 bp (98.7%). Similar band positions in the profiles were also observed for Sko17 and CB-6 (related to Pseudomonas stutzeri and to Azotobacter salinestris). As judged by the almost-identical DNA sequences of the selected 16S rRNA gene region (342 of 343 matches), these organisms were identical. Differences in identification of their closest relatives can be explained by the different lengths of the sequences submitted to the databases. A third identity in band position was found for Sko08 and CB-12. The match in sequences was 274 of 275 bp. Thus, it can be concluded that these organisms too were the same, or at least very closely related in their 16S rRNA genes.
DISCUSSION
The quickly changing physicochemical conditions in composting processes, as described in this study, are likely to select for a succession of different microbial communities. It can be expected that temperature and the available substrates, including electron donors and acceptors, are the main factors in this context (36). In this study, we have shown that SSCP analysis of small-subunit rRNA genes amplified from directly extracted compost DNA can be used to visualize such community structures and successions. The SSCP-generated genetic profiles consisted of more than 30 distinguishable products. These products were different in electrophoretic mobility, probably due to sequence differences causing different conformations under nondenaturing conditions. However, the possibility that single products may also have existed in more than one conformation under such conditions and, thus, generated more than one band cannot be ruled out (34). In contrast to another, previously applied protocol for SSCP-based microbial-community analysis (30), our protocol avoids the reannealing of DNA single strands and the formation of heteroduplices by removal of one strand from each PCR product by exonuclease digestion prior to nondenaturing electrophoresis (41). As a further development of our protocol in this study, we have isolated single products from community profiles and identified them by DNA sequencing. With DGGE profiles the same purpose was achieved in other studies (15, 39). In contrast to direct cloning of PCR-amplified rRNA genes from environmental DNA, the profiling technique has the advantage that single products can be preselected before sequencing, and thus redundant information from repetitive isolation of the same or highly similar sequences can be avoided. This advantage of genetic profiling was shown in our study, since each of 12 molecular isolates extracted from a genetic profile generated with eubacterial primers was unique. However, in contrast to directly cloned molecular isolates, where whole RNA operons can be analyzed, the size of products available from genetic profiles is, to our knowledge, still restricted to a maximum length of about 500 bp, whether SSCP or DGGE-TGGE techniques are used for microbial-community analysis. This can limit the accuracy of species identification but may not be crucial for comparative assessment of microbial-community structures in environmental samples (19).
For community profiling, as described in this study, PCR conditions and primer selections are important factors which may influence the outcome of an analysis. It is known that products amplified by PCR may not accurately reflect the microbial diversity in the template mixture due to different small-subunit rRNA gene copy numbers (17) or biases during the amplification process (38, 45). However, since the number of products which could be detected in the profiles was limited to a maximum of approximately 40, it could be anticipated that shifts in the community structure on the basis of eubacterial PCR amplifications would be detected only when quantitatively dominant organisms were affected (32). More-specific detections, which may be suppressed with eubacterial primer amplifications, may be possible if primers are used which select for specific groups of the microbial community. Such primer systems have been designed to characterize the diversity of actinomycetes in soil (22), ammonia-oxidizing Proteobacteria in compost (29), or fungal genes in plant roots or a rhizosphere (28, 43). In our study we used three primer systems in parallel with the same template DNA, and it was therefore possible to characterize community successions from the same samples with three different specificities.
In accordance, SSCP profiles generated with all three primer systems demonstrated increasing microbial diversity during the composting process. Sequence identification indicated the presence of several species known to be involved in composting. Lactobacilli, which are the typical dominant microorganisms in degrading plant material under oxygen limitation, e.g., in silage (10), occurred at the beginning of the composting process, when the substrate was relatively wet. During the heating phase, 5 of 12 molecular isolates which were closely related to the spore-forming aerobic genus Bacillus and one which was related to the anaerobic genus Clostridium were detected. Bacilli are typical microorganisms in hot composting stages (6, 44). Two Bacillus-related isolates could be detected throughout the composting process. One of these isolates showed perfect homology to a known 16S rRNA gene sequence (B. badius), but the other (CB-9) had only 92.3% identity to a known sequence. Possibly this dominant isolate was related to a group of gram-positive bacteria with a low G+C DNA content which have not been characterized yet.
Surprisingly, one sequence was related, not to bacteria, but to yeast. This result can be explained by the fact that both the Com1 and the Com2 primer have homologous regions in the 18S rRNA genes, each with only one mismatch. The primers amplify between positions 556 and 1,117 (20, 33). The size of the sequenced product was in accordance with this assumption. Only one actinomycete-related sequence could be detected with eubacterial primers, but with primers targeting this group more specifically, four of five isolates could be assigned to this group. These results indicate the usefulness of the primers suggested by Heuer et al. (22) for the analysis of indigenous actinomycete populations in the environment. The increase in pattern diversity observed with these primers is in accordance with the general knowledge that actinomycete populations increase during compost maturation (13, 36). Primers selected for monitoring of fungal diversity also amplified plant DNA. By this means, we were able to determine that plant DNA was degraded during the first 4 days of composting. These results confirm the findings of a previous study in which the stability of a recombinant marker gene was monitored under similar composting conditions (27). Two different molecular isolates, both related to the yeast genus Candida, were identified from the SSCP profiles. The closest relatives of these species, C. krusei and Candida tropicalis, are known to be pathogenic for humans (20). DNA similarities do not prove any pathogenic potential. It is, however, remarkable that C. krusei was also accidentally identified as a close relative of CB-3 with eubacterial primers targeting a different gene region. Possibly the yeast originated from the fecal material (horse dung) which was a component of the organic substrate composted in this study. For accurate identification of the molecular yeast isolates, other rRNA gene regions may be more appropriate, since the taxonomic resolution of small-subunit rRNA genes for fungi seems to be lower than that for bacteria (33, 43). Other fungi may have been present in the composting process as well but may have been missed due to inefficient DNA extraction or nonoptimal primer selection.
The diversity of the cultivated isolates was rather low; only 6 species could be identified out of 290 isolates. It is likely that more-appropriate cultivation media and more-sophisticated incubation conditions would have yielded additional species. Cultivation of bacteria was carried out only at the end of the composting process. With pure-culture SSCP analysis, we found three products which had the same electrophoretic mobilities as dominant products in the profiles. As judged by base pair similarities in the selected 16S rRNA gene region, it is highly probable that at least two of these isolates were identical. One was B. badius, which could be detected throughout the composting process. Since the 16S rRNA gene of this organism was predominant and was also identified by cultivation, we assume that it was quantitatively important. The other organisms with high similarities belonged to the γ-Proteobacteria and were related to P. stutzeri and A. salinestris. In community profiles, this product was detectable from 4 days to 12 days, but by cultivation the related organism was found in 18-day-old samples.
The succession of products in combination with increasing and decreasing band intensities during different composting stages, as detected with SSCP profiles in this study, indicates the high potential of this technique to monitor microbial communities and their variation qualitatively and quantitatively. As more gene sequences become available, PCR-SSCP-mediated monitoring of different subgroups or microorganisms, due to optimized primer design, will become even more attractive. The use of genetic profiles reduces the numbers of products which need to be identified by DNA sequencing to those which are assumed to be of specific importance. By this means and in combination with highly efficient automated DNA sequencing, molecular analysis of microbial communities gains new relevance for the monitoring of biotechnological processes and applied microbial ecology.
ACKNOWLEDGMENTS
We thank Katja Lübke and Evelin Schummer for excellent technical assistance and Klaus Grabbe for help in conducting the composting process.
This work was supported by the Federal Environmental Agency of Germany (grant 11201032).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews S A, Lee H, Trevors J T. Bacterial species in raw and cured compost from a large-scale urban composter. J Ind Microbiol. 1994;13:177–182. [Google Scholar]
- 3.Bassam B J, Caetano-Anolles G, Greshoff P M. Fast and sensitive staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 4.Beffa T, Blanc M, Aragno M. Obligately and facultatively autotrophic, sulfur- and hydrogen-oxidizing thermophilic bacteria isolated from hot composts. Arch Microbiol. 1996;165:34–40. [Google Scholar]
- 5.Beffa T, Blanc M, Lyon P F, Vogt G, Marchiani M, Fischer J L, Aragno M. Isolation of Thermus strains from hot composts (60 to 80°C) Appl Environ Microbiol. 1996;62:1723–1727. doi: 10.1128/aem.62.5.1723-1727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc M, Marilley L, Beffa T, Aragno M. Rapid identification of heterotrophic, thermophilic, spore-forming bacteria isolated from hot composts. Int J Syst Bacteriol. 1997;47:1246–1248. doi: 10.1099/00207713-47-4-1246. [DOI] [PubMed] [Google Scholar]
- 7.Blanc M, Marilley L, Beffa T, Aragno M. Thermophilic bacterial communities in hot composts as revealed by most probable number counts and molecular (16S rDNA) methods. FEMS Microbiol Ecol. 1999;28:141–149. [Google Scholar]
- 8.Bruns M A, Stephen J R, Kowalchuk G A, Prosser J I, Paul E A. Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled, and successional soils. Appl Environ Microbiol. 1999;65:2994–3000. doi: 10.1128/aem.65.7.2994-3000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter-Boggs L, Kennedy A C, Reganold J P. Use of phospholipid fatty acids and carbon source utilization patterns to track microbial community succession in developing compost. Appl Environ Microbiol. 1998;64:4062–4064. doi: 10.1128/aem.64.10.4062-4064.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocconcelli P S, Triban E, Basso M, Botazzi V. Use of DNA probes in the study of silage colonization by Lactobacillus and Pediococcus strains. J Appl Bacteriol. 1991;71:296–301. doi: 10.1111/j.1365-2672.1991.tb03793.x. [DOI] [PubMed] [Google Scholar]
- 11.Derikx P J L, de Jong G A H, op den Camp H J M, van der Drift C, van Griensven L J L D, Vogels G D. Isolation and characterization of thermophilic methanogenic bacteria from mushroom compost. FEMS Microbiol Ecol. 1989;62:251–258. [Google Scholar]
- 12.Duineveld B M, Rosado A S, van Elsas J D, van Veen J A. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl Environ Microbiol. 1998;64:4950–4957. doi: 10.1128/aem.64.12.4950-4957.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards C. Compost—a natural substrate for studies of the diversity of thermophilic actinomycetes and monitoring the fate of genetically modified species. In: Debabov V G, editor. The biology of the actinomycetes '94: proceedings of the Ninth Symposium on the biology of the actinomycetes. Biotechnologiya 7/8. 1995. pp. 229–233. [Google Scholar]
- 14.Fermor T R, Smith J F, Spencer D M. The microflora of experimental mushroom composts. J Hortic Sci. 1979;54:137–147. [Google Scholar]
- 15.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finstein M S, Morris M L. Microbiology of municipal solid waste management. In: Mitchell R, editor. Environmental microbiology. New York, N.Y: John Wiley & Sons; 1975. pp. 355–374. [Google Scholar]
- 17.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi K. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1991;1:34–38. doi: 10.1101/gr.1.1.34. [DOI] [PubMed] [Google Scholar]
- 19.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 20.Hendriks L, Goris A, van der Peer Y, Neefs J M, Vancanneyt M, Kersters M, Hennebert G L, de Wachter R. Phylogenetic analysis of five medically important Candida species as deduced on the basis of small ribosomal subunit RNA sequences. J Gen Microbiol. 1991;137:1223–1230. doi: 10.1099/00221287-137-5-1223. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann R F, Shann J F. Microbial community changes during composting of municipal soil waste. Microb Ecol. 1997;33:78–85. doi: 10.1007/s002489900010. [DOI] [PubMed] [Google Scholar]
- 22.Heuer H, Krsek M, Baker P, Smalla K, Wellington E M H. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann A, Thimm T, Droge M, Moore E R B, Munch J C, Tebbe C C. Intergeneric transfer of conjugative and mobilizable plasmids harbored by Escherichia coli in the gut of the soil microarthropod Folsomia candida (Collembola) Appl Environ Microbiol. 1998;64:2652–2659. doi: 10.1128/aem.64.7.2652-2659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Insam H, Amor K, Renner M, Crepaz C. Changes in functional abilities of the microbial community during composting of manure. Microb Ecol. 1996;31:77–87. doi: 10.1007/BF00175077. [DOI] [PubMed] [Google Scholar]
- 25.Klamer M, Baath E. Microbial community dynamics during composting of straw material studied using phospholipid fatty acid analysis. FEMS Microbiol Ecol. 1998;27:9–20. [Google Scholar]
- 26.Koschinsky S, Peters S, Schwieger F, Tebbe C C. Applying molecular techniques to monitor microbial communities in composting processes. In: Bell C, editor. Progress in microbial ecology. Proceedings of the International Symposium on Microbial Ecology—8, in press. 2000. [Google Scholar]
- 27.Koschinsky S, Schwieger F, Peters S, Grabbe K, Tebbe C C. Characterizing microbial communities of composts at the DNA level. Med Fac Landouww Univ Gent. 1998;63/4b:1725–1732. [Google Scholar]
- 28.Kowalchuk G A, Gerards S, Woldendorp J W. Detection and characterization of fungal infections of Ammophila arenaria (marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S rDNA. Appl Environ Microbiol. 1997;63:3858–3865. doi: 10.1128/aem.63.10.3858-3865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalchuk G A, Naoumenko Z S, Derikx P J L, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D-H, Zu Y-G, Kim S-J. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single strand conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik M, Kain J, Pettigrew C, Ogram A. Purification and molecular analysis of microbial DNA from compost. J Microbiol Methods. 1994;20:183–196. [Google Scholar]
- 32.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neefs J-M, Van der Peer Y, De Rejk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekyia T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Nat Acad Sci USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page A L, Miller R H, Keeney D R. Methods of soil analysis. Part 2. Chemical and microbiological properties. Madison, Wis: American Society of Agronomy, Inc. and Soil Science Society of America, Inc.; 1982. [Google Scholar]
- 36.Paul E A, Clark F E. Soil microbiology and biochemistry. 2nd ed. San Diego, Calif: Academic Press; 1996. [Google Scholar]
- 37.Pfaller S L, Vesper S J, Moreno H. The use of PCR to detect a pathogen in compost. Compost Sci Utiliz. 1994;2:48–54. [Google Scholar]
- 38.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rölleke S, Muyzer G, Wawer C, Wanner G, Lubitz W. Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1996;62:2059–2065. doi: 10.1128/aem.62.6.2059-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schwieger F, Tebbe C C. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol. 1998;64:4870–4876. doi: 10.1128/aem.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma S, Rangger A, Insam H. Effects of decomposing maize litter on community level physiological profiles of soil bacteria. Microb Ecol. 1998;35:301–310. doi: 10.1007/s002489900085. [DOI] [PubMed] [Google Scholar]
- 43.Smit E, Leeflang P, Glandorf B, van Elsas J D, Wernars K. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:2614–2621. doi: 10.1128/aem.65.6.2614-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strom P F. Identification of thermophilic bacteria in solid-waste composting. Appl Environ Microbiol. 1985;50:906–913. doi: 10.1128/aem.50.4.906-913.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vancanneyt M, De Vos P, Kersters K, De Ley J. Isolation, characterization and identification of strictly anaerobic, thermophilic, ethanol producing bacteria from compost. Syst Appl Microbiol. 1987;9:293–298. [Google Scholar]
- 48.Vaneechoutte M, Roussau R, De Vos P, Gillis M, Janssens D, Paepe N, De Rouck A, Fiers T, Claeys G, Kesters K. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA) FEMS Microbiol Lett. 1992;93:227–234. doi: 10.1111/j.1574-6968.1992.tb05102.x. [DOI] [PubMed] [Google Scholar]
- 49.White T J, Burns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal genes for phylogenetics. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. A guide to methods and applications. San Diego, Calif: Academic Press; 1990. pp. 315–322. [Google Scholar]