Abstract
Given the growing number of prediction algorithms developed to predict COVID-19 mortality, we evaluated the transportability of a mortality prediction algorithm using a multi-national network of healthcare systems. We predicted COVID-19 mortality using baseline commonly measured laboratory values and standard demographic and clinical covariates across healthcare systems, countries, and continents. Specifically, we trained a Cox regression model with nine measured laboratory test values, standard demographics at admission, and comorbidity burden pre-admission. These models were compared at site, country, and continent level. Of the 39,969 hospitalized patients with COVID-19 (68.6% male), 5717 (14.3%) died. In the Cox model, age, albumin, AST, creatine, CRP, and white blood cell count are most predictive of mortality. The baseline covariates are more predictive of mortality during the early days of COVID-19 hospitalization. Models trained at healthcare systems with larger cohort size largely retain good transportability performance when porting to different sites. The combination of routine laboratory test values at admission along with basic demographic features can predict mortality in patients hospitalized with COVID-19. Importantly, this potentially deployable model differs from prior work by demonstrating not only consistent performance but also reliable transportability across healthcare systems in the US and Europe, highlighting the generalizability of this model and the overall approach.
Subject terms: Viral infection, Data mining
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused millions of cases of coronavirus disease 2019 (COVID-19) in nearly every country. While most patients with COVID-19 have a mild form of viral pneumonia, an appreciable subgroup develops rapid onset of severe disease. Several large national studies have demonstrated that a variable and potentially significant proportion (ranging from 5% to 70%)1–3 of hospitalized patients with COVID-19 develop cardiorespiratory failure, require mechanical ventilation and hemodynamic support, and may ultimately die. The early identification of patients at high risk for death can improve triage and resource allocation, particularly when numbers of COVID-19 cases overwhelm health systems4.
Numerous studies have reported models using clinical data, including laboratory values, to predict patients at high risk of death for COVID-192. However, most models have not been tested across hospital systems and countries to determine generalizability. Few studies have included patients from multi-national cohorts. The international nature of this disease begs the question of whether models derived using data from one site or one country can be used in another. Is transportability possible if the experience of one site or country could help another make better decisions?
We formed the 4CE Consortium5 as an international research collaborative of nearly 300 hospitals from four countries in order to collect standardized patient-level electronic health record (EHR) data to examine the epidemiology, pathophysiology, management, and healthcare system dynamics of COVID-19. Using the 4CE data, we examined the relationship between pre-selected laboratory values6 and mortality across institutions and countries. We compared prediction models using single laboratory values at admission to a prediction model containing multiple laboratory values. Across all models, we evaluated geographical differences (national and continental) among the outcome prediction models to better understand if models trained on data from one country and institution can be used elsewhere.
Results
Characteristics of the study population
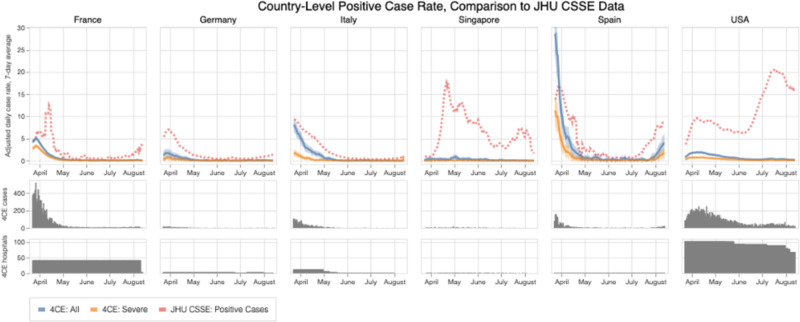
In this study population of 39,969 patients, the incidence of hospitalization for COVID-19 largely tracked with population dynamics of COVID-19 cases7 across different countries during the initial pandemic period (Fig. 1). Both the COVID-19 case rate and the COVID-19 hospitalization rate dropped significantly from the first peak in April 2020. While hospitalization rates remained relatively low for all countries, case rates increased in France, Germany, Spain and United States after June 2020.
Fig. 1. Comparison of National Hospitalization Rates by Data Source.
Adjusted 7-day average new hospitalization rate and rate of ever-severe disease per 100,000 people by country based on 4CE contributors along with 95% confidence intervals compared with 7-day average new case rates collected by Johns Hopkins Center for Systems Science and Engineering (JHU CSSE).
Consistent with prior studies4,8, the study population of patients hospitalized with COVID-19 showed a higher prevalence of men and older populations. See Supplementary Fig. 1 for demographic characteristics and percentages among age group, race/ethnicity, and sex. International comparisons were consistent and showed across three countries that most patients (79.6%) were 50 years of age or older and male (68.6%).
International comparisons of individual laboratory tests at admission for mortality risk prediction
The prediction performances of individual laboratory test across all sites, at country level and continent level were summarized using random-effects meta-analysis. On average, albumin, creatinine, neutrophil count, CRP and white blood cell were stronger predictors of mortality than the other labs (Supplementary Fig. 2). The predictiveness of the laboratory tests for mortality within the next few days after admission tends to be slightly higher than for 1 or 2-week mortality although the decrease in predictiveness over time was moderate. The predictiveness of the labs varies substantially across sites. Albumin has low predictiveness in European sites but higher in the US, CRP appears to be slightly more predictive in Europe than in US, while other labs performed similarly in the US and in Europe on average.
International comparisons of mortality risk prediction model
The estimated log hazard ratios for demographic, nine laboratory tests and Charlson comorbidity index from a comprehensive Cox model are largely consistent across different healthcare systems with respect to their directions and magnitudes (Supplementary Fig. 3). The estimated log hazard ratios across all sites and at country level were summarized using random-effects meta-analysis. The risk models indicate that age, albumin, AST, creatine, CRP, and white blood cell are most predictive of mortality. For example, the risk model predicts a protective effect against mortality from those who are <50 years old, report higher albumin values and lymphocyte count values, and report lower AST, creatinine and CRP values. The average AUC of the full risk model is about 0.80, 0.79 and 0.77 for predicting both 3-day, 1-week, or 2-week mortality (Fig. 2). While the performance of the locally trained site-level models varies across healthcare systems, the average performance of the full model is similar in the US versus Europe.
Fig. 2. Risk Model Performance Across Countries and Continents.
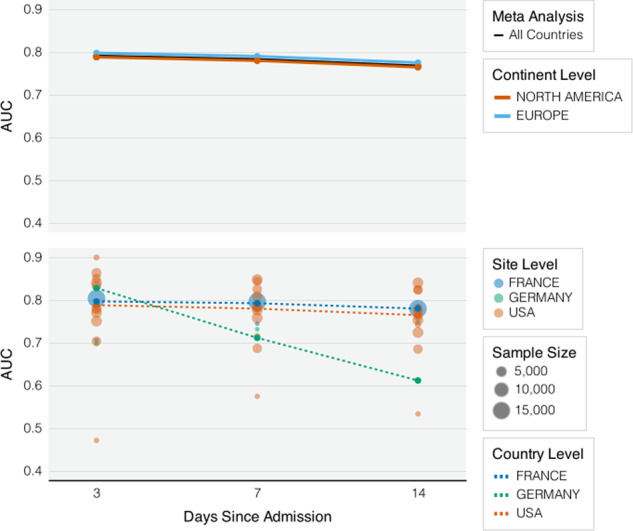
AUCs of cox regression models with nine common laboratory tests (missing rate <30%) in predicting death adjusting for demographic variables and Charlson comorbidity index.
Portability of mortality algorithms across sites, countries, and continents
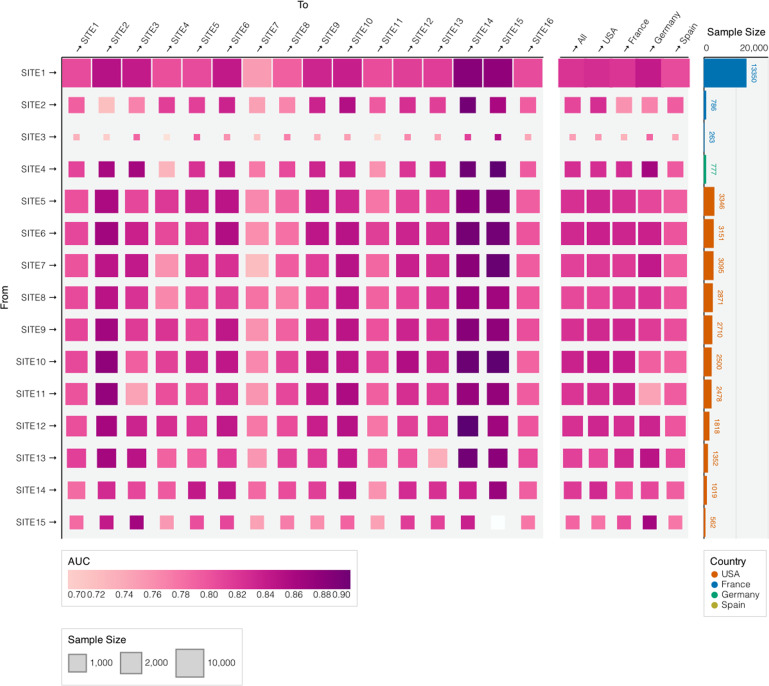
The AUCs of the locally trained mortality risk models for 1-week mortality when porting to external sites were summarized in Fig. 3 (refer to Supplementary Table 4 for numerical results). The averaged AUCs across all sites and at country level were summarized using random-effects meta-analysis. The algorithms trained from sites with large cohort size tend to have better performance both locally and when transported to other sites. For example, the AUCs of the model trained at SITE1 (France) are always close to or higher than the those of the local trained model. We additionally compared the portability performance across continents. In general, when porting to North America sites, the algorithms trained at both continents perform equally well. For example, when porting to SITE5 (US), the maximum AUC was 0.842 and 0.847 for algorithms trained at North America sites and at European sites, respectively, which are very close to the maximum AUC of the local SITE5 algorithm. On the other hand, when porting to Europe sites, the algorithms trained at North America sites perform slightly better than those trained at Europe sites, due to the relatively smaller sample size of the Europe sites. For example, when porting to SITE1 (France), the maximum AUC was 0.813 and 0.791 for algorithms trained at North America sites and at European sites, respectively.
Fig. 3. Transportability of the Mortality Prediction Model Across Sites and Countries.
Heatmap of transportability of the Cox regression model across different sites and countries. Each part of the figure represents performance when the model is trained at one site and evaluated at another.
Discussion
In this large-scale multi-national study, we reported a mortality prediction model for patients hospitalized with COVID-19 that retained accuracy across healthcare systems and countries. Building on the growing literature of COVID-19 mortality prediction, our study is unique in leveraging international cohorts to validate the generalizability of the prediction model, which has the following specific features. First, a predictive model containing nine commonly measured laboratory test values performed better than the model containing 17 laboratory test values: CRP, creatinine, white blood cell count, lymphocyte count, AST, ALT, total bilirubin, neutrophil count, and albumin. From a list of 17 laboratory tests associated with worse outcomes in patients with COVID-19 based on prior reports6, we selected the subset of nine tests based on their low rate of missing data in our data set. Second, we identified albumin, CRP, creatinine, neutrophil count, and white blood cell count as better individual predictors than other individual laboratory tests. Third, a comprehensive model containing the nine commonly measured laboratory tests as well as baseline demographic features and comorbidity burden indicates that age, albumin, AST, CRP, creatine, and white blood cell count are most predictive of mortality. Interestingly, the baseline covariates are more predictive of mortality in the early days after admission for COVID-19, likely because other features gain importance as hospital course prolongs. Finally, when comparing prediction models between North American and European sites, the final model showed crucial consistency across international sites, highlighting its potential generalizable application.
The study has several strengths. Chief among them is the international consortium with a federated data sharing approach that facilitated the pooling of laboratory values across 283 hospitals with diverse healthcare practices and populations, enabling the examination of model transportability. Second, while the accuracy (AUC) of individual laboratory test in predicting mortality after hospital admission for COVID-19 varies substantially cross countries, the accuracy of the mortality risk prediction model is remarkably consistent between US and Europe. Further, the estimated log hazard ratios from the best-performing Cox model are largely consistent across different healthcare systems with respect to their directions and magnitudes. Third, the mortality prediction model using commonly measured laboratory tests and baseline demographic and comorbidity burden trained at healthcare systems performs well both locally and externally when transported to other sites. Interestingly, the transportability does not appear to depend on the continent or country. Taken together, the key innovation of our study that differs from prior studies is the transportability and the potential generalizability of the COVID-19 mortality prediction model that seems independent of the specific healthcare system.
The study also has several limitations that we took measures to mitigate. First, EHR data have variable degree of intrinsic noise, missing data, and available documentation due to differences in clinical practice that contribute to differences among healthcare systems. Indeed, we found healthcare system-level (within-healthcare system and between-healthcare system) differences were greater than country-level differences. By leveraging our federated system of common EHR data elements and capturing healthcare system-level heterogeneity, the 4CE consortium is uniquely positioned to identify international differences in patient characteristics and outcomes as well as to test model transportability. To mitigate the quality issue of EHR data, we performed extensive and iterative quality controls at each participating healthcare system with local collaborators and centrally to address potential imprecision due to healthcare system-specific variations in data extraction and incompleteness of datasets (e.g., incomplete mapping of local EHR codes to desired data elements). These critical quality control steps, which are often underappreciated in multi-center EHR data research, further differentiate the 4CE research efforts from other COVID-19 research efforts. Second, we observed a significant level of heterogeneity in the predictiveness of individual laboratory tests and the locally trained mortality risk models across the participating healthcare systems. The heterogeneity could result from differences in patient population, clinical practice and EHR system. To address this concern, we performed random-effects meta-analyses to account for the heterogeneity across sites. Importantly, the best-performing model showed evidence of good transportability despite of the heterogeneity.
As the pandemic persists and new SARS-CoV-2 variants emerge, two clinically relevant questions remain unanswered: (1) does the mortality prediction model continue to perform well across healthcare systems and countries? (2) can the prediction model predict long-term mortality after COVID-19 hospitalization? To address these questions, we are planning future analyses using patient-level data at each participating healthcare system to assess the temporal trends of the model performance throughout the pandemic waves and at individual patient-level over longer period. We will revise and adapt to temporal changes in clinical scenarios. In this study, we observed that AUCs are generally consistent across genders. Since age is a significant risk factor for mortality, conditioning on the age group, the model performance for distinguishing high-risk vs. low-risk patients within the age group is expected to be lower than the overall accuracy. Further developing age-specific risk prediction models warrants further research. Beyond mortality prediction, the 4CE consortium has established a platform of harmonized data capture through its federated system with iterative and methodical expansion of data elements to enable the clinical investigation of a wide range of domains pertaining to COVID-19 such as coagulopathy and thrombotic events, acute renal failure, pediatric manifestation, neurological complications as well as the post-acute sequelae syndrome (i.e., long-hauler). We will apply the approach from this study to assess other prediction model transportability within our international network of participating healthcare systems.
We make several noteworthy observations of clinical relevance. First, the laboratory tests predictive of mortality in patients hospitalized for COVID-19 represent the combination of acute inflammatory response (as indicated by CRP, white blood cell, lymphocyte, and neutrophil count) and underlying physiological function as well as the acute response of critical organ systems (general nutritional status as indicated by albumin, renal function as indicated by creatinine, and hepatic function as indicated by AST, ALT, and bilirubin). These routinely collected laboratory indicators of systemic response to the SARS-CoV-2 viral infection in conjunction with easily ascertainable baseline demographic and comorbidity burden formulate a clinically deployable prediction tool of mortality risk following hospital admission for COVID-19. Second, the relatively modest accuracy of individual laboratory values in predicting mortality is likely due to its large variation within each participating healthcare system. This combination of commonly measured clinical laboratory tests dramatically improved the prediction performance over individual laboratory tests, and performed better than a larger panel of clinical laboratory tests. A key clinical insight is that clinical laboratory tests beyond the commonly measured routine tests may not inform mortality, which is the most important clinical outcome. Third, the performance of the final model was relatively stable over the hospital course and did not improve beyond the initial hospital days. This finding suggests that additional factors contribute to mortality as the hospital course for COVID-19 patients prolongs. Of particular clinical relevance, it supports the utility of commonly measured routine clinical laboratory test values (and other routine clinical and demographic features) at admission to identify patients at high risk for mortality who would warrant early and aggressive intervention as well as close monitoring, particularly in the setting of limited healthcare resources.
Methods
Cohort identification
We included all patients hospitalized at participating 4CE sites with an admission date from 7 days before to 14 days after the date of their first reverse transcription polymerase chain reaction (PCR)-confirmed SARS-CoV-2 positive test result. The first admission date within this 21-day time window was considered the index admission date. Throughout this work, “days since admission” refers to this index date.
Participating sites
Data were available from 39,969 patients from 284 hospitals (affiliated with 16 sites) across four countries: France, Germany, Spain, and the United States. See Supplementary Table 2 for details about participating sites. Several sites collected data from multiple hospitals. In the United States, 170 medical centers of the US Department of Veterans Affairs were grouped into five regional divisions called Veterans Integrated Service Networks.
Patient and public involvement
Patients and the public were not involved in the design, conduct, or reporting, or dissemination plans of the research.
Outcome
We consider death as the main COVID-19 outcome. Death was identified via standard coding and discharge data aggregation from each site. Each partner institution used local criteria to identify in-hospital mortality.
Local data collection
Patient-level data
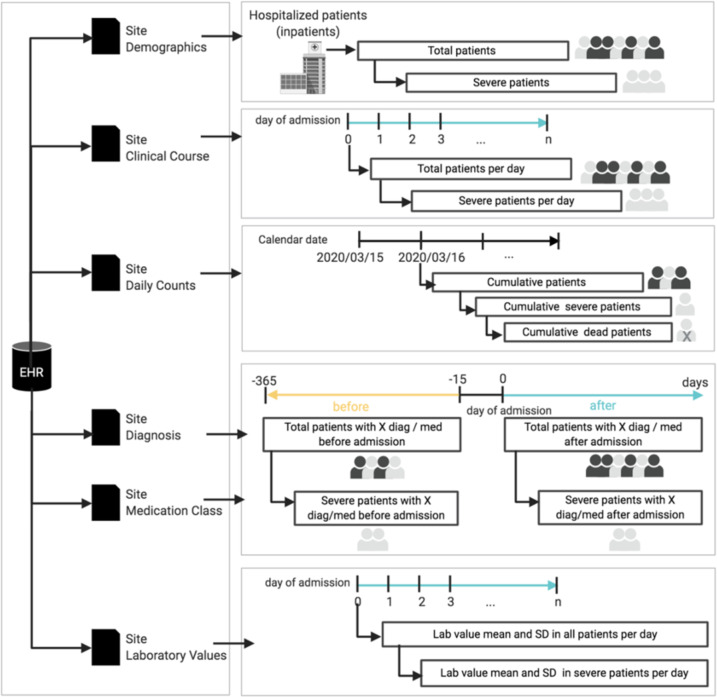
Sixteen sites representing 284 Hospitals assembled patient-level data for detailed analyses, including twelve US sites, and four international sites. Individual healthcare systems then ran separate analyses using the patient-level data within their local firewall and only reported the final analytic results to the central institution for meta-analysis. A schematic of our workflow is presented in Fig. 4, and further details of collected data are reported in Supplementary Table 3.
Fig. 4. Schematic of the federated EHR-based study involving healthcare systems from three countries.
Each site generated three data tables (comma-separated files) containing patient level data: 1) local patient clinical course indicates which days the patient was in the hospital and when the patient died; 2) local patient observation includes first three-character ICD9/10 diagnosis code and laboratory tests, where laboratory test has a numerical value; 3) local patient summary contains demographic variables including age, sex and race. Sites then conduct analysis using these individual level data within their firewall (see Methods).
Software platform
Most sites used the open source i2b2 (Informatics for Integrating Biology and the Bedside) software platform to obtain the data. More than 200 organizations worldwide use i2b2 for purposes that include identifying participants for clinical trials, drug safety monitoring, and clinical and epidemiological research. Those 4CE sites with i2b2 used database scripts to directly query their i2b2 repository, calculate the counts and statistics, and export the data files. The 4CE sites without i2b2 used the Observational Medical Outcomes Partnership (OMOP) Common Data Model or their own clinical data warehouse solutions (e.g., Epic Caboodle) and querying tools to create the required files.
Selection of laboratory tests
We focused on nine laboratory tests that are commonly measured (missing rate <30% at most sites) and associated with mortality in patients with COVID-19 based on prior reports6, We provided each site with a single standard Logical Objects, Identifiers, Names and Codes (LOINC) identifier for each test, but sites often needed to map tests to additional LOINC or custom codes within their EHR. We addressed barriers that arose during initial efforts to extract these laboratory values by stratifying region-specific laboratory test types to reduce extraction errors and enable standardization.
Quality control
We conducted site-specific quality control. Each site ran an R script for the following additional quality control checks: consistency of the total counts of total cases across all datasets within each site, consistency between the 3-digit diagnosis codes and the ICD dictionary, and consistency of the range of laboratory data from each site with the normal range observed from all sites. Sites checked and fixed the data if their laboratory values were consistently lower or higher than the other sites or otherwise implausible.
Statistical analysis
We estimated the country-level daily incidence of new patients hospitalized with COVID-19 during the study period from March 1, 2020 to September 30, 2020. Specifically, for each country, we summed the daily incidence of new patients hospitalized with COVID-19 at each site within that country per 100,000 people of the country and multiplied this by an adjustment factor, defined as the ratio between the country’s overall inpatient discharge rate and the overall inpatient discharge rate of all 4CE sites in that country irrespective of COVID-19 status. We then reported the adjusted 7-day average incidence of new COVID-19 hospitalizations per 100,000 of the country population.
We divided our analysis into two parts: (1) prediction of mortality using individual laboratory values and a comprehensive algorithm derived from multiple laboratory values, comorbid conditions, and demographics available at each site and (2) comparison of these models across sites, countries, and continents.
We evaluated the ability of a biomarker and demographics-based algorithm to predict mortality using admission data. We removed patients who died at admission. We developed mortality risk prediction models using a set of nine common laboratory tests with missing rates <30% at most sites, adjusting for demographic variables and the Charlson comorbidity index. We derived the risk models by fitting penalized Cox proportional hazards model. We evaluated the accuracy of the risk models for predicting mortality by t-days since admission based on the time-specific AUC9. We used the 10-fold cross-validation to estimate the AUC when evaluating the model performance within each local site. The mortality risk prediction model was not trained at Spain because the data were not available at the time when we collected the model training results. To assess the transportability of the mortality risk prediction models across different sites, we validated the algorithm trained at local individual healthcare centers using independent dataset from remaining external sites including the healthcare center from Spain. We used random effects meta-analysis on the prediction performance measures across sites to summarize country level, continent level, and overall average performances.
IRB Approval was obtained at Assistance Publique—Hôpitaux de Paris, Beth Israel Deaconess Medical Center, Bordeaux University Hospital, Hospital Universitario 12 de Octubre, Massachusetts General Brigham, Northwestern University, Medical Center, University of Freiburg, University of Pittsburgh, VA North Atlantic, VA Southwest, VA Midwest, VA Continental, and VA Pacific. An exempt determination was made by the IRB at University of California Los Angeles, University of Michigan, and University of Pennsylvania.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
G.W. reports funding from NCATS UL1TR002541, NCATS UL1TR000005, and NLM R01LM013345. S.M. and J.K. report funding from NCATS 5UL1TR001857-05 and NHGRI 5R01HG009174-04. Z.X. reports funding from NINDS R01NS098023. G.O. reports funding from NIH grants NIEHS P30ES017885 and NCI U24CA210967. S.V. reports funding from NLM R01LM012095 and NCATS UL1TR001857. A.S. reports funding from NHLBI K23HL148394 and L40HL148910, and NCATS UL1TR001420. B.A. reports funding from NHLBI U24 HL148865. D.B. and R.F. report funding from NCATS UL1TR001881. T.G. and T.G. report funding from 01ZZ1801E German Federal Ministry of Education and Research. D.H. reports funding from NCATS UL1TR002240. M.K. reports funding from NHGRI 5T32HG002295-18. D.K. reports funding from MIRACUM Consortium grant 01ZZ1801A. Y.L. reports funding from NLM R01LM01333. J.M. reports funding from NCATS UL1TR001878. D.M. reports funding from NCATS UL1-TR001878 Institutional Clinical and Translational Science Award (University of Pennsylvania). L.P. reports funding from NCATS CTSA Award #UL1TR002366.
Author contributions
G.M.W., C.H., N.P.P., P.A., S.N.M., A.G.S., G.S.O., J.G.K., R.B., M.A., B.J.A., D.S.B., F.T.B., K.C., A.D., J.H.M., I.S.K., T.C., and G.A.B. contributed to design and conceptualization of the study. G.M.W., Z.X., N.P.P., P.A., S.N.M., A.S.L., A.N., S.V., J.G.K., A.M.S., N.H.W.L., M.C., B.K.B.J., R.B., G.A., M.A., D.S.B., V.B., L.C., K.C., A.D., S.L.D., N.G.B., D.A.H., Y.L.H., J.H.H., R.W.I., Y.L., K.E.L., S.E.M., A.M., K.D.M., C.M., M.E.M., J.H.M., J.S.M., M.M., D.L.M., K.Y.N., L.P.P., M.P.J., R.B.R., E.R.S., P.S., P.S.B., A.S., A.L.M.T., B.W.L.T., V.T., C.T., and E.M.T. contributed to data collection. G.M.W., C.H., N.P.P., P.A., S.L., M.S.K., S.N.M., A.G.S., C.L.B., G.S.O., S.V., J.G.K., A.M.S., M.C., B.K.B.J., G.A., M.A., B.J.A., D.S.B., F.T.B., A.D., S.L.D., D.A.H., J.H.H., M.L., Y.L., S.E.M., K.D.M., C.M., M.E.M., J.S.M., M.M., L.P.P., A.L.M.T., C.T., E.M.T., X.W., I.S.K., T.C., and G.A.B. contributed to data analysis and interpretation. All authors contributed to drafting and revision of the manuscript and approved the final manuscript. All authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Data availability
Only aggregate data was shared by sites for this study. All aggregate data in a de-identified fashion can be found and downloaded at www.covidclinical.net.
Code availability
The SQL and R scripts used in this work can be found and downloaded at https://github.com/covidclinical.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Griffin M. Weber, Chuan Hong, Zongqi Xia.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Tianxi Cai, Email: tcai@hsph.harvard.edu.
Gabriel A. Brat, Email: gabriel_brat@hms.harvard.edu
The Consortium for Clinical Characterization of COVID-19 by EHR (4CE):
James R. Aaron, Adem Albayrak, Giuseppe Albi, Anna Alloni, Danilo F. Amendola, François Angoulvant, Li L. L. J. Anthony, Fatima Ashraf, Andrew Atz, Paul Avillach, Paula S. Azevedo, James Balshi, Brett K. Beaulieu-Jones, Antonio Bellasi, Vincent Benoit, Michele Beraghi, José Luis Bernal-Sobrino, Mélodie Bernaux, Romain Bey, Surbhi Bhatnagar, Alvar Blanco-Martínez, Martin Boeker, John Booth, Silvano Bosari, Robert L. Bradford, Gabriel A. Brat, Stéphane Bréant, Nicholas W. Brown, Raffaele Bruno, William A. Bryant, Mauro Bucalo, Emily Bucholz, Anita Burgun, Tianxi Cai, Aldo Carmona, Charlotte Caucheteux, Julien Champ, Krista Y. Chen, Jin Chen, Lorenzo Chiudinelli, Kelly Cho, James J. Cimino, Tiago K. Colicchio, Sylvie Cormont, Sébastien Cossin, Jean B. Craig, Juan Luis Cruz-Bermúdez, Jaime Cruz-Rojo, Mohamad Daniar, Christel Daniel, Priyam Das, Batsal Devkota, Audrey Dionne, Rui Duan, Julien Dubiel, Loic Esteve, Hossein Estiri, Shirley Fan, Robert W. Follett, Thomas Ganslandt, Noelia García-Barrio, Lana X. Garmire, Nils Gehlenborg, Emily J. Getzen, Alon Geva, Tobias Gradinger, Alexandre Gramfort, Romain Griffier, Nicolas Griffon, Olivier Grisel, Alba Gutiérrez-Sacristán, Larry Han, David A. Hanauer, Christian Haverkamp, Derek Y. Hazard, Bing He, Darren W. Henderson, Martin Hilka, Kenneth M. Huling, Meghan R. Hutch, Richard W. Issitt, Anne Sophie Jannot, Vianney Jouhet, Ramakanth Kavuluru, Chris J. Kennedy, Kate F. Kernan, Daniel A. Key, Katie Kirchoff, Jeffrey G. Klann, Isaac S. Kohane, Ian D. Krantz, Detlef Kraska, Ashok K. Krishnamurthy, Trang T. Le, Judith Leblanc, Guillaume Lemaitre, Leslie Lenert, Damien Leprovost, Molei Liu, Qi Long, Sara Lozano-Zahonero, Sadiqa Mahmood, Sarah E. Maidlow, Adeline Makoudjou, Anupama Maram, Patricia Martel, Marcelo R. Martins, Jayson S. Marwaha, Aaron J. Masino, Maria Mazzitelli, Arthur Mensch, Marianna Milano, Marcos F. Minicucci, Bertrand Moal, Taha Mohseni Ahooyi, Jason H. Moore, Cinta Moraleda, Jeffrey S. Morris, Karyn L. Moshal, Sajad Mousavi, Douglas A. Murad, Shawn N. Murphy, Thomas P. Naughton, Carlos Tadeu Breda Neto, Jane Newburger, Kee Yuan Ngiam, Wanjiku F. M. Njoroge, James B. Norman, Jihad Obeid, Marina P. Okoshi, Karen L. Olson, Gilbert S. Omenn, Nina Orlova, Brian D. Ostasiewski, Nathan P. Palmer, Nicolas Paris, Lav P. Patel, Miguel Pedrera-Jiménez, Ashley C. Pfaff, Emily R. Pfaff, Danielle Pillion, Sara Pizzimenti, Hans U. Prokosch, Robson A. Prudente, Andrea Prunotto, Víctor Quirós-González, Rachel B. Ramoni, Maryna Raskin, Siegbert Rieg, Gustavo Roig-Domínguez, Pablo Rojo, Paula Rubio-Mayo, Paolo Sacchi, Carlos Sáez, Elisa Salamanca, Malarkodi Jebathilagam Samayamuthu, L. Nelson Sanchez-Pinto, Arnaud Sandrin, Nandhini Santhanam, Janaina C. C. Santos, Fernando J. Sanz Vidorreta, Maria Savino, Juergen Schuettler, Luigia Scudeller, Neil J. Sebire, Pablo Serrano-Balazote, Patricia Serre, Arnaud Serret-Larmande, Mohsin Shah, Zahra Shakeri Hossein Abad, Domenick Silvio, Piotr Sliz, Jiyeon Son, Charles Sonday, Andrew M. South, Francesca Sperotto, Zachary H. Strasser, Amelia L. M. Tan, Bryce W. Q. Tan, Suzana E. Tanni, Deanne M. Taylor, Ana I. Terriza-Torres, Patric Tippmann, Emma M. S. Toh, Yi-Ju Tseng, Andrew K. Vallejos, Gael Varoquaux, Margaret E. Vella, Guillaume Verdy, Jill-Jênn Vie, Shyam Visweswaran, Michele Vitacca, Kavishwar B. Wagholikar, Lemuel R. Waitman, Demian Wassermann, Griffin M. Weber, Martin Wolkewitz, Scott Wong, Zongqi Xia, Xin Xiong, Ye Ye, Nadir Yehya, William Yuan, Alberto Zambelli, Harrison G. Zhang, Daniela Zöller, Valentina Zuccaro, and Chiara Zucco
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-022-00601-0.
References
- 1.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 2.Wynants L, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goyal P, et al. Clinical characteristics of Covid-19 in New York City. N. Engl. J. Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fried, M. W. et al. Patient characteristics and outcomes of 11,721 patients with COVID19 hospitalized across the United States. Clin. Infect. Dis. 10.1093/cid/ciaa1268 (2020). [DOI] [PMC free article] [PubMed]
- 5.Brat GA, et al. International electronic health record-derived COVID-19 clinical course profiles: the 4CE consortium. NPJ Digit. Med. 2020;3:109. doi: 10.1038/s41746-020-00308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lippi, G. & Plebani, M. Laboratory abnormalities in patients with COVID-2019 infection. Clin. Chem. Lab. Med.10.1515/cclm-2020-0198 (2020). [DOI] [PubMed]
- 7.COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) (Johns Hopkins University (JHU), accessed 7 October 2020); https://www.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6.
- 8.Guan, W. -J. et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 10.1056/NEJMoa2002032 (2020). [DOI] [PMC free article] [PubMed]
- 9.Uno H, Cai T, Tian L, Wei LJ. Evaluating prediction rules for t-year survivors with censored regression models. J. Am. Stat. Assoc. 2007;102:527–537. doi: 10.1198/016214507000000149. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Only aggregate data was shared by sites for this study. All aggregate data in a de-identified fashion can be found and downloaded at www.covidclinical.net.
The SQL and R scripts used in this work can be found and downloaded at https://github.com/covidclinical.