Abstract
It is generally accepted for Escherichia coli that (i) the level of OmpC increases with increased osmolarity when cells are growing in neutral and alkaline media, whereas the level of OmpF decreases at high osmolarity, and that (ii) the two-component system composed of OmpR (regulator) and EnvZ (sensor) regulates porin expression. In this study, we found that OmpC was expressed at low osmolarity in medium of pH below 6 and that the expression was repressed when medium osmolarity was increased. In contrast, the expression of ompF at acidic pH was essentially the same as that at alkaline pH. Neither OmpC nor OmpF was detectable in an ompR mutant at both acid and alkaline pH values. However, OmpC and OmpF were well expressed at acid pH in a mutant envZ strain, and their expression was regulated by medium osmolarity. Thus, it appears that E. coli has a different mechanism for porin expression at acid pH. A mutant deficient in ompR grew slower than its parent strain in low-osmolarity medium at acid pH (below 5.5). The same growth diminution was observed when ompC and ompF were deleted, suggesting that both OmpF and OmpC are required for optimal growth under hypoosmosis at acid pH.
The regulatory mechanism for porin expression has been well defined in both Escherichia coli and Salmonella enterica serovar Typhimurium (for reviews, see references 3, 15, and 18). Gene expression of ompC and ompF is regulated by medium osmolarity; OmpC and OmpF are synthesized at high and low osmolarities, respectively. The regulation is mediated via the OmpR/EnvZ two-component system. OmpR is the regulator and is phosphorylated by the sensor protein EnvZ.
Expression of outer membrane proteins is also affected by medium pH; growth at low pH increases OmpC expression and decreases the level of OmpF (8, 24). Neither OmpC nor OmpF was detectable in an ompR mutant at acid pH, and mutation of envZ reduced the expression of ompC- and ompF-lacZ fusion genes (8). The induction of the ompC-lacZ fusion gene was greatly stimulated in rich medium but was very low in minimal medium in S. enterica serovar Typhimurium at acid pH (6). Thus, the mechanism for pH-dependent porin expression is still enigmatic.
Two possible mechanisms for the regulation at acid pH can be postulated: (i) the activity of this two-component regulatory system is modified by pH, and (ii) E. coli uses different systems at different pH values for porin expression, as proposed previously for Na+ extrusion systems (21). In this study, we found that OmpC was synthesized in medium of low osmolarity at acid pH and that the level decreased under hyperosmosis. In contrast, the expression of OmpF was essentially unchanged at both high and low pHs. Therefore, both OmpF and OmpC are expressed under hypoosmosis at acid pH. In agreement with these observations, the expression of OmpF and OmpC increased the growth rate in low-osmolarity medium at low pH, but no difference in growth was observed when medium osmolarity was increased. Neither OmpF nor OmpC was detectable in an ompR mutant at low pH or at high pH. The expression of OmpF and OmpC requires envZ at alkaline pH, but both proteins were expressed at low pH in an envZ mutant, suggesting that porin expression is regulated in a different manner by medium osmolarity at a low pH.
MATERIALS AND METHODS
E. coli strains and growth media.
The E. coli strains used are listed in Table 1. The following three media were used. Medium A contained 5 mM K2HPO4, 20 mM NH4Cl, and 1% glucose. Medium B contained 1 mM K2HPO4, 10 mM NH4Cl, and 0.1% glucose. Medium C contained 1 mM K2HPO4, 1 mM NH4Cl, and 0.1% glucose. All media contained 1 mM MgCl2, 0.1 mM CaCl2, 0.1 mM FeSO4, and 20 μg of thiamine/ml. Fifty millimolar N-Tris(hydroxymethyl)-methylglycine (Tricine) for media of pH 8.5 and 2-(N-morpholino)ethanesulfonic acid monohydrate (MES) for media of pHs 5.5 and 5.0 were used. Medium C contained 5 mM concentrations of these buffers instead of 50 mM concentrations. The medium pH was adjusted by the addition of KOH. Growth was monitored by measuring the absorbance of the medium at 600 nm when cells were cultured in media A and B. The viable cells in medium C were counted on L broth (10) agar plates.
TABLE 1.
E. coli strains used in this study
| Strain | Relevant genotype | Source (reference) |
|---|---|---|
| JB100 | ompR::Tn10 | K. Nishimura (2) |
| AT142 | MC4100 envZ::Kana | T. Mizuno (14) |
| MH1160 | MC4100 ompR101 | T. J. Silhavy (7) |
| W3110 | Wild type | Laboratory stock |
| MSR31 | W3110 ompR::Tn10 | This study; W3110×P1(JB100) |
| MSZ31 | W3110 envZ::Kan | This study; W3110×P1(AT142) |
| PS2209 | W3110 Δ(arg-lac)U169 | R. G. Matthews (4) |
| MH225 | MC4100 malQ7 Φ(ompC-lacZ)10-25 ompC::Tn5 | R. G. Matthews (4) |
| MKC5.4 | PS2209 Φ(ompC-lacZ)10-25 | This study; PS2209×P1(MH225) |
| MKCZ5.4 | PS2209 Φ(ompC-lacZ)10-25 envZ::Kan | This study; MKC5.4×P1 (AT142) |
| MKCR5.4 | PS2209 Φ(ompC-lacZ)10-25 ompR::Tn10 | This study; MFC5.4×P1(JB100) |
| MF8.1 | PS2209 Φ(ompF-lacZ)16-13 ompF+ | R. G. Matthews (4) |
| MSR6 | PS2209 Φ(ompF-lacZ)16-13 ompF+ ompR::Tn10 | This study; MF8.1×P1(JB100) |
| MSZ6 | PS2209 Φ(ompF-lacZ)16-13 ompF+ envZ::Kan | This study; MF8.1×P1(AT142) |
| D3 | W3110 recD::Tn10 ompC::Kan | This study; see Materials and Methods |
| CAG12094 | MG1655 zcb-3059::Tn10 OmpF+ | K. Nishimura (22) |
| MH621 | MH20 Φ(ompF-lacZ)16-21(Hyb) ompF ompC+ | R. G. Matthews (4) |
| MKCF36 | MH20 Φ(ompF-lacZ)16-21(Hyb) ompF ompC::Kan | This study; MH621×P1(D3) |
| MKW505 | MH20 Φ(ompF-lacZ)16-21(Hyb) ompF+ ompC+ | This study; MH621×P1(CAG12094) |
| MKC505 | MH20 Φ(ompF-lacZ)16-21(Hyb) ompF+ ompC::Kan | This study; MKW505×P1(D3) |
Kan, gene conferring kanamycin resistance.
Construction of ompC::Kan.
The chromosomal ompC of W3110 recD::Tn10 was replaced by the nonfunctional ompC::Kan with pBSompC::Kan (1) cut with KpnI and EcoRI, and the resulting strain was designated DK3. Plasmid pBSompC::Kan was kindly supplied by K. Igarashi (Chiba University, Chiba, Japan).
Isolation of outer membrane proteins and analysis.
Outer membrane proteins were prepared as described previously (12) with modifications. Cells washed with 10 mM Tris-HCl (pH 8.0) were suspended in the same buffer containing 20% sucrose, and the suspension was kept on ice after the addition of 1 mM EDTA and lysozyme (0.1 mg/ml). Cells were disrupted by sonication. After centrifugation at 2,500 × g for 5 min, cell envelopes were collected by centrifugation of the supernatant at 25,000 × g for 10 min, washed twice with 10 mM Tris-HCl (pH 8.0), and then suspended in the same buffer containing 2% Triton X-100. The suspension was incubated at room temperature for 30 min and centrifuged at 100,000 × g for 30 min. The resulting pellet was stored at −20°C before use. The pellet was heated in a 1.25% sodium dodecyl sulfate (SDS) solution containing β-mercaptoethanol (1.25%) at 100°C for 5 min. The proteins (15 μg) were separated by urea-SDS–polyacrylamide gel electrophoresis as described previously (26) and stained with Coomassie brilliant blue R250. The stained gel was scanned with ScanJet IIC (Hewlett-Packard), and densitometric analysis was performed using NIH Image (version 1.61).
Other methods and chemicals.
Measurement of β-galactosidase activity (13), protein determination (11), and P1 transduction (10) were carried out as described previously. Reagents used were of analytical grade.
RESULTS
Effect of medium osmolarity on gene expression of OmpC and OmpF at low pH.
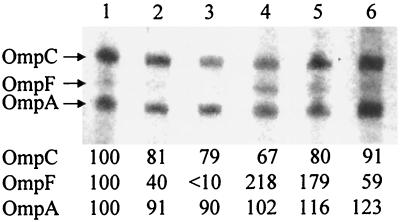
OmpF was expressed in low-osmolarity medium of initial pH 8.5, and the expression of OmpC was stimulated at high osmolarity under our growth conditions (Fig. 1). In contrast to what was observed at alkaline pH, both OmpC and OmpF were expressed in cells growing in low-osmolarity medium of initial pH 5.5, and expression of both proteins was repressed by the addition of NaCl (Fig. 1). The medium pH decreases rapidly when glucose is used as an energy source. In our present study, when cells were harvested at an absorbance of 0.3 to 0.4, the medium pH decreased to 5.1 to 5.3 and 7.9 to 8.1 in media with initial pHs of 5.5 and 8.5, respectively.
FIG. 1.
Urea-SDS–polyacrylamide gel electrophoresis of outer membrane proteins. W3110 (wild type) was grown in medium A of initial pH 5.5 (lanes 1 to 3) and pH 8.5 (lanes 4 to 6) and harvested at an absorbance of 0.3 to 0.4 as described in Materials and Methods. When cells were harvested, the medium pH values were 5.1 to 5.3 and 7.9 to 8.1, respectively. Densities of lane 1 were taken as 100, and relative densities are represented. Lanes: 1 and 4, cells grown without the addition of NaCl; 2 and 5, cells grown in the presence of 100 mM NaCl; 3 and 6, cells grown in the presence of 200 mM NaCl.
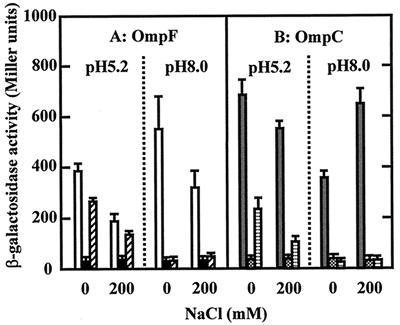
In order to measure the expression more quantitatively, a fusion gene with lacZ was used. The expression of OmpF decreased in high-osmolarity medium at both pH values (Fig. 2A), whereas the expression of OmpC decreased with the increase in medium osmolarity at acid pH (Fig. 2B). When 0.3 M NaCl was added, the OmpC expression decreased to approximately 50% of the expression without the addition of NaCl in medium of initial pH 5.5 (data not shown). Thus, it appears that the effect of osmolarity on OmpF expression at acid pH is the same as that at alkaline pH but that the effect on OmpC expression is reversed.
FIG. 2.
Expression of OmpC and OmpF at low pH. Cells were grown in medium A of initial pHs 5.5 and 8.5 with and without the addition of 200 mM NaCl. Cells were harvested as described in the legend of Fig. 1. One Miller unit of β-galactosidase activity was defined as described previously (13). Each bar represents the mean ± standard deviation of four independent experiments. (A) MF8.1 (ompF-lacZ; open bars), MSR6 (ompF-lacZ ompR::Tn10; solid bars), and MSZ6 (ompF-lacZ envZ::kan; hatched bars). (B) MKC5.4 (ompC-lacZ; gray bars), MKCR5.4 (ompC-lacZ ompR::Tn10; dotted bars), and MKCZ5.4 (ompC-lacZ envZ::kan; horizontally striped bars).
Role of OmpR and EnvZ in porin expression at low pH.
The above results suggested the possibility that E. coli uses different mechanisms for porin gene expression at different pH values. To elucidate this possibility, we constructed mutants containing a disrupted ompR gene by the insertion of Tn10. Neither OmpC nor OmpF was detected by electrophoresis of outer membrane proteins from MSR31 (W3110 ompR::Tn10; data not shown). No significant expression of ompC- and ompF-lacZ fusion genes in MKCR5.4 (ompC-lacZ ompR::Tn10) and MSR6 (ompF-lacZ ompR::Tn10) grown in acid and alkaline media was observed (Fig. 2). EnvZ may not have been expressed when Tn10 was inserted into ompR, as envZ and ompR are in an operon with the promoter located upstream of ompR (23). Next, we used the ompR101 mutation (7), an in-frame deletion of 57 bp (17). The expression levels of OmpC and OmpF in the outer membrane fraction from MH1160 (ompR101; Fig. 3) were very low, suggesting that ompR is essential for gene expression of OmpC and OmpF at low as well as at alkaline pHs.
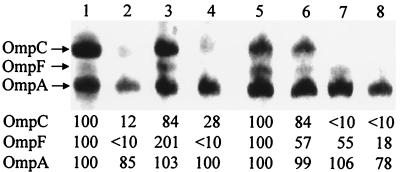
FIG. 3.
Urea-SDS–polyacrylamide gel electrophoresis of outer membrane proteins. Cells grown in medium A of initial pH 5.5 (lanes 1, 2, 5, and 6) and pH 8.5 (lanes 3, 4, 7, and 8) were harvested as described in the legend of Fig. 1. Densities of lanes 1 and 5 were taken as 100, and relative densities are represented. Lanes: 1 and 3, W3110 (wild type); 2 and 4, MH1160 (ompR101); 5 and 7, MSZ31 (W3110 envZ) grown without the addition of NaCl; 6 and 8, MSZ31 (W3110 envZ) grown in the presence of 200 mM NaCl.
Mizuno and Mizushima (14) showed that an envZ deletion mutant produced small amounts of OmpC and OmpF and that their expression was regulated by osmolarity. Our data demonstrate a weak expression of OmpC and OmpF at alkaline pH (Fig. 3). In contrast to what was observed at alkaline pH, both OmpC and OmpF were well expressed in envZ mutants at acid pH (Fig. 3). Expression of OmpC and OmpF was repressed by the addition of NaCl to the growth medium at acid pH (Fig. 3). Both ompC- and ompF-lacZ fusion genes were expressed at acid pH even if envZ was deleted, and the expression was again regulated by medium osmolarity (Fig. 2). These results suggest that E. coli has an EnvZ-independent system for porin expression functioning at acid pH.
Effect of OmpF and OmpC on growth at acid pH.
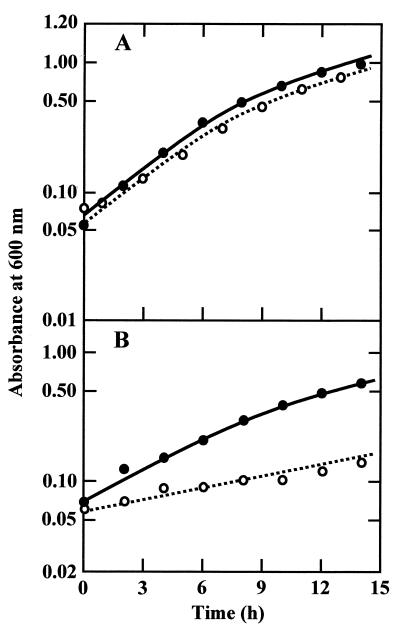
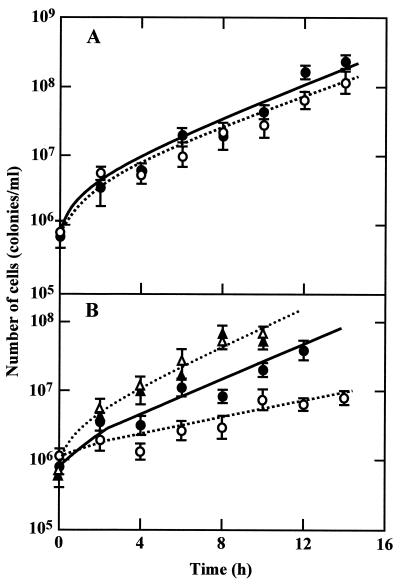
As shown in Fig. 4, growth of MSR31 (W3110 ompR::Tn10) was nearly the same as that of its parent strain, W3110, in medium A (approximately 0.20 osM) of initial pH 5.5 but slower than that of W3110 in medium B (approximately 0.12 osM). The difference in growth rate at low pH was enhanced in medium C, whose osmolarity was approximately 0.02 osM (Fig. 5B). Since medium C contained a low concentration of MES buffer, the medium pH dropped rapidly at a high cell density. Therefore, we started the cell culture at a low density of 0.5 × 106 to 1.1 × 106 cells/ml, and growth was monitored by determining the viable cell count. When growth in medium A was measured in terms of viable cell count, it was found that the cell number increased in proportion to the absorbance of the culture medium in both strains. MSR31 (W3110 ompR::Tn10) grew in medium C containing 200 mM NaCl at the same rate as W3110 (Fig. 5B), suggesting that the low growth rate of the ompR mutant is due to a low osmolarity but not to a low concentration of some nutrients of this medium. In contrast to the results for growth at acid pH, no significant difference in the growth rates of the two strains was observed at alkaline pH in medium C (Fig. 5A) and medium A containing 200 mM NaCl (data not shown).
FIG. 4.
Growth of W3110 and MSR31. W3110 (wild type; solid symbols) and MSR31 (W3110 ompR::Tn10; open symbols) were grown in media A (A) and B (B) of initial pH 5.5. Growth was monitored by measuring the absorbance of the medium.
FIG. 5.
Growth of W3110 and MSR in low-osmolarity medium. W3110 (wild type; solid symbols) and MSR31 (W3110 ompR::Tn10; open symbols) were grown in medium C of initial pHs 8.5 (A) and 5.5 (B). Growth was monitored by determining the viable cell count. Symbols: circles, no addition of NaCl; triangles, 200 mM NaCl added.
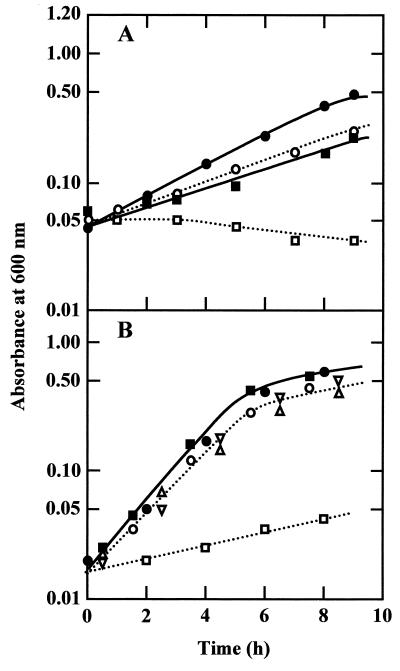
The difference in growth rates between W3110 (wild type) and MSR31 (W3110 ompR::Tn10) was enhanced in medium of initial pH 5.0 (Fig. 6A). It is quite likely that growth diminution is due the absence of porin proteins. To confirm this idea, we constructed a mutant deficient in ompC and ompF. MKCF36 (ompC ompF) grew slower than MKW505 (ompC+ ompF+) in medium B of initial pH 5.0, and no difference in the growth rate was observed when 200 mM NaCl was added to the medium (Fig. 6B). The same results were obtained in strains deficient in ompC and ompF constructed separately by the same procedure. MKW505 (ompC+ ompF+) grew faster than W3110, probably due to a different genetic background.
FIG. 6.
Growth of mutants deficient in OmpC and OmpF in medium of initial pH 5.0. Cells were grown in medium B of initial pH 5.0 (open symbols) and the same medium containing 200 mM NaCl (solid symbols). Growth was monitored by measuring the absorbance of the medium. (A) ● and ○, W3110 (ompR+); ■ and □, MSR31 (ompR). (B) ● and ○, MKW505 (ompC+ ompF+); ■ and □, MKCF36 (ompC ompF); Δ, MKC505 (ompC ompF+); ▿, MH621 (ompC+ ompF).
The growth rates of MKC505 (ompC) and MH621 (ompF) were the same as that of MKW505 (ompC+ ompF+) in medium B of initial pH 5.0 (Fig. 6B) as well as in the same medium containing 200 mM NaCl (data not shown). MKFC36 (ompC ompF) grew at the same rate as MKW505 (ompC+ ompF+), MKC505 (ompC), and MH621 (ompF) in medium B of initial pH 8.5 with and without the addition of 200 mM NaCl (data not shown). The growth yield was low in medium B of initial pH 5.0 (Fig. 6B). Since the buffering capacity of MES decreases rapidly at a pH below 5, the low yield is probably due to the decrease in medium pH. In fact, the medium pH dropped to approximately 4 when growth stopped.
DISCUSSION
In agreement with previous reports (8, 24), our data showed that the level of OmpC was high at low pH when cells were cultured without the addition of NaCl. However, the OmpC level was low in cells grown in high-osmolarity medium at acid pH, as the OmpC expression is down-regulated by medium osmolarity at low pH. Thomas and Booth (24) have reported that OmpF expression is repressed in medium of pH 6.0 when glucose is used. However, OmpF was expressed in cells growing in glucose medium of initial pH 5.5 under our experimental conditions (Fig. 1 and 2). As described in Results, the medium pH decreases rapidly when glucose is used as an energy source. Therefore, the discrepancy might be due to a different pH value when the cells were harvested.
Our present observations lead us to conclude that E. coli has two kinds of mechanisms for porin expression. One is working at neutral and alkaline pH values, and the other is for acid pH. OmpR participates in both mechanisms, but EnvZ is involved only in the former system. OmpC was expressed at pH 5.5 in an S. enterica serovar Typhimurium mutant deficient in ompR (6). Thus, the regulation at low pH might be different between the two strains.
Why does E. coli have multiple regulatory systems for porin expression?
Expression of OmpF in envZ mutants is slightly lower than that in the wild type at pH 5.2, suggesting that the activity of the mechanism mediated by EnvZ is low at acid pH. Thus, the most likely explanation would be that EnvZ has a low activity at acid pH. It is also possible that the expression of EnvZ decreases at low pH. In both cases, a mechanism independent from EnvZ may be required in acid environments. An unknown factor might function as a sensor instead of EnvZ at acid pH. In agreement with the previous report by Mizuno and Mizushima (14), weak expression of OmpF and OmpC was observed at alkaline pH in our envZ mutant (Fig. 2 and 3). These results suggest that the activity of the system independent from EnvZ proposed here is very low at pH around 8.0 but detectable.
How does the environmental pH affect the activity of the OmpR/EnvZ system?
Since EnvZ is a membrane protein, a domain located outside of the cytoplasmic membranes might function as a pH sensor. Alternatively, the regulatory system may be affected by cytoplasmic pH. Mugikura et al. (16) demonstrated that the cytoplasmic pH is kept constant within the range of medium pH from 6 to 8 and that the cytoplasm is acidified in acid medium below pH 6. Since porin expression at pH 6.5 was similar to that at pH 8.0 (data not shown), the latter possibility is more likely. Heyde and Portalier (8) have reported that the porin expression was mediated via EnvZ in Luria-Bertani medium of initial pH 5.6 without glucose. The medium they used is alkalinized rapidly at a high cell density. Therefore, if the medium pH was above 6 when cells were harvested (absorbance of 0.6 to 0.8), expression of OmpC could be mediated via EnvZ.
To the best of our knowledge, this is the first report to demonstrate the effect of OmpC or OmpF on growth of E. coli. It has been shown in this study that the expression of either OmpC or OmpF is required for growth in low-osmolarity medium at acid pH (Fig. 6). The decrease in the OmpC expression due to NaCl was only about 50% (Fig. 2). However, the above results suggest that the higher expression of OmpC and OmpF is essential for growth under hypoosmosis at acid pH. It is an open question why these porins are required under such conditions. It can be argued that the active accumulation of nutrients decreases at acid pH because of the low activity of energy metabolism. Therefore, an acceleration of the movement of nutrients across the outer membranes through porins would be especially important at low pH when the surroundings contain a low concentration of nutrients.
When E. coli cells were transferred from acid medium of pH 5.5 to alkaline medium, cells became sensitive to alkalinity (19, 20). The sensitivity to alkalinity was reduced by ompC lesion (20), and our results show that OmpC is expressed at low pH. These observations suggest that the presence of OmpC reduces the ability to survive at alkaline pH. The pore size of OmpC is regulated by pH; an increase in pH induced the structural change from a small-sized channel conformation to a large-sized one (25). Kennedy (9) reported that membrane-derived oligosaccharides (MDO) were accumulated in the periplasmic space at low osmolarity. The deletion of MDO synthesis enhanced OmpC expression (5). Thus, porins might participate in the maintenance of osmolytes in the periplasmic space under hypoosmosis.
ACKNOWLEDGMENTS
We thank K. Igarashi (Chiba University) R. G. Matthews (University of Michigan, Ann Arbor, Mich.), T. Mizuno (Nagoya University, Nagoya, Japan), K. Nishimura (National Institute of Genetics, Mishima, Japan), and T. J. Silhavy (Princeton University, Princeton, N.J.) for gifts of strains and plasmids. We are grateful to R. L. Kaspar for his help with the preparation of the manuscript and valuable suggestions.
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Apirakaramwong A, Fukuchi J, Kashiwagi K, Kakinuma Y, Ito E, Ishihama A, Igarashi K. Enhancement of cell death due to decrease in Mg2+ uptake by OmpC (cation-selective porin) deficiency in ribosome modulation factor-deficient mutant. Biochem Biophys Res Commun. 1998;251:482–487. doi: 10.1006/bbrc.1998.9494. [DOI] [PubMed] [Google Scholar]
- 2.Brass J M, Bauer K, Ehmann U, Boos W. Maltose-binding protein does not modulate the activity of maltoporin as a general porin in Escherichia coli. J Bacteriol. 1985;161:720–726. doi: 10.1128/jb.161.2.720-726.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csonka L N. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrario M, Ernsting B R, Borst D W, Wiese II D E, Blumenthal R M, Matthews R G. The leucine-responsive regulatory protein of Escherichia coli negatively regulates transcription of ompC and micF and positively regulates translation of ompF. J Bacteriol. 1995;177:103–113. doi: 10.1128/jb.177.1.103-113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiedler W, Rotering H. Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J Biol Chem. 1988;263:14684–14689. [PubMed] [Google Scholar]
- 6.Foster J W, Park Y K, Bang I S, Karem K, Betts H, Hall H K, Shaw E. Regulatory circuits involved with pH-regulated gene expression in Salmonella typhimurium. Microbiology. 1994;140:341–352. doi: 10.1099/13500872-140-2-341. [DOI] [PubMed] [Google Scholar]
- 7.Hall M N, Silhavy T J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K-12. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 8.Heyde M, Portalier R. Regulation of major outer membrane porin proteins of Escherichia coli K12 by pH. Mol Gen Genet. 1987;208:511–517. doi: 10.1007/BF00328148. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy E P. Osmotic regulation and the biosynthesis of membrane-derived oligosaccharides in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:1092–1095. doi: 10.1073/pnas.79.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 11.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 12.Matsuyama S, Inokuchi K, Mizushima S. Promoter exchange between ompF and ompC, genes for osmoregulated major outer membrane proteins of Escherichia coli K-12. J Bacteriol. 1984;158:1041–1047. doi: 10.1128/jb.158.3.1041-1047.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 14.Mizuno T, Mizushima S. Isolation and characterization of deletion mutants of ompR and envZ, regulatory genes for expression of the outer membrane proteins OmpC and OmpF in Escherichia coli. J Biochem. 1987;101:387–396. doi: 10.1093/oxfordjournals.jbchem.a121923. [DOI] [PubMed] [Google Scholar]
- 15.Mizuno T, Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol. 1990;4:1077–1082. doi: 10.1111/j.1365-2958.1990.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 16.Mugikura S, Nishikawa M, Igarashi K, Kobayashi H. Maintenance of a neutral cytoplasmic pH is not obligatory for growth of Escherichia coli and Streptococcus faecalis at an alkaline pH. J Biochem. 1990;108:86–91. doi: 10.1093/oxfordjournals.jbchem.a123168. [DOI] [PubMed] [Google Scholar]
- 17.Nara F, Matsuyama S, Mizuno T, Mizushima S. Molecular analysis of mutant ompR genes exhibiting different phenotypes as to osmoregulation of the ompF and ompC genes of Escherichia coli. Mol Gen Genet. 1986;202:194–199. doi: 10.1007/BF00331636. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowbury R J. Regulatory components, including integration host factor, CysB and H-NS, that influence pH responses in Escherichia coli. Lett Appl Microbiol. 1997;24:319–328. doi: 10.1046/j.1472-765x.1997.00065.x. [DOI] [PubMed] [Google Scholar]
- 20.Rowbury R J, Hussain N H. Exposure of Escherichia coli to acid habituation conditions sensitizes it to alkaline stress. Lett Appl Microbiol. 1996;22:57–61. doi: 10.1111/j.1472-765x.1996.tb01108.x. [DOI] [PubMed] [Google Scholar]
- 21.Sakuma T, Yamada N, Saito H, Kakegawa T, Kobayashi H. pH dependence of the function of sodium ion extrusion systems in Escherichia coli. Biochim Biophys Acta. 1998;1363:231–237. doi: 10.1016/s0005-2728(97)00102-3. [DOI] [PubMed] [Google Scholar]
- 22.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slauch J M, Garrett S, Jackson D E, Silhavy T J. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol. 1988;170:439–441. doi: 10.1128/jb.170.1.439-441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas A D, Booth I R. The regulation of expression of the porin gene ompC by acid pH. J Gen Microbiol. 1992;138:1829–1835. doi: 10.1099/00221287-138-9-1829. [DOI] [PubMed] [Google Scholar]
- 25.Todt J C, McGroarty E J. Acid pH decreases OmpF and OmpC channel size in vivo. Biochem Biophys Res Commun. 1992;189:1498–1502. doi: 10.1016/0006-291x(92)90244-f. [DOI] [PubMed] [Google Scholar]
- 26.Uemura J, Mizushima S. Isolation of outer membrane proteins of Escherichia coli and their characterization on polyacrylamide gel. Biochim Biophys Acta. 1975;413:163–176. doi: 10.1016/0005-2736(75)90101-7. [DOI] [PubMed] [Google Scholar]