Abstract
A Pseudomonas 2,4-diacetylphloroglucinol (DAPG)-producing population that occurred naturally on the roots, in rhizosphere soil of Zea mays and in the nonrhizosphere soil was investigated in order to assess the microbial diversity at five stages of plant growth. A total of 1,716 isolates were obtained, and 188 of these isolates were able to produce DAPG. DAPG producers were isolated at each stage of plant growth, indicating that the maize rhizosphere is colonized by natural DAPG producers throughout development. The frequency of DAPG producers was very low in the first stage of plant growth and increased over time. An analysis of the level of biodiversity of the DAPG producers at the species level was performed by comparing the AluI restriction patterns of the 16S ribosomal DNAs (rDNAs) amplified by PCR from 167 isolates. This comparison allowed us to cluster the isolates into four amplified rDNA restriction analysis (ARDRA) groups, and the main group (ARDRA group 1) contained 89.8% of the isolates. The diversity of the 150 isolates belonging to ARDRA group 1 was analyzed by the random amplified polymorphic DNA (RAPD) technique. An analysis of RAPD patterns by a molecular variance method revealed that there was a high level of genetic diversity in this population and that the genetic diversity was related to plant age. Finally, we found that some of the DAPG producers, which originated from all stages of plant growth, had the same genotype. These DAPG producers could be exploited in future screening programs for biocontrol agents.
Some rhizobacteria, which are commonly called plant growth-promoting rhizobacteria, interact with plant roots and protect the roots against pathogenic microorganisms (26). Pseudomonads are common members of the plant growth-promoting rhizobacterial microflora in the rhizospheres of protected plants (13, 25, 42). The ability of pseudomonads to suppress soilborne fungal pathogens depends on their ability to produce antibiotic metabolites, such as pyoluteorin, pyrrolnitrin, phenazine-1-carboxylic acid, and 2,4-diacetylphloroglucinol (DAPG) (14, 32, 38, 47, 49, 55, 57). One of these metabolites, DAPG, is a major factor in the biological control of a range of plant pathogens (7, 23, 24, 57). The antibiotic DAPG is produced by pseudomonads of worldwide origin, and its biosynthetic locus is conserved in pseudomonads obtained from diverse geographic locations (25, 40). Bacteria that produce DAPG play a key role in agricultural environments, and their potential for use in sustainable agriculture is promising. However, introduction of these bacteria in the field often fails because the organisms are not able to recolonize the roots or colonize the roots but do not produce antibiotic compounds in the new environment (11, 41).
It is well known that root exudates are sources of nutrition for rhizosphere microorganisms (6, 43). The composition of root exudates is affected by the stage of plant development (18), which results in changes in the patterns and activities of rhizobacterial populations (9, 18, 33). Thus, the stage of plant development results in selection of a bacterial genotype. Understanding the relationship between the rhizosphere environment and the genetic diversity of local microbial populations seems to be required for evaluating the effect of microbial inoculation on the preexisting balance in indigenous populations. Therefore, analysis of the genetic structure and activity of a microbial population has practical importance, as the results can be used to estimate the fate of released strains and their impact on pathogenic resident microbial communities due to the production of antibiotics.
The aim of this work was to analyze the genetic diversity of rhizobacterial DAPG-producing populations isolated from maize roots at different stages of development. Such an analysis could result in identification of strains which could be successfully reinoculated into fields and protect maize.
In the present work, identification and analysis of the genetic polymorphism of DAPG producers isolated from maize roots were carried out by using a combination of phenotypic and PCR-based molecular techniques which have been used previously to study bacterial populations isolated from different natural environments (2, 5, 8–10). The strategy used included the following steps. (i) DAPG-producing strains isolated from maize roots were first grouped into clusters on the basis of an analysis of the restriction patterns of 16S ribosomal DNAs (rDNAs) amplified by PCR (amplified rDNA restriction analysis [ARDRA]). This technique has been used previously to distinguish species in different genera (17, 22, 27, 28, 31, 51–53). DNA digestion was performed by using the tetrameric restriction enzyme AluI, which very often generates species-specific restriction patterns, as shown previously (2, 5, 8–10, 16). (ii) Biolog fingerprinting and nucleotide sequencing of the 16S rDNAs of some representatives of the main ARDRA groups were carried out in order to identify the organisms. The biodiversity of strains belonging to the main ARDRA groups was examined by using the random amplified polymorphic DNA (RAPD) technique (58, 60), and the data obtained were analyzed by the analysis of molecular variance (AMOVA) technique (12).
MATERIALS AND METHODS
Bacterial strains.
The four reference strains used are Pseudomonas fluorescens 2-79 (49, 56) and Q2-87 (39), Pseudomonas putida M.3.1 (Digat, INRA, Angers, France), and Pseudomonas aureofaciens PGS12 (14).
Isolation of bacteria from roots of maize and from soil.
Maize plants (Zea mays cv. DEA; Pioneer France Maïs) were cultivated in pots (height, 30 cm; width, 20 cm) containing 5,000 g of soil in a growth chamber by using a 16-h photoperiod, a nighttime temperature of 19°C, a daytime temperature of 22°C, and a relative humidity of 65%. The soil used had not been used previously to grow maize. The soil composition was as follows: sand, 35%; clay, 27%; silt, 38%; organic C, 1.5% (wt/wt); pH 6.8.
Plants were collected after 27 (end of germination), 55 (tassel appearance), 72 (end of flowering), 92, and 106 (physiological maturation) days of growth. At each sampling time, three plants were harvested, the roots were excised, and nonadhering soil was removed. Rhizosphere soil was separated from the roots by vigorously shaking the roots in 100 ml of sterile water for 5 min by using a Vortex mixer. Washed roots and rhizosphere soil were separately blended with a Waring blender in sterile water. Serial dilutions of both suspensions were plated onto S1 medium, which is known to be selective for the recovery of fluorescent pseudomonads (15). The plates were incubated at 27°C, and numbers of CFU were determined after 48 h. Bacteria were also isolated from three samples of nonrhizosphere soil by using the protocol used for the rhizosphere soil. At each sampling time 52 randomly selected isolates were picked and transferred to new S1 medium plates. A total of 1,716 bacterial colonies were collected in this way (52 × 5 stages of plant growth × 3 repetitions = 780 isolates for roots and 780 isolates for rhizosphere soil; 52 × 3 repetitions = 156 isolates for nonrhizosphere soil). Isolates that harbored the phlD gene were selected by colony hybridization and high-performance liquid chromatography (HPLC) analysis.
Colony hybridization.
Bacterial colonies were transferred to nylon membranes (Boehringer Mannheim) by standard methods (45). The membranes were baked for 1 h at 80°C and then were treated with a 2-mg/ml proteinase K solution (0.5 ml for a membrane that was 82 mm in diameter) for 1 h at 37°C. To remove bacterial cell debris from colony blots, the membranes were blotted between pieces of filter paper wetted with distilled water, and pressure was applied by passing a ruler over the area. The hybridization buffer used contained 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium lauroylsarcosine, 0.02% sodium dodecyl sulfate (SDS), and 2% blocking reagent (Boehringer Mannheim). Prehybridization was performed for 1 h at 42°C, and hybridization was performed overnight at 42°C. The membranes were washed twice for 5 min at room temperature in 2× SSC–0.1% SDS and twice for 15 min at 68°C in 0.5× SSC–0.1% SDS.
The probe used was a 745-bp DNA fragment obtained by amplifying the DNA from DAPG-producing strain P. fluorescens Q2-87 (55) with primers Phl2a and Phl2b (40), which were designed on the basis of the sequence of the phlD gene, one of the six clustered genes involved in DAPG biosynthesis (1). Amplification reactions were carried out by using the protocol described below. The nucleotide sequence of the PCR product was verified (Genome Express) before we labeled the probe, which was done by using the nonradioactive digoxigenin system in accordance with the protocol recommended by the manufacturer (Boehringer Mannheim). The hybridized probe was immunodetected with anti-digoxigenin-alkaline phosphatase-Fab fragments and was visualized with the colorimetric substrate nitroblue tetrazolium salt and 5-bromo-4-chloro-3-indolylphosphate, as described in the protocols provided by the supplier. The specificity of the probe was verified previously by Raaijmakers et al. (40) and in this study by hybridizing the probe to the genomic DNAs of five DAPG-producing Pseudomonas reference strains and to the DNAs of three DAPG-negative strains. Isolates harboring the phlD gene were preserved at −80°C in 40% glycerol.
Detection of antibiotic production by HPLC.
Production of DAPG, monoacetylphloroglucinol (MAPG) (the precursor of DAPG), and pyoluteorin was verified by using analytical HPLC methods described by Keel et al. (24). First, bacteria were inoculated into 50 ml of liquid S1 medium and incubated for 24 h at 27°C. The bacterial cultures were centrifuged at 7,000 × g for 20 min at 4°C, and the supernatants were extracted with an equal volume of ethyl acetate for 2 h by using a rotary shaker. The ethyl acetate extracts were dried in a vacuum at 35°C and were dissolved in 1.5 ml of 65% methanol. Aliquots were filtered (pore size, 0.2 μm; (Sartorius) and analyzed with a Beckman liquid chromatograph. A C18 reverse-phase column (catalog no. LiChroCART 250-4; Merck) was thermostatically controlled at 45°C. The samples were eluted with a three-step gradient consisting of methanol in 0.43% o-phosphoric acid; from zero time to 5 min the solution contained 18 to 23% methanol, from 5 to 6 min the solution contained 23 to 53% methanol, and from 6 to 26.8 min the solution contained 53 to 61% methanol. The column was then washed for 10 min with pure methanol, and before each analysis it was conditioned 5 min with the first elution gradient. The flow rate was 1 ml/min. DAPG and MAPG were detected at a wavelength of 270 nm; their retention times were 22 and 12 min, respectively. Pyoluteorin was detected at a wavelength of 313 nm; its retention time was 14 min.
Sample preparation prior to PCR amplification.
Bacterial isolates were grown overnight at 27°C on YPGA, which contained (per liter) 5 g of yeast extract, 5 g of peptone, 10 g of glucose, and 15 g of agar. A single colony of each strain was picked with a sterile toothpick, resuspended in 20 μl of sterile distilled water, and heated at 95°C for 10 min to lyse the cells; the lysate was then cooled in ice, briefly centrifuged with a microcentrifuge, and used for PCR amplification.
ARDRA.
The DNA coding for the 16S rRNA of each isolate was amplified with primers P0 (5′-GAGAGTTTGATCCTGGCTCAG) and P6 (5′-CTACGGCTACCTTGTTACGA). These primers were designed (16) on the basis of the conserved bacterial sequences at the 5′ and 3′ ends of the 16S rRNA gene (positions 27f and 1495r, respectively, on Escherichia coli rDNA), which allowed amplification of almost the entire gene (16). Amplification was performed as previously described (9); each mixture contained 2 μl of lysed cell suspension in 20 μl of Polymed Taq buffer containing 1.5 mM MgCl2, 150 ng of each primer, each deoxynucleoside triphosphate at a concentration of 250 μM, and 0.5 U of Taq DNA polymerase (Polymed). The reaction mixtures were incubated in a thermocycler (model 9600; Perkin-Elmer) at 95°C for 1.5 min and then subjected to 35 cycles consisting of 95°C for 30 s, the annealing temperature for 30 s, and 72°C for 4 min. The annealing temperature was 60°C for the first 5 cycles, 55°C for the next 5 cycles, and 50°C for the last 25 cycles. Finally, the mixtures were incubated at 72°C for 10 min and then at 60°C for 10 min. Two microliters of each amplification mixture was analyzed by agarose (1.2%, wt/vol) gel electrophoresis in Tris-acetate-EDTA (TAE) buffer containing 0.5 μg of ethidium bromide per ml.
A 5-μl aliquot of each PCR mixture containing approximately 1.5 μg of amplified 16S rDNA was digested with 3 U of restriction enzyme AluI (Boehringer Mannheim) in a total volume of 20 μl at 37°C for 3 h. The enzyme was inactivated by heating the preparations at 65°C for 15 min, and the reaction products were analyzed by agarose (2.5%, wt/vol) gel electrophoresis in TAE buffer containing 0.5 μg of ethidium bromide per ml.
Sequencing 16S rDNA.
Amplified 16S rDNA was purified from each reaction mixture by agarose (1.2%, wt/vol) gel electrophoresis in TAE buffer containing 0.5 μg of ethidium bromide per ml. A small agarose slice containing the band of interest (observed under long-wavelength [312-nm] UV light) was excised from the gel and purified by using a QIAquick gel extraction kit (Qiagen) according to the supplier's instructions. The 16S rDNA nucleotide sequence was determined by cycle sequencing by using the dye terminator method and an automatic device for sequencing. Almost complete 16S rDNA sequences were determined for isolates 3-1 (1,454 bp), 3-45 (1,456 bp), and 16-31 (1,456 bp); partial (717-bp) sequences were determined for isolates 11-19, 22-27, 23-27, and 37-27.
Analysis of sequence data.
The 16S rDNA nucleotide sequences obtained were aligned with the most similar sequences in the Ribosomal Database Project database (30) by using Ribosomal Database Project utilities. The alignment was checked manually, corrected, and then analyzed by using the neighbor-joining method according to the models of Jukes and Cantor with TREECON 2.2 (50). The robustness of the inferred trees was evaluated by 100 bootstrap resamplings.
RAPD-PCR analysis.
Amplification reactions were performed by using 25 μl of platinum buffer containing 3 mM MgCl2, 2 μl of a lysed cell suspension, 500 ng of primer AP12 (5′-CGGCCCCTGC-3′) (which has been shown previously to give reproducible and informative results [9, 10, 34]), each deoxynucleoside triphosphate at a concentration of 200 μM, and 0.625 U of platinum Taq polymerase (Gibco BRL). The reaction mixtures were incubated in a thermocycler (model 9600; Perkin-Elmer) at 94°C for 2 min. They were then subjected to 45 cycles consisting of 95°C for 30 s, 36°C for 1 min, and 72°C for 2 min. The final extension step consisted of 75°C for 10 min and then 60°C for 10 min.
A 5-μl sample of each reaction mixture was electrophoresed on a 2% (wt/vol) agarose gel in TAE buffer containing 0.5 μg of ethidium bromide per ml. The amplification patterns were analyzed with a scanner-densitometer (model GDS2000; Ultra-Violet Product Ltd.).
The reproducibility of the unique RAPD-PCR patterns produced by single isolates was tested by performing three independent experiments, and no differences in the RAPD patterns were found under the standard conditions used (data not shown); the same results were obtained for several randomly chosen strains that produced the same pattern.
AMOVA of amplification products.
The AMOVA procedure was used to estimate the variance components for RAPD patterns by partitioning the variations among samplings times and/or locations. The AMOVA technique is a method for analyzing molecular variance that produces estimates of variance components which reflect the correlation of haplotypic diversity at different levels of a hierarchical subdivision. The significance of the variance components is tested by a permutational approach (12). The vectors for the presence of RAPD markers (1 for the presence of each band on a gel; 0 for the absence of each band on a gel) for each strain were used to compute the genetic distance for each pair of strains. The parameter used was the Euclidean metric measurement (E) of Excoffier et al. (12), as defined by Huff et al. (20) as follows: E = ɛ2xy = n(1 − 2nxy/2n), where 2nxy is the number of markers shared by two strains and n is the total number of polymorphic sites.
All analyses were performed with the Arlequin program (45a), which is used in several scientific fields (microbiology, medicine, population genetics) and is available at the following URL: http://anthropologie.unige.ch/arlequin/.
Identification and characterization of the isolates.
Some isolates were analyzed by performing the tests described below.
(i) PCR analysis with specific phlD primers.
Amplification was performed with 1 μl of each heat-lysed bacterial suspension in 50 μl of Pro-HA DNA polymerase buffer (Eurogentec, Seraing, Belgium) containing 1.5 mM MgCl2, 0.2 μM primer Phl2a, 0.2 μM primer Phl2b, each deoxynucleoside triphosphate at a concentration of 200 μM, and 0.5 U of Pro-HA DNA polymerase. The reaction mixtures were incubated in a thermocycler (model Gene Cycler; Bio-Rad) at 95°C for 10 min and then subjected to 35 cycles consisting of: 92°C for 1 min, 62°C for 1 min, and 72°C for 1 min. Finally, the mixtures were incubated at 72°C for 2 min. PCR-amplified DNAs were detected by using a horizontal 1.5% (wt/vol) agarose gel electrophoresed in TAE buffer at 10 V · cm−1 for 1 h. The gel was stained with ethidium bromide for 30 min, and the PCR products were visualized with a UV transilluminator.
(ii) Phenotypic characterization.
Isolates were characterized by using the Biolog system (Biolog, Hayward, Calif.), which is based on the differential utilization of a large number of organic compounds (3). Biolog GN microplates were inoculated as recommended by the manufacturer and were incubated at 27°C for 24 h. Formazan accumulation in bacterial cells was measured by determining the optical density at 550 nm with an automatic microplate reader. The isolates were identified on the basis of their patterns of utilization of 95 substrates by using the Biolog Microlog software.
Nucleotide sequence accession numbers.
The 16S rDNA nucleotide sequences obtained were deposited in the GenBank database under the following accession numbers: strain 3-1, AF126101; strain 3-45, AF126102; strain 16-31, AF126103; strain 11-19, AF177474; strain 22-27, AF177467; strain 23-27, AF177468; and strain 37-27, AF177469.
RESULTS
Isolation of DAPG-producing bacterial strains.
Bacteria isolated on S1 medium were enumerated 48 h after plating for each of the five sampling times and locations examined during plant development. Bacteria were isolated from nonrhizosphere soil, from rhizosphere soil, and from washed roots (Table 1) at densities ranging from 4.9 to 7.2 log CFU/g (dry weight). A total of 1,716 colonies (156 colonies from nonrhizosphere soil, 780 colonies from rhizosphere soil, and 780 colonies from washed roots) were randomly picked and tested for hybridization to the phlD probe. The DNAs of 188 isolates (75 isolates from rhizosphere soil and 113 isolates from roots) gave hybridization signals with the phlD gene; below these isolates are referred to as PhlD+ isolates. In contrast, no PhlD+ isolate was obtained from nonrhizosphere soil, indicating that the natural level of DAPG producers was less than 2.6 × 102 cells per g (dry weight) of soil. Depending on the age of the plant, the percentages of PhlD+ isolates ranged from 1.9 to 25.6% (Table 1). The percentage of root PhlD+ isolates was very low at the first sampling time (27 days), but it dramatically increased and remained constant for the next three sampling times (55, 72, and 92 days) and then decreased at the last sampling time (106 days). Very similar results were obtained with the rhizosphere soil isolates, but the increase in the percentage of isolates harboring phlD began at 72 days instead of 55 days (Table 1).
TABLE 1.
Hybridizing isolates and ARDRA analysis
| Location | Day | No. of bacteria (log CFU/g [dry wt])a
|
Presence of PhlD+ isolates
|
No. of strains in:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Cultivable bacteria |
DAPG-producing bacteria |
Fractionb | % | ARDRA group 1 | ARDRA group 2 | ARDRA group 3 | ARDRA group 4 | ||
| Rhizosphere soil | 27 | 5.8 | 4.0 | 3/156 | 1.9 | 2 | |||
| 55 | 5.4 | 4.8 | 40/156 | 25.6 | 28 | 4 | 1 | ||
| 72 | 7.2 | 6.4 | 27/156 | 17.3 | 22 | ||||
| 92 | 6.2 | 5.5 | 33/156 | 21.1 | 27 | 3 | |||
| 106 | 6 | 4.8 | 10/156 | 6.4 | 7 | 1 | |||
| Total | 6.12 | 5.10 | 113/780 | 14.5 | 86 | 8 | 0 | 1 | |
| Washed roots | 27 | 5.4 | 3.6 | 3/156 | 1.9 | 2 | |||
| 55 | 6.4 | 4.8 | 4/156 | 2.5 | 3 | 1 | |||
| 72 | 5.8 | 4.9 | 22/156 | 14.1 | 19 | 1 | 1 | ||
| 92 | 4.9 | 4.2 | 35/156 | 22.4 | 31 | 3 | 1 | ||
| 106 | 5.3 | 4.1 | 11/156 | 7.0 | 9 | 1 | |||
| Total | 5.56 | 4.32 | 75/780 | 9.6 | 64 | 4 | 3 | 1 | |
| Total | 5.84 | 4.71 | 188/1,560 | 12 | 150 | 12 | 3 | 2 | |
The values are means based on three samples. Samples were plated on S1 medium.
Number of bacteria that hybridized to the phlD probe/total number of bacteria tested for hybridization.
HPLC analysis showed that all of the hybridizing isolates were able to produce DAPG. The amount of DAPG produced was at least 1 μg/ml of culture, which is enough to inhibit pathogenic fungi in vitro (54). In addition, all of these isolates produced MAPG but no pyoluteorin (data not shown). PCR amplification of DNAs from 10 randomly chosen isolates belonging to ARDRA group 1 and from all of the isolates belonging to the other ARDRA groups (see below) (Fig. 1) by using primers specifically designed by using the phlD gene gave an amplification fragment of the expected size (745 bp) (data not shown).
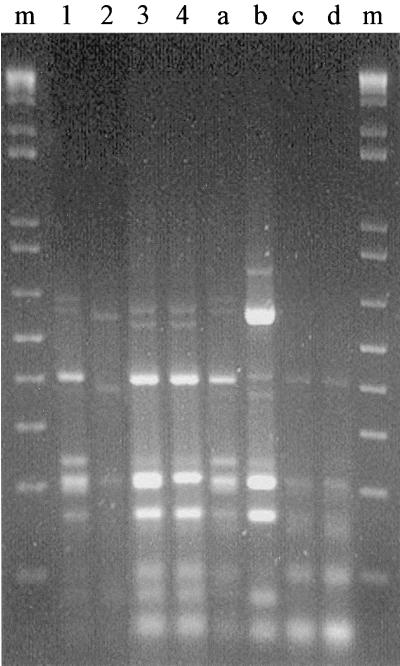
FIG. 1.
Agarose gel electrophoresis of amplified 16S rDNAs digested with endonuclease AluI from 167 PhlD+ bacteria isolated from maize roots and rhizosphere soil (lanes 1 through 4, corresponding to patterns 1 through 4, respectively) and from reference strains (lane a, pattern 1 strain Q2-87; lane b, pattern 6 strain 2-79; lane c, pattern 5 strain PGS12; lane d, pattern 5 strain M.3.1). Lanes m contained a 100-bp molecular size marker ladder. The slow faint bands present in some ARDRA patterns might be attributable to sequence heterogeneities between the different 16S rRNA gene copies present in the genome of the same strain.
Identification of the PhlD+ isolates.
The 16S rDNAs of the four reference strains and of 167 of 188 hybridizing isolates (72 and 95 isolates from rhizosphere soil and roots, respectively) were amplified (data not shown). The remaining 21 isolates were not investigated further since they did not survive the cryoconservation in glycerol. Restriction analysis of the amplified 16S rDNA of each sample with AluI revealed six ARDRA patterns (Fig. 1). The isolates produced four different patterns (Table 1). Most of the isolates (150 of 167 isolates [89.8%]) produced pattern 1, which was identical to the pattern obtained with the 16S rDNA of P. fluorescens reference strain Q2-87. Patterns 2, 3, and 4 were produced by 12 (7.2%), 3, and 2 isolates, respectively, and were different from the patterns obtained with the 16S rDNAs of the four reference strains used. Patterns 5 and 6 were produced by only reference strains; pattern 5 was produced by P. aureofaciens PGS12 and P. putida M3.1, and pattern 6 was produced by P. fluorescens 2-79.
The nucleotide sequences of 16S rDNAs from three isolates, ARDRA group 1 strain 3-1 and ARDRA group 2 strains 3-45 and 16-31, were determined and used to construct the phylogenetic trees shown in Fig. 2.
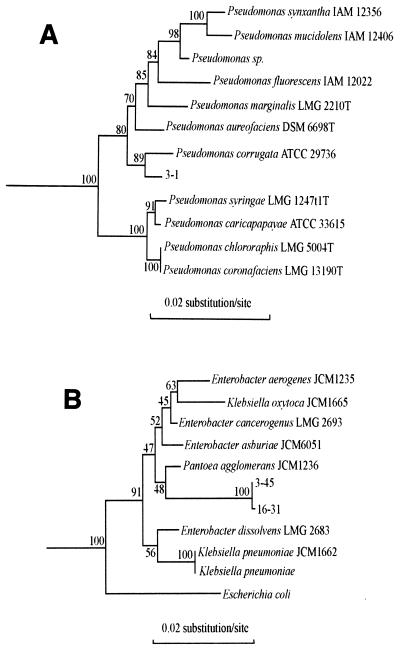
FIG. 2.
Phylogenetic trees showing the relationships among the 16S rDNA sequences of three PhlD+ isolates and reference 16S rDNA sequences of organisms belonging to the gamma subclass of the class Proteobacteria. (A) Relationships of isolate 3-1 and fluorescent Pseudomonas spp. The outgroup sequence used was the sequence of A. calcoaceticus ATCC 33604, (accession no. M34139). (B) Relationships of isolates 3-45 and 16-31 and members of the family Enterobacteriaceae. The outgroup sequence used was the sequence of P. putida JCM 6156 (accession no. D37924). The sequence positions used for alignment were positions 48 to 1468 (A) and 37 to 1370 (B) (Escherichia coli numbering).
Isolate 3-1 was placed in the Pseudomonas cluster, and its 16S rDNA exhibited the highest level of sequence similarity (99.4%) with the 16S rDNA of Pseudomonas corrugata ATCC 29736. The partial nucleotide sequences of the 16S rDNAs of four other ARDRA group 1 isolates (isolates 11-19, 22-27, 23-27, and 37-27) were also determined. The four sequences obtained (positions 185 to 900) were identical to the corresponding region of the 16S rDNA sequence of isolate 3-1. The data obtained suggested that ARDRA group 1 was a rather homogeneous cluster of isolates affiliated with fluorescent Pseudomonas spp. This finding was confirmed by the results of Biolog tests, which were performed with 20 isolates belonging to ARDRA group 1 and showed that 18 of them should be assigned to P. fluorescens with similarity levels of at least 65% and 2 of them (including isolate 3-1) should be assigned to P. corrugata (69% similarity for isolate 3-1 and 80% similarity for the other strain).
Isolates 3-45 and 16-31, which belonged to ARDRA group 2, were placed in the family Enterobacteriaceae, very close to Pantoea agglomerans (accession no. AB004691). Biolog tests performed with all of the ARDRA group 2 isolates showed that most of them should be assigned to P. corrugata (60 to 80% similarity with P. corrugata); the only exception was isolate 3-45, which was assigned to Enterobacter cloacae with a level of similarity of 80%. In contrast, isolate 16-31 was assigned to P. corrugata (69% similarity).
RAPD analysis.
The DNAs of lysed cell suspensions of the DAPG-producing isolates were amplified by the RAPD technique with the 10-mer oligonucleotide AP12.
Isolates belonging to ARDRA groups 2 through 4 were not distinguished by the RAPD method. RAPD amplification of DNAs from the 150 ARDRA group 1 isolates resulted in 31 bands, whose sizes ranged from 200 to 2,200 bp. The RAPD patterns obtained for these isolates were defined by the presence or absence of 31 RAPD markers. Each RAPD pattern was compared with the other patterns, and a Euclidean distance matrix (E) was calculated (data not shown). An important level of genetic diversity was found in ARDRA group 1, whose members had 64 genotypes. In particular, we obtained 33 different genotypes from rhizosphere soil and 44 genotypes from roots; the two habitats shared 13 genotypes. To analyze the RAPD variation in the 150 ARDRA group 1 isolates, we performed an AMOVA with the Euclidean distance matrix. First, rhizosphere soil and roots were considered two different groups. The AMOVA data revealed that 5.93% of genetic variability was associated with differences among samples (P = 0.00006). The level of genetic diversity found within sampling locations was high (93.14%) and very significant (P = 0.00001).
Second, RAPD variation within roots or rhizosphere soil was analyzed separately (Table 2). Sampling times that yielded only two or three isolates (e.g., 27 days) were not included in this analysis. When the root isolates were examined, the AMOVA data showed that most genetic variability (97.21%) at a significant P value (P = 0.03) occurred within sampling times; only 2.9% of the genetic variability was associated with differences among sampling times. To investigate whether this genetic diversity was related to divergence among strains and/or sampling times, we analyzed all of the possible combinations of two, three, and four sampling times. The data obtained (Table 2) clearly showed that in all of the combinations most of the total molecular variance between sampling times was attributable to divergence among strains.
TABLE 2.
AMOVA 150 Pseudomonas isolates determined with 31 RAPD markers at four sampling times
| Location | Variance componenta | Total variance | % of variance among sampling times | % of variance within sampling times | Pb |
|---|---|---|---|---|---|
| Roots | I vs II vs III vs IV | 2.03826 | 2.79 | 97.21 | 0.03213 |
| I vs II vs III | 2.02884 | 3.76 | 96.24 | 0.00869 | |
| I vs II vs IV | 2.06942 | 3.88 | 96.12 | 0.04768 | |
| I vs III vs IV | 1.98945c | 2.55 | 97.45 | 0.09005 | |
| II vs III vs IV | 2.06091 | 0.02 | 99.98 | 0.42176 | |
| I vs II | 2.07221 | 6.13 | 93.87 | 0.00987 | |
| I vs III | 1.97668 | 4.23 | 95.77 | 0.02018 | |
| I vs IV | 1.92857 | 0.62 | 99.38 | 0.35352 | |
| II vs III | 2.04228 | 0.82 | 99.18 | 0.27740 | |
| II vs IV | 2.13518 | −0.76 | 100.76 | 0.45707 | |
| III vs IV | 1.99780 | −1.85 | 101.85 | 0.58218 | |
| Rhizosphere soil | I vs II vs III vs IV | 1.94041 | 11.13 | 88.87 | 0.00044 |
| I vs II vs III | 1.78320 | 11.25 | 88.75 | 0.00081 | |
| I vs II vs IV | 2.39099 | 2.55 | 97.45 | 0.21941 | |
| I vs III vs IV | 1.81214 | 19.53 | 80.47 | 0.00362 | |
| II vs III vs IV | 1.94979 | 10.84 | 89.16 | 0.00019 | |
| I vs II | 2.24820 | 1.36 | 98.64 | 0.31746 | |
| I vs III | 1.59599 | 26.69 | 73.31 | 0.01619 | |
| I vs IV | 2.39383 | 1.60 | 98.40 | 0.32915 | |
| II vs III | 1.77407 | 10.21 | 89.79 | 0.00069 | |
| II vs IV | 2.48141 | 3.01 | 96.99 | 0.17042 | |
| III vs IV | 1.82303 | 19.65 | 80.35 | 0.00175 |
I, samples obtained on day 55; II, samples obtained on day 72; III, samples obtained on day 92; IV, samples obtained on day 106.
P, probability of a more extreme variance distribution.
Values in boldface type are not significant.
Similar analyses carried out with the rhizosphere soil isolates (Table 2) showed that most of the genetic variability occurred within sampling times, but an important fraction was associated with differences among sampling times (up to 26.69% for sampling times I and III).
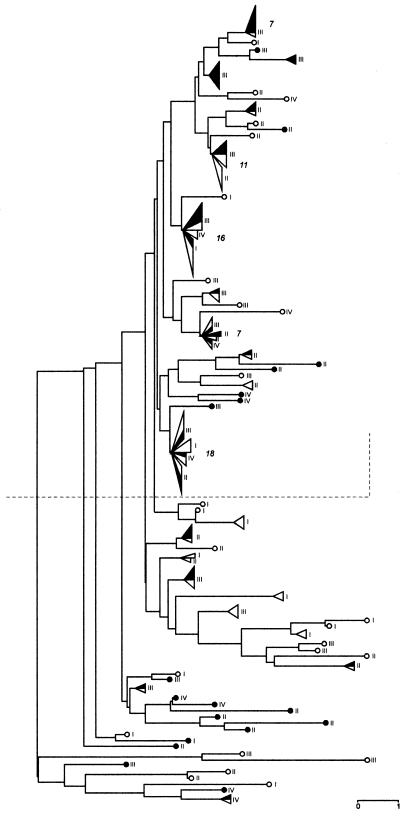
The relationships among the 150 isolates examined were represented as a dendrogram (Fig. 3) by using E and the neighbor-joining method (44). As previously reported for soil bacterial populations with frequent recombinations (9, 59), the unrooted dendrogram looks more like a bush than a tree. Moreover, the lower part of the dendrogram appears to be more variable than the upper part. The upper part of the dendrogram includes (i) most isolates collected from rhizosphere soil at sampling time III (92 days) and (ii) at least five main genotypes, which were expressed by 7, 11, 16, 7, and 18 isolates (Fig. 3), representing about 40% of the total population analyzed. In addition, these genotypes were found in both root and rhizosphere soil and very often at different sampling times; moreover, they appeared to be closely related.
FIG. 3.
Dendrogram showing genetic relationships among 150 isolates belonging to ARDRA group 1, based on RAPD patterns produced with primer AP12. Genetic distances were calculated by using E (12). Solid symbols, isolates from rhizosphere soil; open symbols, isolates from roots; circles, single isolates, triangles, genotypes isolated from roots and/or rhizosphere soil at the same/or different sampling times (sampling times I through IV). Each triangle's base is proportional to the number of isolates. The arabic numerals indicate the numbers of isolates that have the same genotype.
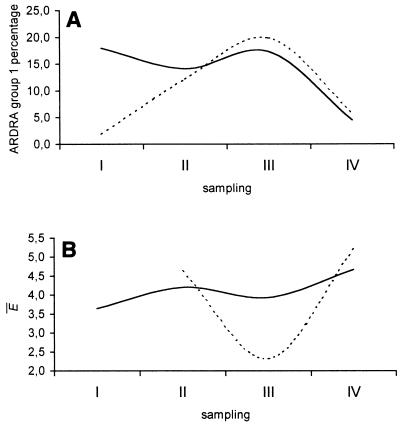
We also determined the E within each sampling time for root and rhizosphere soil populations. The results shown in Fig. 4 raise at least one salient point. The genetic diversity of rhizosphere soil isolates decreased significantly at sampling time III (92 days) and increased at sampling time IV (106 days); similar, although much more pronounced, behavior was observed for root isolates.
FIG. 4.
(A) Percentages of isolates belonging to ARDRA group 1 collected from roots and rhizosphere soil at different sampling times (sampling times I, II, III, and IV, corresponding to 55, 72, 92, and 106 days, respectively). (B) Mean E values determined for isolates within each sampling time. Mean E values were not calculated for the sampling time at which only two or three isolates were obtained (i.e., 27 days). Solid lines, roots; dashed lines, soil.
DISCUSSION
Production of DAPG has been shown to play a major role in suppression of soilborne plant pathogens (7, 23, 25, 57). Furthermore, the DAPG biosynthetic locus is conserved in DAPG producers of worldwide origin (25, 40). An analysis of metabolite patterns and molecular typing showed that there is considerable diversity among DAPG-producing pseudomonads depending on the plant species and soil origin (25). However, to our knowledge, the frequency and diversity of rhizosphere DAPG producers during one growing season have not been described previously. In the present study we showed that DAPG producers were present in the maize rhizosphere during all the plant development and that the DAPG producer population was genetically very diverse.
Fluctuations in the number of DAPG producers and the low frequency of these organisms (<0.65%) in nonrhizosphere soil could be explained by assuming that there was a spatial and temporal selection gradient for the DAPG producers resulting from the root exudates, which started earlier in roots than in rhizosphere soil and was absent in nonrhizosphere soil. Even if root exudates of young immature plants allowed DAPG producers to grow, they did not appear to exert specific selection for this type of bacteria. An alternative explanation for this finding is that biological activity of the natural DAPG producers is not induced by the exudates of young plant roots but is induced by exudates of older plants, which results in selective pressure against other rhizosphere microorganisms. Indeed, it is well known that carbon medium composition significantly affects antibiotic production (21, 37, 47), and thus variations in the carbon composition of root exudates should also affect DAPG production.
Therefore, the presence of DAPG-producing bacteria in the rhizosphere of maize was significantly affected by plant development.
The level of diversity of DAPG producers as determined by ARDRA resulted in recognition of four ARDRA patterns. On the basis of Biolog and 16S rDNA sequence analyses some strains belonging to ARDRA group 1 (89.8% of the isolates) were assigned to P. fluorescens, whereas other strains were assigned to P. corrugata. This is consistent with the taxonomic heterogeneity of P. fluorescens and with the previous finding of Laguerre et al. (28), who included in the same AluI ARDRA pattern some P. fluorescens strains, as well as strains belonging to Pseudomonas chlororaphis, Pseudomonas syringae, and P. putida biovar C. No P. corrugata strain was included in the analysis of Laguerre et al., but the 16S rDNA sequence of P. corrugata ATTC 29736 (accession no. D84012) exhibited a high degree of sequence identity with strain 3-1 (99.4%) and produced the same AluI ARDRA pattern as strain 3-1.
The same analyses revealed heterogeneity among strains that produced pattern 2 (7.2% of the isolates), which were assigned to P. corrugata or to the family Enterobacteriaceae close to P. agglomerans and/or E. cloacae. Strains belonging to the latter species are widely distributed in nature and are often associated with a variety of plant and animal species (4). Some of them, which are isolated from seeds, hypocotyl tissue, or the rhizosphere, are effective agents for biological control of plant-pathogenic fungi (19, 35); in fact, they are able to suppress fungus growth by producing of a siderophore or by competing for root exudates (19, 29, 36). The identification of strains belonging to the family Enterobacteriaceae that are able to produce DAPG indicated that this ability is not confined to the genus Pseudomonas. It is possible that genes involved in DAPG biosynthesis might have been horizontally transferred between members of the genus Pseudomonas and members of other bacterial genera. This possibility was suggested by the results of a preliminary analysis of the nucleotide sequence of the 745-bp phlD fragment obtained by PCR from some strains belonging to ARDRA groups 1 and 2. We found that there were extremely high levels of sequence identity (>99.8%) between the sequences of phlD DNA fragments of isolates belonging to ARDRA group 2, including isolates 3-45 and 16-31, and the homologous sequences obtained from strains belonging to ARDRA group 1 (including isolate 3-1). The lack of divergence between phlD sequences from isolates belonging to different bacterial genera supports the hypothesis that lateral gene transfer occurs, even though we cannot a priori rule out any other possible explanation.
The ARDRA results confirmed the high levels of genetic heterogeneity among fluorescent pseudomonads observed by Laguerre et al. (28) and the fact that several species could be grouped together on the basis of having the same AluI ARDRA pattern. This is in agreement with previous findings which showed that there are several groups of organisms which have almost identical 16S rRNA sequences (and therefore produce identical ARDRA patterns) but for which the levels of DNA hybridization are significantly less than 70%, indicating that they represent individual species (48).
The genetic diversity of isolates belonging to ARDRA group 1 was checked by performing a RAPD analysis and was analyzed by AMOVA. Significant genetic variability between root and rhizosphere soil isolates was not found, which was consistent with the finding that 13 genotypes were found in both locations. When we analyzed the two locations separately, we found that plant development did not have the same effect on biodiversity in the two populations. The genetic variability of the root isolates did not change significantly over time except for the last sampling time (106 days), at which time the genetic diversity increased. The lowest level of genetic variability for DAPG producers was found when maize roots were highly colonized, suggesting that selection of particular strains occurred when roots produced large amount of exudates.
The genetic diversity of rhizosphere soil isolates was much greater (range, 10.21 to 26.69%) than the genetic diversity of root isolates, suggesting that an important part of genetic variability may be attributed to divergence among strains obtained at different stages of plant growth.
In conclusion, the stage of plant growth appeared to significantly affect the level of genetic diversity of the Pseudomonas DAPG-producing population and seemed to be related to the level of root exudate production. Our data are consistent with previous data which showed that Burkholderia cepacia (9, 33) and Paenibacillus azotofixans (46) populations in maize rhizospheres changed while plants grew. Moreover, Seldin et al. (46) also showed that P. azotofixans populations isolated from rhizosphere soil, roots, and non-root-associated soil of maize were statistically different.
Finally, the results of this study have practical importance in the context of using biocontrol agents to protect crops against rhizosphere pathogens. Indeed, we found that in pot experiments DAPG producers were present in the maize rhizosphere during all stages of plant development but were present at a very low density during the first stage of maize growth. Since the maize root systems at the first three sampling times (27, 55, and 72 days) did not saturate the pot soil (data not shown), it is likely that the changes in the DAPG-producing population during this time were similar to changes that occur under field conditions. However, at the last two sampling times (92 and 106 days) very large root systems filled the pots, and therefore it would be interesting to compare this part of our results with field data. It is known that in the field maize is more sensitive to rhizosphere pathogens during the first stage of growth, which corresponds to the end of germination. The low densities of DAPG producers in the rhizosphere could explain this sensitive period during maize growth. Thus, releasing a good biocontrol agent during the first stage of plant development should be very useful. Furthermore, our data indicate that similar populations of DAPG producers are present in the maize rhizosphere throughout the growing season and that some of these strains have the same genotype. As these strains are very similar, we hypothesize that if they were included in an inoculum, they would efficiently colonize the maize rhizosphere and protect it throughout the growing season. Thus, the type of DAPG producers described here could be used in future screening programs for biocontrol agents.
ACKNOWLEDGMENTS
We thank Philippe Kieffer for technical assistance, Wafa Achouak for help with the Biolog analysis and for helpful discussions, Donatella Paffetti for useful suggestions and comments, and Marco Bosco for scientific discussions. We are also deeply indebted to two anonymous reviewers for their many helpful comments and suggestions which improved the manuscript.
REFERENCES
- 1.Bangera M G, Thomashow L S. Characterization of a genomic locus required for synthesis of the antibiotic 2,4-diacetylphloroglucinol by the biological control agent Pseudomonas fluorescens Q2-87. Mol Plant-Microbe Interact. 1996;9:83–90. doi: 10.1094/mpmi-9-0083. [DOI] [PubMed] [Google Scholar]
- 2.Barberio C, Fani R. Biodiversity of an Acinetobacter population isolated from activated sludge. Res Microbiol. 1998;149:665–673. doi: 10.1016/s0923-2508(99)80014-x. [DOI] [PubMed] [Google Scholar]
- 3.Bochner B R. “Breathprints” at the microbial level. ASM News. 1989;55:536–539. [Google Scholar]
- 4.Brenner D J. The genus Enterobacter. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Vol. 2. Berlin, Germany: Springer-Verlag; 1981. pp. 1173–1180. [Google Scholar]
- 5.Caccamo D, Di Cello F, Giugliandolo C, Fani R, Maugeri T. Polyphasic approach to the characterisation of marine luminous Vibrionaceae. Res Microbiol. 1999;50:221–230. doi: 10.1016/s0923-2508(99)80039-4. [DOI] [PubMed] [Google Scholar]
- 6.Chiarini L, Bevivino A, Tabacchioni S. Proceedings of the Third International Workshop on Plant Growth-Promoting Rhizobacteria. Adelaide, Australia: CSIRO Australia; 1994. Factors affecting the competitive ability in rhizosphere colonization of plant-growth promoting strains of Burkholderia cepacia; pp. 204–206. [Google Scholar]
- 7.Défago G. 2,4-Diacetylphloroglucinol, a promising compound in biocontrol. Plant Pathol. 1993;42:311–312. [Google Scholar]
- 8.Di Cello F, Fani R. A molecular strategy for the study of natural bacterial communities by PCR-based techniques. Minerva Biotecnol. 1996;8:126–134. [Google Scholar]
- 9.Di Cello F, Bevivino A, Chiarini L, Fani R, Paffetti D, Tabacchioni S, Dalmastri C. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl Environ Microbiol. 1997;63:4485–4493. doi: 10.1128/aem.63.11.4485-4493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cello F, Pepi M, Baldi F, Fani R. Molecular characterization of a n-alkane degrading bacterial community and identification of a new species, Acinetobacter venetianus. Res Microbiol. 1997;148:237–249. doi: 10.1016/S0923-2508(97)85244-8. [DOI] [PubMed] [Google Scholar]
- 11.Elsherif M, Grossmann F. Comparative investigations on the antagonistic activity of fluorescent pseudomonads against Gaeumannomyces graminis var. tritici in vitro and in vivo. Microbiol Res. 1994;149:371–377. [Google Scholar]
- 12.Excoffier L, Smouse P E, Quattro J M. Analysis of molecular variance inferred from metric distances among DNA genotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenton A, Stephens P, Crowley J, O'Callaghan M, O'Gara F. Exploitation of gene(s) involved in 2,4-diacetylphloroglucinol biosynthesis to confer a new biocontrol capability to a Pseudomonas strain. Appl Environ Microbiol. 1992;58:3873–3878. doi: 10.1128/aem.58.12.3873-3878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgakopoulos D, Hendson M, Panopoulos N, Schroth M. Cloning of a phenazine biosynthetic locus of Pseudomonas aureofaciens PGS12 and analysis of its expression in vitro with the ice nucleation reporter gene. Appl Environ Microbiol. 1994;60:2931–2938. doi: 10.1128/aem.60.8.2931-2938.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould W, Hagedorn C, Bardinelli T, Zablotowicz R. New selective media for enumeration and recovery of fluorescent pseudomonads from various habitats. Appl Environ Microbiol. 1985;49:28–32. doi: 10.1128/aem.49.1.28-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grifoni A, Bazzicalupo M, Di Serio C, Fancelli S, Fani R. Identification of Azospirillum strains by restriction fragment length polymorphism of the 16S rDNA and the histidine operon. FEMS Microbiol Lett. 1995;127:85–91. doi: 10.1111/j.1574-6968.1995.tb07454.x. [DOI] [PubMed] [Google Scholar]
- 17.Gurtler V, Wilson V, Mayall B. Classification of medically important clostridia using restriction endonuclease site differences of PCR-amplified 16S rDNA. J Gen Microbiol. 1991;137:2673–2679. doi: 10.1099/00221287-137-11-2673. [DOI] [PubMed] [Google Scholar]
- 18.Hamlen R, Lukezic F, Bloom J. Influence of age and stage of development on the neutral carbohydrate components in root exudates from alfalfa plants grown in a gnotobiotic environment. Can J Plant Sci. 1972;52:633–642. [Google Scholar]
- 19.Howell C R, Beier R C, Stipanovic R D. Production of ammonia by Enterobacter cloacae and its possible role in the biological control of Pythium preemergence damping-off by the bacterium. Phytopathology. 1988;78:1075–1078. [Google Scholar]
- 20.Huff D R, Peakall R, Smouse P E. RAPD variation among natural populations of outcrossing buffalograss [Buchloe dactyloides (Nutt.) Engelm.] Theor Appl Genet. 1993;86:927–934. doi: 10.1007/BF00211043. [DOI] [PubMed] [Google Scholar]
- 21.James D W, Gutterson N I. Multiple antibiotics produced by Pseudomonas fluorescens HV37a and their differential regulation by glucose. Appl Environ Microbiol. 1986;52:1183–1198. doi: 10.1128/aem.52.5.1183-1189.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaunet T, Laguerre G, Lemanceau P, Frutos R, Notteghem J. Diversity of Pseudomonas fuscovaginae and other fluorescent pseudomonads isolated from diseased rice. Phytopathology. 1995;85:1534–1541. [Google Scholar]
- 23.Keel C, Wirthner P, Oberhänsli T, Voisard C, Burger M, Haas D, Défago G. Pseudomonas as antagonists of plant pathogens in the rhizosphere: role of the antibiotic 2,4-diacetylphloroglucinol in the suppression of black rot of tobacco. Symbiosis. 1990;9:327–341. [Google Scholar]
- 24.Keel C, Schnider U, Maurhofer M, Voisard C, Burger M, Wirthner P, Haas D, Défago G. Suppression of root disease by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant-Microbe Interact. 1992;5:4–13. [Google Scholar]
- 25.Keel C, Weller D, Natsch A, Défago G, Cook R, Thomashow L. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62:552–563. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloepper J, Schrot M. Plant growth-promoting rhizobacteria on radishes. Proc Int Conf Plant Pathog Bact. 1978;2:879–882. [Google Scholar]
- 27.Laguerre G, Allard M, Revoy F, Amarger N. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rDNA genes. Appl Environ Microbiol. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laguerre G, Rigottier-Gois L, Lemanceau P. Fluorescent Pseudomonas species categorized by using polymerase chain reaction (PCR)/restriction fragment analysis of 16S rDNA. Mol Ecol. 1994b;3:479–487. doi: 10.1111/j.1365-294x.1994.tb00126.x. [DOI] [PubMed] [Google Scholar]
- 29.Loper J E, Ishimaru C A, Carnegie S R, Vanavichit A. Cloning and characterization of aerobactin biosynthesis genes of the biological control agent Enterobacter cloacae. Appl Environ Microbiol. 1993;59:4189–4197. doi: 10.1128/aem.59.12.4189-4197.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Murcia A, Acinas S, Rodriguez-Valera F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Lett. 1995;17:247–256. [Google Scholar]
- 32.Maurhofer M, Keel C, Haas D, Défago G. Influence of plant species on disease suppression by Pseudomonas fluorescens strain CHA0 with enhanced antibiotic production. Plant Pathol. 1995;44:40–50. [Google Scholar]
- 33.McArthur J V, Kovacic D A, Smith M H. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc Natl Acad Sci USA. 1988;85:9621–9624. doi: 10.1073/pnas.85.24.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori E, Lio' P, Daly S, Damiani G, Perito B, Fani R. Molecular nature of RAPD markers amplified from Haemophilus influenzae Rd genome. Res Microbiol. 1999;150:83–93. doi: 10.1016/s0923-2508(99)80026-6. [DOI] [PubMed] [Google Scholar]
- 35.Nelson E B. Biological control of Pythium seed rot and preemergence damping-off of cotton with Enterobacter cloacae and Erwinia herbicola applied as seed treatments. Plant Dis. 1988;72:140–142. [Google Scholar]
- 36.Nelson E B. Exudate molecules initiating fungal responses to seeds and roots. Plant Soil. 1990;129:61–73. [Google Scholar]
- 37.Novak-Thompson B, Gould S J, Kraus J, Loper J E. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf5. Can J Microbiol. 1995;40:1064–1066. [Google Scholar]
- 38.O'Sullivan D, O'Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierson E A, Weller D M. Use of mixtures of fluorescent pseudomonads to suppress take-all and improve the growth of wheat. Phytopathology. 1994;84:940–947. [Google Scholar]
- 40.Raaijmakers J, Weller D, Thomashow L. Frequency of antibiotic-producing Pseudomonas spp. in natural environments. Appl Environ Microbiol. 1997;63:881–887. doi: 10.1128/aem.63.3.881-887.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy M S, Hynes R K, Lazarovits G. Relationship between in vitro growth inhibition of pathogens and suppression of preemergence damping-off and postemergence root rot of white bean seedlings in the greenhouse by bacteria. Can J Microbiol. 1993;40:113–119. [Google Scholar]
- 42.Rosales A, Thomashow L, Cook R, Mew T. Isolation and identification of antifungal metabolites produced by rice-associated antagonistic Pseudomonas spp. Phytopathology. 1995;85:1028–1032. [Google Scholar]
- 43.Rovira A. Plant root exudates and their influence upon soil micro-organisms. In: Baker K F, Snyder W C, editors. Ecology of soil-borne plant pathogens—prelude to biological control. Berkeley: University of California Press; 1965. p. 170. [Google Scholar]
- 44.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45a.Schneider S, Kueffer J-M, Roessli D, Excoffier L. Arlequin ver. 1.1: a software for population genetic data analysis. Geneva, Switzerland: Genetics and Biometry Laboratory, University of Geneva; 1997. [Google Scholar]
- 46.Seldin L, Rosado A S, da Cruz D W, Nobrega A, van Elsas J D, Paiva E. Comparison of Paenibacillus azotofixans strains isolated from rhizoplane, rhizosphere, and non-root-associated soil from maize planted in two different Brazilian soils. Appl Environ Microbiol. 1998;64:3860–3868. doi: 10.1128/aem.64.10.3860-3868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanahan P, O'Sullivan D, Simpson P, Glennon J, O'Gara F. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl Environ Microbiol. 1992;58:353–358. doi: 10.1128/aem.58.1.353-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rDNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 49.Thomashow L, Weller D. Role of phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van der Peer Y, De Wacher R. TREECON: a software package for the construction and drawing of evolutionary trees. Comput Applic Biosci. 1993;9:177–182. doi: 10.1093/bioinformatics/9.2.177. [DOI] [PubMed] [Google Scholar]
- 51.Vaneechoutte M, Rossau R, De Vos P, Gillis M, Janssen D, Paepe N, De Rouck A, Fiers T, Claeys G, Kersters K. Rapid identification of bacteria of the Comamonadaceae with amplified ribosomal DNA-restriction analysis (ARDRA) FEMS Microbiol Lett. 1992;93:227–234. doi: 10.1111/j.1574-6968.1992.tb05102.x. [DOI] [PubMed] [Google Scholar]
- 52.Vaneechoutte M, De Beenhouwer H, Claeys G, Verschraegen G M, De Rouck A, Paepe N, Eilachouni A, Portaels F. Identification of Mycobacterium species with amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1993;31:2061–2065. doi: 10.1128/jcm.31.8.2061-2065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaneechoutte M, Dijkshoorn I, Tjernberg A, Eilachouni A, De Vos P, Claeys G, Verschraegen G. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J Clin Microbiol. 1995;33:11–15. doi: 10.1128/jcm.33.1.11-15.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ventura M, Picard C, Gallet A, Benizri E, Guckert A. Production of 2,4-diacetylphloroglucinol by a Pseudomonas strain in the rhizosphere of maize: implication of this compound in the biological control of Fusarium graminearum. Int Org Biol Integrated Control Bull. 1997;21(9):27–31. [Google Scholar]
- 55.Vincent M, Harrison L, Brackin J, Kovacevich P, Murkerji P, Weller D, Pierson E. Genetic analysis of the antifungal activity of a soilborne Pseudomonas aureofaciens strain. Appl Environ Microbiol. 1991;57:2928–2934. doi: 10.1128/aem.57.10.2928-2934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weller D M, Cook R J. Suppression of take-all of wheat by seed treatment with fluorescent pseudomonads. Phytopathology. 1983;73:463–469. [Google Scholar]
- 57.Weller D M, Thomashow L S. Use of rhizobacteria for biocontrol. Curr Opin Biotechnol. 1993;4:306–311. [Google Scholar]
- 58.Welsh J, McClelland M. Fingerprinting genome using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whittam T S. Sex in the soil. Curr Biol. 1992;2:676–678. doi: 10.1016/0960-9822(92)90140-6. [DOI] [PubMed] [Google Scholar]
- 60.Williams J G K, Kubelick A R, Livak K J, Rafalski J A, Tingey S V. Polymorphism generated by arbitrarily primed PCR in the mouse: application to strain identification and genetic mapping. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/19.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]