Summary
Huanglongbing (HLB) is a devastating disease of citrus, which is mostly caused by Candidatus Liberibacter asiaticus (CLas). To realize the specific application of nano-transgenic technology in HLB, AuNPs-PEI (Gold Nanoparticles-Polyethylenimine) was used to carry foreign genes into the leaves of periwinkle (Catharanthus roseus) by infiltration. Here, we demonstrated that NPR1-GFP protein expression was observed from the 12th hour to the 10th day after infiltrating AuNPs-PEI-pNPR1 (Arabidopsis thaliana nonexpressor of pathogenesis-related gene 1)-GFP. Fluorescence of mCherry was observed 6 h after AuNPs-PEI-pNLS (nuclear localization signal sequence)-mCherry infiltration and fluorescence of FAM was observed in the nucleus 4 h after AuNPs-PEI-FAM-siRNANPR1 infiltration. In addition, NPR1-GFP expression in CLas-infected periwinkle leaves was significantly higher than that in healthy periwinkle leaves after infiltration. Our work confirmed that the expression of exogenous NPR1-GFP could reduce the CLas titers by promoting the expression of PR (pathogenesis related) genes and ICS (isochorismate synthase) gene.
Subject areas: Biological sciences, Biomaterials, Biotechnology, Materials in biotechnology
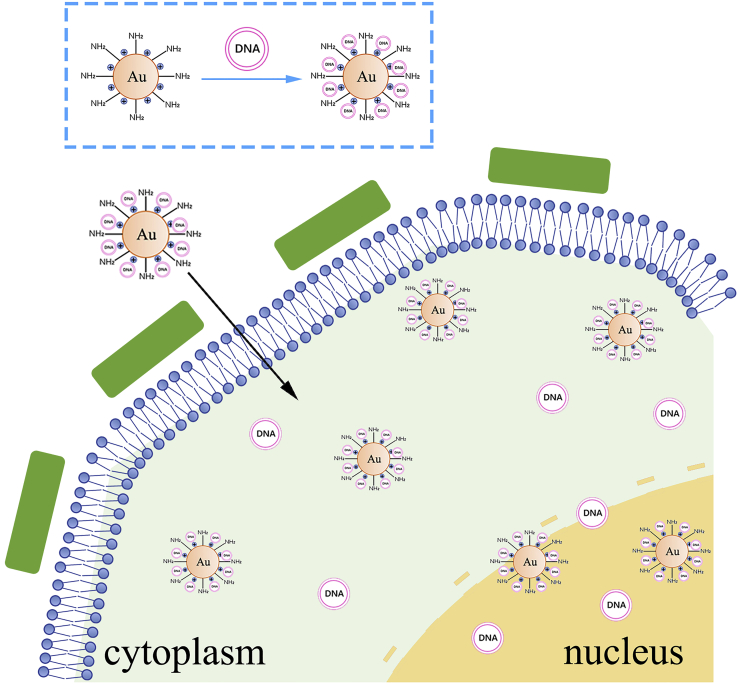
Graphical abstract
Highlights
-
•
AuNPs-PEI-FAM-siRNANPR1 entered the nucleus within 4 h after infiltration
-
•
AuNPs-PEI-pNLS-mCherry expressed the corresponding protein within 6 h
-
•
AuNPs-PEI-pNPR1-GFP continued to express the corresponding protein for 14 days
-
•
After AuNPs-PEI-pNPR1-GFP infiltration for 2 days, CLas titer decreased significantly
Biological sciences; Biomaterials; Biotechnology; Materials in biotechnology
Introduction
Citrus huanglongbing (HLB) is the most serious bacterial disease in the citrus, causing immeasurable losses to the citrus industry around the world every year. CLas (Candidatus Liberibacter asiaticus) is the most widespread, destructive citrus HLB pathogen around the world. Periwinkle (Catharanthus roseus) is the host of CLas, which can be quickly infected by grafting and dodder transmission. Compared with citrus, periwinkle has more obvious symptoms and higher bacterial titers after being infected by CLas. This allows periwinkle to be widely used as an excellent model plant for exploring citrus HLB (Bové and Barros, 2006). Overexpression of Arabidopsis thaliana nonexpressor of pathogenesis-related gene 1 (NPR1) in citrus has the effect of resisting CLas invasion (Qiu et al., 2020). However, there are no reports showing the effect of exogenous gene expression on CLas in periwinkle. Nanoparticle has been extensively studied as a nucleic acid carrier in mammalian cell. But due to its limited permeability against the cell wall, it has limited application in plant cell. At present, only GFP transfection has been done using nanoparticle in plant cell (Demirer et al., 2019).
AuNPs (Aurum or gold nanoparticles), as one of the most widely used metal nanomaterials, have excellent chemical stability, enzyme stability, low cytotoxicity, and other physical and chemical properties (Connor et al., 2005; Yeh et al., 2012). The high surface area and adjustability of AuNPs provide excellent support for the attachment of nucleic acids and other biological molecules (Castañeda et al., 2007). PEI (polyethylenimine) is widely used as a gene transfection vector for animal cells (Godbey et al., 2001; Xu et al., 2009). According to proton sponge effect, PEI is used to carry plasmids into cells and then release them inside (Godbey et al., 1999, 2001). Controlling the ratio of PEI to DNA, PEI can protect and help DNA enter cells (González-Domínguez et al., 2019). However, PEI is highly cytotoxic (MÁRton et al., 1979).
AuNPs-PEI can be obtained using PEI reduction (Brondani et al., 2013; Li et al., 2019b). The free PEI in solution can be removed by centrifugation, thus reducing the toxicity of PEI to cells. The positively charged AuNPs-PEI is easier to bind to negatively charged DNA (Elfinger et al., 2009). AuNPs-PEI has a higher capability at delivering plasmid DNA (pDNA) into cells through the proton sponge effect, which is 15 times more efficient than its counterpart without PEI modification (Thomas and Klibanov, 2003).
It has been previously confirmed that AuNPs-PEI carrying exogenous siRNANPR1 successfully silenced the NPR1 gene with high specificity (Lei et al., 2020). However, the timing of AuNPs-PEI entry into the cytoplasm and nucleus has not been further investigated. So far, there are no reports on the entry of AuNPs-PEI into plant cell nucleus and the use of AuNPs-PEI to deliver pDNA with functional genes to plant cells and to make them expressed. By infiltrating AuNPs-PEI-FAM-siRNANPR1 and AuNPs-PEI-pDNA into periwinkle leaves in separate experiments, we became the first research team to use AuNPs-PEI to transfer functional foreign genes into plants and quantify their expression with confocal imaging and western blot.
In this study, we utilized AuNPs-PEI to carry three plasmids: NLS-mCherry (pBI221 vector contains a nuclear localization signal sequence -mCherry gene, pNLS-mCherry), GFP (pBI221 vector contains GFP gene, pGFP), and AtNPR1-GFP (pCB302 vector contains Arabidopsis thaliana nonexpressor of pathogenesis-related gene 1 -GFP gene, pNPR1-GFP). We not only observed their expression in plant cells and the process of AuNPs-PEI entering cell nucleus but also found that the expression of exogenous NPR1-GFP significantly inhibited the growth of CLas in periwinkle.
Results
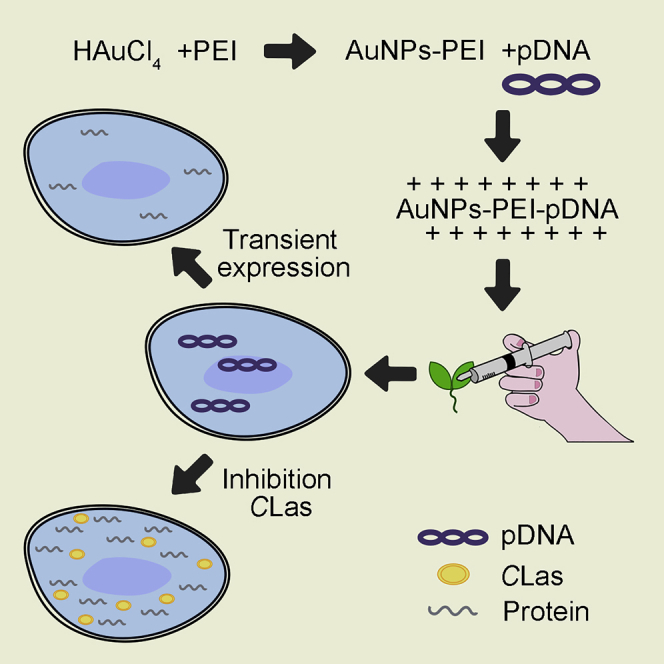
Schematic diagram of AuNPs-PEI-pDNA entering plant cells
The positively charged AuNPs-PEI-pDNA complexes attach to the cell through electrostatic adsorption and enter into the cell wall via pits of the cell walls (Lakshmanan et al., 2012). Then, through endocytosis of the cell membrane, they enter the cytoplasm and further into the nucleus (Choi et al., 2013; Mohamed et al., 2015; Panzarini et al., 2018). Owing to the proton spongy effect of PEI (Pollard et al., 1998; Graczyk et al., 2020), AuNPs-PEI release pDNA, which is transcribed and translated to express corresponding proteins (Figure 1).
Figure 1.
Schematic diagram of AuNPs-PEI-pDNA complexes entering and expressing in cells
AuNPs-PEI-pDNA complex could attach around the cell through electrostatic adsorption and entered the cell wall through pores. Subsequently, it entered the cytoplasm through endocytosis of cell membrane, and further entered the nucleus. Owing to the proton sponge effect of PEI, AuNPs-PEI released pDNA they carried.
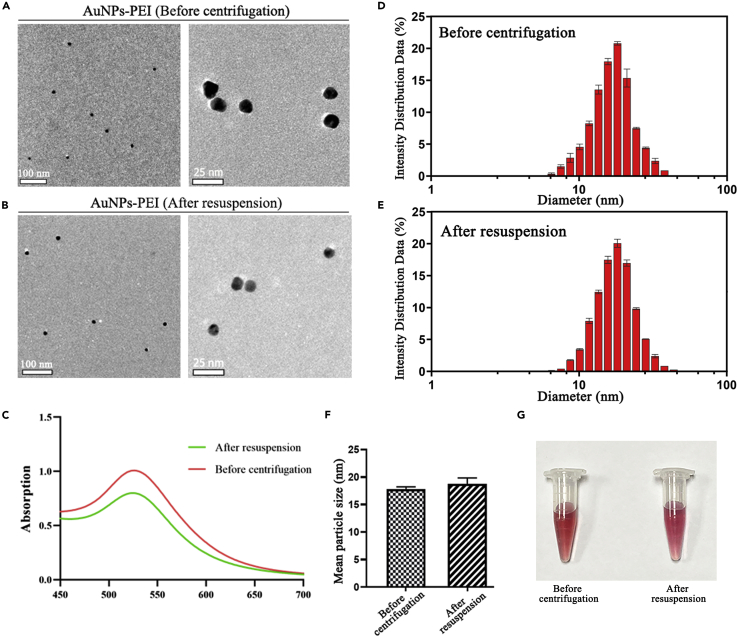
Synthesis and characterization of AuNPs-PEI
The results of TEM and particle size measurement mutually confirmed that the particle size of AuNPs-PEI was mainly distributed in 17–18 nm (Figures 2A, 2D, and 2F). The range of 6–200 nm is theoretical size of nanoparticles that can enter the cells (Graczyk et al., 2020). The AuNPs-PEI in this subject adopt the PEI reduction method (Brondani et al., 2013; Li et al., 2019b), and the gold glue solution prepared was ruby red (Figure 2G). Generally, the nano-gold with a particle size between 10 and 20 nm for optical colorimetric analysis has an absorption peak at about 530 nm (Figure 2C). To obtain materials with smaller and uniform particle size and to remove free PEI in the solution, we continued with the centrifugation for 15 min, carefully aspirated the supernatant, and then resuspended the pellet with 200 μL ddH2O. The particle size of the solution had no significant change after resuspension (Figures 2B, 2E, and 2F). The UV absorption peak was not shifted (Figure 2C) and the color of the solution remained stable at ruby red (Figure 2G).
Figure 2.
Characterization of AuNPs-PEI
(A) TEM images of AuNPs-PEI before centrifugation.
(B) TEM images of AuNPs-PEI after resuspension. It was observed that the AuNPs-PEI had uniform particle size and good dispersibility after resuspension. The centrifugal resuspension operation did not cause its agglomeration.
(C) UV absorption peaks of AuNPs-PEI did not shift significantly before centrifugation and after resuspension.
(D) The particle size distribution of AuNPs-PEI before centrifugation. Data were represented as means ± SD from three repeats.
(E) The particle size distribution of AuNPs-PEI after resuspension. Data were represented as means ± SD from three repeats.
(F) The average particle size of AuNPs-PEI before centrifugation and after resuspension. Data were represented as means ± SD from three repeats.
(G) The color change of AuNPs-PEI solution before centrifugation (left) and after resuspension (right). The AuNPs-PEI resuspended after centrifugation and the color was still ruby red, indicating that no coagulation had occurred.
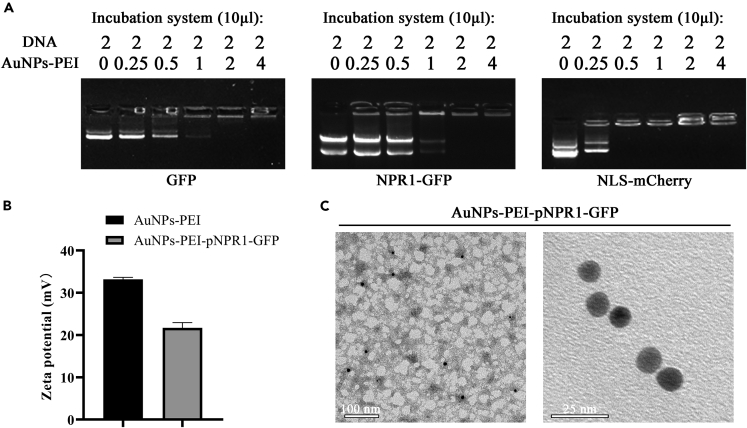
Preparation of AuNPs-PEI-pDNA
The AuNPs-PEI synthesized above was mixed with a volume of 0, 0.25, 0.5, 1, 2, 4, and the 2 μL plasmid (600 ng/μL) respectively, then incubated at 25°C for 15 min. 1% agarose gel was used to explore the best binding ratio of AuNPs-PEI and plasmid by electrophoresis. Cationic polymer vectors bind to pDNA via electrostatic interaction. The negative charge of pDNA could be partially or completely neutralized by the vector, consequently resulting in its retardation upon gel electrophoresis. The bands visible in the gel are free pDNA bands (Shan et al., 2012; Demirer et al., 2019). As the proportion of AuNPs-PEI in the system increases, more pDNA binds to AuNPs-PEI to form AuNPs-PEI-pDNA. Compared with the positive control in the first lane, the fluorescence intensity of the bands in the gel gradually decreases. When 2 μL of plasmid and 2 μL of previously synthesized AuNPs-PEI were added, the two could be completely combined (Figure 3A).
Figure 3.
Preparation and characterization of AuNPs-PEI-pDNA
(A) Gel retardation assay after AuNPs-PEI combining with pBI121-GFP, pCB302-NPR1-GFP, and pBI221-NLS-mCherry, respectively. As the concentration of AuNPs-PEI increased, the DNA band gradually weakened.
(B) Changes in zeta potential before and after AuNPs-PEI and pNPR1-GFP binding. Owing to the cancellation of the positive and negative potentials, the zeta potential of the AuNPs-PEI surface dropped by about 12mV after binding. Data were represented as means ± SD from three repeats.
(C) TEM images of AuNPs-PEI-pNPR1-GFP. The particle size of AuNPs-PEI did not change significantly after binding the plasmid.
After binding to the plasmid, the zeta potential of AuNPs-PEI-pNPR1-GFP was decreased from +32 mV to +20 mV (Figure 3B), which not only showed the combination of the two but also showed that AuNPs-PEI-pDNA still had good dispersibility. Although the hydrated particle size of AuNPs-PEI-pNPR1-GFP measured was about 64 nm (Figure S1), the results of TEM confirmed that the particle size of AuNPs-PEI did not change significantly after binding the plasmid (Figure 3C) and precipitation or obvious color change had not been seen during the whole process.
Expression of AuNPs-PEI-pGFP in A. thaliana and periwinkle leaves
In order to measure the gene delivering capacity of AuNPs-PEI, we first used it to transfect plasmid GFP into plant cells. Two days after infiltrating the AuNPs-PEI-pGFP complex into the Arabidopsis thaliana leaves, GFP fluorescence appeared in mesophyll and stomata cells (Figure S2A), which indicated that GFP protein was successfully expressed through transcription and translation. GFP fluorescence did not been observed in the leaves infiltrated only with AuNPs-PEI (Figure S2A). Western blotting results confirmed that after infiltration, GFP protein was expressed on the day 1, day 3, and day 5 in Arabidopsis thaliana leaves (Figure S2B). It reminded us that AuNPs-PEI-pGFP had entered the nucleus and expressed protein within one day.
Arabidopsis thaliana was the basic model plant, and we wanted to know whether AuNPs-PEI-pGFP could be applied to other plants. Our laboratory had been committed to the prevention and treatment of citrus HLB. Periwinkle was the host of CLas. Two days after infiltrating the AuNPs-PEI-pGFP complex into the periwinkle leaves, GFP fluorescence was observed in periwinkle leaves under confocal microscopy (Figure S2C). In contrast, no GFP fluorescence was observed in the control periwinkle leaves infiltrated with AuNPs-PEI alone (Figure S2C). The western blot results also showed that GFP protein was indeed expressed (Figure S2D).
Next, we wanted to further explore whether pNPR1-GFP carried by AuNPs-PEI could be expressed in periwinkle leaves and whether it had the function of inhibiting CLas.
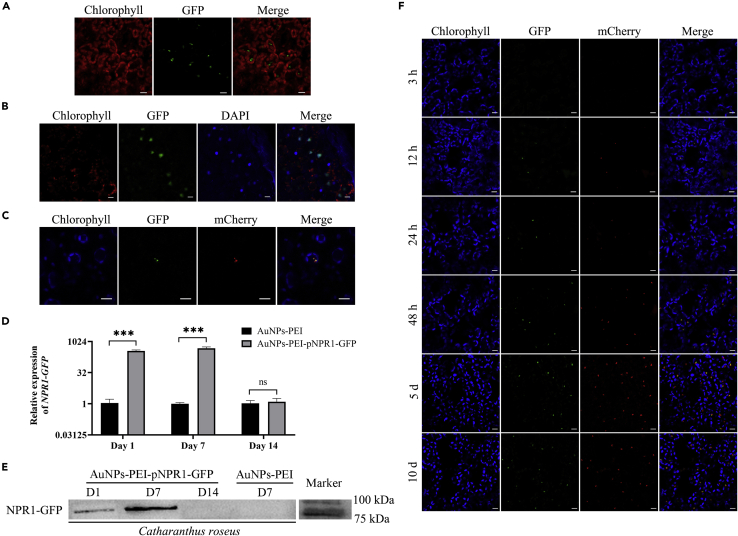
Expression and nuclear positioning of AuNPs-PEI-pNPR1-GFP in healthy periwinkle leaves
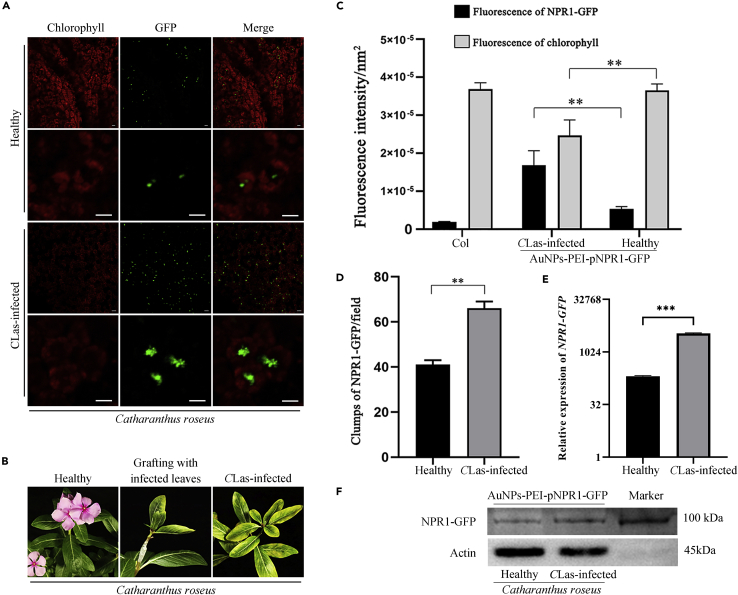
First, we infiltrated AuNPs-PEI-pNPR1-GFP into healthy (CLas-free) periwinkle leaves. Two days later, we observed a large amount of GFP fluorescence. The fluorescence appeared in the form of small green particles in mesophyll cells, and the small particles in each cell gathered together (Figures 4A and 6A).
Figure 4.
Expression of AuNPs-PEI-pNPR1-GFP in healthy periwinkle leaves
(A) NPR1-GFP fluorescence was accumulated in healthy periwinkle mesophyll cells 2 days after infiltrating. The green fluorescence was the fluorescence of NPR1-GFP. The red fluorescence was the autofluorescence of chlorophyll. Scale bars, 10 μm.
(B) NPR1-GFP fluorescence co-localized with the nucleus stained by DAPI. The green fluorescence was the fluorescence of NPR1-GFP. The red fluorescence was the autofluorescence of chlorophyll. The blue fluorescence was the fluorescence of DAPI. Scale bars, 10 μm.
(C) NPR1-GFP fluorescence co-localized with NLS (Nuclear localization signal sequence)-mCherry fluorescence which located in the nucleus. The green fluorescence was the fluorescence of NPR1-GFP. The blue fluorescence was the autofluorescence of chlorophyll. The red fluorescence was the fluorescence of NLS-mCherry. Scale bars, 10 μm.
(D) Relative expression levels of NPR1-GFP mRNA in healthy periwinkle leaves after infiltration with AuNPs-PEI-pNPR1-GFP. Data were represented as means ± SD from three biological repeats. Statistical analyses were performed using Student’s t-test (∗∗∗, p < 0.001 and ns indicated no significant difference).
(E) Western blot for NPR1-GFP at different time points after AuNPs-PEI-pNPR1-GFP infiltration of healthy periwinkle leaves. Corresponding bands could be observed at day 1 and day 7. The molecular weight of NPR1-GFP was 93 kDa. Irrelevant or superfluous lanes had been eliminated from the blot.
(F) Confocal observation of healthy periwinkle leaves after AuNPs-PEI-pNPR1-GFP and AuNPs-PEI-pNLS-mCherry infiltration at different time points. The green fluorescence was the fluorescence of NPR1-GFP. The blue fluorescence was the autofluorescence of chlorophyll. The red fluorescence was the fluorescence of NLS-mCherry. Scale bars, 10 μm.
Figure 6.
The expression of AuNPs-PEI-pNPR1-GFP in CLas-infected periwinkle leaves
(A) More NPR1-GFP fluorescence was observed in CLas-infected periwinkle compared to healthy leaves 2 days after infiltrating. The green fluorescence was the fluorescence of NPR1-GFP. The red fluorescence was the autofluorescence of chlorophyll. Scale bars, 10 μm.
(B) CLas-infected periwinkle leaves were grafted on to healthy periwinkle. The symptom shown in the figure appeared 50 days after grafting.
(C) Statistical fluorescence comparison of NPR1-GFP fluorescence and chlorophyll autofluorescence changes in CLas-infected and healthy leaves mesophyll cells. NPR1-GFP fluorescence of the CLas-infected periwinkle leaves was stronger than healthy, and the chlorophyll autofluorescence was weaker. Data were represented as means ± SD from three repeats. Statistical analyses were performed using Student’s t-test (∗∗, p < 0.01).
(D) Quantitative analysis of NPR1-GFP fluorescence in confocal images. There was more fluorescence of NPR1-GFP in the mesophyll cells of the CLas-infected periwinkle leaves. Data were represented as means ± SD from three repeats. Statistical analyses were performed using Student’s t-test (∗∗, p < 0.01). Statistical graphs came from (A). ZEN software (Carl Zeiss) was used for the fluorescence intensity and quantitative analysis of NPR1-GFP fluorescence and chlorophyll autofluorescence.
(E) Relative expression levels of NPR1-GFP mRNA in healthy and CLas-infected periwinkle leaves after 2 days of infiltration. Data were represented as means ± SD from three biological repeats. Statistical analyses were performed using Student’s t-test (∗∗∗, p < 0.001).
(F) Western blot for NPR1-GFP in healthy and CLas-infected periwinkle leaves 2 days after infiltration. The molecular weight of NPR1-GFP was 93 kDa and the molecular weight of actin was 42 kDa.
Because NPR1 performs its function in the nucleus (Qiu et al., 2020), we came up with a reasonable guess: those gathered particles were NPR1-GFP expressed in the nucleus. Two days after infiltrating AuNPs-PEI-pNPR1-GFP, the periwinkle leaves were directly immersed in the DAPI staining solution for 90 min before confocal observation. NPR1-GFP fluorescence was overlapped with DAPI fluorescence (Figure 4B).
The nucleus-localized plasmid pNLS-mCherry was incubated with AuNPs-PEI and then co-infiltrated with AuNPs-PEI-pNPR1-GFP in the leaves of healthy periwinkle. After two days, co-localization of GFP fluorescence and mCherry fluorescence was observed, confirming that the pNPR1-GFP was expressed in the nucleus (Figure 4C).
Real-time fluorescence quantitative PCR analysis and western blot experiment were used to probe the duration of NPR1-GFP expression in periwinkle leaves. Compared with control leaves infiltrated with AuNPs-PEI, the relative expression level of NPR1-GFP mRNA reached more than 300-fold on days 1 and 7 after infiltration with AuNPs-PEI-pNPR1-GFP, which decreased to no significant difference by day 14 (Figure 4D). Similarly, the results of western blot experiment confirmed the successful expression of pNPR1-GFP on day 1 and day 7 after infiltration, while there was no significant band on day 14 (Figure 4E).
To further determine the timing of expression of pNPR1-GFP and pNLS-mCherry, confocal observation was performed at 1, 2, 3, 12, 24, 48 h, day 5, and day 10 after infiltrating, respectively. We found that GFP fluorescence was co-localized with mCherry fluorescence, which started to express at 12 h, and had more expression at 24 and 48 h. The expressions were still not significantly weakened on day 5 and day 10 (Figure 4F). These results suggested that AuNPs-PEI carried pNPR1-GFP and pNLS-mCherry into the nucleus within 12 h.
The process of AuNPs-PEI-pDNA entering the nucleus
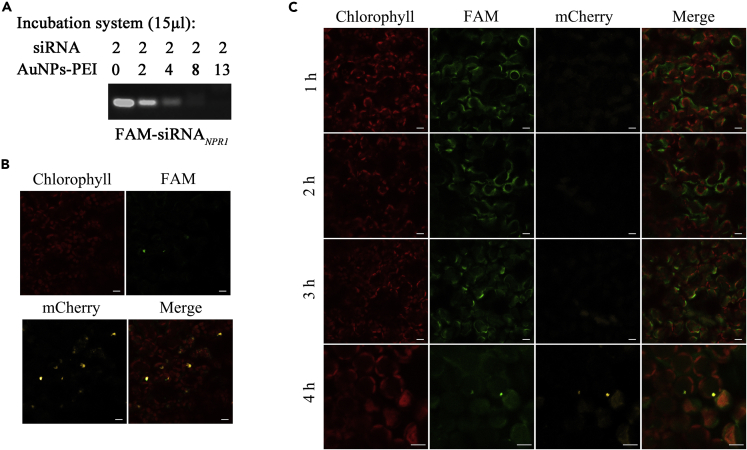
Our previous reports revealed that AuNPs-PEI-FAM-siRNANPR1 successfully silenced the function of NPR1 (Lei et al., 2020). Here, we infiltrated healthy periwinkle leaves with AuNPs-PEI-FAM-siRNANPR1 and used the fluorescence of FAM-siRNANPR1 to follow the entry of AuNPs-PEI into the nucleus. Gel hysteresis assays of AuNPs-PEI and different concentrations of FAM-siRNANPR1 were performed. When 2 μL of FAM-siRNANPR1 (0.6 μM) and 13 μL of previously synthesized AuNPs-PEI were added, the two could be completely combined (Figure 5A).
Figure 5.
The process of AuNPs-PEI-FAM-siRNANPR1 entry into the nucleus
(A) Gel retardation assay after AuNPs-PEI combining with FAM-siRNANPR1. As the concentration of AuNPs-PEI increased, the RNA band gradually weakened.
(B) Co-localization of FAM-siRNANPR1 fluorescence and NLS-mCherry fluorescence in healthy periwinkle leaves 2 days after AuNPs-PEI-FAM-siRNANPR1 infiltration. The green fluorescence was the fluorescence of FAM-siRNANPR1. The red fluorescence was the autofluorescence of chlorophyll. The yellow fluorescence was the fluorescence of NLS-mCherry. Scale bars, 10 μm.
(C) Confocal observation of healthy periwinkle leaves after AuNPs-PEI-FAM-siRNANPR1 infiltration at different time points. AuNPs-PEI-pNLS-mCherry was infiltrated 2 h in advance. The green fluorescence was the fluorescence of FAM-siRNANPR1. The red fluorescence was the autofluorescence of chlorophyll. The yellow fluorescence was the fluorescence of NLS-mCherry. Scale bars, 10 μm.
The periwinkle leaves were infiltrated with AuNPs-PEI-pNLS-mCherry 2 h in advance, and then the same leaves were infiltrated with AuNPs-PEI-FAM-siRNANPR1. Two days after infiltrating AuNPs-PEI-FAM-siRNANPR1, co-localization of FAM fluorescence and mCherry fluorescence was observed, implying that within two days AuNPs-PEI-FAM-siRNANPR1 had entered the nucleus (Figure 5B).
To further investigate the process of AuNPs-PEI entry into cells, we recorded our observation at 1, 2, 3, and 4 h after infiltrating AuNPs-PEI-FAM-siRNANPR1. After infiltration of AuNPs-PEI-FAM-siRNANPR1 for 1 h and 2 h, FAM fluorescence was observed in the extracellular space. 3 h after infiltration, bright spot-like FAM fluorescence was observed around the cells, which might be that AuNPs-PEI-FAM-siRNANPR1 were entering the cells through endocytosis. 4 h after infiltration, co-localization of FAM fluorescence and mCherry fluorescence was observed (Figure 5C). When FAM-siRNANPR1 alone was used to infiltrate periwinkle leaves as a control, punctate FAM fluorescence was observed in the stomata at 1 h; 2 h after infiltration, a large amount of diffuse FAM fluorescence was observed around the cells; 3–4 h after infiltration, only a bit diffuse FAM fluorescence could be observed. No FAM fluorescence was observed in the mesophyll cells (Figure S3). These results indicated that AuNPs-PEI indeed carried nucleic acid into the nucleus within 4 h, and pNLS-mCherry was expressed within 6 h.
There had been reported that overexpression of NPR1 could enhance the resistance against HLB in citrus (Qiu et al., 2020). These results inspired us to further explore the expression of pNPR1-GFP in CLas-infected periwinkle leaves and whether it performed its functions.
NPR1-GFP expression in CLas-infected periwinkle leaves
CLas-infected periwinkle plants were obtained by grafting (Figure 6B). It was relatively easy to infiltrate AuNPs-PEI-pNPR1-GFP complex into the CLas-infected periwinkle leaves 1 month after grafting. After two days of infiltration, the fluorescence intensity of NPR1-GFP in the CLas-infected periwinkle leaves was significantly stronger than that in the healthy leaves (Figure 6A). A large amount of NPR1-GFP fluorescence was clustered in the form of small particles, and the fluorescence was even dispersed (Figure 6A). The chloroplast autofluorescence of infected leaves also decreased to a certain extent relative to the mesophyll cells in healthy leaves (Figure 6A). The statistical results showed that the fluorescence quantity and intensity of NPR1-GFP was higher in CLas-infected periwinkle leaves compared with healthy periwinkle leaves (Figures 6C and 6D), while the chloroplast autofluorescence intensity was significantly lower (Figure 6C).
Further, we examined the relative expression of NPR1-GFP mRNA in healthy and CLas-infected periwinkle leaves two days after infiltration with AuNPs-PEI-pNPR1-GFP. The relative expression of NPR1-GFP in CLas-infected periwinkle reached more than 3000-fold, which was significantly higher than that in healthy periwinkle (Figure 6E). It was also confirmed by western blotting experiments of healthy and CLas-infected periwinkle leaves two days after infiltration with AuNPs-PEI-pNPR1-GFP (Figure 6F).
A large amount of expression of NPR1-GFP in CLas-infected periwinkle leaves might be related to its corresponding functions. The first thing we thought about was whether a reliable research method could be established using CLas-infected periwinkle leaves to explore the role of exogenous substances on CLas.
AuNPs-PEI-pNPR1-GFP had inhibitory effect on CLas in periwinkle leaves
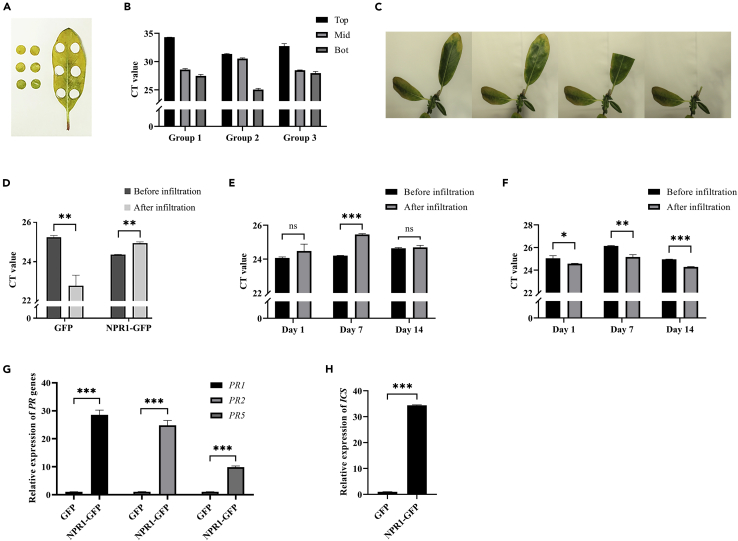
It had been reported that CLas was mainly distributed in the citrus leaf veins (Li et al., 2009; Kunta et al., 2014). The distribution of CLas in the periwinkle leaves was very uneven. Our results showed that the CLas infection degree in a leaf was up < middle < bottom (Figures 7A and 7B). It indicated that CLas titers in the lower half of a leaf were higher than that in the upper half. So, this regularity of CLas in leaves could be used for scientific detection of the inhibitory/promoting effect of foreign genes or drugs on CLas. In the morning, when the stomata opened, the AuNPs-PEI-pDNA complex (or other materials or chemicals) was infiltrated into CLas-infected periwinkle leaves. The upper part of the leaves was cut to determine CLas titers, and the lower part of the leaves was cut to determine CLas titers a few days later (Figure 7C). If the CT value increased after infiltrating the complex, it indicated that the CLas titers in periwinkle leaves decreased, proving that the complex significantly inhibited the growth of CLas.
Figure 7.
AuNPs-PEI-pNPR1-GFP inhibited CLas in periwinkle leaves
(A) Top, mid, and bottom location of CLas-infected periwinkle leaf diagram.
(B) The CT value in top, mid, and bottom of CLas-infected periwinkle leaves was different. The accumulation of CLas in the lower half of a leaf was significantly higher. Data were represented as means ± SD from three repeats.
(C) Sampling diagram for studying the effects of infiltration of AuNPs-PEI-pNPR1-GFP (or other reagent solution) on CLas. Half of the CLas-infected periwinkle leaves were cut as a control immediately after infiltration, and the other half was cut 2 days later.
(D) The changes of CLas were measured 2 days after infiltrating AuNPs-PEI-pNPR1-GFP, and the infiltration of AuNPs-PEI-pGFP was used as a control. Data were represented as means ± SD from three biological repeats. Statistical analyses were performed using Student’s t-test (∗∗, p < 0.01).
(E) Changes in CT values in CLas-infected periwinkle leaves at different time points after AuNPs-PEI-pNPR1-GFP infiltration. There was a significant increase at day 7. Data were represented as means ± SD from three biological repeats. Statistical analyses were performed using Student’s t-test (∗∗∗, p < 0.001 and ns indicated no significant differences).
(F) Changes in CT values in CLas-infected periwinkle leaves at different time points after AuNPs-PEI-pGFP infiltration. As a control, the CT values in the lower half of the leaves were consistently significantly lower than those in the upper half. Data were represented as means ± SD from three biological repeats. Statistical analyses were performed using Student’s t-test (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001).
(G) Relative expression levels of PR genes mRNA in CLas-infected periwinkle leaves after 2 days of infiltration. Leaves infiltrated with AuNPs-PEI-GFP were used as controls. Data were represented as means ± SD from three biological repeats. Statistical analyses were performed using Student’s t-test (∗∗∗, p < 0.001).
(H) Relative expression levels of ICS gene mRNA in CLas-infected periwinkle leaves after 2 days of infiltration. Leaves infiltrated with AuNPs-PEI-GFP were used as controls. Data were represented as means ± SD from three biological repeats. Statistical analyses were performed using Student’s t-test (∗∗∗, p < 0.001).
We infiltrated AuNPs-PEI-pNPR1-GFP into CLas-infected periwinkle leaves, taking the upper half first and then the remaining half of the leaves 2 days later (Figure 7C). The CLas titers of both were measured separately. The results showed that the CT values of CLas were higher in the lower half of the leaves than in the upper half, while the opposite was true in the leaves infiltrated with AuNPs-PEI-pGFP as a control (Figure 7D). It indicated that the expression of the exogenous pNPR1-GFP had indeed inhibited the reproduction of CLas in periwinkle leaves. Also, it confirmed that the method of studying whether CLas was inhibited was feasible.
Furthermore, there was a significant increase in the CT values of CLas-infected periwinkle leaves at day 7 after infiltration with AuNPs-PEI-pNPR1-GFP (Figure 7E). On days 1 and 14, there was no significant difference between the CT values of the upper part of the leaves and the lower part. However, the CLas titers were higher in the lower half of the leaves than in the upper half. Thus, the expression of NPR1-GFP still significantly inhibited the growth of CLas at days 1 and 14. In control leaves infiltrated with AuNPs-PEI-pGFP, CT values were consistently significantly lower in the lower half of the leaves than in the upper half (Figure 7F).
The relative expression levels of pathogenesis-related (PR) genes were determined in CLas-infected periwinkle leaves. Compared with control leaves infiltrated with AuNPs-PEI-pGFP, the relative expression levels of PR1, PR2, and PR5 genes were significantly upregulated two days after infiltration with AuNPs-PEI-pNPR1-GFP (Figure 7G).
Isochorismate has been identified as a precursor for synthesizing SA in Nicotiana benthamiana and tomato (Uppalapati et al., 2007; Catinot et al., 2008). We have determined the relative expression of isochorismate synthase (ICS) genes to assess SA levels in periwinkle leaves. Compared to control leaves infiltrated with AuNPs-PEI-pGFP, the ICS mRNA was significantly upregulated in CLas-infected periwinkle leaves two days after infiltration with AuNPs-PEI-pNPR1-GFP (Figure 7H). It implied that the infiltration of AuNPs-PEI-pNPR1-GFP might have promoted the increase of SA level in the leaves.
In conclusion, it was hypothesized that the expression of NPR1-GFP significantly increased the resistance of periwinkle to CLas.
Discussion
At present, two common genetic transformation methods, the agrobacterium-mediated and the gene bombardment, have obvious limitations and drawbacks (MÁRton et al., 1979; Hayashimoto et al., 1990). Therefore, developing efficient gene delivery vectors and feasible new methods of genetic transformation are great significance for innovation in plant genetic engineering. It had been reported that carbon nanotubes entered the nucleus within 3 h (Serag et al., 2011). Since the plant cell wall is a transport barrier, which limits the efficiency of plant genetic engineering, it is difficult to deliver exogenous biomolecules into plant cells. Our results showed that AuNPs-PEI accumulated on certain areas of the cell surface after 3 h of infiltration and entered the nucleus after 4 h (Figure 5). It was reported that the size exclusion limit of the plant cell wall was 20 nm (Wang et al., 2016; Schwab et al., 2016), while the particle size of the synthesized AuNPs-PEI was less than 20 nm (Figure 2). Free PEI was removed by centrifugation, which greatly reduces the toxicity of PEI. Also, the transfection efficiency of AuNPs-PEI was significantly higher than that of AuNPs without PEI modification (Thomas and Klibanov, 2003).
For the first time, our study confirmed the expression of exogenous NLS-mcherry, GFP, and NPR1-GFP genes in periwinkle leaves using confocal and western blot experiments after infiltrating AuNPs-PEI-pDNA. Our results proved that the mCherry fluorescence in the nucleus was observed after infiltrating AuNPs-PEI-pNLS-mCherry for 6 h. Green fluorescence was observed in the nucleus under confocal after 12 h of infiltration of AuNPs-PEI-pNPR1-GFP, while more fluorescence was observed at 24 and 48 h. Significant fluorescence could still be observed at day 5 and day 10 after infiltration.
CLas-infected periwinkle had more obvious symptoms and higher bacterial titers, which made it widely used as an excellent material for exploring HLB (Bové and Barros, 2006). Since the CLas titers in the half of the leaf near the petiole was higher than the CLas titers in the half of the leaf near the tip, we found a reliable system for studying the effects of drugs or genes on CLas-infected periwinkle leave. The CLas-infected periwinkle selected came from 1 month after grafting because at this time it was easier to infiltrating liquid substances into the leaves. Used this system, we discover that the transient overexpression of NPR1-GFP in 2 days significantly inhibited the growth of CLas (Figure 7).
Confocal results showed that NPR1-GFP was accumulated in cells in the CLas-infected periwinkle leaves (Figure 6). It has been reported that NPR1 promotes cell survival by forming salicylic acid-induced NPR1 condensates (SINCs). Relying on the agrobacterium-mediated transient expression system, SA treatment resulted in significant nuclear accumulation of NPR1-GFP (Zavaliev et al., 2020). We observed that NPR1-GFP expression was higher in the CLas-infected periwinkle leaves, which might be due to the increase of endogenous SA levels caused by the invasion of CLas, lead to the expression of NPR1-GFP protein in periwinkle leaves cells forming SINCs-like described above, and our results also showed that SA synthesis key enzyme ICS was significantly upregulated (Figure 7H). For this reason, we suggest that CLas-induced NPR1 condensates may be caused by the increase in SA in vivo.
AuNPs-PEI-pNPR1-GFP could enter and express in plant cells only by infiltration, which greatly reduced the cost and time consuming of experiments. Citrus stably expressing NPR1 have been proven to reduce the damage of HLB (Qiu et al., 2020). However, the construction of stable transgenic plants may take a long time, a lot of work, and the success rate is low. Using AuNPs-PEI-pNPR1-GFP to treat CLas-infected periwinkle can allow NPR1 to be expressed in a short period of time and reduce the titer of CLas. It not only verifies the resistance effect of NPR1 against CLas but also shortens the experimental period. The application of this system provides a new method for exploring the disease resistance genes of CLas.
In conclusion, we used the PEI reduction method to synthetic AuNPs-PEI, which could form a complex with nucleic acid molecules. AuNPs-PEI enabled the expression of three plasmids in the chloroplasts of periwinkle leaves. By confocal microscopy, AuNPs-PEI-FAM-siRNA was observed to enter the nucleus within 4 h. NLS-mcherry was expressed in 6 h after infiltrating AuNPs-PEI-pNLS-mcherry complex. We found that brighter NPR1-GFP fluorescence was observed in CLas-infected periwinkle leaves 2 days after infiltration with AuNPs-PEI-pNPR1-GFP and CLas was significantly inhibited.
In the future, we will use the periwinkle leaves system to further explore the relationship between NPR1 and CLas. At the same time, we will further explore the possibility of obtaining stable transgenic plants and make our own contribution to the biological control of citrus HLB.
Limitations of the study
Huanglongbing is a disease on citrus. Currently, restricted by the presence of wax on the surface of citrus leaves, we were unable to infiltrate AuNPs-PEI-pDNA into citrus leaves by syringe. In future studies, we hope to use this system on citrus leaves through methodological improvements to more directly verify the effect of a gene on citrus huanglongbing.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GFP (JL-8 Monoclonal Antibody) | Invitrogen | CAT#A-6455; RRID: AB_221570 |
| anti-ACTIN | Sigma | CAT#A0480; RRID:AB_476670 |
| Goat anti-mouse IgG-HRP | Absin | CAT#abs20001; RRID: AB_2716555 |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethylenimine (PEI) | Shyuanye | CAT#S24467-25mL |
| Gold chloride trihydrate | Aladdin | CAT#G141105-1g |
| Critical commercial assays | ||
| Trizol | Takara | CAT#9109 |
| Hifair® Ⅱ 1st Strand cDNA Synthesis Kit | Yeasen Biotechnology | CAT#11139ES60 |
| Hieff® qPCR SYBR Green Master Mix (Low Rox) | Yeasen Biotechnology | CAT#11202ES08 |
| EZ-10 Spin Column Plant Genomic DNA Purification Kit | Sangon | CAT#B518261-0050 |
| 2X TaqMan Fast qPCR Master Mix | Sangon | CAT#B639274-0005 |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana ecotype Col-0 | N/A | N/A |
| Catharanthus roseus | N/A | N/A |
| Recombinant DNA | ||
| pBI221-NLS-mCherry | Li et al. (2019a) | N/A |
| pBI121-GFP | Li et al. (2019a) | N/A |
| pCB302-AtNPR1-GFP | Chen et al. (2019) | N/A |
| Software and algorithms | ||
| Graphpad Prism | Graphpad | N/A |
| Excel | Microsoft | N/A |
| ZEN | Carl Zeiss | N/A |
Resource availability
Lead contact
Further information and requests for reagents may be directed to and will be fulfilled by the corresponding author Wenli Chen (chenwl@scnu.edu.cn).
Materials availability
Plasmids and seeds used in this study are available with a completed Materials Transfer Agreement Request for these materials by submitting to Wenli Chen (chenwl@scnu.edu.cn).
Experimental model and subject details
Arabidopsis thaliana
The wild-type of Arabidopsis thaliana (Col-0) came from our own laboratory. The cultivation conditions of Arabidopsis thaliana were light for 16 h (6 am to 10 pm), darkness for 8 h (10 pm to 6 pm at night). And the ambient temperature was controlled at 23°C, the relative humidity of the environment was 82%. Soil-grown Arabidopsis thaliana plants were used at three-week-old.
Catharanthus roseus
Healthy (CLas-free) periwinkle (Catharanthus roseus) plants were purchased from Guangzhou flower market and maintained in the greenhouse of our laboratory. The experimental site had a subtropical monsoon climate with an average annual temperature of 22.1°C. The annual average sunshine hours were 1608 h, and the annual average relative humidity was 78%.
CLas-infected periwinkle (Catharanthus roseus) was first provided by Professor Xiaolin Deng of South China Agricultural University and get more CLas-infected periwinkle through grafting. The obtained CLas-infected periwinkle was identified with specific primers HLBas, HLBr, and HLBp (Li et al., 2006; Zhou et al., 2011; Duan et al., 2021). After grafting, all the plants were kept for a week in a cool and ventilated room (26°C) for 7 days, and then moved to the greenhouse to cultivate under natural weather conditions (Zhang et al., 2010; Ya et al., 2018).
Method details
AuNPs-PEI synthesis
Prepare 14 mL PEI (branched polyethylenimine, 25KD, Shyuanye, Shanghai, China) solution (final concentration 0.2 mM) and 1 mL chloroauric acid (HAuCl4, Aladdin, Shanghai, China) solution (final concentration 2 mM) with double distilled water (ddH2O) respectively. Added PEI solution to HAuCl4 solution and slowly heated to 85°C until the solution was ruby red in color. Continued to stir until the solution cooled to room temperature to obtain AuNPs-PEI (Li et al., 2019b). Centrifuged at 7040×g for 15 min to remove the free PEI from the supernatant and resuspended the pellet in 200 μL ddH2O.
Plasmids and AuNPs-PEI-pDNA preparation
The plasmids carrying GFP gene (pBI121-GFP) and NLS (Nuclear localization signal sequence) gene (pBI221-NLS-mCherry) were provided by Professor Caiji-Gao of South China Normal University (Li et al., 2019a). The plasmid carrying AtNPR1-GFP (Arabidopsis thaliana NPR1-GFP) gene was provided by Professor Zhengqing-Fu of the University of South Carolina (Chen et al., 2019). In the following text, the complexes of AuNPs-PEI and pBI121-GFP are expressed in the form of AuNPs-PEI-pGFP, and the complexes of AuNPs-PEI and pBI221-NLS-mCherry are expressed in the form of AuNPs-PEI-pNLS-mCherry. The complexes of AuNPs-PEI and pCB302-AtNPR1-GFP are expressed in the form of AuNPs-PEI-pNPR1-GFP.
Particle size and zeta potential detection
800 μL of AuNPs-PEI or AuNPs-PEI-pNPR1-GFP ten-fold dilution were tested by Zetasizer Nano ZS90 (Malvern Instruments, Malvern, USA) (Xu et al., 2019; Liao et al., 2019).
Ultraviolet visible detection
The solution of AuNPs-PEI was examined using the UV-Vis Spectrometer (Shimadzu, Japan).
Transmission electron microscopy
The morphology and size of the AuNPs-PEI and AuNPs-PEI-pDNA nanoparticles were analyzed by transmission electron microscopy (TEM) (Producer: JEOL, Tokyo, Japan. Model: JEM-1400 PLUS) at an acceleration voltage of 120 kV (Thomas and Klibanov, 2003; Chen et al., 2014).
Gel retardation assay
2 μL plasmid (600 ng/μL) was mixed with the above synthetic AuNPs-PEI 0, 0.25, 0.5, 1, 2, 4 μL separately, incubated for 15 min at room temperature, diluted with ddH2O to 10 μl. These mixtures were loaded onto 1% agarose gels and electrophoresed for 15 min using the agarose gel electrophoresis apparatus (BG power 600K, China) at 100 V.
Mixed the AuNPs-PEI with a volume of 0, 2, 4, 8, 13 μL and the 2 μL FAM-siRNANPR1 (0.6 μM) respectively, incubated at room temperature for 15 min, and diluted to 15 μl with ddH2O. These mixtures were loaded onto 1.2% agarose gels and electrophoresed for 15 min using the same instrument as above at 100 V.
Infiltrating plant leaves
Leaves were infiltrated at 8–10 am. The criterion for completion of the infiltration was that the entire leaf was moistened (Figure S4).
The plasmid DNA (600 ng/μL) was mixed with the AuNPs-PEI prepared above in a 1:4 volume ratio and incubated for 15 min at room temperature before being diluted 10 times with the infiltration buffer (10 mM MgCl2 and 10 mM MES (2-(N-morpholino) ethanesulfonic acid) pH 5.7) for infiltration.
FAM-siRNANPR1 (20 μM) was mixed with the AuNPs-PEI prepared above in a 1:9 volume ratio, diluted 3 times with the infiltration buffer and incubated at room temperature for 30 min with 60 rpm shaking, then diluted 5 times before infiltration.
qPCR experiments for gene expression
Two-step qPCR was performed to quantify NPR1-GFP gene, PR genes, and ICS gene expression in periwinkle leaves with the following commercially available kits: Trizol (Takara, Tokyo, Japan) for total RNA extraction from leaves, Hifair® Ⅱ 1st Strand cDNA Synthesis Kit (Yeasen Biotechnology, Shanghai, China) to reverse transcribe total RNA into cDNA, and Hieff® qPCR SYBR Green Master Mix (Low Rox) (Yeasen Biotechnology, Shanghai, China) for qPCR. The 18S rDNA gene was used as a reference gene in all qPCR experiments. The sequences of the primers used were collated in Table S1. The qPCR conditions were as follows: 94°C for 2 min, followed by 30 cycles of 94°C for 20 s, 60°C for 20 s, and 72°C for 20 s. The qPCR data were analyzed by the ddCt method to obtain the normalized gene expression fold change with respect to the 18S rDNA housekeeping gene and control sample (Li et al., 2019c; Demirer et al., 2019). For each sample, qPCR was performed as three technical replicates (three reactions from the same isolated RNA batch), and the entire experiment consisting of independent infiltrations and RNA extractions from different plants was repeated three times (three biological replicates).
CT values detection
Leaves samples, stored at −80°C after weighing, were rapidly ground in a mortar using liquid nitrogen. Total genomic DNA was extracted using EZ-10 Spin Column Plant Genomic DNA Purification Kit (Sangon, Shanghai, China) following the manufacturer’s instructions. Adjusted the concentration of nucleic acid to the same 200 ng/μL, which was used as templates for real-time qPCR to get CT (Cycle threshold) values directly. The lower the CT value, the higher the CLas titer (concentration). The reaction mixture was performed in a total volume of 20 μL and contained 0.4 μL primers HLBas (Sangon, Shanghai, China), 0.4 μL primers HLBr (Sangon, Shanghai, China), 0.4 μL probe HLBp (Takara, Tokyo, Japan), 10 μL 2X TaqMan Fast qPCR Master Mix (Sangon, Shanghai, China), 1 μL DNA template, and 7.8 μL water. The sequences of the primers and probes used are collated in Table S1. The amplification protocol was 95°C for 5 min followed by 40 cycles at 95°C for 30 s and 60°C for 30 s (Duan et al., 2021).
DAPI stained plant leaves
DAPI (4′,6-diamidino-2-phenylindole) (Genview, America) staining solution at 0.5 mg/mL was prepared using 0.1 mol/L PBS (pH = 7.4). Two days after infiltrating AuNPs-PEI-pNPR1-GFP, the periwinkle leaves were directly immersed in the staining solution for 90 min, then washed three times with ddH2O for 1 min each, and then the leaves were placed under the confocal microscope for observation.
Confocal microscopy imaging
Confocal images were obtained in LSM 880 with Airyscan confocal laser scanning microscope (SH120). FAM, GFP, mCherry, DAPI and Chlorophyll fluorescence wave range was selected on the confocal microscope.
Confocal images reported in Figures S2, S3, 4, 5, and 6 were representative images chosen from 10 replicates (10 different fields of view from the same leaf per sample). ZEN software (Carl Zeiss) was used for the fluorescence intensity and quantitative analysis of GFP and Chlorophyll fluorescence.
Western blot
Leaves were collected at different time points after AuNPs-PEI-pGFP and AuNPs-PEI-pNPR1-GFP were infiltrated. 0.4 g of leaf tissue was ground in liquid nitrogen before adding extraction buffer. The extracts were centrifuged, and the supernatant protein was collected to be denatured in the SDS sample buffer at 100°C for 10 min. Total proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) with a constant current of 100 mA and then transferred onto PVDF (polyvinylidene difluoride) membranes (Merck, Darmstadt, Germany) at a constant voltage 100 V (Bio-Rad, CA, USA). Skimmed milk powder (5% w/v) was dissolved in TBST (Tris-buffered Saline Tween 20) buffer and used to block the membrane. The antibody was diluted in TBST at a ratio of 1:2,000 and incubated with the membrane overnight at 4°C. The membrane was then washed three times with TBST for 10 min. Goat anti-mouse IgG-HRP (Absin, abs 20001) was then incubated with the membrane for 2 hours at room temperature. The membrane was washed twice for 10 minutes and another one time for 30 minutes with TBST. Clarity Western Substrate (BIO-RAD) was used for chemiluminescent detection of the GFP protein. The detection was performed using a LI-COR Odyssey Infrared Imaging System (Tanon, 5,200, China) (An et al., 2022).
Quantification and statistical analysis
All experiments were repeated at least three times or three parallel replicate samples, and the results were processed by Excel and GraphPad Prism 8. ZEN software was used for the processing of confocal images and the quantitative analysis of fluorescence intensity. Data were expressed as mean ± SD. Statistical analyses were performed using Student's t-test (∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001 and ns indicated no significant differences).
Acknowledgments
This research was funded by the National Natural Science Foundation of China (Grants 31570256) and a grant from the science and technology project of Guangzhou (Grant No.201805010002). The authors thank the anonymous reviewers for helpful comments on the manuscript.
Author contributions
J.Z. conducted the experiments and wrote the manuscript. W.L. contributed to the experiment designing and data analysis. Y.M. and B.Z. collected the previous literature and summarized the contents. C.Z., J.Y., M.W., and D.X. helped to revise the manuscript. X.M. supervised the project, reviewed the manuscript, and provided funding support. W.C. supervised the project, proofread the manuscript, and provided funding support.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104479.
Contributor Information
Xiangchun Meng, Email: mengxiangchun@gdaas.cn.
Wenli Chen, Email: chenwl@scnu.edu.cn.
Supplemental information
Data and code availability
All data produced in this study are included in the published article and its supplemental information, or are available from the lead contact upon request. No unique code was generated. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- An Z., Cao B., Zhang J., Zhang B., Zhou C., Hu X., Chen W. Efficient transient expression of plasmid DNA using poly (2-(N, N-dimethylamino) ethyl methacrylate) in plant cells. Front. Bioeng. Biotechnol. 2022;10:805996. doi: 10.3389/fbioe.2022.805996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bové J., Barros A. Huanglongbing: a destructive, newly emerging, century-old disease of citrus. J. Plant Pathol. 2006;88:7–37. [Google Scholar]
- Brondani D., De Souza B., S Souza B., Neves A., C Vieira I. PEI-coated gold nanoparticles decorated with laccase: a new platform for direct electrochemistry of enzymes and biosensing applications. Biosens. Bioelectron. 2013;42:242–247. doi: 10.1016/j.bios.2012.10.087. [DOI] [PubMed] [Google Scholar]
- Castañeda M., Alegret S., Merkoçi A. Electrochemical sensing of DNA using gold nanoparticles. Electroanalysis. 2007;19:743–753. doi: 10.1002/elan.200603784. [DOI] [Google Scholar]
- Catinot J., Buchala A., Abou-Mansour E., Métraux J.P. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS. Lett. 2008;582:473–478. doi: 10.1016/j.febslet.2007.12.039. [DOI] [PubMed] [Google Scholar]
- Chen J., Mohan R., Zhang Y., Li M., Chen H., Palmer I.A., Chang M., Qi G., Spoel S.H., Mengiste T., et al. NPR1 promotes its own and target gene expression in plant defense by recruiting CDK8. Plant Physiol. 2019;181:289–304. doi: 10.1104/pp.19.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang L., He Y., Li Y. Sandwich-type Au-PEI/DNA/PEI-Dexa nanocomplex for nucleus-targeted gene delivery in vitro and in vivo. Acs Appl. Mater. Interfaces. 2014;6:14196–14206. doi: 10.1021/am503483w. [DOI] [PubMed] [Google Scholar]
- Choi C.H.J., Hao L., Narayan S.P., Auyeung E., Mirkin C.A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U S A. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor E.E., Mwamuka J., Gole A., Murphy C.J., Wyatt M.D. Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small. 2005;1:325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- Demirer G.S., Zhang H., Matos J.L., Goh N.S., Cunningham F.J., Sung Y., Chang R., Aditham A.J., Chio L., Cho M.-J., et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019;14:456–464. doi: 10.1038/s41565-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J., Li X., Zhang J., Cheng B., Liu S., Li H., Zhou Q., Chen W. Cocktail therapy of fosthiazate and cupric-ammoniun complex for citrus huanglongbing. Front. Plant Sci. 2021;12:643971. doi: 10.3389/fpls.2021.643971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfinger M., Pfeifer C., Uezguen S., Golas M.M., Sander B., Maucksch C., Stark H., Aneja M.K., Rudolph C. Self-assembly of ternary insulin-polyethylenimine (PEI)-DNA nanoparticles for enhanced gene delivery and expression in alveolar epithelial cells. Biomacromolecules. 2009;10:2912–2920. doi: 10.1021/bm900707j. [DOI] [PubMed] [Google Scholar]
- Godbey W., Wu K.K., Mikos A.G. Poly (ethylenimine) and its role in gene delivery. J. Contr. Release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- Godbey W.T., Wu K.K., Mikos A.G. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22:471–480. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- González-Domínguez I., Grimaldi N., Cervera L., Ventosa N., Gòdia F. Impact of physicochemical properties of DNA/PEI complexes on transient transfection of mammalian cells. New Biotechnol. 2019;49:88–97. doi: 10.1016/j.nbt.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Graczyk A., Pawlowska R., Jedrzejczyk D., Chworos A. Gold nanoparticles in conjunction with nucleic acids as a modern molecular system for cellular delivery. Molecules. 2020;25:204. doi: 10.3390/molecules25010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashimoto A., Li Z., Murai N. A polyethylene glycol-mediated protoplast transformation system for production of fertile transgenic rice plants. Plant. Physiol. 1990;93:857–863. doi: 10.1104/pp.93.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunta M., da Graça J.V., Malik N.S., Louzada E.S., Sétamou M. Quantitative distribution of Candidatus Liberibacter asiaticus in the aerial parts of the huanglongbing-infected citrus trees in Texas. Hortscience. 2014;49:65–68. doi: 10.21273/hortsci.49.1.65. [DOI] [Google Scholar]
- Lakshmanan M., Kodama Y., Yoshizumi T., Sudesh K., Numata K. Rapid and efficient gene delivery into plant cells using designed peptide carriers. Biomacromolecules. 2012;14:10–16. doi: 10.1021/bm301275g. [DOI] [PubMed] [Google Scholar]
- Lei W.X., An Z.S., Zhang B.H., Wu Q., Gong W.J., Li J.M., Chen W.L. Construction of gold-siRNA NPR1 nanoparticles for effective and quick silencing of NPR1 in Arabidopsis thaliana. RSC Adv. 2020;10:19300–19308. doi: 10.1039/d0ra02156c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li Y., Zhao Q., Li T., Wei J., Li B., Shen W., Yang C., Zeng Y., Rodriguez P.L., et al. The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nature plants. 2019;5:512–524. doi: 10.1038/s41477-019-0400-5. [DOI] [PubMed] [Google Scholar]
- Li M., Guo Z., Yang H., Liu Y., Tong Y., Kong J. Electrochemical sensor based on polyethyleneimine-aunps- anthraquinone-2-carboxylic acid nanocomposite for cysteine detection. Int. J. Electrochem. Sci. 2019;14:943–956. doi: 10.20964/2019.01.72. [DOI] [Google Scholar]
- Li W., Hartung J.S., Levy L. Quantitative real-time PCR for detection and identification of candidatus liberibacter species associated with citrus huanglongbing. J. Microbiol. Methods. 2006;66:104–115. doi: 10.1016/j.mimet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Li W., Levy L., Hartung J.S. Quantitative distribution of ‘Candidatus Liberibacter asiaticus’ in citrus plants with citrus huanglongbing. Phytopathology. 2009;99:139–144. doi: 10.1094/phyto-99-2-0139. [DOI] [PubMed] [Google Scholar]
- Li Y., Yu Q., Wang B., Chen L. Differential expression of Isochorismate synthase in Catharanthus roseus during ‘Candidatus Liberibacter asiaticus’ infection. Trop. Plant Pathol. 2019;44:363–370. doi: 10.1007/s40858-019-00287-y. [DOI] [Google Scholar]
- Liao S., Zhang Y., Pan X., Zhu F., Jiang C., Liu Q., Cheng Z., Dai G., Wu G., Wang L., Chen L. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant <em>Pseudomonas aeruginosa</em>. Int. J. Nanomed. 2019;14:1469–1487. doi: 10.2147/ijn.s191340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márton L., Wullems G.J., Molendijk L., Schilperoort R.A. In vitro transformation of cultured cells from Nicotiana tabacum by Agrobacterium tumefaciens. Nature. 1979;277:129–131. doi: 10.1038/277129a0. [DOI] [Google Scholar]
- Kodiha M., Wang Y.M., Hutter E., Maysinger D., Stochaj U. Off to the organelles - killing cancer cells with targeted gold nanoparticles. Theranostics. 2015;5:357–370. doi: 10.7150/thno.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzarini E., Mariano S., Carata E., Mura F., Rossi M., Dini L. Intracellular transport of silver and gold nanoparticles and biological responses: an update. Int. J. Mol. Sci. 2018;19:1305. doi: 10.3390/ijms19051305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard H., Remy J.-S., Loussouarn G., Demolombe S., Behr J.-P., Escande D. Polyethylenimine but not cationic lipids promotes transgene delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- Qiu W., Soares J., Pang Z., Huang Y., Sun Z., Wang N., Grosser J., Dutt M. Potential mechanisms of AtNPR1 mediated resistance against Huanglongbing (HLB) in citrus. Int. J. Mol. Sci. 2020;21:2009. doi: 10.3390/ijms21062009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab F., Zhai G., Kern M., Turner A., Schnoor J.L., Wiesner M.R. Barriers, pathways and processes for uptake, translocation and accumulation of nanomaterials in plants--Critical review. Nanotoxicology. 2016;10:257–278. doi: 10.3109/17435390.2015.1048326. [DOI] [PubMed] [Google Scholar]
- Serag M.F., Kaji N., Gaillard C., Okamoto Y., Terasaka K., Jabasini M., Tokeshi M., Mizukami H., Bianco A., Baba Y. Trafficking and subcellular localization of multiwalled carbon nanotubes in plant cells. ACS Nano. 2011;5:493–499. doi: 10.1021/nn102344t. [DOI] [PubMed] [Google Scholar]
- Shan Y., Luo T., Peng C., Sheng R., Cao A., Cao X., Shen M., Guo R., Tomás H., Shi X. Gene delivery using dendrimer-entrapped gold nanoparticles as nonviral vectors. Biomaterials. 2012;33:3025–3035. doi: 10.1016/j.biomaterials.2011.12.045. [DOI] [PubMed] [Google Scholar]
- Thomas M., Klibanov A.M. Conjugation to gold nanoparticles enhances polyethylenimine's transfer of plasmid DNA into mammalian cells. Proc. Natl. Acad. Sci. U S A. 2003;100:9138–9143. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati S.R., Ishiga Y., Wangdi T., Kunkel B.N., Anand A., Mysore K.S., Bender C.L. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 2007;20:955–965. doi: 10.1094/mpmi-20-8-0955. [DOI] [PubMed] [Google Scholar]
- Wang P., Lombi E., Zhao F.J., Kopittke P.M. Nanotechnology: a new opportunity in plant sciences. Trends Plant Sci. 2016;21:699–712. doi: 10.1016/j.tplants.2016.04.005. [DOI] [PubMed] [Google Scholar]
- Xu P., Quick G.K., Yeo Y. Gene delivery through the use of a hyaluronate-associated intracellularly degradable crosslinked polyethyleneimine. Biomaterials. 2009;30:5834–5843. doi: 10.1016/j.biomaterials.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Zhang S., Zhou Q., Chen W. VHPKQHR peptide modified magnetic mesoporous nanoparticles for MRI detection of atherosclerosis lesions. Artificial Cells. 2019;47:2440–2448. doi: 10.1080/21691401.2019.1626411. [DOI] [PubMed] [Google Scholar]
- Li Y., Xu M.r., Dai Z.H., Deng X.L. Distribution pattern and titer of Candidatus Liberibacter asiaticus in periwinkle (Catharanthus roseus) J. Integr. Agric. 2018;17:2501–2508. doi: 10.1016/s2095-3119(18)61918-5. [DOI] [Google Scholar]
- Yeh Y.C., Creran B., Rotello V.M. Gold nanoparticles: preparation, properties, and applications in bionanotechnology. Nanoscale. 2012;4:1871–1880. doi: 10.1039/c1nr11188d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavaliev R., Mohan R., Chen T., Dong X. Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell. 2020;182:1093–1108.e18. doi: 10.1016/j.cell.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Duan Y., Zhou L., Turechek W.W., Stover E., Powell C.A. Screening molecules for control of citrus huanglongbing using an optimized regeneration system for 'Candidatus Liberibacter asiaticus'-infected periwinkle (Catharanthus roseus) cuttings. Phytopathology. 2010;100:239–245. doi: 10.1094/phyto-100-3-0239. [DOI] [PubMed] [Google Scholar]
- Zhou L., Powell C.A., Hoffman M.T., Li W., Fan G., Liu B., Lin H., Duan Y. Diversity and plasticity of the intracellular plant pathogen and insect symbiont “Candidatus Liberibacter asiaticus” as revealed by hypervariable prophage genes with intragenic tandem repeats. Appl. Environ. Microbiol. 2011;77:6663–6673. doi: 10.1128/aem.05111-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data produced in this study are included in the published article and its supplemental information, or are available from the lead contact upon request. No unique code was generated. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.