Abstract
Despite intensive studies of microbial-community diversity, the questions of which kinds of microbial populations are associated with changes in community diversity have not yet been fully solved by molecular approaches. In this study, to investigate the impact of livestock wastewater on changes in the bacterial communities in groundwater, bacterial communities in subsurface aquifers were analyzed by characterizing their 16S rDNA sequences. The similarity coefficients of restriction fragment length polymorphism (RFLP) patterns of the cloned 16S ribosomal DNAs showed that the bacterial communities in livestock wastewater samples were more closely related to those in contaminated aquifer samples. In addition, calculations of community diversity clearly showed that bacterial communities in the livestock wastewater and the contaminated aquifer were much more diverse than those in the uncontaminated aquifer. Thus, the increase in bacterial-community diversity in the contaminated aquifer was assumed to be due to the infiltration of livestock wastewater, containing high concentrations of diverse microbial flora, into the aquifer. Phylogenetic analysis of the sequences from a subset of the RFLP patterns showed that the Cytophaga-Flexibacter-Bacteroides and low-G+C gram-positive groups originating from livestock wastewater were responsible for the change in the bacterial community in groundwater. This was evidenced by the occurrence of rumen-related sequences not only in the livestock wastewater samples but also in the contaminated-groundwater samples. Rumen-related sequences, therefore, can be used as indicator sequences for fecal contamination of groundwater, particularly from livestock.
Stock farming in Korea has increased considerably over the last decade due to changes in eating habits. Consequently, attention has been paid recently to pollution in the soil, surface water, and groundwater caused by intensive stock farming. The application of animal manure and livestock wastewater to surface soil results in a rapid increase in microbial biomass and nitrogenous nutrients (9, 21, 29). As animal manure and wastewater are important sources of nitrogen compounds, studies have been carried out mainly to analyze the community structure of microorganisms responsible for nitrogen transformations (25, 33). Also, a few papers (23, 62) have been dedicated to the study of the effect of livestock waste on groundwater quality in the context of various nutrients and some indicator bacteria. However, the effect of livestock manure and wastewater on bacterial-community structure in environments receiving livestock waste is currently poorly understood. In our recent study (12), subsurface aquifers in the Wonju stock-farming area (Fig. 1) were found to be heavily contaminated with nitrate and pollution indicator bacteria due to the infiltration of livestock wastewater. This study indicates that the composition and diversity of the bacterial community in groundwater may change dramatically through the introduction of livestock wastewater into aquifers. Merely by measuring the indicator bacterial abundance using culture-dependent methods, however, it could not be precisely determined which kinds of bacteria were responsible for the change in the bacterial community and which types of organisms could be used as pollution indicators, because most bacteria in the environment have not been cultivated (11, 43). The analysis of bacterial communities, including noncultivated bacteria, therefore, can provide more-precise information on the bacterial populations responsible for rapid changes in the bacterial community and the pollution indicators of fecal contamination in groundwater ecosystems.
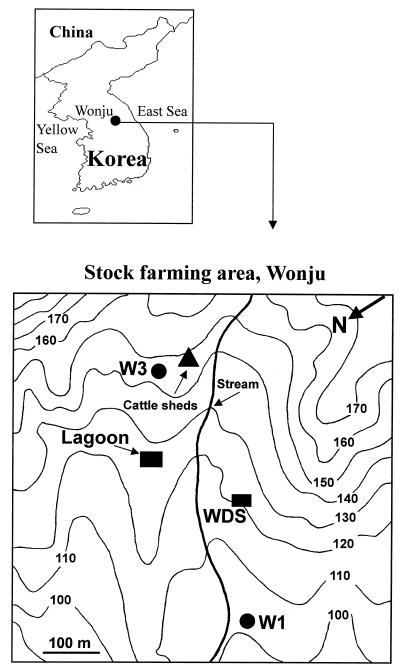
FIG. 1.
Locations of sampling sites and boreholes drilled at a livestock farming area in Wonju, Korea. Numbers on the contours show altitudes above sea level (in meters). W3, used as an uncontaminated control, is a 120-m-deep borehole located adjacent to the cattle sheds. Borehole W1 is located downstream from the lagoon and the livestock waste disposal sites and is contaminated with livestock wastewater. Livestock wastewater samples were taken from a livestock wastewater storage lagoon (Lagoon) and an animal waste dump site (WDS).
A variety of molecular methods have been used to determine the species composition of bacterial communities without enrichment culture (18, 26). Many of these methods rely on 16S ribosomal DNA (rDNA) sequences, including in situ hybridization (2, 3), direct amplification of 16S rDNA, and further analysis using community fingerprinting such as denaturing gradient gel electrophoresis (DGGE) (32, 45), temperature gradient gel electrophoresis (TGGE) (17, 19), single-strand-conformation polymorphism (SSCP) (37, 51), restriction fragment length polymorphism (RFLP) (41), terminal RFLP (13), or 16S rDNA cloning-sequencing (20, 42, 61). Although 16S rDNA cloning-sequencing requires much time and effort and hence cannot provide an immediate overview of the community structure, it is a useful method for detecting infrequent sequences in the environment. Studies using this method have shown the presence of several novel genera or divisions of microorganisms such as the Acidobacterium, the Holophaga, and the Verrucomicrobium groups (5, 20, 39, 61).
Numerous studies using 16S rDNA technologies have reported higher molecular diversity of uncultivated microorganisms in various environmental samples (15, 27, 47). However, evaluation of microbial diversity in quantitative terms using molecular approaches had not been carried out until recent reports (16, 42, 46) on the quantitative estimation of bacterial diversity in soils and microbial mats were published. Also, the questions of how bacterial communities are changed by the large input of nutrients and allochthonous microorganisms and which kinds of bacterial populations are responsible for changes in the community have not yet been fully solved using molecular approaches.
The purposes of this study are to show the impact of livestock wastewater on change in the bacterial community, to define the bacterial populations responsible for the shift of bacterial community, and to determine which types of organisms can be used as indicators of fecal contamination in groundwater. We studied subsurface aquifers and a livestock wastewater-stabilizing lagoon in the Wonju stock farming area, where intensive stock farming has been carried out for 8 years. The 16S rDNAs from an aquifer contaminated with livestock wastewater, an uncontaminated aquifer, and a livestock wastewater-stabilizing lagoon were amplified, and RFLP analysis of the cloned sequences was performed; then various diversity indices were calculated based on the RFLP patterns in five clone libraries. Further phylogenetic analyses of sequences of representative RFLP patterns were conducted to define the bacterial populations responsible for the change in bacterial community.
MATERIALS AND METHODS
Site description and sample collection.
Subsurface groundwater and livestock wastewater samples were recovered from boreholes drilled and from the livestock wastewater-stabilizing lagoon, respectively, in a stock-farming area. The sampling site was located in a stock-farming area, Buron-myun, approximately 20 km southwest of Wonju, Korea (Fig. 1). The history of stock farming and the sampling locations were described in a previous study (12). Briefly, livestock wastewater (slurry) had been removed from the lagoon about bimonthly since 1992, and ca. 25 m3 of this slurry had been spread onto the surrounding land surface. Consequently, livestock wastewater had been introduced directly or indirectly into the aquifers. Borehole W3, an uncontaminated control site located adjacent to the cattle sheds, is a 120-m-deep domestic well. Borehole W1 is located ca. 300 m downstream from the lagoon and ca. 200 m downstream from the livestock waste disposal sites and is contaminated with livestock wastewater. Groundwater samples were collected from borehole W1 and borehole W3 with a suction-lift pump in March (designated W1-R and W3-R, respectively) and May (designated W1-Y and W3-Y, respectively) of 1998 after each borehole was flushed by pumping until at least 3 well volumes of water had been evacuated. In addition, livestock wastewater samples (LW) were taken from the wastewater-stabilizing lagoon and the livestock wastewater dump site on May 1998 and pooled.
Collection of bacterial community and DNA extraction.
Two liters of groundwater samples was filtered with a stirred ultrafiltration cell (Amicon Co., Beverly, Mass.) and YM100 disk membranes (molecular weight cutoff, 100,000; Amicon Co.) under N2 gas at constant pressure (20 lb in−2). To detach the bacteria from the membrane, the filter was soaked with 2 ml of sterile STE buffer (0.1 M NaCl, 10 mM Tris, 1 mM EDTA [pH 7.6]) for 1 h at 4°C. The resuspended cells were centrifuged (at 10,000 × g for 15 min at 4°C), and the supernatant was removed gently. This detaching step was repeated four times, and the products were used for DNA extraction. Two milliliters of livestock slurry samples was centrifuged (at 10,000 × g for 15 min at 4°C) and used likewise for DNA extraction. Total nucleic acids were extracted from each sample by using lysozymes, freeze-thawing, and phenol-chloroform as previously described (37). Extracted nucleic acids were purified by electrophoresis through a 0.75% low-melting-point agarose gel (SeaPlaque GTG; FMC Bioproducts, Rockland, Maine). Total DNAs were purified from the excised gel slice with a DNA purification kit (DNA PrepMate; Bioneer Co., Chungbuk, Korea), eluted in 30 μl of sterile Tris-EDTA (TE) buffer, and used as templates for PCR.
PCR, cloning, and sequencing.
Bacterial 16S rDNA was amplified with the two bacterial universal primers, 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGYTACCTTGTTACGACTT-3′) (36). PCR amplification was carried out with a thermal cycler (PE2400; Perkin-Elmer Co., Norwalk, Conn.) under the following conditions: 94°C for 3 min; 35 cycles of denaturation at 94°C for 1.5 min, annealing at 50°C for 1.5 min, and extension at 72°C for 2.0 min; and 72°C for 30 min. Reaction mixtures (final volume, 50 μl) contained 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.8 at 25°C), 50 mM KCl, 0.1% Triton X-100, 200 ng of bovine serum albumin μl−1, 200 μM each deoxynucleoside triphosphate, 0.2 μM each oligonucleotide primer, and 2 U of Dynazyme (Finnzymes Oy., Espoo, Finland). Approximately 50 ng of DNA template was added to each PCR tube. The amplification products were visualized by electrophoresis through a 1.2% low-melting-point agarose gel (SeaPlaque GTG). After triplicate amplified products were pooled, the band corresponding to the correctly sized product (∼1.5 kb) was excised, purified, and concentrated. The 16S rDNA library was generated with the purified 16S rDNAs from each sample by each ligation into the pGEM-T vector (Promega Co., Madison, Wis.). Ligation, transformation into Escherichia coli DH5α competent cells, and blue/white screening were performed according to the manufacturer's instructions. Plasmid inserts were directly amplified from the transformant cells by PCR with pGEM-T primers T7 and SP6 for 16S rDNA screening. The composition of the reaction mixtures and PCR conditions were the same as described above. To define RFLP patterns of cloned 16S rDNA sequences, aliquots (5.0 μl) of reamplified PCR products were digested for 3 h at 37°C with 2 U of HaeIII (Promega Co.). The resulting fragments were separated by gel electrophoresis in 4% NuSieve 3:1 agarose (FMC Bioproducts). RFLP patterns for each library were grouped visually, and representative clones were selected for sequencing. Plasmid preparations for DNA sequencing were made with Wizard Mini-Preps (Promega Co.). A total of 109 16S rDNA clones were sequenced by the chain termination method on an ALFexpress DNA autosequencer (Pharmacia Biotech, Uppsala, Sweden) using the 27F primer and the Cy5 AutoRead sequencing kit.
Phylogenetic analyses.
Initially, all sequences of approximately 500 bases were compared with sequences available in the EMBL/GenBank database by using BLAST network services (1) and with sequences in the Ribosomal Database Project II (RDP) database (7) by using the SEQUENCE_MATCH (version 2.7) function to determine their approximate phylogenetic affiliations. Sequences were initially aligned with the CLUSTAL W program (55), visually examined, and relocated to allow maximal alignment by referring to representative bacterial sequences from the RDP. Sequences were also checked for chimeric properties by using CHIMERA_CHECK from the RDP. Among the 109 16S rDNA sequences from samples analyzed, 3 clones appeared to be chimeras and were excluded in the phylogenetic analyses. Phylogenetic trees were constructed using DNADIST with the Jukes-Cantor model (30) and NEIGHBOR with the neighbor-joining method (49) in the programs of PHYLIP (phylogeny inference package), version 3.5 (J. Felsenstein, Department of Genetics, University of Washington, Seattle). Two hundred bootstrapped replicate resampling data sets were generated with SEQBOOT (PHYLIP, version 3.5). The similarity values between sequences were calculated from DNADIST matrices by reversing the Jukes-Cantor distance formula (30). Phylogenetic assignments were made from both the constructed phylogenetic trees by using PHYLIP and the HTML TREE by using SEQUENCE_MATCH in the RDP database.
Calculation of community diversity.
To determine the similarities of bacterial populations present in five clone libraries, pairwise comparisons of the RFLP patterns between clone libraries were performed, and a matrix (presence or absence of each RFLP pattern) was constructed. The matrix was computed by the unweighted pair group method using arithmetic averages (UPGMA) algorithm using the NTSYS program (version 1.8). Various diversity indices were calculated from the RFLP patterns of five clone libraries. Species richness, which represents the total number of species or operational taxonomic units (OTUs), was calculated by rarefaction (28, 50) with the online Rarefaction Calculator (34; http://gause.biology.ualberta.ca/jbrzusto/rarefact.html). Bacterial diversity was calculated on the basis of RFLP patterns by using the Shannon-Weaver index (H), Pielou's evenness index (e), equitability (J), and Simpson's dominance index (c) (48).
Nucleotide sequence accession numbers.
The partial sequences determined in this study have been deposited in the GenBank database under accession no. AF175579 to AF175674.
RESULTS
RFLP patterns and similarity coefficients.
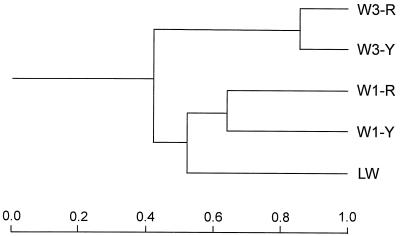
Bacterial diversity in subsurface aquifers and livestock wastewater was analyzed by RFLP patterns obtained by HaeIII-digested 16S rDNA fragments. About 250 clones containing 16S rDNA in each clone library were analyzed by reamplification with T7 and SP6 primers. After exclusion of partially inserted 16S rDNA clones, in the five clone libraries taken together, the analysis of HaeIII-digested RFLP types of a total of 1,160 clones resulted in 222 RFLP patterns (Table 1). In clone libraries W3-R and W3-Y from the uncontaminated borehole, W3, 38 and 42 RFLP patterns, respectively, were detected, while in LW and borehole-W1 samples, 91 to 127 RFLP patterns were detected. In this analysis, each RFLP pattern was taken as an OTU. Therefore, the similarity values among the clone libraries could be calculated by the pairwise comparison of RFLP patterns. In this way a similarity coefficient was obtained for every pair of clone libraries and a similarity UPGMA dendrogram was generated (Fig. 2). The dendrogram showed the similarity levels in the samples analyzed. Clone libraries from two borehole-W3 samples (W3-R and W3-Y; similarity coefficient, 0.829) were more similar to each other than those from two borehole-W1 samples (W1-R and W1-Y; similarity coefficient, 0.613). Interestingly, the RFLP patterns from LW were more closely related to those from the borehole-W1 samples than to those from the borehole-W3 samples. The similarity coefficients between the LW and W1-R and between the LW and W1-Y clone libraries were 0.523 and 0.586, respectively, while those between the LW and W3-R and between the LW and W3-Y clone libraries were 0.356 and 0.419, respectively.
TABLE 1.
Numbers of clones analyzed and a variety of diversity indices obtained based on RFLP patterns in 16S rDNA clone libraries from Wonju groundwater and livestock wastewater
| Parameter | Clone library
|
||||
|---|---|---|---|---|---|
| W-3R | W-3Y | W-1R | W-1Y | LW | |
| Total no. of clones | 225 | 216 | 242 | 227 | 250 |
| No. of RFLP patterns | 38 | 42 | 127 | 91 | 111 |
| RFLP pattern richnessa | 36.9 | 41.3 | 116.8 | 88.2 | 100.6 |
| Shannon-Weaver diversity (H)b | 2.589 | 2.538 | 4.585 | 4.207 | 4.381 |
| Evenness (e)c | 0.712 | 0.679 | 0.947 | 0.933 | 0.930 |
| Equitability (J)d | 0.478 | 0.472 | 0.835 | 0.776 | 0.793 |
| Simpson's dominance (c)e | 0.147 | 0.152 | 0.014 | 0.021 | 0.018 |
Calculated by rarefaction for a standard sample size of 210 clones for each clone library (28, 34).
Calculated as H = −Σ(Pi log Pi), where Pi is the proportion for each RFLP pattern.
Calculated from H as follows: e = H/log S, where S is the total number of RFLP patterns.
Calculated from H as follows: J = H/Hmax, where Hmax is the theoretical maximal Shannon-Weaver diversity index for the clone libraries.
Calculated as c = Σ(Pi)2 where Pi is the proportion for each RFLP pattern.
FIG. 2.
Dendrogram of 16S rDNA RFLP similarities among five clone libraries calculated on the basis of similarity coefficients with the clustering algorithm of UPGMA. See Materials and Methods for designations.
Diversity indices.
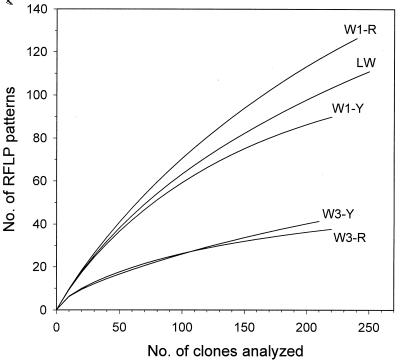
The molecular bacterial diversity of groundwater and livestock wastewater in each clone library was calculated using five diversity indices in the context of two components of diversity, richness (the number of RFLP patterns), and evenness (relative abundance of each RFLP pattern), and results are given in Table 1. For the calculation of richness, the numbers of RFLP patterns according to a sample size of 210 clones were estimated by rarefaction. The estimated values of richness in the W1-R (116.8), W1-Y (88.2), and LW (100.6) clone libraries were much higher than those in the W3-R (36.9) and W3-Y (41.3) clone libraries. The rarefaction curves in Fig. 3 show different patterns of richness among the clone libraries. The slopes of the RFLP pattern richness curves for the W1 and LW clone libraries were much steeper than those for the W3 clone libraries. As shown in Table 1, the W1 and LW clone libraries had higher values on diversity indices such as richness, the Shannon-Weaver diversity index, evenness, and equitability than the W3 clone libraries. Otherwise, the Simpson's dominance index value for the W3 clone libraries was much higher than those for the W1 and LW clone libraries, which means that the W3 clone libraries were dominated by a few RFLP patterns. Overall, the calculated diversity index values showed that the bacterial communities of the contaminated borehole, W1, and of livestock wastewater were more diverse than those of the uncontaminated borehole, W3.
FIG. 3.
RFLP pattern richness curves of the five clone libraries obtained by 16S rDNA PCR-RFLP patterns. Sampling curves were calculated by rarefaction (28, 34). For designations, see Materials and Methods.
Analysis of 16S rDNA sequences.
In the five clone libraries taken together, a total of 109 clones (3 clones were chimeras) were partially (approximately 500 bp at the 5′ end) sequenced. Fourteen clones, which were composed of two to six clones from each of four selected RFLP patterns (WJGRT-1, WJGRT-2, WJGRT-6, and WJGRT-20), were sequenced to determine the 16S rDNA sequence similarity of clones representing the same RFLP patterns. The ranges of similarity values of 16S rDNA sequences for WJGRT-1, WJGRT-2, WJGRT-6, and WJGRT-20 were 97.5 to 100% (average, 99.8%), 96.3 to 99.7% (average, 98.4%), 98.2 to 99.8% (average, 99.5%), and 98.6%, respectively. After the 3 chimeras and 10 clones among the 14 clones used for determining sequence similarity were excluded, the resulting 96 clones representing the unique RFLP patterns were used for the phylogenetic analysis. These 96 RFLP patterns were chosen from the total of 222 RFLP patterns as those most commonly occurring on the basis of comparisons between the W3-R and W3-Y, W1-R and W1-Y, W3 and W1, W3 and LW, and W1 and LW clone libraries and among the W3, W1, and LW clone libraries.
Phylogenetic assignment of 96 clones to bacterial divisions was carried out using the RDP database. As shown in Table 2, most clones in each clone library, except the W3-R and W3-Y clone libraries, were assigned to four well-characterized divisions (the Proteobacteria, the low-G+C gram-positive group, the high-G+C gram-positive group, and the Cytophaga-Flexibacter-Bacteroides group). Two clusters including eight clones, however, were not assigned to any of the known bacterial divisions or candidate divisions (27) in the phylogenetic analysis (see Fig. 6). In this study, therefore, these clusters were designated WJG groups 1 and 2. In the groundwater clone libraries (W3 and W1), clones belonging to the β-Proteobacteria were most abundant, while in the LW clone library, clones belonging to the low-G+C gram-positive group were more abundant. The molecular bacterial diversities in clone libraries were highly different from each other. The relative abundance of clones assigned to the β-Proteobacteria, the low-G+C gram-positive group, the Cytophaga-Flexibacter-Bacteroides group, and WJG group 1 clearly made this point. In the W3 clone libraries, sequences of the β-Proteobacteria and WJG group 1 comprised 43.2 and 24.3% (pooled data set), respectively, and sequences of the gram-positive bacteria and the Cytophaga-Flexibacter-Bacteroides group were rare. In the W1 clone libraries, however, the relative abundances of the low-G+C gram-positive group and the Cytophaga-Flexibacter-Bacteroides group were 19.5 and 17.8%, respectively (pooled data set), and WJG group 1 was rare. In the LW clone library, as with W1, the relative abundances of the low-G+C gram-positive group and the Cytophaga-Flexibacter-Bacteroides group were as high as 29.8 and 22.8%, respectively, and no sequences belonging to WJG group 1 were found. From these results, it could be inferred that borehole W1 was affected by the input of livestock wastewater.
TABLE 2.
Relative abundances of clones related to various bacterial phylogenetic divisions in clone libraries from Wonju groundwater and livestock wastewater
| Phylogenetic group | Relative clone abundance (%) in:
|
||||||
|---|---|---|---|---|---|---|---|
| Clone libraries of groundwater and livestock wastewater
|
Pooled data set of groundwater
|
||||||
| W3-R | W3-Y | W1-R | W1-Y | LW | W3a | W1b | |
| α-Proteobacteria | 5.6 | 10.5 | 7.7 | 7.5 | 7.0 | 8.1 | 7.6 |
| β-Proteobacteria | 44.4 | 42.1 | 32.3 | 28.3 | 21.1 | 43.2 | 30.5 |
| γ-Proteobacteria | 5.6 | 10.5 | 9.2 | 7.5 | 8.8 | 8.1 | 8.5 |
| δ-Proteobacteria | 0.0 | 0.0 | 1.5 | 1.9 | 1.8 | 0.0 | 1.7 |
| Low-G+C gram-positive group | 0.0 | 5.3 | 20.0 | 18.9 | 29.8 | 2.7 | 19.5 |
| High-G+C gram-positive group | 5.6 | 5.3 | 3.1 | 5.7 | 3.5 | 5.4 | 4.2 |
| Cytophaga-Flexibacter-Bacteroides | 0.0 | 5.3 | 15.4 | 20.8 | 22.8 | 2.7 | 17.8 |
| Cyanobacteria | 0.0 | 0.0 | 1.5 | 0.0 | 0.0 | 0.0 | 0.8 |
| Nitrospira | 0.0 | 0.0 | 1.5 | 1.9 | 0.0 | 0.0 | 1.7 |
| Verrucomicrobia | 0.0 | 0.0 | 1.5 | 1.9 | 1.8 | 0.0 | 1.7 |
| Spirochaetales | 0.0 | 0.0 | 1.5 | 1.9 | 1.8 | 0.0 | 1.7 |
| Acidobacterium | 5.6 | 5.3 | 0.0 | 1.9 | 0.0 | 5.4 | 0.8 |
| WJG group 1c | 33.3 | 15.8 | 3.1 | 0.0 | 0.0 | 24.3 | 1.7 |
| WJG group 2c | 0.0 | 0.0 | 1.5 | 1.9 | 1.8 | 0.0 | 1.7 |
W3-R and W3-Y.
W1-R and W1-Y.
The unclassified groups, WJG groups 1 and 2, are defined in Fig. 6.
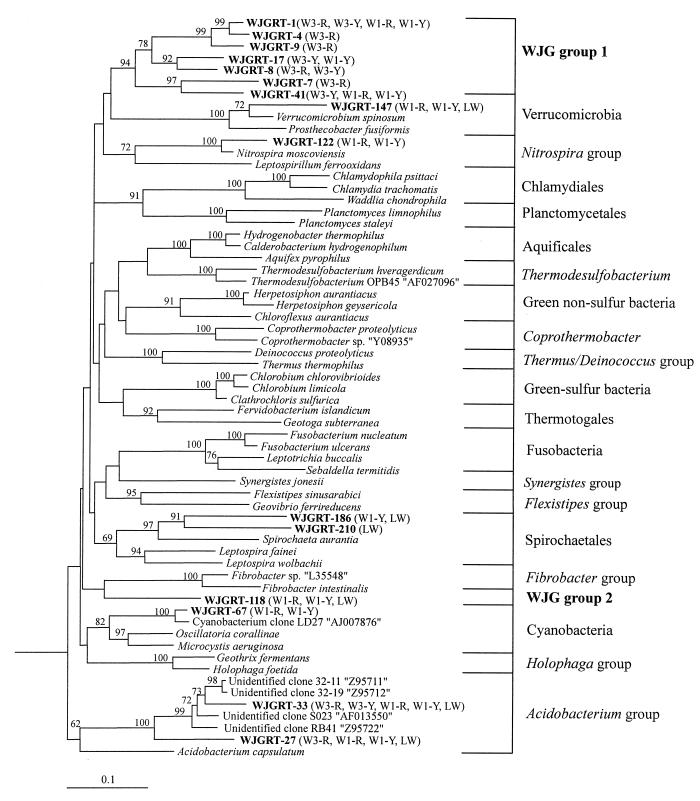
FIG. 6.
Phylogenetic tree generated by the neighbor-joining method showing the phylogenetic relationships among Wonju groundwater and livestock wastewater clones within the Bacteria based on analysis of ca. 400 bases of aligned 16S rDNA sequences. Only sequences that are not assigned to the Proteobacteria, the high-G+C gram-positive group, the low-G+C gram-positive group, or the Cytophaga-Flexibacter-Bacteroides group are represented in the tree. For sequence nomenclature, clone libraries in parentheses, accession numbers in quotation marks, and support for branch points, see the legend to Fig. 4. Methanobacterium thermoflexum in the Archaea served as the outgroup organism. The scale bar indicates 0.1 change per nucleotide.
Phylogenetic distribution of the bacterial community.
The Wonju groundwater and livestock wastewater sequences analyzed, their occurrence in the libraries, and their phylogenetic affiliations are shown in Fig. 4 to Fig. 6. Among 96 sequences of cloned 16S rDNA, 33 sequences (34.3%) had similarities greater than 95.0% with sequences obtained from the RDP and the GenBank database. Thirty-four sequences (35.4%) had similarity values lower than 90.0%, and the sequences with similarity values lower than 85.0% comprised 21.2%.
FIG. 4.
Phylogenetic tree generated by the neighbor-joining method showing the phylogenetic relationships among Wonju groundwater and livestock wastewater clones within the Proteobacteria based on analysis of ca. 400 bases of aligned 16S rDNA sequences. Bootstrap values are shown for each node that had >50% support in a bootstrap analysis of 200 replicates. Sequences obtained from samples are designated in boldface by the prefix WJGRT, followed by replicate numbers (WJGRT-1, WJGRT-2, etc.). The clone libraries from which the individual 16S rDNA clone sequences came are given in parenthesis. GenBank accession numbers of environmental clones or unclassified cultural isolates are given in quotation marks. Actinomyces bovis in the high-G+C gram-positive group served as the outgroup organism. The scale bar indicates 0.1 change per nucleotide.
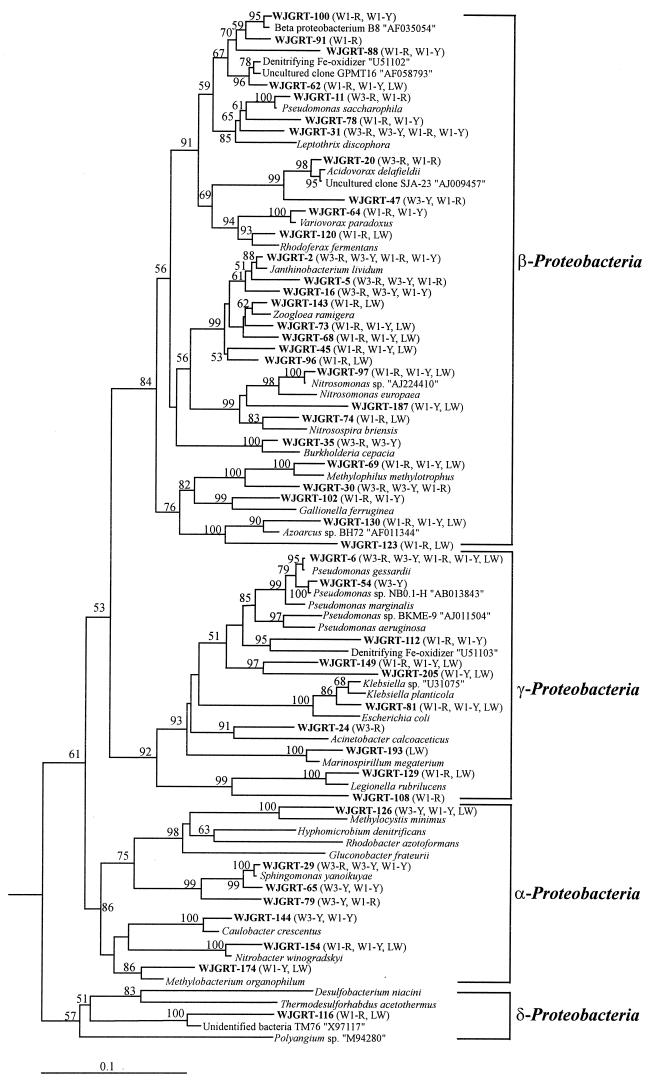
The sequences assigned to the Proteobacteria are represented in Fig. 4. The α-proteobacterial sequences were closely related to the cultured bacteria with strong support in the bootstrap analysis and had relatively high similarities (92.2 to 99.8%; mean, 96.1%) to the reference sequences. Five sequences assigned to the β-Proteobacteria had as their nearest neighbors unclassified or unidentified bacterial clones and were regarded as unclassified β-proteobacterial clones. A total of nine clones were assigned to the γ-Proteobacteria with a bootstrap value of 92%. Clone WJGRT-116 was taken as an unclassified δ-proteobacterium, although there was no strong support (bootstrap value, 57%) for this relationship.
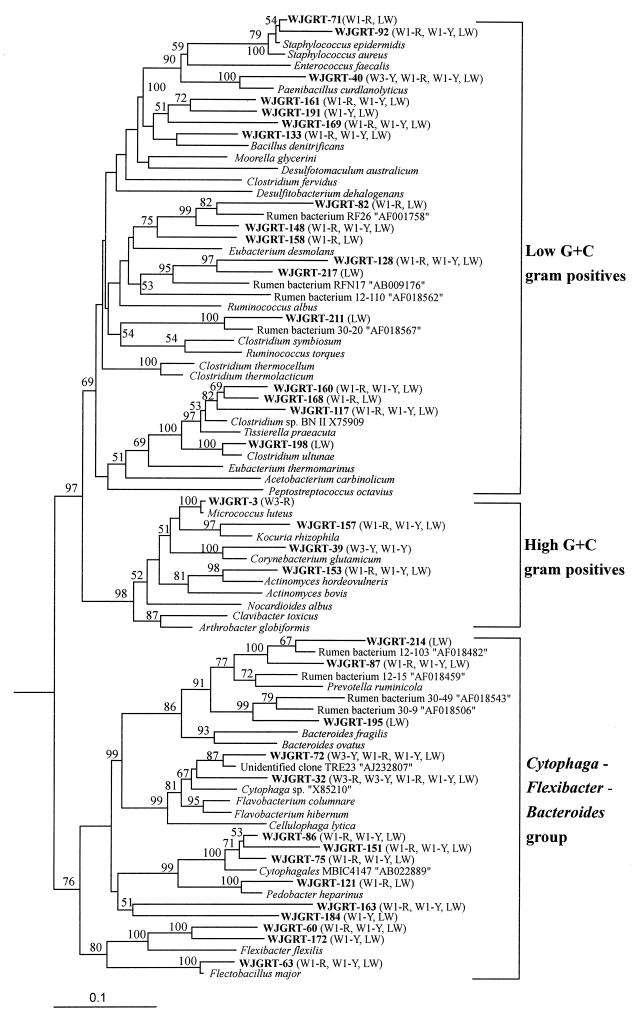
The sequences assigned to the low-G+C gram-positive group, the high-G+C gram-positive bacteria, and the Cytophaga-Flexibacter-Bacteroides group are shown in Fig. 5. Most of the sequences assigned to the low-G+C gram-positive group and the Cytophaga-Flexibacter-Bacteroides group were found only in the W1 and LW clone libraries. Six clones assigned to the low-G+C gram-positive group were not related to any cultivated bacterial sequences; however, interestingly, these sequences were related to the unidentified rumen bacteria RF26 (GenBank accession no. AF001758), RFN17 (AF009176), and 30-1205 (AF018567) obtained from recent studies (54, 59) on rumen microbiota. As with low-G+C gram-positive group, sequences related to unidentified rumen bacteria were also found in the Cytophaga-Flexibacter-Bacteroides group. Three clones, WJGRT-87, WJGRT-195, and WJGRT-214, assigned to the Cytophaga-Flexibacter-Bacteroides group were related to the unidentified rumen bacteria 12-15 (AF018459), 12-103 (AF018482), and 30-9 (AF018506) obtained from the rumen fluid of cattle (59). Clones related to rumen bacterial sequences did not occur in the W3 clone libraries from uncontaminated groundwater, while they occurred in both the W1 and LW clone libraries.
FIG. 5.
Phylogenetic tree generated by the neighbor-joining method showing the phylogenetic relationships among Wonju groundwater and livestock wastewater clones within the high-G+C gram-positive group, the low-G+C gram-positive group, and the Cytophaga-Flexibacter-Bacteroides group based on analysis of ca. 400 bases of aligned 16S rDNA sequences. For sequence nomenclature, clone libraries in parentheses, accession numbers in quotation marks, and support for branch points, see the legend to Fig. 4. E. coli in the γ-Proteobacteria served as the outgroup organism. The scale bar indicates 0.1 change per nucleotide.
The results of phylogenetic analysis of 15 clones that were not represented in Fig. 4 or 5 are shown in Fig. 6. Each group of the Verrucomicrobia, Nitrospira, and Cyanobacteria had only one sequence. Two clones, WJGRT-27 and WJGRT-33, were assigned to the recently defined Acidobacterium group (5, 39), and they are related to environmental sequences from other studies (35, 39). Eight clones which were not assigned to any of the defined bacterial divisions comprised two novel groups designated WJG groups 1 and 2. WJG group 1 contained seven clones that comprised a single group with a bootstrap value of 94% and also comprised three strongly supported lineages.
DISCUSSION
In this study, the molecular diversity of the bacterial community in the Wonju stock-farming area was analyzed to show the effect of livestock wastewater on changes in the bacterial community in groundwater. The comparison of the bacterial communities among the five clone libraries was carried out by three approaches using cloned 16S rDNA RFLP patterns: calculation of the similarity coefficients between the distribution of RFLP patterns in each clone library, calculation and comparison of various diversity indices, and phylogenetic analysis by sequencing of the representative RFLP patterns.
All the approaches described above were based on RFLP patterns of 16S rDNA sequences as units of biodiversity. Because the current bacteriological species concepts are not equivalent to those of macrobiology (53, 58), there is a basic difficulty in applying diversity indices based on the species concept directly to bacterial communities. Therefore, various taxonomic levels used as operational units have been preferred in the field of microbiology. These have included the morphotypes (14, 46), sequence similarity between isolates or clones (8, 42), the heterogeneity of total community DNA (56), RFLP patterns of amplified 16S rDNA (16, 44), bands in community fingerprints (17, 41), and various biochemical markers (6, 31). RFLP patterns as OTUs were proposed by Moyer et al. (44), who estimated the number of RFLP patterns and the abundance of each RFLP pattern. Similarly, a recent study by Dunbar et al. (16) calculated various diversity indices based on RFLP patterns. The amplification of 16S rDNA, cloning, and RFLP typing are known to have biases such as efficiency and selectivity of DNA extraction, differential PCR amplification, rrn operon heterogeneity, and chimera formation that can distort the perception of the bacterial community (26, 60). However, in spite of shortcomings in estimating bacterial-community diversity based on the above methods, the bacterial-community diversity in each clone library could be compared with that in every other, since the biases might operate uniformly for identically treated environmental samples. In addition, the triplicate PCR products were pooled before cloning to minimize the effect of PCR drift.
Similarity analysis by the pairwise comparison of RFLP patterns between clone libraries showed remarkable relationships between clone libraries. Although the boreholes studied are located in the same geographical area, the similarities (0.42 ± 0.08) between the bacterial communities from these boreholes (W1 and W3) were lower than those between bacterial communities from livestock wastewater and borehole W1 (0.56 ± 0.04). In our previous study (12), borehole W1 was found to be heavily contaminated with nitrate and various pollution indicator bacteria due to the infiltration of livestock wastewater after rainfall. These results indicate that the bacterial community of the aquifer located downstream from a livestock wastewater dump site has been influenced by the input of livestock wastewater into the aquifer, while borehole W3 as an uncontaminated control located upstream from a livestock wastewater dump site was not influenced by it. However, by estimating only similarity values between clone libraries, it is impossible to determine whether the input of livestock wastewater caused an increase in bacterial-community diversity or a decrease. Therefore, diversity index values were calculated for each clone library and compared among clone libraries.
Calculated RFLP pattern richness, Shannon-Weaver diversity, evenness, equitability, and Simpson's dominance index based on 16S rDNA RFLP patterns showed distinct patterns among the clone libraries. The overall patterns of estimated diversity in bacterial communities showed clearly that the bacterial communities of the livestock wastewater and borehole-W1 samples were much more diverse than those of the borehole-W3 samples in terms of richness, evenness, and the Shannon-Weaver index. The lower diversity observed in borehole W3 was due to the higher dominance of three RFLP types (WJGRT-1, WJGRT-2, and WJGRT-6) which comprise 60.5% of the total clones analyzed. Therefore, the Simpson's dominance index value in the W3 clone libraries was much higher than those in the W1 and LW clone libraries. It can be deduced from these results that the bacterial community of the aquifer became more diverse as livestock wastewater containing diverse microbial flora and a high concentration of nutrients infiltrated into the aquifer.
Subsurface bacteria show limited physiological and biochemical diversity (24), and hence the microbial community in a subsurface environment is less complex than those in near-surface environments, showing the strong selectivity of physically controlled ecosystems (10). In contrast, animal manure itself contains a large number of microorganisms that originate from gut microflora or rumen microflora, which have great diversity (21, 52, 63). Bacterial-community diversity generally is low under stressed conditions, as in soils polluted by chemical contaminants (4). Also, Torsvik et al. (57) suggested that agricultural management, fish farming, and pollution may lead to profound changes in community structures and reduction in bacterial diversity. However, it was assumed that the infiltration of livestock wastewater into an aquifer did not act as a stress because a greater increase in community diversity was found in an aquifer contaminated with livestock wastewater.
In aquifers into which livestock wastewater, with its diverse microflora, is introduced, the diversity of the bacterial community may greatly increase. Bacteria can colonize aquifer environments by active or passive migration via percolation from the surface soil and lateral movement from the recharge areas (22, 40). Relatively rapid migration of allochthonous bacterial populations occurs in large fractures and fissures. The aquifers studied in this research contained a large number of fractures distributed rather uniformly and had a high hydraulic conductivity (38); therefore, bacterial populations originating from livestock wastewater can migrate rapidly into the aquifer. It remains open, however, whether bacterial populations introduced into the aquifer successfully adapt to the groundwater ecosystems and colonize them or are defeated in competition and die off. Because 16S rDNAs were amplified from total community DNA, RFLP patterns in clone libraries themselves do not represent sequences of live cells but rather sequences of total cells, including dead cells and even DNA fragments released from dead cells. Some introduced aerobic bacterial populations might adapt well to aerobic aquifers; however, that rumen-related bacteria would so adapt is doubtful, since these are strictly anaerobic organisms, found almost exclusively in the guts of warm-blooded animals. Therefore, the detection of sequences of rumen-related bacteria in the groundwater was likely due to persistence of DNA rather than adaptation.
Finally, phylogenetic analyses of the cloned sequences were carried out to define bacterial populations responsible for changes in bacterial-community diversity in groundwater. This is the first study, to our knowledge, to define the bacterial populations responsible for the contamination of groundwater and the shift in community diversity by using molecular biological methods in a pollution study. This approach clearly showed that mainly the Cytophaga-Flexibacter-Bacteroides group and the low-G+C gram-positive group are associated with the changes in the bacterial community of the aquifer contaminated with livestock wastewater in the Wonju stock-farming area. There were significant (P < 0.001 by t-test) differences in the abundances of the Cytophaga-Flexibacter-Bacteroides group, the low-G+C gram-positive group, and WJG group 1 between the contaminated borehole (W1) and the uncontaminated borehole (W3). Introduction of the Cytophaga-Flexibacter-Bacteroides group and low-G+C gram-positive group into the groundwater ecosystem was apparent from the result that livestock wastewater and borehole-W1 samples showed higher abundances of the Cytophaga-Flexibacter-Bacteroides group and the low-G+C gram-positive group, while borehole-W3 samples showed lower abundances. More obviously, six of the clones assigned to the Cytophaga-Flexibacter-Bacteroides group and three of the clones assigned to the low-G+C gram-positive group were related to unidentified rumen bacterial sequences obtained from rumen fluid (54, 59) (Fig. 6). In addition, these sequences were not found in the uncontaminated borehole, W3, while they were found in borehole W1 and livestock wastewater. Members of the Cytophaga-Flexibacter-Bacteroides and low-G+C gram-positive groups are often reported as being among the most numerous culturable or uncultivated microbes present in the rumen or gut. Therefore, sequences related to the rumen bacteria found in borehole W1 must have originated from livestock wastewater containing a variety of rumen microorganisms. From this perspective, rumen-related sequences can be used as correct indicator sequences of fecal contamination of groundwater, particularly from livestock.
Besides the Cytophaga-Flexibacter-Bacteroides and low-G+C gram-positive groups, sequences related to nitrogen cycling were found only in W1 and/or LW clone libraries; they were not found in W3 clone libraries. In the α-Proteobacteria subdivision, sequences related to nitrite oxidizers (Nitrobacter winogradskyi) were found. In the β-Proteobacteria subdivision, sequences related to the β-group ammonium oxidizers (Nitrosomonas spp. and Nitrosospira spp.) and Fe-oxidizing denitrifying bacteria (U51102) were found. In addition, a sequence related to the Nitrospira moscoviensis subgroup was found in the W1 clone libraries. Livestock wastewater contains high concentrations of inorganic and organic nitrogen compounds. It has been reported that ammonium N is oxidized to nitrate N in septic tanks, wastewater stabilization ponds, and a cow slurry storage lagoon (23, 62). Also, our previous study on the flux of nitrogen compounds suggested that nitrification of ammonium N actively occurred in a livestock wastewater-stabilizing lagoon and that denitrification occurred in an aquifer as a change to anoxic conditions following the introduction of livestock wastewater containing high concentrations of carbon substrates (12). The occurrence of sequences related to nitrogen cycling in the borehole-W1 and livestock wastewater samples also supports our previous findings.
In summary, comparative study of 16S rDNA sequences using diversity indices and phylogenetic analysis showed that the diversity of the bacterial community in groundwater increased significantly following the input of livestock wastewater into an aquifer and that this increase of diversity was mainly due to the Cytophaga-Flexibacter-Bacteroides and low-G+C gram-positive groups originating from animals. Especially, this study showed that rumen-related sequences could be indicator sequences of fecal contamination of groundwater by livestock wastewater. Further studies are required to directly detect indicator sequences in groundwater and to trace the survival or adaptation of microorganisms introduced into aquifers.
ACKNOWLEDGMENTS
We are grateful to the Rural Development Corporation (RDC) of Korea for allowing us to use the well field in Wonju and to Daniel R. Noguera (University of Wisconsin, Madison) for very helpful comments. We thank Hack Sung Jung for generous hospitality, providing us with the opportunity to use laboratory facilities during the study, and Kyu-Ho Lee, Dong-Hun Lee, Myeong-Woon Kim, and Soon-Gyu Hong for particularly helpful discussions.
This work was supported by the G-7 Projects Grant from the Ministry of Environment of the Republic of Korea.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer K-H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlas R M, Horowitz A, Krichevsky M, Bej A K. Response of microbial populations to environmental disturbance. Microb Ecol. 1991;22:249–256. doi: 10.1007/BF02540227. [DOI] [PubMed] [Google Scholar]
- 5.Barns S M, Takala S L, Kuske C R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berardesco G, Dyhrman S, Gallagher E, Shiaris M P. Spatial and temporal variation of phenanthrene-degrading bacteria in intertidal sediments. Appl Environ Microbiol. 1998;64:2560–2565. doi: 10.1128/aem.64.7.2560-2565.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnie L M, Cole J R, Parker C T, Garrity J G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman J P, McCammon S A, Brown M V, Nichols D S, McMeekin T A. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol. 1997;63:3068–3078. doi: 10.1128/aem.63.8.3068-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter-Boggs L, Kennedy A C, Reganold J P. Use of phospholipid fatty acids and carbon source utilization patterns to track microbial community succession in developing compost. Appl Environ Microbiol. 1998;64:4062–4064. doi: 10.1128/aem.64.10.4062-4064.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler D P, Li S, Spadoni C M, Drake G R, Balkwill D L, Fredrickson J K, Brockman F J. A molecular comparison of culturable aerobic heterotrophic bacteria and 16S rDNA clones derived from a deep subsurface sediment. FEMS Microbiol Ecol. 1997;23:131–144. [Google Scholar]
- 11.Cho J-C, Kim S-J. Viable, but non-culturable, state of a green fluorescence protein-tagged environmental isolate of Salmonella typhi in groundwater and pond water. FEMS Microbiol Lett. 1999;170:257–264. doi: 10.1111/j.1574-6968.1999.tb13382.x. [DOI] [PubMed] [Google Scholar]
- 12.Cho, J.-C., H. B. Cho, and S.-J. Kim. Heavy contamination of a subsurface aquifer and a stream by livestock wastewater in a stock farming area, Wonju, Korea. Environ. Pollut., in press. [DOI] [PubMed]
- 13.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 14.Del Val C, Barea J M, Azcón-Aguilar C. Diversity of arbuscular mycorrhizal fungus populations in heavy-metal-contaminated soils. Appl Environ Microbiol. 1999;65:718–723. doi: 10.1128/aem.65.2.718-723.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunbar J, Takala S, Barns S M, Davis J A, Kuske C R. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol. 1999;65:1662–1669. doi: 10.1128/aem.65.4.1662-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichner C A, Erb R W, Timmis K N, Wagner-Döbler I. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl Environ Microbiol. 1999;65:102–109. doi: 10.1128/aem.65.1.102-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Embley T M, Stackebrandt E. The use of 16S rRNA sequences in microbial ecology. In: Pickup W, Saunders J R, editors. Molecular approaches in environmental microbiology. London, United Kingdom: Ellis Horwood; 1996. pp. 39–62. [Google Scholar]
- 19.Felske A, Akkermans A D L, DeVos W M. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl Environ Microbiol. 1998;64:4581–4587. doi: 10.1128/aem.64.11.4581-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felske A, Wolterink A, Lis R V, Akkermans A D L. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands) Appl Environ Microbiol. 1998;64:871–879. doi: 10.1128/aem.64.3.871-879.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frostegård Å, Petersen S O, Bååth E, Nielsen T H. Dynamics of a microbial community associated with manure hot spots as revealed by phospholipid fatty acid analyses. Appl Environ Microbiol. 1997;63:2224–2231. doi: 10.1128/aem.63.6.2224-2231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghiorse W C, Wilson J T. Microbial ecology of the terrestrial subsurface. Adv Appl Microbiol. 1988;33:107–172. doi: 10.1016/s0065-2164(08)70206-5. [DOI] [PubMed] [Google Scholar]
- 23.Goody D C, Withers P J A, McDonald H M, Chilton P J. Behaviour and impact of cow slurry beneath a storage lagoon. II. Chemical composition of chalk porewater after 18 years. Water Air Soil Pollut. 1998;107:51–72. [Google Scholar]
- 24.Haldeman D L, Amy P S. Diversity within a colony morphotype: implications for ecological research. Appl Environ Microbiol. 1993;59:933–935. doi: 10.1128/aem.59.3.933-935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastings R C, Ceccherini M T, Miclaus N, Saunders J R, Bazzicalupo M, McCarthy A J. Direct molecular biological analysis of ammonia oxidizing bacteria populations in cultivated soil plots treated with swine manure. FEMS Microbiol Ecol. 1997;23:45–54. [Google Scholar]
- 26.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 27.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurlbert S H. The non-concept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 29.Insam H, Amor K, Renner M, Crepaz C. Changes in functional abilities of the microbial community during composting of manure. Microb Ecol. 1996;31:77–87. doi: 10.1007/BF00175077. [DOI] [PubMed] [Google Scholar]
- 30.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 31.Ka J O, Holben W E, Tiedje J M. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils Appl. Environ Microbiol. 1994;60:1106–1115. doi: 10.1128/aem.60.4.1106-1115.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalchuk G A, Stephen J R, Boer W D, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowalchuk G A, Naoumenko Z S, Derikx P J L, Felske A, Stephen J R, Arkhipchenko I A. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl Environ Microbiol. 1999;65:396–403. doi: 10.1128/aem.65.2.396-403.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krebs C J. Ecological methodology. New York, N.Y: Harper & Row; 1989. [Google Scholar]
- 35.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 37.Lee D-H, Zo Y-G, Kim S-J. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand-conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J-Y, Lee K-K. Analysis of the quality of parameter estimates from repeated pumping and slug tests in a fractured porous aquifer system in Wonju, Korea. Ground Water. 1999;37:692–700. doi: 10.1111/j.1745-6584.1999.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 39.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 40.Madson E L, Ghiorse W C. Groundwater microbiology: subsurface ecosystem process. In: Ford T E, editor. Aquatic microbiology: an ecological approach. Boston, Mass: Blackwell Sciences; 1993. pp. 167–213. [Google Scholar]
- 41.Martinez-Murcis A J, Acinas S G, Rodriguez-Valeria F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol. 1995;14:219–232. [Google Scholar]
- 42.McCaig A E, Glover L A, Prosser J I. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol. 1999;65:1721–1730. doi: 10.1128/aem.65.4.1721-1730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDougald D, Rice S A, Weichart D, Kjelleberg S. Nonculturability: adaptation or debilitation? FEMS Microbiol Ecol. 1998;25:1–9. [Google Scholar]
- 44.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muyzer G, DeWaal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nübel U, Garcia-Pichel F, Kühl M, Muyzer G. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl Environ Microbiol. 1999;65:422–430. doi: 10.1128/aem.65.2.422-430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nüsslein K, Tiedje J M. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl Environ Microbiol. 1998;64:1283–1289. doi: 10.1128/aem.64.4.1283-1289.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odum E P. Basic ecology. Philadelphia, Pa: Saunders College Publishing; 1983. [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Sanders H L. Marine benthic diversity: a comparative study. Am Nat. 1968;102:243–282. [Google Scholar]
- 51.Schwieger F, Tebbe C C. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol. 1998;64:4870–4876. doi: 10.1128/aem.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spoelstra S F. Enumeration and isolation of anaerobic microbiota of piggery wastes. Appl Environ Microbiol. 1978;35:841–846. doi: 10.1128/aem.35.5.841-846.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staley J T. Biodiversity: are microbial species threatened? Curr Opin Biotechnol. 1997;8:340–345. doi: 10.1016/s0958-1669(97)80014-6. [DOI] [PubMed] [Google Scholar]
- 54.Tajima, Aminov K R I, Nagamine T, Ogata K, Nakamura M, Matsui H, Benno Y. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol Ecol. 1999;29:159–169. [Google Scholar]
- 55.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torsvik V, Salte K, Sørheim R, Goksøyr J. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol. 1990;56:776–781. doi: 10.1128/aem.56.3.776-781.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torsvik V, Daae F L, Sandaa R-A, Øvreås L. Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol. 1998;64:53–62. doi: 10.1016/s0168-1656(98)00103-5. [DOI] [PubMed] [Google Scholar]
- 58.Ward D M. A natural species concept for prokaryotes. Curr Opin Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. [DOI] [PubMed] [Google Scholar]
- 59.Whitford M F, Foster R J, Beard C E, Gong J, Teather R M. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe. 1998;4:153–163. doi: 10.1006/anae.1998.0155. [DOI] [PubMed] [Google Scholar]
- 60.Wintzingerode F V, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 61.Wise M G, McArthur J V, Shimkets L J. Bacterial diversity of a Carolina bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl Environ Microbiol. 1997;63:1505–1514. doi: 10.1128/aem.63.4.1505-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Withers P J A, McDonald H M, Smith K A, Chumbley C. Behaviour and impact of cow slurry beneath a storage lagoon. I. Groundwater contamination 1975–1982. Water Air Soil Pollut. 1998;107:35–49. [Google Scholar]
- 63.Wolin M J. The rumen fermentation: a model for interactions in anaerobic ecosystems. Adv Microb Ecol. 1979;3:49–78. [Google Scholar]