Abstract
Thermoanaerobacter ethanolicus is a gram-positive thermophile that produces considerable amounts of ethanol from soluble sugars and polymeric substrates, including starch. Growth on maltose, a product of starch hydrolysis, was associated with the production of a prominent membrane-associated protein that had an apparent molecular weight of 43,800 and was not detected in cells grown on xylose or glucose. Filter-binding assays revealed that cell membranes bound maltose with high affinity. Metabolic labeling of T. ethanolicus maltose-grown cells with [14C]palmitic acid showed that this protein was posttranslationally acylated. A maltose-binding protein was purified by using an amylose resin affinity column, and the binding constant was 270 nM. Since maltase activity was found only in the cytosol of fractionated cells and unlabeled glucose did not compete with radiolabeled maltose for uptake in whole cells, it appeared that maltose was transported intact. In whole-cell transport assays, the affinity for maltose was approximately 40 nM. Maltotriose and α-trehalose competitively inhibited maltose uptake in transport assays, whereas glucose, cellobiose, and a range of disaccharides had little effect. Based on these results, it appears that T. ethanolicus possesses a high-affinity, ABC type transport system that is specific for maltose, maltotriose, and α-trehalose.
Thermoanaerobacter ethanolicus is a gram-positive anaerobic thermophile (11) that produces considerable amounts of ethanol from a wide range of polymeric and soluble carbohydrates (15, 24, 25). The physiology of T. ethanolicus type strain JW200 and strain 39E has been studied in some detail, and the high specific rate of ethanol production makes this species an attractive candidate for use in bioconversion processes (7–9). Starch is a potentially useful substrate for biomass conversion because of its availability and relatively low cost (26). The physiology (24) and enzymology (7) of starch breakdown in T. ethanolicus have been studied previously. However, there are still many gaps in our knowledge concerning fundamental processes, such as substrate transport. T. ethanolicus grows more slowly on maltose, cellobiose, or starch than on glucose (16, 23, 25) and apparently lacks disaccharide phosphorylase activities possessed by other thermophiles (16); it was therefore hypothesized that in this organism a glucose permease is responsible for uptake of monosaccharides derived from extracellularly degraded maltose and starch (7). However, there has been no systematic comparison of the substrate transport systems of T. ethanolicus cultures grown on glucose, maltose, or starch, and an alternative hypothesis is that the differences in growth rates observed are due to discrete sugar transport systems for mono- and disaccharides.
Maltose transport systems in mesophilic bacteria have been widely studied (2), and several systems in thermophiles have been characterized to different degrees (12, 19, 27). The thermophilic transporters that have been examined in thermophiles are binding-protein-dependent systems, which constitute a subfamily within the ABC transporter superfamily (5). Each of these systems is composed of a periplasmic or membrane-associated binding protein that transfers the substrate to integral membrane components which mediate ATP-dependent uptake. Substrate specificity is mediated in large measure by the binding protein (2). Since little information concerning sugar transport mechanisms in T. ethanolicus is available, we decided to determine whether T. ethanolicus possesses a discrete uptake system for maltose. In this paper we describe biochemical characterization of a discrete maltose/maltotriose-binding protein and an associated transport system that recognizes α-linked oligomers in T. ethanolicus.
MATERIALS AND METHODS
Cell growth.
T. ethanolicus 39E (= ATCC 33323) was obtained from the American Type Culture Collection (Rockville, Md.) and was grown in a basal medium as previously described (4) at 70°C with a carbon source supplied at a concentration of 4 g/liter unless otherwise noted. Cell growth was measured spectrophometrically at 600 nm.
Analyses.
Maltose and glucose concentrations in culture medium were determined enzymatically as previously described (13). The protein contents of samples were determined by the method of Lowry et al. (14) after the samples were boiled in 0.1 N NaOH for 20 min. Bovine serum albumin (catalog no. A4503; Sigma Chemical Co., St. Louis, Mo.) was used as the standard. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed by standard procedures (10).
Whole-cell transport assays.
Cultures (10 ml) were harvested anaerobically by centrifugation (10,000 × g, 5 min, 25°C) during mid-exponential growth and were washed twice with anaerobically prepared 50 mM potassium phosphate (pH 7.0) containing 10 mM MgCl2. The cells were resuspended in the same buffer to a protein concentration of approximately 6 μg/ml. Individual lots of cells (100 μl) were prewarmed by incubating them at 70°C for 1 min, and sugar uptake was initiated by adding radiolabeled [14C]maltose (360 mCi/mmol). Uptake was terminated after 5, 10, and 15 s by diluting the cells with 2 ml of cold 100 mM LiCl and immediately filtering the preparations through 0.45-μm-pore-size nitrocellulose membrane filters. The filters were dried at 105°C for 20 min, and radioactivity was determined by liquid scintillation counting (Econo-Safe; RPI Corp., Mount Prospect, Ill.). All uptake values were calculated after subtraction of the value for a control reaction that was immediately terminated after the radioisotope was added. Preliminary experiments established that uptake was linear for 10 s when radiolabeled maltose was provided at concentrations ranging from 2.5 to 100 nM and that the adherence of radiolabeled maltose to nitrocellulose filters in the absence of cells was negligible. Uptake values were transformed by using a Lineweaver-Burk double-reciprocal plot to calculate the apparent transport affinity (Km) and Vmax for maltose transport. Competition studies were performed by providing radiolabeled maltose at concentrations of 5, 10, 25, 50, 100, and 200 nM and measuring the initial rates of uptake in the presence of each of the following unlabeled sugars at concentrations between 40 and 200 nM: α-trehalose, maltotriose, maltotetraose, and maltopentaose. The apparent inhibition constant (KI) was calculated for each unlabeled saccharide from a plot of (Km/Vmax)×(1 + I/KI) versus I (where I is the unlabeled sugar concentration).
Enzyme assays.
Maltase activity was measured in a discontinuous fashion by using the same fractions that were used for partial purification of the binding protein. Each initial reaction mixture contained approximately 20 μg of protein, 50 mM piperazine-N,N′-bis-2(ethanesulfonic acid) (PIPES) (pH 6.8), and 20 mM maltose. The reaction mixture (1 ml) was incubated at 70°C for 2 min, and samples (250 μl) were removed at various times. The glucose concentrations in the samples were then determined by determining NADPH formation in an end point spectrophotometric assay (340 nm) at 37°C; each assay mixture contained 50 mM PIPES (pH 6.8), 3 mM MgCl2, 1 mM ATP, 1 mM NADP, 5 U of hexokinase (EC 2.7.1.1), and 5 U of glucose-6-phosphate dehydrogenase (EC 1.1.1.49) (13). The control reaction mixtures lacked substrate or crude extract. Since maltase activity was not detected in the culture supernatant of cells growing on maltose, the extracellular protein present in the supernatant of an 80-ml culture was concentrated. Approximately 100 μg of protein was precipitated with a saturated ammonium sulfate solution at 4°C for 1 h, pelleted by centrifugation (32,000 × g, 20 min, 4°C), and then resuspended in 100 μl of 50 mM Tris-HCl (pH 7.5). The sample was then dialyzed overnight against 5 liters of 50 mM Tris-HCl (pH 7.5) at 4°C before the assay was performed.
Purification of binding protein.
Cells (5 liters) were harvested by centrifugation (10,000 × g, 20 min, 4°C) during late-exponential growth (optical density at 600 nm [OD600], 0.6), washed twice with 50 mM Tris-HCl (pH 7.5), resuspended in the same buffer to an OD600 of 50, and then frozen at −80°C. Cells prepared and stored in this manner retained binding activity for at least 6 months (data not shown). A 2-ml sample of concentrated cells (approximately 30 mg of protein) was thawed and passed twice through a chilled French pressure cell (1,200 kg/cm2). Whole cells and cellular debris were removed by centrifugation (32,000 × g, 30 min, 4°C), which resulted in a crude extract fraction.
The crude extract fraction was used for localization and initial identification of the maltose-binding protein. It was fractionated by centrifugation (105,000 × g, 60 min, 4°C) into cytosolic and membrane fractions. The membrane pellet was washed twice with 50 mM Tris-HCl (pH 7.5) and resuspended in 700 μl of the same buffer containing 1% octyl-β-glucoside. The membranes were extracted by gentle agitation for 75 min at 4°C. The resulting mixture was centrifuged (105,000 × g, 60 min, 4°C), and the supernatant fraction containing the solubilized binding protein was retained and used for the assay.
The maltose-binding protein was purified by performing affinity chromatography with an amylose resin column (New England Biolabs, Beverly, Mass.). The column (15 ml of resin) was prepared, chromatographed, and regenerated as recommended by the manufacturer. All steps were carried out at 4°C. Our initial attempts to purify the maltose-binding protein from the membrane fractions were unsuccessful (data not shown). However, preliminary experiments revealed that the maltose-binding activity present in crude extracts bound to the amylose resin. Therefore, freshly prepared crude extract (170 mg of protein) was loaded onto the affinity column, and proteins that did not bind to the amylose resin were eluted with 12 column volumes of 20 mM Tris-HCl (pH 7.4) containing 200 mM NaCl and 1 mM EDTA. Maltose-binding activity was then eluted with the same buffer supplemented with 10 mM maltose. The fractions that contained the most binding protein (as estimated by SDS-PAGE) were pooled and concentrated by using a centrifugal filter (10,000-molecular-weight cutoff; MSI, Westboro, Mass.). The concentrated fraction was dialyzed to reduce the maltose concentration to less than 10 nM prior to reapplication to the amylose affinity resin. The fractions containing the most binding activity after elution were again pooled, concentrated, and dialyzed.
Maltose-binding assay.
The maltose-binding activities in various cell fractions were determined essentially as described previously (17). Crude extract, cytosol, or membrane fractions (100 μl) and pooled, concentrated, and dialyzed fractions obtained from the affinity chromatography procedure (2 μl) were prewarmed in 50 mM Tris-HCl (pH 7.5) for 30 s at 70°C before radiolabeled d-[14C]maltose (final concentration, 0.69 μM; 360 mCi/mmol) was added. The reaction mixtures were then placed in an ice bath and diluted with 2 ml of ice-cold 50 mM Tris-HCl (pH 7.5) saturated with ammonium sulfate. After 10 min of incubation, samples were passed through a 0.45-μm-pore-size nitrocellulose filter and washed with an additional 2 ml of the Tris-ammonium sulfate solution. The filters were then dried for 20 min at 105°C, and the radioactivity in each sample was determined by scintillation counting. Preliminary experiments revealed that there was linearity with respect to protein concentration, and there was negligible nonspecific adsorption (<5%) of radiolabeled maltose to filters in the absence of protein.
Detergent-extracted membrane samples were used to investigate the effect of temperature and pH on maltose binding. In these experiments, reaction mixtures were incubated at the different temperatures or pH values for 5 min before radiolabeled sugar was added. Sugar competition studies were performed with the extracted membranes by adding a 50-fold excess (34.5 μM) of unlabeled sugar to the reaction mixture 5 s before radiolabeled maltose was added. Maltose and maltooligomers were obtained from Sigma Chemical Co. The purity of the maltooligomers was sufficiently high that any contaminating maltose did not significantly affect the competition experiments.
Equilibrium dialysis.
An assay based on the retention of ligand by binding protein (20) was used to determine the binding affinity (Kd) of the maltose-binding protein. Purified binding protein from the affinity chromatography experiments was diluted with 50 mM Tris-HCl (pH 7.5) to a concentration of 0.67 μM. A 100-μl sample of the resulting solution was pipetted into a 10,000-molecular-weight cutoff Slide-a-Lyzer Mini dialysis unit (Pierce Chemical Co., Rockford, Ill.). An identical control unit contained only 50 mM Tris-HCl (pH 7.5). Both units were equilibrated for 16 h at 4°C against a 40-ml Tris-HCl solution containing [14C]maltose (594 mCi/mmol) at a final concentration of 198 nM. The dialysis units were then removed and placed in separate flasks containing 1 liter of 50 mM Tris-HCl (pH 7.5) that was gently stirred. Samples (10 μl) were removed from both units over time, and radioactivity was determined by liquid scintillation counting. The half-life of retention of ligand in the unit containing binding protein was theoretically greater by a factor of 1 + ([P]/Kd) (where [P] is the molar concentration of ligand) than the half-life of retention of ligand in the unit that did not contain any binding protein; Kd was expressed as a nanomolar concentration (20).
Labeling with [1-14C]palmitate.
Ten milliliters of medium containing either 0.4% maltose or glucose was inoculated to obtain an initial OD600 of approximately 0.1. The cultures were grown to an OD600 of 0.25, and then 10 μCi of [1-14C]palmitic acid (52 mCi/mmol) was added. Growth was allowed to continue until the OD600 reached 0.65. Cells were then harvested by centrifugation (10,000 × g, 20 min, 4°C), washed twice with 50 mM Tris-HCl (pH 7.5), and resuspended with 100 μl of lysis solution containing 50 mM Tris-HCl (pH 7.5), 2% SDS, and 0.1% 2-mercaptoethanol. Samples were incubated at 70°C for 15 min and centrifuged at 15,000 × g, 20 min, 25°C), and a sample (approximately 15 μg of protein in 25 μl) of each resulting supernatant was loaded onto a 10% polyacrylamide SDS-PAGE gel. Proteins were visualized by staining the gels with Coomassie brilliant blue and were fixed with 10% methanol before the gels were dried. Radiolabeled proteins were visualized by autoradiography after the gels were exposed to Kodak XOMAT LS film for 2 weeks.
Protein sequence analysis.
The proteins present in detergent-extracted membranes from maltose-grown cells were resolved by SDS-PAGE and then were electroblotted onto a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.). Protein alkylation, enzymatic cleavage, peptide purification, and peptide sequence analysis were performed at the Macromolecular Structure Analysis Facility at the University of Kentucky. Briefly, the putative maltose-binding protein (approximately 40 μg on a polyvinylidene difluoride membrane) was alkylated and then cleaved with 0.1 μg of endopeptidase LysC (Boehringer Mannheim, Indianapolis, Ind.) at 37°C for 18 h. The resulting peptides were separated by reverse-phase chromatography by using a model 1050 high-performance liquid chromatograph (Hewlett-Packard, Palo Alto, Calif.) equipped with a Vydec column (Vydec, Hesperia, Calif.). Absorbing peaks (214 nm) were collected manually, and one peak was used for amino acid sequence analysis. A peptide sequence analysis was performed with a model 477A peptide sequencer (Applied Biosystems, Foster City, Calif.).
RESULTS
Maltase activity in cell fractions.
Maltase activity (3.1 nmol/min/mg of protein) was detected in crude extracts derived from maltose-grown cells, but activity was not detected in glucose-grown cell extracts. After further fractionation, it was established that maltase activity (3.9 nmol/min/mg of protein) was present only in the cytosol. Activity was not detected in either the membrane samples or the extracellular culture fluid of maltose-grown cells. In addition, there was little glucose accumulation (<20 μM) in the extracellular culture fluid of T. ethanolicus logarithmic- or stationary-phase cells supplied with 11 mM maltose. Together, these data suggested that rather than maltose being cleaved to glucose by extracellular enzymes and then monomers being internalized via a glucose permease, as had been hypothesized previously (7), the disaccharide was actually transported intact across the cell membrane prior to intracellular cleavage.
Maltose transport by whole cells.
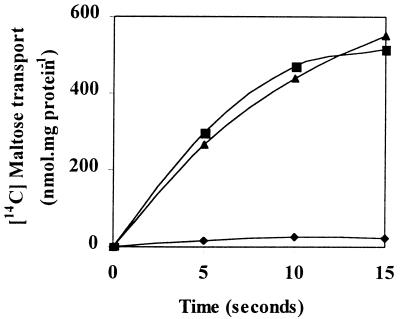
Given the lack of extracellular maltase activity, we hypothesized that T. ethanolicus possesses a transport system(s) that is specific for maltose uptake. To test this hypothesis, mid-logarithmic-growth-phase cells grown on maltose were used in uptake studies. These cells transported the disaccharide (Fig. 1), and no effect on radiolabeled maltose uptake was observed if a 50-fold excess of unlabeled glucose, galactose, sucrose, or lactose (data not shown) was added to the incubation mixture. In addition, there was virtually no maltose uptake in glucose-grown cells. Since 15 min of incubation of cells at 70°C prior to transport abolished 95% of the original activity, uptake appeared to be an energy-dependent process and not simply the result of maltose binding to the external surfaces of the cells.
FIG. 1.
Radiolabeled maltose uptake in maltose-grown (■) and glucose grown (⧫) cells. Uptake of the disaccharide by maltose-grown cells in the presence of a 50-fold excess of unlabeled glucose was also examined (▴). Uptake experiments were performed at 70°C with 100-μl reaction mixtures containing 25 nM radiolabeled maltose.
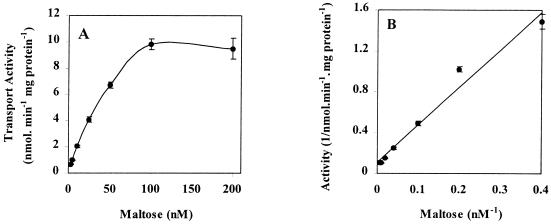
Incubation of cells with maltose at concentrations ranging from 2.5 to 200 nM revealed that the kinetics of uptake were saturable (Fig. 2A); it appeared that T. ethanolicus possessed a high-affinity (Km, 40 nM) system for maltose uptake (Fig. 2B). The results of an Eadie-Hofstee transformation suggested that there was a single maltose transport system in this species (data not shown). Sugar competition studies showed that both maltotriose and α-trehalose strongly inhibited maltose uptake in a competitive manner (data not shown), and the KI values for these sugars were 48 and 51 nM, respectively. In contrast, the kinetic studies revealed mixed competition for maltotetraose and maltopentaose, whose KI values were 470 and 1,650 nM, respectively. This is the first quantitative data which indicates the relative affinities of maltose, malto-oligomers, and α-trehalose for a maltose transport system in a thermophile.
FIG. 2.
Kinetics of maltose transport in T. ethanolicus 39E. (A) Uptake experiments were performed at 70°C with 100-μl reaction mixtures containing radiolabeled maltose at concentrations ranging from 2.5 to 200 nM. (B) Double-reciprocal plot of the initial rate of radiolabeled maltose uptake versus maltose concentration in T. ethanolicus maltose-grown cells. The error bars indicate standard deviations based on three independent observations.
Purification of maltose-binding activity.
In general, bacterial and archaeal high-affinity transport systems, such as the maltose transport system in T. ethanolicus, achieve their high affinity in large measure due to binding proteins which are specific for the substrate and either are present in the periplasm or are associated with the cell surface. Therefore, we performed experiments to determine whether T. ethanolicus possesses a d-maltose-binding protein as one of the components of its high-affinity maltose transport system. Radiolabeled maltose was bound by crude extracts of cells grown on the disaccharide (Table 1), and this activity did not appear to be caused by nonspecific binding since extracts obtained from glucose-grown cells bound less than 5% as much maltose as extracts obtained from maltose-grown cells. In addition, maltose-binding activity was not detected in membrane fractions of cultures grown on glucose, xylose, sucrose, cellobiose, lactose, or mannitol (data not shown).
TABLE 1.
Localization and purification of a maltose-binding protein from T. ethanolicus
| Fraction | Amt of protein (mg) | Amt of maltose bound (nmol)a | Sp act (nmol/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Localization | |||||
| Crude extract | 15.0 | 2.2 | 0.15 | 1 | 100 |
| Cytosol | 11.9 | 0.8 | 0.07 | ||
| Membrane | 2.2 | 1.8 | 0.82 | 5.5 | 82 |
| Extracted membraneb | 1.4 | 1.3 | 0.93 | 6.2 | 59 |
| Purification | |||||
| Crude extract | 172 | 24.1 | 0.14 | 1 | 100 |
| Affinity fraction | 0.01 | 0.1 | 10.0 | 71.4 | 0.42 |
Binding activity was determined by a filter-binding assay. Fractions were prewarmed for 30 s at 70°C before [14C]maltose (final concentration, 0.69 μM; 360 mCi/mmol) was added.
Membranes were extracted with 1% octyl-β-glucoside for 75 min at 4°C.
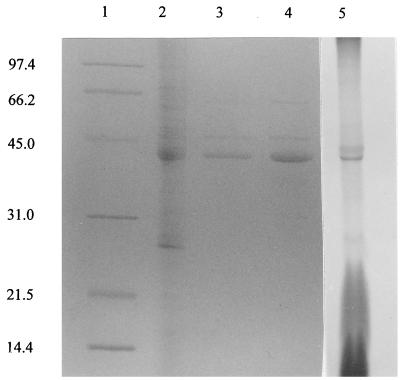
When extracts of maltose-grown cells were partitioned into cytosolic and membrane fractions, more than 80% of the total binding activity was associated with the membrane fraction (data not shown). This activity could be solubilized by 1% octyl-β-glucoside, and the detergent-extracted material appeared to be enriched for a protein with an apparent molecular weight of 43,800 (Fig. 3, lane 4). However, the specific activity for maltose binding was only sixfold greater than the specific activity of the crude extract (Table 1).
FIG. 3.
SDS-PAGE of T. ethanolicus cell fractions. Lane 1, molecular weight standards; lane 2, maltose-grown cell crude extract; lane 3, affinity-purified maltose-binding protein; lane 4, membranes derived from maltose-grown cells; lane 5, autoradiograph of crude cell lysate grown on maltose in the presence of [14C]palmitic acid. Lanes 2 and 4 contained approximately 20 μg of protein, and lane 3 contained approximately 7 μg of protein.
In an effort to purify the maltose-binding protein, detergent-extracted membrane fractions were passed over amylose affinity resin. However, the binding activity of these fractions was not retained by the resin (data not shown). In contrast, we found that the maltose-binding activity of crude extracts selectively bound to the amylose resin. Indeed, after two passes over the column, the maltose-binding specific activity of the preparation was more than 70-fold greater than that of the crude extract fraction (Table 1 and Fig. 3, lane 3).
Metabolic labeling and peptide sequencing.
Since T. ethanolicus has a gram-positive cell envelope and possesses a membrane-bound lipoprotein for xylose binding (4), it seemed reasonable to hypothesize that the putative maltose-binding protein is also a lipoprotein. Therefore, T. ethanolicus cells grown on either maltose or glucose were incubated with [14C]palmitate to label any lipoproteins. The results indicated that the prominent 43.8-kDa membrane-associated protein in maltose-grown cells (Fig. 3, lane 4) incorporated radiolabeled palmitate (Fig. 3, lane 5) and that this labeled protein was not present in membrane samples from glucose-grown cells (data not shown). Thus, the putative maltose-binding protein appeared to be lipidated and located on the T. ethanolicus cell surface.
Since the 43.8-kDa protein appeared in the membrane fraction of maltose-grown cells and since its presence was correlated with maltose-binding activity, it was partially sequenced to determine its identity. Since this protein was apparently lipidated as shown by the metabolic labeling experiment, most likely at the N terminus (21), it was probably resistant to N-terminal Edman degradation. Therefore, peptides were derived by cleavage with LysC and were subsequently purified by high-performance liquid chromatography. One of the resulting products was sequenced (Fig. 4), but no matches were found when this peptide was compared to sequences in the GenBank database by using the BLAST algorithim (1). This was not entirely unexpected since the query sequence was only 17 amino acids long and the maltose-binding proteins currently available from the GenBank database exhibit considerable sequence diversity (data not shown). However, when the peptide was individually aligned with several thermophilic maltose-binding proteins using ClustalW (22), we found that it exhibited 41% sequence identity and 82% similarity (Fig. 4) with a portion of the C-terminal region of Thermococcus litoralis MalE (gi2828820 [6]) and was less similar to maltose-binding proteins from Thermoanaerobacterium thermosulfurigenes (gi565382 [19]) and Thermotoga maritima (gi1850900 [12]) (data not shown). In fact, the C-terminal portion of the T. litoralis α-trehalose/maltose-binding protein exhibited a higher level of identity to the T. ethanolicus peptide sequence than to any other maltose-binding protein. This sequence comparison provided further evidence that the 43.8-kDa protein was probably a maltose-binding protein.
FIG. 4.
Alignment of the sequence of a peptide derived from digestion of the T. ethanolicus putative maltose-binding protein with the sequence of a portion of the C-terminal region of T. litoralis MalE (gi2828820). The alignment was generated by using ClustalW with a Blosum residue substitution matrix (22). An asterisk indicates that the amino acids in the two peptides are identical. A colon indicates residue conservation.
Biochemical characteristics.
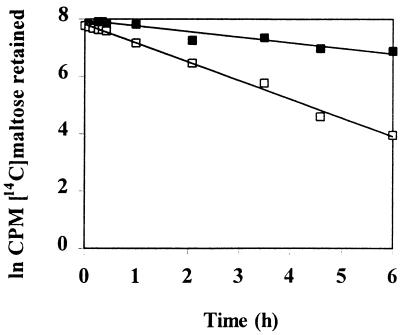
Equilibrium dialysis (20) was used to determine the binding affinity of the purified T. ethanolicus maltose-binding protein. In this assay, the rates at which a radiolabeled substrate left a dialysis membrane in the presence and in the absence of binding protein were compared (Fig. 5), and the Kd for maltose was 270 nM.
FIG. 5.
Determination of the binding affinity constant of the T. ethanolicus 39E maltose-binding protein, as measured by equilibrium dialysis. A solution containing 198 nM [14C]maltose (594 mCi/mmol) was used in the equilibration step. Exit of radiolabel from a 10,000-molecular-weight cutoff dialysis unit containing either purified binding protein at a concentration of 0.67 μM (■) or buffer alone (□) is shown.
Maltose-binding activity was also characterized by using the detergent-extracted membrane fraction and a membrane filter assay. As expected, a 50-fold excess of unlabeled maltose completely eliminated radiolabeled maltose binding (Table 2). In addition, a similar excess of the trisaccharide maltotriose or the disaccharide α-trehalose decreased binding of labeled maltose by more than 85%. These data are consistent with results obtained in the whole-cell uptake studies. Maltose binding was also decreased by adding excess maltotetraose and maltopentaose. However, binding activity was largely unaffected by unlabeled glucose, cellobiose, or β-trehalose.
TABLE 2.
Sugar specificity of T. ethanolicus maltose-binding protein
| Competing sugara | Relative maltose binding (%)b |
|---|---|
| Control | 100 |
| d-Maltose | 4 |
| α-Maltotriose | 12 |
| α-Maltotetraose | 60 |
| α-Maltopentaose | 52 |
| α-Trehalose | 14 |
| β-Trehalose | 105 |
| Glucose | 91 |
| Cellobiose | 95 |
Fractions were prewarmed at 70°C for 30 s before [14C]maltose (final concentration, 0.69 μM; 360 mCi/mmol) was added. A 50-fold excess of unlabeled sugar (34.5 μM) was provided.
The binding activity of the control was 0.87 nmol · mg of protein−1.
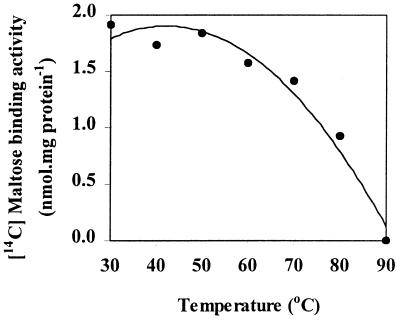
Using detergent-extracted membranes, we found that maltose-binding activity was relatively resistant to changes in pH and temperature. Binding activity varied less than 20% over a pH range from 4.5 to 8.5 (data not shown). Binding activity was constant at temperatures ranging from 30 to 70°C, but binding activity was dramatically decreased when extracts were incubated at temperatures greater than 70°C (Fig. 6). Thus, the maltose-binding activity in T. ethanolicus was less thermostable than the xylose-binding activity in the same organism (4).
FIG. 6.
Effect of temperature on maltose binding by detergent-extracted membranes derived from maltose-grown T. ethanolicus. Fractions were prewarmed at the appropriate temperature for 5 min before [14C]maltose (final concentration, 0.69 μM; 360 mCi/mmol) was added.
DISCUSSION
The elucidation of mechanisms by which thermophilic bacteria utilize maltose during starch degradation is of interest with regard to the use of these microorganisms in production of vendable chemicals (15, 26). The amylosaccharide metabolism pathways in T. ethanolicus 39E (formerly Clostridium thermohydrosulfuricum) were investigated in detail by Hyun et al. (7). These authors concluded that maltose and other soluble dextrins derived from starch are hydrolyzed to glucose by an array of extracellular enzymes before the monosaccharide is taken up via a glucose permease. This hypothesis apparently was confirmed by the observation that glucose accumulated in the extracellular culture fluid of cells grown in the presence of high concentrations of maltose or starch. However, since all enzyme activities in the study of Hyun et al. were measured by using crude cell extracts, the cellular locations of the activities could not be definitively established. In addition, there was no attempt to determine whether T. ethanolicus actually possessed only a glucose transport system or whether there were also mechanisms for transporting oligomers that arise from starch degradation.
In contrast to the previous studies of maltose utilization by T. ethanolicus, we found no evidence that glucose accumulates in the extracellular culture fluid of cells growing on maltose. Additionally, maltase activity was not detected either in membranes of maltose-grown T. ethanolicus cells or in the extracellular culture fluid; maltase activity was found only in the cytosolic fraction. Finally, radiolabeled maltose uptake was specifically associated with maltose-grown cells, and uptake was not affected by glucose. Therefore, our results support the hypothesis that maltose is transported intact across the cell membrane prior to cleavage by cytoplasmic maltase activity. The observations described above show that a thorough analysis of both the biochemistry and the genetics of maltose uptake is particularly important in developing a more complete understanding of the starch bioconversion process in T. ethanolicus.
As appears to be the case in T. ethanolicus, maltose transport is mediated by binding-protein-dependent systems in the thermophiles T. litoralis (27) and T. thermosulfurigenes (19). The affinity of the T. ethanolicus transport system for maltose is very similar to the affinity of the maltose-binding protein of T. litoralis (40 and 20 nM, respectively). In addition, α-trehalose is a common substrate for each transporter in these two organisms. Accumulation of α-trehalose has been mentioned as a possible osmoprotective mechanism when an organism encounters high salt conditions (6). However, in other respects, the T. ethanolicus system appears to be different from the systems in other thermophiles. Unlike the T. ethanolicus system, maltotriose does not appear to be a substrate for binding or transport by the T. litoralis MalEFG transport system (6, 27). Maltotriose has been reported to be a possible substrate for the T. thermosulfurigenes maltose transporter, but no kinetic data was presented (19). Maltose was also transported with a higher affinity in T. ethanolicus than in T. thermosulfurigenes (40 nM versus 7 μM). In contrast to the kinetics of transport of other sugars in T. thermosulfurigenes (19), Thermoanaerobacter thermosulfuricus (3), and T. ethanolicus (4), the kinetics of maltose transport in T. ethanolicus are monophasic. The substrate specificity of the T. ethanolicus maltose transport system appears to be more similar to the substrate specificities of mesophilic systems exemplified by Escherichia coli, in which the maltose-binding protein binds both maltose and maltotriose with similar affinities, albeit in the micromolar range (2). This is the first study in which the relative affinities of substrates for a maltose transport system in a thermophile were quantified.
Since the Km of whole-cell transport was 40 nM, it was somewhat surprising that the binding constant of the purified protein was 270 nM. However, an eightfold difference between whole-cell transport affinity and the binding constant of T. litoralis MalE was noted by Horlacher et al. (6). This difference was attributed to the nature of the interaction of the binding protein with the membrane components of the maltose transport system (6). The 43.8-kDa membrane protein specifically associated with maltose-grown cells undergoes fatty acyl modification in the presence of radiolabeled palmitate, and a peptide derived from the protein was found to have a sequence very similar to the sequence of a portion of the C-terminal region of the T. litoralis α-trehalose/maltose-binding protein (6).
Together, our results provide presumptive evidence that the maltose-binding protein in T. ethanolicus is covalently attached to the cell membrane, as are other binding proteins of gram-positive organisms that have been characterized, including binding proteins of T. ethanolicus (4, 18, 20). Maltose-binding activity in T. ethanolicus was remarkably insensitive to changes in temperature. In contrast, in T. litoralis there was no significant maltose-binding activity at room temperature (6). Future crystallographic studies should provide interesting comparative data for the active sites and overall structures of maltose-binding proteins from mesophilic and thermophilic organisms.
Our description of maltose transport and a binding protein in T. ethanolicus is the first detailed characterization of a maltose/maltotriose transport system in a thermophile, and we found that T. ethanolicus possesses a functional ATP-dependent transport system for maltose. Short, degenerate oligonucleotide probes obtained from reverse translation of the known sequence of the T. ethanolicus maltose-binding protein and degenerate PCR primers based on alignment of the conserved sequences of MalG, one of the membrane-bound components of the maltose ABC transport system (2, 5), are currently being used to clone and analyze the components that encode the maltose ABC transporter in T. ethanolicus.
ACKNOWLEDGMENTS
This work was supported by Hatch project KY007007. C.R.J. was supported by a University of Kentucky Presidential Graduate Fellowship.
Footnotes
Published with the approval of the Director of the Kentucky Agricultural Experiment Station as Journal article no. 00-07-7.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boos W, Shuman H. Maltose/maltodextrin system of Escherichia coli: transport, metabolism and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook G M, Janssen P H, Russell J B, Morgan H W. Dual mechanisms of xylose uptake in the thermophilic bacterium Thermoanaerobacter thermosulfuricus. FEMS Microbiol Rev. 1994;116:257–262. [Google Scholar]
- 4.Erbeznik M, Dawson K A, Strobel H J, Jones C R. d-Xylose-binding protein, XylF, from Thermoanaerobacter ethanolicus 39E: cloning, molecular analysis, and expression of structural gene. J Bacteriol. 1998;180:3570–3577. doi: 10.1128/jb.180.14.3570-3577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 6.Horlacher R, Xavier K B, Santos S, DiRuggiero J, Kossman M, Boos W. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1998;180:680–689. doi: 10.1128/jb.180.3.680-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyun H H, Shen G-J, Zeikus J G. Differential amylosaccharide metabolism of Clostridium thermosulfurigenes and Clostridium thermohydrosulfuricum. J Bacteriol. 1985;164:1153–1161. doi: 10.1128/jb.164.3.1153-1161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lacis L S, Lawford H G. Thermoanaerobacter ethanolicus in a comparison of the growth efficiencies of thermophilic and mesophilic anaerobes. J Bacteriol. 1985;163:1275–1278. doi: 10.1128/jb.163.3.1275-1278.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacis L S, Lawford H G. Strain selection in carbon-limited chemostats affects reproducibility of Thermoanaerobacter ethanolicus fermentations. Appl Environ Microbiol. 1992;58:761–764. doi: 10.1128/aem.58.2.761-764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemelli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y-E, Jain M K, Lee C, Lowe S E, Zeikus J G. Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurigenes, and Clostridium thermohydrosulfuricum E100-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int J Syst Bacteriol. 1993;43:41–51. [Google Scholar]
- 12.Liebl W, Stemplinger I, Ruile P. Properties and gene structure of the Thermotoga maritima alpha-amylase AmyA, a putative lipoprotein of a hyperthermophilic bacterium. J Bacteriol. 1997;179:941–948. doi: 10.1128/jb.179.3.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lou J, Dawson K A, Strobel H J. Glycogen biosynthesis via UDP-glucose in the ruminal bacterium Prevotella bryantii B14. Appl Environ Microbiol. 1997;63:4355–4359. doi: 10.1128/aem.63.11.4355-4359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 15.Lynd L R. Production of ethanol from lignocellulosic materials using thermophilic bacteria: critical evaluation of potential and review. Adv Biochem Eng. 1989;38:1–52. [Google Scholar]
- 16.Ng T K, Zeikus J G. Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum. J Bacteriol. 1982;150:1391–1399. doi: 10.1128/jb.150.3.1391-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richarme G, Kepes A. Study of binding protein-ligand interactions by ammonium sulfate-assisted adsorption on cellulose ester filters. Biochim Biophys Acta. 1983;742:16–24. doi: 10.1016/0167-4838(83)90353-9. [DOI] [PubMed] [Google Scholar]
- 18.Russell R R, Aduse-Opoku J, Sutcliffe I C, Tao L, Ferretti J J. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem. 1992;267:4631–4637. [PubMed] [Google Scholar]
- 19.Sahm K, Matuschek M, Muller H, Mitchell W J, Bahl H. Molecular analysis of the amy gene locus of Thermoanaerobacterium thermosulfurigenes EM1 encoding starch-degrading enzymes and a binding protein-dependent maltose transport system. J Bacteriol. 1996;178:1039–1046. doi: 10.1128/jb.178.4.1039-1046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silhavy T J, Boos W, Szmelcman S, Schwartz M. On the significance of retention of ligand by protein. Proc Natl Acad Sci USA. 1975;72:2120–2124. doi: 10.1073/pnas.72.6.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J D, Higgins D G, Gibson T J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiegel J. Formation of ethanol by bacteria. A pledge for the use of extreme thermophilic bacteria in industrial ethanol fermentation processes. Experientia. 1980;36:1434–1446. [Google Scholar]
- 24.Wiegel J, Carriera L H, Mothershed C P, Ljungdahl L G. Production of ethanol from biopolymers by anaerobic, thermophilic and extreme thermophilic bacteria. II. Thermoanaerobacter ethanolicus JW200 and its mutants in batch cultures and resting cell experiments. Biotechnol Bioeng Symp. 1983;13:193–205. [Google Scholar]
- 25.Wiegel J, Ljungdahl L G. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new, extreme thermophilic, anaerobic bacterium. Arch Microbiol. 1981;128:343–348. [Google Scholar]
- 26.Wyman C E, Goodman B J. Biotechnology for production of fuels, chemicals, and materials from biomass. Appl Biochem Biotechnol. 1993;39:41–59. [Google Scholar]
- 27.Xavier K B, Martins L O, Peist R, Kossmann M, Boos W, Santos H. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1996;178:4773–4777. doi: 10.1128/jb.178.16.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]