Abstract
Numerous agents can damage the DNA of prokaryotes in the environment (e.g., reactive oxygen species, irradiation, and secondary metabolites such as antibiotics, enzymes, starvation, etc.). The large number of potential DNA-damaging agents, as well as their diverse modes of action, precludes a simple test of DNA damage based on detection of nucleic acid breakdown products. In this study, free 3′-OH DNA ends, produced by either direct damage or excision DNA repair, were used to assess DNA damage. Terminal deoxyribonucleotide transferase (TdT)-mediated dUTP nick end labeling (TUNEL) is a procedure in which 3′-OH DNA ends are enzymatically labeled with dUTP-fluorescein isothiocyanate using TdT. Cells labeled by this method can be detected using fluorescence microscopy or flow cytometry. TUNEL was used to measure hydrogen peroxide-induced DNA damage in the archaeon Haloferax volcanii and the bacterium Escherichia coli. DNA repair systems were implicated in the hydrogen peroxide-dependent generation of 3′-OH DNA ends by the finding that the protein synthesis inhibitors chloramphenicol and diphtheria toxin blocked TUNEL labeling of E. coli and H. volcanii, respectively. DNA damage induced by UV light and bacteriophage infection was also measured using TUNEL. This methodology should be useful in applications where DNA damage and repair are of interest, including mutant screening and monitoring of DNA damage in the environment.
DNA damage is a ubiquitous phenomenon experienced by microbes in the environment. Potential DNA-damaging agents include enzymes (e.g., restriction enzymes encoded by addiction modules or bacteriophage) (1, 23, 31), as well as nonenzymatic attacks via physical (e.g., irradiation) (18) and chemical (e.g., secondary metabolites such as antibiotics and reactive oxygen species [ROS]) agents (13). Additionally, it is probable that during differentiation and/or under ecological pressures, microbes undergo programmed cell death involving DNA fragmentation (1, 12, 16). Identifying conditions in which DNA damage occurs is important to understanding microbes in their natural environments.
DNA damage is a serious threat to survival. Consequently, all organisms have developed intricate and extensive DNA repair systems (6, 9, 15, 20, 30). There are two general categories of DNA repair systems: direct and indirect (20). Direct DNA repair systems are rare and involve the chemical reversal of damage (e.g., photoreactivation of pyrimidine dimers) (9, 20). Indirect DNA repair systems require the recognition and removal of DNA damage (i.e., excision) and the synthesis of new DNA (9). Indirect DNA repair systems are responsible for fixing most forms of DNA damage (9, 15). The excision step of indirect DNA repair systems results in single-strand DNA breaks, regardless of the type of original DNA-damaging agent (9, 15). For example, hydrogen peroxide treatment of cells results in DNA damage mainly through the modification of DNA bases by the attack of hydroxyl radicals (13). DNA repair is initiated when the modified bases are recognized and removed by a DNA glycosylase (reviewed in reference 13). Removal of the base forms an apurinic or apyrimidinic (AP) site. AP endonucleases then nick the strand at the AP site, thereby creating a 3′-OH DNA end, which is used as a substrate for synthesis of a new DNA strand to replace the damaged area. The strand is then closed by DNA ligases. Hypothetically, any DNA damage that is recognized by a cell's DNA repair system should result in production of 3′-OH DNA ends. Identifying cells with an excess of 3′-OH DNA ends would indicate that the cell has experienced DNA damage and is undergoing DNA repair.
Previous methods to measure biologically relevant microbial DNA damage in the environment have included (i) the killing rate of DNA-repair-negative Escherichia coli (19), (ii) DNA- and bacteriophage (phage)-containing dosimeters (25), (iii) bacterial activity (2, 10, 14, 29), (iv) antibodies to damaged DNA (17, 22), and (v) expression of RecA (M. G. Booth and R. V. Miller, presentation at Limnology and Oceanography: Navigating into the Next Century, 1999, Santa Fe, N.Mex.). In this study, we tested the hypothesis that DNA damage in prokaryotes can be measured in individual cells by monitoring the formation of 3′-OH DNA. This was done by adapting the TUNEL (terminal deoxyribonucleotide transferase [TdT]- mediated dUTP nick end labeling) method to prokaryotes (11). TUNEL is a procedure in which 3′-OH DNA ends are enzymatically labeled with dUTP-fluorescein isothiocyanate (dUTP · FITC) using TdT. TUNEL is routinely used to monitor the DNA fragmentation associated with apoptosis in eukaryotes (27). DNA damage caused by hydrogen peroxide treatment of the bacterium E. coli and the archaeon Haloferax volcanii was readily detectable using TUNEL. Similarly, UV-induced DNA damage and phage-induced DNA breaks could be detected using this procedure. TUNEL is simple to perform, and samples can be stored for at least 3 weeks before analysis, making it useful for field studies of DNA damage in prokaryotes.
MATERIALS AND METHODS
Culturing conditions and treatments.
E. coli strain K37 [W3110 galK rpsL(Strr)] was grown in Luria-Bertani medium (LB) at 37°C with constant shaking (26). H. volcanii was grown in H. volcanii medium (3.5 M NaCl, 150 mM MgSO4 · 7H2O, 50 mM KCl, 127.5 μg of MnCl2 liter−1, 50 mM Tris · Cl [pH 7.2], 0.05% CaCl2 · 2H2O, 0.003% Bacto yeast extract, 0.005% Bacto tryptone) at 42°C with constant shaking (8).
For the hydrogen peroxide treatments, 100 μl of an overnight E. coli culture or 500 μl of a late-log–early-stationary-phase H. volcanii culture was inoculated into 10 ml of fresh medium. Aliquots were removed from the new cultures at various times, and the optical density at 600 nm was measured. Cells from exponentially growing cultures were treated with 0.2% hydrogen peroxide (≈86 mM) or an equivalent volume of water (i.e., control) and incubated at room temperature for 30 min. For the protein inhibitor studies, the exponentially growing cells were pretreated for 10 min with chloramphenicol (E. coli; 100 μg ml−1) or diphtheria toxin (DT) (H. volcanii; 1 μg ml−1; Calbiochem-Novabiochem Corp., San Diego, Calif.) before the 30-min hydrogen peroxide treatment. Stationary E. coli cells were obtained by pelleting 1 ml of an overnight culture, resuspending it in 10 ml of M-9 minimal medium (MM) (26), and incubating it at 37°C for 2 h. One-milliliter aliquots were treated with 0.4% hydrogen peroxide or an equivalent volume of water for 30 min and then analyzed by TUNEL. A third aliquot of the stationary cells was fixed and permeabilized, and then the DNA was digested with the restriction enzyme SmaI. The fixed cells were washed three times with 1× NEBuffer 4 (New England Biolabs, Beverly, Mass.), resuspended in 100 μl of 1× NEBuffer 4 supplemented with 100 U of SmaI (New England Biolabs), and incubated at 37°C for 15 min. The SmaI-treated cells were then washed two times with 1× phosphate-buffered saline (PBS) (10× PBS = 1.4 M NaCl, 27 mM KCl, 15 mM KH2PO4, 96 mM Na2HPO4, pH 7.3) and analyzed via TUNEL.
For the UV experiments, a 10-ml E. coli culture was rinsed one time with MM supplemented with 1.5% glucose (26), resuspended in 10 ml of MM-glucose, and incubated at 37°C for 1 h. Five milliliters of the resuspended culture was then transferred into a small petri dish (50 by 9 mm; Fisher Scientific) and placed 20 cm under a Spectrolin XX-15F germicidal lamp (Spectronics Corporation, Westbury, N.Y.) without a cover. The lamp contained a 120-V, 60-Hz, 0.7-A bulb outputting a nearly monochromatic band at 255 nm (i.e., UV-C). The control culture was treated identically, except that the petri dish was covered with aluminum foil during UV exposure. After the 30-s UV irradiation, one-half of the culture was removed and fixed for TUNEL analysis. The rest of the culture was allowed to recover at 37°C for 15 min before fixation and TUNEL analysis (i.e., recovered samples).
For the phage experiments, 100 μl of an overnight E. coli culture was transferred to LB supplemented with maltose and MgCl2 (26) and monitored using spectrophotometry. Lambda phage cI857 was added to exponentially growing cells at a multiplicity of infection of 1:1 (phage/bacterium ratio) and incubated at 37°C for 1 h before TUNEL analysis. Similarly, an exponentially growing culture of Roseobacter strain SIO67 grown in ZoBell 2216E medium (5 g of peptone and 1 g of yeast extract, in 0.45-μm-pore-size-filtered seawater) was exposed to Roseophage SIO1 (multiplicity of infection of 1) for 1 h before fixing and TUNEL analysis. The phage titers were determined using standard top agar techniques (26).
Cell fixation and permeabilization.
After appropriate treatments, cells were pelleted by centrifugation at 13,000 × g for 2 min in a Hermle Z 231 M microcentrifuge. The supernatant was decanted, and the E. coli cells were resuspended into 1 ml of ice-cold E. coli fixing solution (4% paraformaldehyde [Sigma, St. Louis, Mo.] in 1× PBS). H. volcanii cells were resuspended in H. volcanii fixing solution, which was made by combining 1 part E. coli fixing solution with 1 part 1× H. volcanii wash buffer (HVWB; 1× HVWB is 3.5 M NaCl, 150 mM MgSO4 · 7H2O, 50 mM KCl, 127.5 μg of MnCl2 liter−1, 50 mM Tris-Cl [pH 7.2], and 0.05% CaCl2 · 2H2O). Cells were incubated in their appropriate fixing solutions for 30 min at room temperature. At the end of the incubation, cells were again pelleted and the fixing solution was removed. The pellets were resuspended in 500 μl of ice-cold permeabilization solution (0.1% Triton X-100 and 0.1% sodium citrate) and incubated on ice for 2 min (in situ cell death detection kit, fluorescein, booklet, 1998; Roche Molecular Biochemicals). Again cells were pelleted and washed once in PBS (E. coli) or HVWB (H. volcanii).
Direct TUNEL with dUTP · FITC.
The fixed and permeabilized cells were resuspended in 100 μl of TUNEL reaction mix from the in situ cell death detection kit, fluorescein, from Roche Molecular Biochemicals (catalog no. 1 684 795; Indianapolis, Ind.). This reaction mix includes dUTP · FITC, TdT, and appropriate buffers. The reaction mixture was then incubated at 37°C in the dark for 1 h. Samples were washed one time with PBS or HVWB and then resuspended in 1 ml of PBS or HVWB for storage and analysis. The PBS and HVWB used for the final resuspension were filtered through a 0.2-μm-pore-size Acrodisc syringe filter (Gelman Sciences, Ann Arbor, Mich.).
Indirect TUNEL with dUTP · biotin.
Biotin-16-dUTP was added to DNA breaks by first washing the cells one time with PBS and then resuspending them in 100 μl of elongation buffer (1× Roche Molecular Biochemicals TdT buffer, 2.5 mM CoCl2, 0.1 mM biotin-16-dUTP, 5 U of TdT; all reagents were obtained from Roche Molecular Biochemicals) and incubating them at 37°C for 60 min. After elongation, the cells were pelleted and the supernatant was removed. The cells were resuspended in 100 μl of TUNEL staining solution (5× SSC [20× SSC is 3 M NaCl and 0.3 M sodium citrate, pH 7.5], 5% Carnation nonfat dried milk, 0.2 μg of FITC · avidin [Sigma] ml−1, 0.01% Triton X-100) and incubated at room temperature for 30 min in the dark. After staining, the cells were pelleted, the supernatant was removed, and the pellet was resuspended in 100 μl of 1× PBS.
Propidium iodide (PI) staining.
A 500× stock solution was made by dissolving dried PI at 10 mg ml−1 in 1× PBS. This solution was filtered through a 0.2-μm-pore-size syringe filter to remove particulates and stored in the dark at 4°C. Samples were stained by adding PI at a final 1× concentration directly to cells in growth medium or wash buffers.
Flow cytometric analysis.
TUNEL and PI samples were analyzed on a FACSort sorter (Becton Dickinson, San Jose, Calif.).
Unless otherwise noted, reagents were obtained from Sigma.
RESULTS
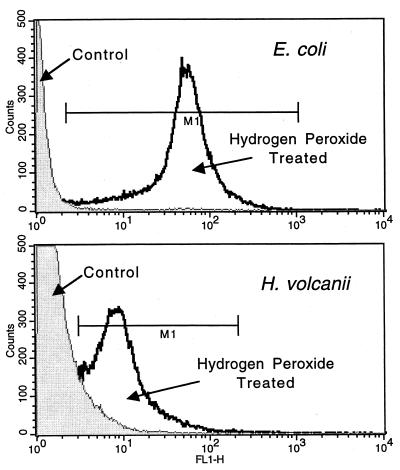
Hydrogen peroxide-induced DNA damage was measured by TUNEL in the bacterium E. coli and the archaeon H. volcanii. Aliquots of exponentially growing cultures were treated with 0.2% hydrogen peroxide or an equivalent volume of water (i.e., control) and incubated at room temperature for 30 min. The cells were then fixed, permeabilized, and analyzed via TUNEL. Figure 1 shows a representative experiment in which hydrogen peroxide treatment caused the vast majority of both populations to become TUNEL positive. Since TdT selectively uses the 3′-OH end of DNA as a substrate for extension, the TUNEL-positive cells in the hydrogen peroxide treatment must have more of these ends (in situ cell death detection kit booklet, Roche Molecular Biochemicals; 26). To quantify this effect, cellular DNA was stained with PI subsequent to the TUNEL procedure. The DNA-containing population (i.e., PI positive) was electronically gated away from non-DNA-containing particles, thereby removing unwanted background. The DNA-containing population was then analyzed for TUNEL staining. For convenience, the gate, marked M1 in Fig. 1, was started at the intersection of the control and hydrogen peroxide treatment histograms. In this experiment, 97.4% of the hydrogen peroxide-treated E. coli cells were TUNEL positive. In comparison, less than 1% of the control E. coli cells were TUNEL positive. Similarly, 84.3% of the hydrogen peroxide-treated H. volcanii cells were TUNEL positive, as opposed to 9.6% of the control cells. Neither the relative size, shape, nor granularity, as determined by forward and side scatter, changed significantly with hydrogen peroxide treatment (data not shown).
FIG. 1.
TUNEL analysis of hydrogen peroxide-induced DNA excision repair in bacteria and archaea. Samples of E. coli and H. volcanii were exposed to 0.2% hydrogen peroxide for 30 min before TUNEL analysis. Control cultures were treated with an equivalent volume of water for 30 min. The x axis (FL1-H) represents the relative FITC fluorescence, and the y axis (counts, or events) is the number of cells.
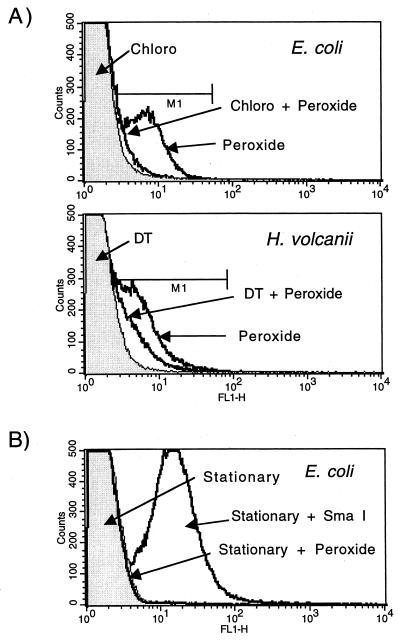
Hydrogen peroxide-induced DNA damage predominately involves the attack of OH* on the various rings of the DNA bases and backbone (13). Hydrogen peroxide has also been reported to induce both double- and single-strand breaks in DNA (13). To determine if the DNA breaks observed upon hydrogen peroxide treatment of E. coli were due to activation of repair enzymes, the effect of the protein synthesis inhibitor chloramphenicol on hydrogen peroxide-induced 3′-OH DNA ends was measured. Since many of the DNA repair systems involve de novo protein synthesis, it was hypothesized that chloramphenicol may block the generation of 3′-OH DNA ends produced by excision repair processes. Conversely, if hydrogen peroxide was directly breaking the DNA, chloramphenicol would not be expected to block the formation of 3′-OH DNA ends detectable by TUNEL. Figure 2A shows that treating the cells with 0.2% hydrogen peroxide for 30 min induces 3′-OH DNA breaks detectable by TUNEL. Analysis of the DNA-containing population showed that 84.1% of the treated E. coli cells were within the gated region (M1). Only 4.2% of the untreated cells were in M1. Pretreating the same cells with chloramphenicol for 10 min before the hydrogen peroxide treatment blocks the generation of TUNEL-positive cells (7.8% in M1). The control population was the same E. coli population treated with chloramphenicol for 40 min. Therefore, the hydrogen peroxide-induced formation of 3′-OH DNA ends was dependent on de novo translation, presumably involved in activating cellular DNA repair systems. Similarly, pretreatment of H. volcanii with DT, a protein synthesis inhibitor in archaea (21), blocked approximately 50% of the hydrogen peroxide-induced 3′-OH DNA ends (percentage of population in M1 for experiment shown in Fig. 2A: DT-alone control, 6.9%; hydrogen peroxide, 78.2%; and hydrogen peroxide plus DT, 31.4%). Finally, it was hypothesized that if hydrogen peroxide was directly breaking DNA, the growth phase of the cells should not have an effect on generation of 3′ ends. As shown in Fig. 2B, stationary E. coli cells treated with hydrogen peroxide did not experience DNA breaks detectable by TUNEL. In this experiment, the cells were treated with 0.4% hydrogen peroxide, double the concentration used in the other experiments. It was a possibility that the cell wall (e.g., peptidoglycan structure) of stationary cells differs significantly from that of exponentially growing cells, thereby excluding the TUNEL reagents and preventing efficient labeling. To address this possibility, stationary cells were fixed, permeabilized, and treated with the restriction enzyme SmaI before TUNEL analysis. A strongly positive population was observed in the SmaI-treated cells, showing that the stationary cells were susceptible to TUNEL labeling (Fig. 2B).
FIG. 2.
(A) The effect of protein synthesis inhibitors on the formation of 3′-OH DNA ends in E. coli and H. volcanii. Three aliquots of an exponentially growing E. coli culture were treated with (i) chloramphenicol (100 μg ml−1) for 40 min (Chloro), (ii) 0.2% hydrogen peroxide for 30 min (Peroxide), and (iii) chloramphenicol for 10 min followed by 0.2% hydrogen peroxide for 30 min (Chloro + Peroxide). DT (1 μg ml−1), a protein synthesis inhibitor in archaea, was used in an identical study of H. volcanii. (B) Stationary E. coli cells do not form 3′-OH DNA ends when treated with hydrogen peroxide. One milliliter of an overnight E. coli culture was pelleted, resuspended in 10 ml of MM, and incubated at 37°C for 2 h. One-milliliter aliquots were treated with 0.4% hydrogen peroxide (Stationary + Peroxide) or an equivalent volume of water (Stationary) for 30 min and then analyzed by TUNEL. A third aliquot of the stationary cells was fixed, permeabilized, treated with the restriction enzyme SmaI, and then analyzed via TUNEL (Stationary + SmaI).
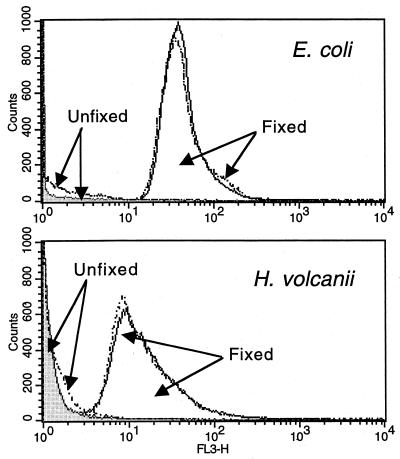
PI is a DNA stain that cannot cross intact cellular membranes (28). This property of PI was exploited to determine if hydrogen peroxide treatment selectively destroyed the membranes of TUNEL-positive cells, thereby exposing them to exogenous DNases, rather than activating internal DNA repair systems. E. coli cells were treated with 0.2% hydrogen peroxide for 30 min. During the last 10 min of this treatment, PI was added to the cells. These cells were washed once with PBS and then analyzed by flow cytometry. Figure 3 shows that the cells excluded PI whether or not they were treated with hydrogen peroxide. Therefore, it was concluded that the inside of the cell was not accessible to exogenous DNases. To show that PI was effective at staining DNA under these conditions, two control populations of E. coli were fixed and permeabilized (as described for the TUNEL procedure) and then resuspended in LB. These samples were then treated exactly like the experimental samples. In both of these control populations, the cells became stained with PI. Similar results were obtained with H. volcanii (Fig. 3) and stationary E. coli cells (data not shown).
FIG. 3.
Hydrogen peroxide treatment does not induce the loss of membrane integrity. Exponentially growing E. coli cells were treated with 0.2% hydrogen peroxide (dashed lines) or water for 30 min (solid lines). During the last 10 min of this treatment, 1× PI was added. After the incubation was complete, the cells were washed once with PBS and analyzed on the flow cytometer (Unfixed). The positive controls were E. coli or H. volcanii previously fixed, permeabilized, and analyzed in parallel with the experimental samples (Fixed).
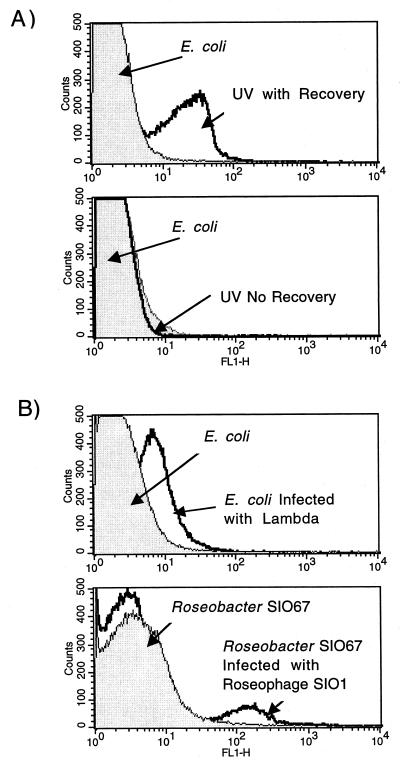
Two other DNA-damaging agents frequently encountered by prokaryotes in the environment are UV and bacteriophage. To determine if DNA damage induced by UV treatment could be detected using TUNEL, E. coli cells were exposed to a germicidal UV lamp (120-V Spectrolin XX-15F with maximal emission at 255 nm) for 30 s, fixed, and analyzed using the assay. No DNA damage could be detected (Fig. 4A). However, when the cells were exposed to the same UV treatment and then allowed to recover for 15 min, a TUNEL-positive population was observed.
FIG. 4.
TUNEL analyses of bacteria treated with UV radiation or infected with bacteriophages. (A) An E. coli culture was exposed to a wide-coverage UV lamp for 30 s. An aliquot of the cells was immediately fixed and analyzed using TUNEL (UV No Recovery). A second aliquot was allowed to recover for 15 min after the UV exposure and then analyzed by TUNEL (UV with Recovery). The control cultures were treated exactly the same as the experimental cultures, except that they were covered with aluminum foil. (B) An E. coli culture was incubated with coliphage λ cI857, and a Roseobacter strain SIO67 culture was exposed to roseophage SIO1 for 1 h and then fixed for TUNEL analyses. The multiplicity of infection was approximately 1 for both samples. The control populations were treated with a heat-killed aliquot of the respective phages.
To determine if TUNEL could detect DNA breaks associated with phage infections, E. coli was exposed to coliphage λ cI857 and Roseobacter strain SIO67 was exposed to roseophage SIO1. Roseobacter strain SIO67 is a marine alpha-proteobacterium susceptible to infection by roseophage SIO1, a marine podovirus genetically related to coliphages T3 and T7 (25a, 31). Previous studies have shown that roseophage SIO1 degrades the host's DNA and incorporates the freed nucleotides into its genome during replication (31). Roseophage SIO1 also encodes an endo-DNase that is closely related to that of T3 and T7. This endo-DNase presumably cuts up the host's chromosome during the phage replication. Therefore, 3′ ends for TUNEL labeling should include both those made by the phage endo-DNase and those made by the production of new phage, each of which have two termini. In contrast, coliphage λ does not degrade the host's DNA during lytic reproduction (3). Therefore, 3′-OH DNA ends available for TUNEL labeling in E. coli infected by coliphage λ should be the result of new phage termini alone. Figure 4B shows that infection of both bacteria, by their respective phages, results in a TUNEL-positive population, indicating that this method can be used to identify cells undergoing a lytic phage infection.
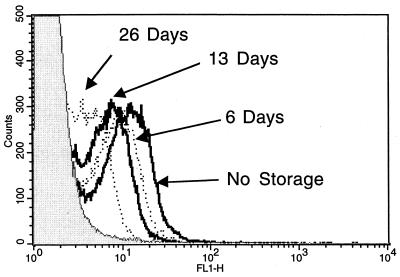
Samples collected in the field must often be stored before analysis; therefore, the effect of storing TUNEL-stained samples was tested. In this assay, E. coli was treated with 0.2% hydrogen peroxide and incubated at room temperature for 1 h. The cells were then permeabilized, fixed, and TUNEL stained. The samples were immediately analyzed on the flow cytometer and then placed at 4°C in the dark. The samples were reanalyzed 6, 13, and 26 days after the original treatment and TUNEL staining. There was a slight loss of mean fluorescence over the first 13 days (Fig. 5). However, this change was minor, and it can be concluded that TUNEL samples can be stored for at least 2 weeks without any special treatment. The effects of storing samples in 4% paraformaldehyde in PBS (fixing solution) or in 70% ethanol after fixing and permeabilization were also tested. These treatments significantly reduced the mean fluorescence of the TUNEL-positive cells (data not shown). Based on these observations, it is suggested that samples be fixed, permeabilized, and TUNEL treated as soon as possible. The cells can then be stored until analysis on a flow cytometer or fluorescence microscope.
FIG. 5.
Effect of storing TUNEL samples. E. coli cells were treated with 0.2% hydrogen peroxide for 60 min, fixed, permeabilized, and TUNEL stained. This sample was then analyzed on the fluorescence-activated cell sorter immediately (No Storage; solid line). After fluorescence-activated cell sorter analysis, the remaining sample was stored in the dark at 4°C. At 6 (6 Days; dotted line), 13 (13 Days; solid line), and 26 (26 Days; dotted line) days, the sample was reanalyzed on the fluorescence-activated cell sorter.
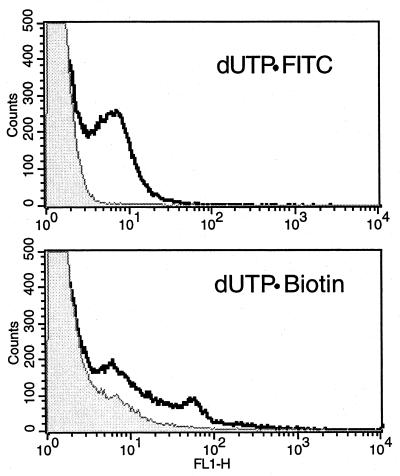
The TUNEL procedure in the previous studies requires FITC as the fluorochrome. There are times, however, when it is desirable to use other fluorochromes (e.g., when analyzing cells expressing green fluorescent protein). We tested a second TUNEL protocol in which dUTP · biotin was used as the substrate for TdT. After the 3′-OH DNA ends had been labeled with the dUTP · biotin, the cells were stained with avidin conjugated to the fluorochrome of choice (in this case, FITC). To compare these two methods, E. coli was treated with 0.2% hydrogen peroxide for 30 min and split into two aliquots. The first aliquot was TUNEL stained using the dUTP · FITC method described above. The second aliquot was TUNEL stained using dUTP · biotin in the elongation step, and then the cells were stained with avidin · FITC. Figure 6 shows that both methods detect 3′-OH DNA ends produced during hydrogen peroxide treatment of E. coli. However, the dUTP · FITC protocol results in a significantly lower background than that for the dUTP · biotin method. Many attempts were made to lower the background in dUTP · biotin samples by changing staining conditions (e.g., increasing milk and/or salt concentrations, addition of bovine serum albumin, etc.). However, none of these treatments significantly changed the background staining. It can be concluded that the dUTP · biotin method will work in prokaryotes, but with less satisfactory results than those of the dUTP · FITC approach.
FIG. 6.
Comparison TUNEL using dUTP · FITC or dUTP · biotin. Exponentially growing E. coli cells were treated with 0.2% hydrogen peroxide for 30 min. The sample was split into two aliquots that were then analyzed using either the dUTP · FITC or the dUTP · biotin protocol.
DISCUSSION
In order to develop a general assay for detecting DNA damage in prokaryotes, the formation of 3′-OH DNA ends was monitored in individual cells using TUNEL. Three types of environmentally relevant DNA-damaging agents (e.g., hydrogen peroxide, UV irradiation, and phage infection) produced 3′-OH DNA ends that were detectable using TUNEL. We believe that 3′-OH ends produced during DNA excision repair, as well as 3′-OH ends caused by directly breaking the DNA backbone, were measured in these studies. First, activation of excision DNA repair mechanisms was implied by the requirement of de novo protein synthesis for the formation of 3′-OH DNA ends in cells treated with hydrogen peroxide. Second, DNA breaks were not produced in stationary E. coli cells treated with hydrogen peroxide, supporting the conclusion that hydrogen peroxide was not directly breaking the DNA backbone (however, this interpretation may be complicated by entry of the stationary cells into a hydrogen peroxide-hyperresistant state [C. Fraley, M. McCann, M. Keyhan, and A. Matin, Abstr. 94th Gen. Meet. Am. Soc. Microbiol. 1994, abstr. H-98, p. 217, 1994]). Third, UV irradiation, a treatment which does not directly induce DNA backbone breaks, produced TUNEL-detectable 3′-OH DNA ends. Finally, we have identified an E. coli mutant strain that does not produce measurable 3′-OH DNA ends upon hydrogen peroxide exposure (i.e., hydrogen peroxide treatment of the mutant does not result in TUNEL-positive population [data not shown; analysis of this mutant will be presented elsewhere]). This finding indicates that a specific genetic component was necessary for the observed phenomenon. Based on these observations, it was concluded that excision repair responses, activated by DNA damage, can be measured using TUNEL. By extension, any DNA-damaging agent that activates DNA excision repair systems should be detectable using this method.
One potential complication associated with using the TUNEL approach to detecting DNA damage would be the Okazaki fragments produced during DNA replication (20). Each of these fragments has a 3′-OH DNA end that should be targeted by TdT in the TUNEL reaction. Therefore, replicating cells should be more brightly labeled than nonreplicating cells. Except for the experiment whose results are shown in Fig. 2B, the cell populations used in this study were exponentially growing cultures and contained replicating cells. In each control population, there was a small percentage (less than 10%) of the cells that may have TUNEL-labeled Okazaki fragments. However, untreated stationary cells (Fig. 2B) and exponentially growing cells had almost the same level of background labeling, suggesting that Okazaki fragments were not major targets for TUNEL labeling.
During the course of these studies, a difference in the mean fluorescence induced by similar ROS treatments was observed. For example, a 30-min 0.2% hydrogen peroxide treatment of E. coli resulted in a mean fluorescence shift of approximately 2 logs in Fig. 1 and 1 log in Fig. 2A. These differences were somewhat surprising, because it was expected that the mean fluorescence should be proportional to the amount of DNA damage. To address this ambiguity, the same cells used for Fig. 2 were also exposed to 0.2% hydrogen peroxide for 60 min (Fig. 5, “No Storage” sample), on the assumption that longer exposure to the ROS would induce more DNA damage. Comparison of these two samples showed that treating E. coli cells with one concentration of hydrogen peroxide for a longer time resulted in more DNA damage. Additionally, treating the cells with varying concentrations of hydrogen peroxide induced changes in the mean fluorescence that were proportional to the hydrogen peroxide concentration (i.e., more peroxide results in more measurable DNA breaks [data not shown]). In retrospect, the differences observed from experiment to experiment were due to variations in the concentration of cells in the sample. Though care was taken to use only exponentially growing cells, the actual concentration of cells per milliliter was not normalized. Later analysis revealed that a higher cell concentration decreased the amount of DNA damage by hydrogen peroxide. It was assumed that this phenomenon was due to the increased concentration of ROS scavenging molecules associated with higher concentrations of cells.
The concentration of hydrogen peroxide used in this study (i.e., 0.2% or ≈86 mM) was approximately 10 times greater than the highest reported hydrogen peroxide concentration in surface and ground freshwater (6) and approximately 105 times higher than the predominant hydrogen peroxide concentrations found in marine systems (e.g., 0.5 to 200 nM) (7) and freshwater systems (e.g., <800 nM) (5). However, microniches (e.g., around algal cells [24]) may have much higher hydrogen peroxide concentrations, thereby complicating the hydrogen peroxide regimens that a cell experiences. At hydrogen peroxide concentrations of <10 μM, we were unable to detect DNA damage in E. coli using TUNEL (data not shown). Therefore, DNA damage to prokaryotes due to exogenous hydrogen peroxide may be relatively limited in the bulk phase of natural environments. Future studies need to more closely address the relationship between cell concentration, ROS scavenging, and DNA damage experienced in natural prokaryotic communities.
To our knowledge, this is the first use of TUNEL in prokaryotes. Based on the observation that both bacteria and archaea show similar responses to hydrogen peroxide treatment, this method should be useful for monitoring DNA damage and excision repair in the majority of prokaryotic groups. This approach has many potential applications, including screening for DNA-damaging agents, determining how effective DNA-damaging agents are in an environmental context, monitoring DNA stress of natural prokaryotic populations in their natural ecosystem, screening of mutants to see if they are more or less sensitive to DNA damage, etc.
ACKNOWLEDGMENTS
We thank Anca Segall for critical readings of the paper and Kelly Bidle for providing H. volcanii and advice on how to grow these cells.
This work was supported by NSF OCE 9900301.
REFERENCES
- 1.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by 3′,5′-bipyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey C A, Neihof R A, Tabor P S. Inhibitory effect of solar radiation on amino acid uptake in Chesapeake Bay bacteria. Appl Environ Microbiol. 1983;46:44–49. doi: 10.1128/aem.46.1.44-49.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. [Google Scholar]
- 4.Collins A, Downes C S, Johnson R T, editors. DNA repair and its inhibition. Vol. 13. Washington, D.C.: IRL Press; 1984. [Google Scholar]
- 5.Cooper W J, Lean D R S. Hydrogen peroxide concentration in a northern lake: photochemical formation and diel variability. Environ Sci Technol. 1989;23:1425–1428. [Google Scholar]
- 6.Cooper W J, Zika R G. Photochemical formation of hydrogen peroxide in surface and ground waters exposed to sunlight. Science. 1983;220:711–712. doi: 10.1126/science.220.4598.711. [DOI] [PubMed] [Google Scholar]
- 7.Cooper W J, Zika R G, Petasne R G, Plane J M C. Photochemical formation of H2O2 in natural waters exposed to sunlight. Environ Sci Technol. 1988;22:1156–1160. doi: 10.1021/es00175a004. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann E M, Place A R, Robb F T, Schreier H J. Protocols for archaebacterial research. Baltimore: University of Maryland; 1991. [Google Scholar]
- 9.Friedberg E C, Hanawalt P C, editors. DNA repair: a laboratory manual of research procedures. New York, N.Y: Marcel Dekker; 1988. [Google Scholar]
- 10.Fujioka R S, Hashimoto H H, Siwak E B, Young R H F. Effect of sunlight on survival of indicator bacteria in seawater. Appl Environ Microbiol. 1981;41:690–696. doi: 10.1128/aem.41.3.690-696.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavrieli Y, Sherman Y, Ben-Sasson S. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman S. Protecting the neighborhood: extreme measures. Proc Natl Acad Sci USA. 1998;95:2731–2732. doi: 10.1073/pnas.95.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halliwell B, Gutteridge J M C. Free radicals in biology and medicine. 2nd ed. Oxford, United Kingdom: Clarendon Press; 1995. [Google Scholar]
- 14.Herndl G J, Muller-Niklas G, Frick J. Major role of ultraviolet-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature. 1993;361:717–719. [Google Scholar]
- 15.Hickson I D, editor. Base excision repair of DNA damage. New York, N.Y: Chapman & Hall; 1997. [Google Scholar]
- 16.Hochman A. Programmed cell death in prokaryotes. Crit Rev Microbiol. 1997;23:207–214. doi: 10.3109/10408419709115136. [DOI] [PubMed] [Google Scholar]
- 17.Jeffrey W H, Pledger R J, Aas P, Hager S, Coffin R B, Haven R V, Mitchell D L. Diel and depth profiles of DNA photodamage in bacterioplankton exposed to ambient solar ultraviolet radiation. Mar Ecol Prog Ser. 1996;137:283–291. [Google Scholar]
- 18.Karentz D, Bothwell M L, Coffin R B, Hansen A, Herndl G J, Kilham S S, Lesser M P, Lindell M, Moeller R E, Morris D P, Neale P J, Sanders R W, Weiler C S, Wetzel R G. Impact of UVB radiation on pelagic freshwater ecosystems: report of working group on bacteria and phytoplankton. Arch Hydrobiol Beih Ergeb Limnol. 1994;43:31–69. [Google Scholar]
- 19.Karentz D, Lutze L H. Evaluation of biologically harmful ultraviolet radiation in Antarctic with a biological dosimeter designed for aquatic environments. Limnol Oceanogr. 1990;35:549–561. [Google Scholar]
- 20.Lewin B. Genes VI. Oxford, United Kingdom: Oxford University Press; 1997. [Google Scholar]
- 21.Madigan M T, Marinko J M, Parker J. Brock biology of microorganisms. 8th ed. Upper Saddle River, N.J: Prentice-Hall, Inc.; 1997. [Google Scholar]
- 22.Mitchell D L, Haepek C A, Clarkson J M. (6-4) Photo-products are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers. Mutat Res. 1985;143:109–112. doi: 10.1016/s0165-7992(85)80018-x. [DOI] [PubMed] [Google Scholar]
- 23.Naito T, Kusano K, Koboyashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 24.Oda T, Nakamura A, Shikayama M, Kawano I, Ishimatsu A, Muramatsu T. Generation of reactive oxygen species by raphidophycean phytoplankton. Biosci Biotechnol Biochem. 1997;61:1658–1662. doi: 10.1271/bbb.61.1658. [DOI] [PubMed] [Google Scholar]
- 25.Regan J D, Carrier W L, Gucinski H, Olla B L, Yoshida H, Fujimura R K, Wicklund R I. DNA as a solar dosimeter in the ocean. Photochem Photobiol. 1992;56:35–42. doi: 10.1111/j.1751-1097.1992.tb09599.x. [DOI] [PubMed] [Google Scholar]
- 25a.Rohwer, F., A. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with non-marine phages. Limnol. Oceanogr., in press.
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sgonc R, Gruber J. Apoptosis detection: an overview. Exp Gerontol. 1998;33:525–533. doi: 10.1016/s0531-5565(98)00031-x. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro H M. Practical flow cytometry. 3rd ed. New York, N.Y: Wiley-Liss; 1995. [Google Scholar]
- 29.Sieracki M E, Seiburth J M. Sunlight-induced growth delay of planktonic marine bacteria in filtered seawater. Mar Ecol Prog Ser. 1986;33:19–27. [Google Scholar]
- 30.Tardiff R G, Lohman P H M, Wogan G N, editors. Methods to assess DNA damage and repair: interspecies comparisons. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 31.Wikner J, Vallino J J, Steward G F, Smith D C, Azam F. Nucleic acids from the host bacterium as a major source of nucleotides for three marine bacteriophages. FEMS Microbiol Ecol. 1993;12:237–248. [Google Scholar]