Summary
Bio-electrochemical systems are based on extracellular electron transfer (EET), whose efficiency relates to the expression level of numerous genes. However, the lack of multi-functional tools for gene activation and repression hampers the enhancement of EET in electroactive microorganisms (EAMs). We thus develop a type I-F CRISPR/PaeCascade-RpoD-mediated activation and inhibition regulation (CRISPR-PAIR) platform in the model EAM, Shewanella oneidensis MR-1. Gene activation is achieved (3.8-fold) through fusing activator RpoD (σ70) to Cas7 when targeting the prioritized loci upstream of the transcription start site. Gene inhibition almost has no position preference when targeting the open reading frame, which makes the design of crRNAs easy and flexible. Then CRISPR-PAIR platform is applied to up-/down-regulate the expression of six endogenous genes, resulting in the improved EET efficiency. Moreover, simultaneous gene activation and inhibition are achieved in S. oneidensis MR-1. CRISPR-PAIR platform offers a programmable methodology for dual regulation, facilitating in-depth EET studies in Shewanella spp.
Subject areas: Biochemistry, Biochemical engineering, Metabolic engineering
Graphical abstract
Highlights
-
•
CRISPR-PAIR platform enables both gene activation and inhibition in Shewanella oneidensis
-
•
An efficient type I-F CRISPR-Cas tool is developed for S. oneidensis
-
•
Transcription regulation of endogenous genes enhances extracellular electron transfer (EET)
Biochemistry; Biochemical engineering; Metabolic engineering
Introduction
Electroactive microorganisms (EAMs) utilize the bidirectional extracellular electron transfer (EET) pathway to exchange electrons with the environment and enable a variety of microbial electrochemical techniques (METs) (Logan et al., 2019). Shewanella oneidensis MR-1 is regarded as an important model EAM; however, the relatively low EET efficiency of wild-type S. oneidensis MR-1 severely limits practical applications (Fredrickson et al., 2008; Logan et al., 2019; Shi et al., 2016), driving it necessary to conduct elaborate genetic engineering. The mechanism of EET is highly sophisticated, including direct EET pathways mainly mediated by outer-membrane cytochromes (OM-cyts) (Shi et al., 2007), and indirect EET pathways that function via self-secreted electron shuttles (Brutinel and Gralnick, 2012; Gralnick and Newman, 2007; Watanabe et al., 2009). In addition, complex metabolic networks and multiple cellular activities also affect the efficiency of EET, such as the anaerobic respiration pathway (Liu et al., 2017), biofilm formation (Sivakumar et al., 2014), endogenous electron shuttle (Mevers et al., 2019; Zou et al., 2017), and carbon source utilization (Li et al., 2017). Many genes associate with these complicated processes and are thus needed to be regulated, either up or down, to match the highly active electron transporting. Hence, multi-level modulation of gene expression is critical to promote EET and broaden the applications of METs in S. oneidensis MR-1 (Meitl et al., 2009).
To date, there have been various tools available to artificially carry out gene regulation. For enhancing gene expression, the current approaches, plasmid-based overexpression and genomic knock-in, are utilized to increase the expression level of the specific genes in S. oneidensis MR-1 (Fan et al., 2021a, 2021b; Min et al., 2017). For repressing gene expression, CRISPRi system efficiently blocks RNA polymerase binding or elongation to achieve transcriptional interference (Cao et al., 2017; Li et al., 2020). However, the above approaches of enhancing and repressing gene expression are two types of totally separate systems. These mono-functional systems could not easily meet the requirements for the modulation of substance and energy metabolism in S. oneidensis MR-1. Therefore, the development of a dual-regulation tool which enables both gene activation and gene inhibition is urgently needed, not only to reduce the time cost of mining the mechanism of EET, but to obtain more engineered strains with high EET efficiency readily.
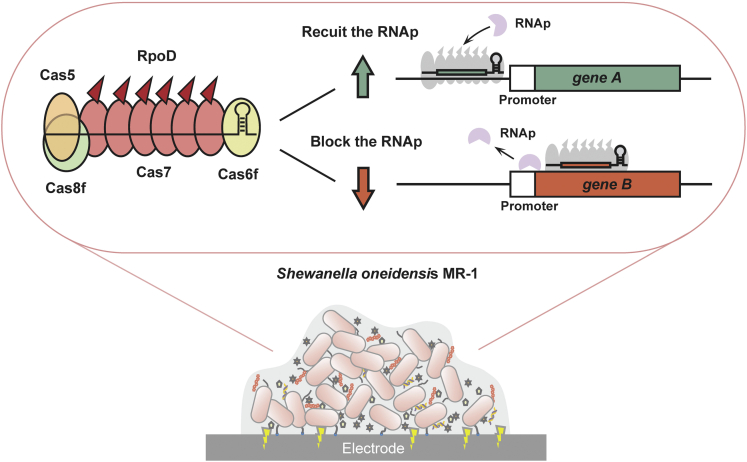
Type I and type II CRISPR-Cas systems have been widely used as the tool for gene manipulation (Chang et al., 2016; Chen et al., 2020; Lian et al., 2017). Despite pioneering work with type II CRISPR-Cas tools for different functions in diverse species (Li et al., 2015; Lian et al., 2017), type I CRISPR-Cas systems have many attractive features as a transcriptional regulation tool for S. oneidensis (Semenova et al., 2015; Zheng et al., 2019). Firstly, type I CRISPR-Cas systems are prevalent in the species close to S. oneidensis, such as Shewanella putrefaciens (Dwarakanath et al., 2015) and Pseudomonas aeruginosa (Xu et al., 2019), elevating the possibility of choosing an appropriate system which would function effectively in S. oneidensis MR-1. Secondly, type I system relies on a cascade for DNA binding and Cas3 for degrading the foreign DNA (Westra et al., 2012). The subunit responsible to bind the protospacer in type I cascade has several copies, unlike type II Cas protein functioning as a single molecule (Xu et al., 2019). In this case, more effectors for activation or inhibition are able to be recruited to this subunit (Figure 1) (Chen et al., 2020). These properties make type I CRISPR-Cas system an ideal two-way regulation tool for large-scale control of the electron flux in EAMs.
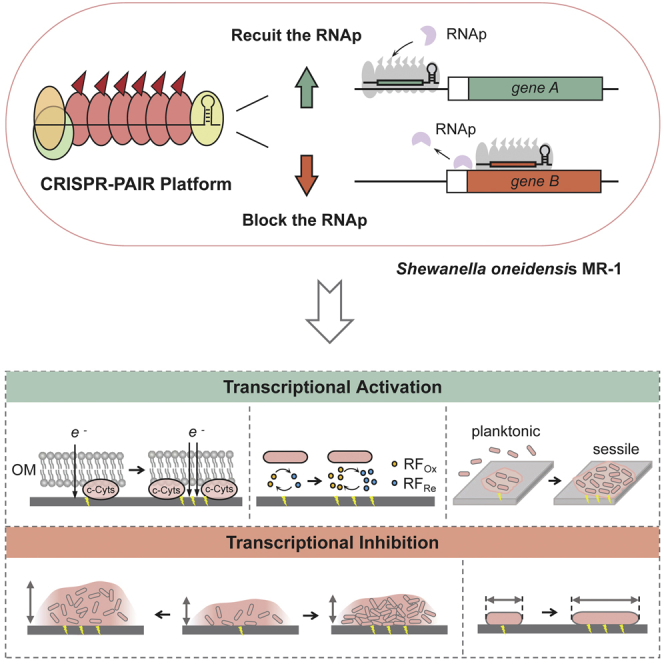
Figure 1.
The programmable type I-F CRISPR/PaeCascade-RpoD-mediated activation and inhibition regulation (CRISPR-PAIR) platform developed in S. oneidensis
In this study, we developed a dual-regulation tool using type I CRISPR-Cas system in S. oneidensis MR-1 (Figure 1). Firstly, PaeCascade (Cascade of type I-F CRISPR-Cas system from P. aeruginosa) was selected from three candidates as a suitable system in S. oneidensis MR-1. Secondly, for gene activation, PaeCascade-RpoD was constructed by fusing activator RpoD with Cas7 (functioning as several copies to bind the protospacer). The prioritized targeting sites for activation were identified and 3.8-fold activation was successfully achieved via type I-F CRISPR-Cas system in S. oneidensis MR-1. For gene inhibition, there was almost no position dependence when targeting the open reading frame (ORF), thus it is quite flexible to design the crRNAs. Thirdly, we utilized PaeCascade-RpoD system to activate 3 genes and repress 3 genes, related to biofilm formation, outer-membrane cytochrome, and so forth. The EET efficiency of all regulated strains was improved, and corresponding phenotype changes including thicker biofilm and longer cell morphology were observed. Finally, the feasibility of simultaneous gene activation and inhibition was verified in S. oneidensis MR-1. In sum, type I-F CRISPR/PaeCascade-RpoD-mediated activation and inhibition regulation (namely CRISPR-PAIR) platform provides a programmable and facile methodology for dual modulation, which would facilitate comprehensive EET studies and multi-dimensional MET applications in Shewanella spp.
Results
To screen and characterize type I CRISPR-Cas systems in S. oneidensis
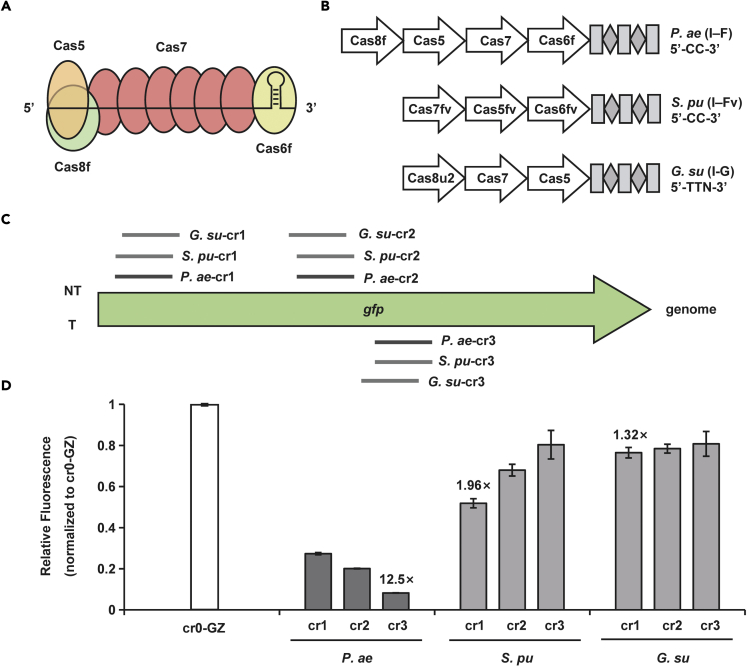
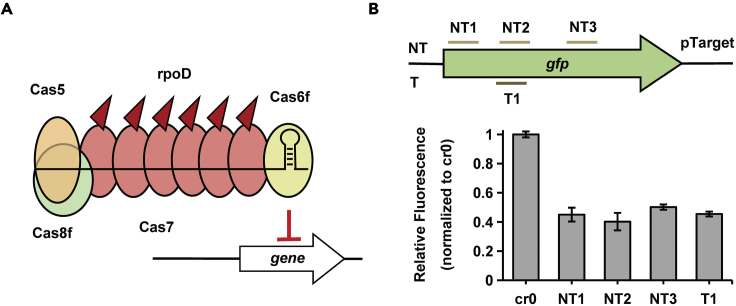
Type I CRISPR-Cas systems have the effector modules consisting of multiple Cas proteins which function together in binding and processing the target (Luo et al., 2015). Taking Pseudomonas aeruginosa type I-F CRISPR-Cas system (PaeCascade) as an example, PaeCascade complex is composed of 4 kinds of Cas protein subunits, Cas8f, Cas5, Cas7, and Cas6f (Chowdhury et al., 2017; Wiedenheft et al., 2011) (Figure 2A). Cas8f is responsible for recognizing PAM (5’-CC-3’) at the 5’ end of the protospacer and Cas5 binds to the 5’ handle of the crRNA. Cas6f combines with crRNA 3’ hairpin structure and mediates pre-crRNA maturation. Cas7 functioning as several copies binds to the protospacer, serving as the backbone of the cascade complex (Gleditzsch et al., 2016; Gu et al., 2019). Compared to the standalone dCas9, more than one effector for activation or inhibition fused to Cas7 can be recruited to the targets, allowing stronger transcriptional regulation (Figure 1) (Cady et al., 2012; Chen et al., 2020).
Figure 2.
To screen type I CRISPR-Cas systems in S. oneidensis
(A) The structure of PaeCascade-crRNA complex. Cas5 binds to the 5′ handle of the crRNA and Cas8f is responsible for PAM recognition (5′-CC-3′) at the 5′ end of the protospacer. Cas7 proteins function as several copies bind to the protospacer, serving as the backbone of the cascade complex. Cas6f combines with crRNA 3′ hairpin structure and mediates pre-crRNA maturation.
(B) The schematic diagram of the P. aeruginosa type I-F (P. ae I-F), S. putrefaciens type I-Fv (S. pu I-Fv), and G. sulfurreducens type I-G (G. su I-G) CRISPR-Cas systems used in this study. Cas proteins are presented with arrows. CRISPR repeats and spacers are indicated with squares and diamonds, respectively.
(C) Targeting sites of designed crRNAs binding different positions of gfp in the genome of the strain S. oneidensis GZ for three type I CRISPR-Cas systems. Three crRNAs were designed for each system.
(D) The inhibition efficiency of three type I CRISPR-Cas systems targeting gfp. The values above the bar indicate the highest repression folds of three crRNAs for each system. Median GFP levels are normalized to the control strain cr0-GZ. Values and error bars indicate mean ± standard error of mean (SEM) of three replicates.
Several type I CRISPR-Cas systems have been discovered, driving it necessary to find a suitable one for application in S. oneidensis MR-1. We thus chose three candidates from close relative species of S. oneidensis, PaeCascade (Pseudomonas aeruginosa type I-F CRISPR-Cascade) (Luo et al., 2015), SpuCascade (Shewanella putrefaciens type I-F variant CRISPR-Cascade) (Gleditzsch et al., 2016) and GsuCascade (Geobacteraceae sulfurreducens type I-G CRISPR-Cascade) (Makarova et al., 2020) (Figure 2B).To determine the ability of transcriptional regulation of these three systems in S. oneidensis, we adopted gfp as a reporter in the genome (S. oneidensis GZ) (Li et al., 2020) for transcriptional interference. crRNAs (cr1, cr2, cr3) either targeting the template strand (T) or the non-template strand (NT) were designed to repress the expression of gfp (Figure 2C). As shown in Figure 2D, after 24-h incubation, the inhibition efficiency of PaeCascade for gfp expression was up to 12.5-fold in S. oneidensis. However, the highest repression efficiencies of SpuCascade and GsuCascade were only 1.96-and 1.32-fold, respectively. Such differences reaffirm the notion that different cascade complexes have distinct properties from one another (Zheng et al., 2020). Hence, in the following sections, we will focus on type I-F PaeCascade system and investigate how to utilize it efficiently to modulate transcriptional level.
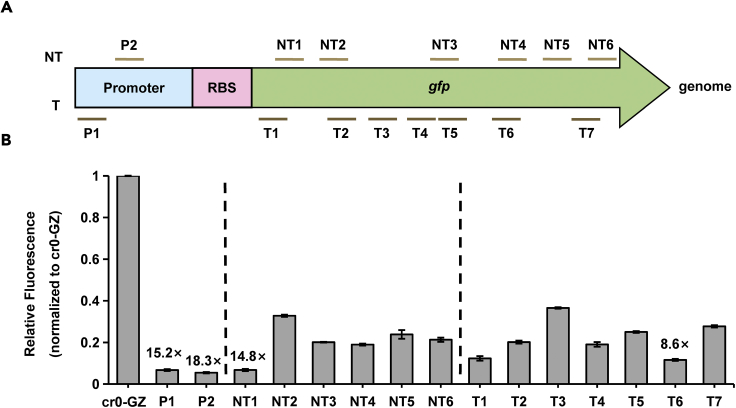
We next identified the correlation between PaeCascade-mediated repression and the targeted sites. crRNAs were designed complementary to different regions of the gfp sequence (NT1∼NT6, and T1∼T7), or promoter (P1 and P2) (Figure 3A). Spacers of crRNA were constructed through the Golden Gate assembly method (Engler et al., 2008; Fang et al., 2021). The strains harboring the plasmid with PaeCascade and corresponding crRNA were incubated for 24 h, and the fluorescence intensities were detected. In comparison to the strain with spacer-free plasmid (the strain termed cr0 hereafter), strains targeting P1, P2 and NT1 demonstrated high repression efficiency, which was ∼15.2-fold, ∼18.3-fold, and ∼14.8-fold, respectively (Figure 3B). Besides, interference levels were similar when targeting other sites, from 2.5-fold to 5-fold, no matter if the targeting sites were adjacent to or far from the initiation codon, on template or non-template strand. (Figure 3B). The phenomena were different from what were observed in type II CRISPRi system, in which sgRNAs ought to be designed close to the initiation codon and target the non-template strand (Cao et al., 2017). We speculated the difference was because for PaeCascade, the whole cascade, containing multiple Cas7 proteins, was responsible to block transcription, while dCas9 functioned as the blocker alone in the type II CRISPRi system (Burstein et al., 2017; Ghavami and Pandi, 2021; Zheng et al., 2020). In this way, PaeCascade led to increased space steric hindrance, resulting in almost no position preference. As there was almost no limitation for the design of crRNA in the PaeCascade CRISPRi system, more spacers could be obtained than in type II CRISPRi. Therefore, it is easy to design effective crRNAs, which makes PaeCascade an outstanding tool for transcriptional inhibition in S. oneidensis MR-1.
Figure 3.
To characterize optimal targeting sites of PaeCascade-mediated CRISPRi in S. oneidensis
(A) Targeting sites of designed crRNAs binding different regions of gfp in the genome of the strain S. oneidensis GZ by type I-F PaeCascade-mediated system.
(B) The inhibition efficiency of type I-F PaeCascade system targeting different regions of gfp. The values above the bar indicate the corresponding repression folds. Median GFP levels are normalized to the control strain cr0-GZ. Values and error bars indicate mean ± SEM of three replicates.
Transcription activation via PaeCascade-RpoD in S. oneidensis MR-1
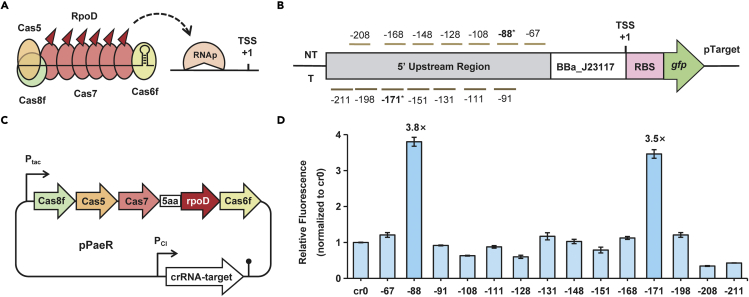
We then aimed to employ PaeCascade as a transcriptional activation tool. It has been proven that RpoD (σ70) is able to bind to the transcriptional regulatory region and recruit the core RNA polymerase (ɑ2ββ′ω) to enable gene activation in bacteria (Dong et al., 2018). We thus selected RpoD (encoded by rpoD of S. oneidensis MR-1) as the activator. In addition, it has been reported that Cas7-activator fusion protein showed the highest transcription-activation dominance at targeted loci, compared to fusing with Cas5, Cas6f, or Cas8f (Luo et al., 2015). Therefore, the RpoD was C-terminally fused with Cas7 through five amino acid linkers (Figure 4A). To testify whether CRIPSR/PaeCascade-Cas7-RpoD system (hereafter PaeCascade-RpoD) could enable transcription activation in S. oneidensis MR-1, we adopted gfp in the plasmid pTarget as the reporter gene. Besides, to find the effective sites for upregulation, a well-designed sequence was applied with several PAM (5′-CC-3′) sites located in the upstream region of gfp as in the previous study (Dong et al., 2018). Then we designed a panel of crRNAs to target these sites from −211 bp to −67 bp upstream of the transcription start site (TSS) (Figure 4B). In our CRISPRa system, polycistronique cascade fused with RpoD was controlled by IPTG-induced promoter Ptac, and crRNA was driven by constructive promoter PCI in the plasmid pPaeR (Figure 4C). We noticed that significantly increased GFP expression occurred for sites at 88 bp (NT) and 171 bp (T) upstream of TSS, and the activation efficiency was 3.8-fold and 3.5-fold, respectively (Figure 4D). In the prior reports of the bacterial CRISPRa system, the efficiency of transcription activation depended on the distance of the crRNA targets from the TSS (Bikard et al., 2013). Similarly, this phenomenon of position dependence was also observed for PaeCascade-RpoD in S. oneidensis MR-1. Furthermore, we then attempted to promote the activation efficiency by adding a promoter Ptac in the forepart of Cas7-RpoD, allowing a higher expression level (Figure S1). However, the fluorescence intensity of GFP was not activated when targeting all these six sites (Figure S1). We thus employed the PaeCascade-RpoD without an additional promoter. Given the strong position dependence on upregulation, we recommended that the prioritized sites could be settled around 88 bp (NT) and 171 bp (T) upstream of TSS.
Figure 4.
Transcription activation via PaeCascade-RpoD in S. oneidensis MR-1
(A) The schematic diagram and mechanism of PaeCascade-RpoD to recruit RNA polymerase (RNAp). RpoD (σ70) binds to transcriptional regulatory region and recruits the core RNAp (ɑ2ββ′ω) to enable gene activation. RpoD is fused to Cas7, responsible for binding protospacer.
(B) Location sites of designed crRNAs targeting TSS upstream of gfp in the plasmid pTarget.
(C) Plasmid map of PaeCascade-RpoD for transcriptional activation. The plasmid pPaeR is consisted of Cascade fused with RpoD proteins and a designed crRNA cassette controlled by Ptac and PCI promoter, respectively.
(D) The activation efficiency of type I-F PaeCascade-RpoD system targeting gfp. The values above the bar indicate the activation folds. Median GFP levels are normalized to the cr0 control strain. Values and error bars indicate mean ± SEM of three replicates.
See also Figure S1.
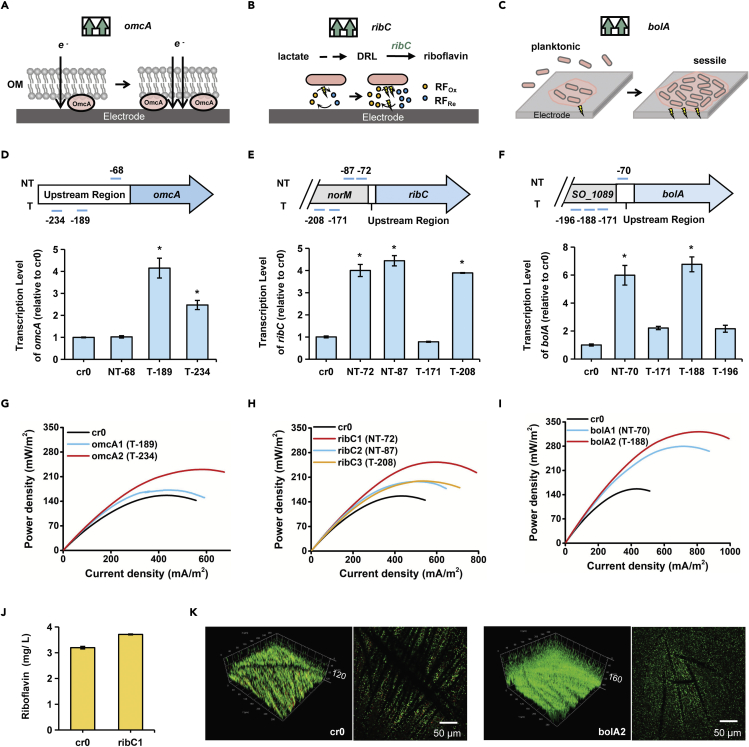
Activation of cytochrome, electron shuttle, cell motility-related gene expression by PaeCascade-RpoD for enhancing extracellular electron transfer efficiency
To demonstrate the application of PaeCascade-RpoD-mediated transcription activation for enhancing EET efficiency, the expression of three genes, omcA, ribC, and bolA were activated individually in S. oneidensis MR-1. OmcA, the outer membrane c-type cytochrome (OM c-Cyts), has been identified as a central player in transferring electrons from periplasm to electron acceptor (Figure 5A) (Meitl et al., 2009; Reardon et al., 2010). Riboflavin (RF) is the main electron shuttle of Shewanella, and RibC catalyzes the last step to synthesize RF from lactate (Figure 5B) (Yong et al., 2013). BolA is a motile/adhesive transcriptional switch, involved in the transition between the planktonic and the attachment stage of strains in the process of biofilm formation (Dressaire et al., 2015). Overexpression of bolA led to increased electroactive biomass, which is a crucial factor for electron transfer (Figure 5C) (Silva et al., 2020).
Figure 5.
Activation of cytochrome, electron shuttle, cell motility-related gene expression by PaeCascade-RpoD for enhancing EET efficiency in S. oneidensis MR-1
(A–C) Schematic representation of (A) promoting OM c-Cyts by activating the expression of omcA, (B) modulating RF biosynthesis pathway to produce more RF by activating the expression of ribC, (C) transiting engineered strains from planktonic to the attachment stage by activating the expression of bolA to enhance the EET efficiency.
(D–F) Targeting sites and corresponding transcriptional activation efficiency of (D) omcA, (E) ribC, and (F) bolA by qRT-PCR. norM and SO_1089 are the adjacent upstream genes of ribC and bolA, respectively. Transcription levels are normalized to the control strain cr0. Values and error bars indicate mean ± SEM of three replicates. Asterisks mean the strains selected for the bio-electrochemical analysis in microbial fuel cells.
(G–I) The power density output curves obtained by linear sweep voltammetry (LSV) with a scan rate of 0.1 mV/s of (G) omcA, (H) ribC, and (I) bolA-activated strains. cr0 is the control strain. See also Figure S2.
(J) The production of riboflavin (RF) of the control strain cr0 and engineered strain ribC1. Values and error bars indicate mean ± SEM of three replicates.
(K) Bio-image of strain cr0 and the activated strain bolA2 embedded on anode carbon cloth by Confocal Laser Scanning Microscope (CLSM). The number on the right of the biofilm means the bio-image thickness. All scale bars are 50 μm.
To activate the expression of omcA, ribC and bolA, we designed three to four crRNAs to target each gene. As shown in Figures 5D–5F, the qRT-PCR results demonstrated that the highest activation fold changes were 4.1 (T-189), 4.4 (NT-87), and 6.8 (T-188) of genes omcA, ribC, and bolA, respectively, compared with the no-targeting control strain cr0. Noteworthy, as the upstream regulatory regions of ribC and bolA are short, the designed crRNAs were in the coding sequence of their adjacent upstream genes. In spite of this, the transcription level of ribC and bolA were still successfully activated (Figures 5E and 5F). Then to identify the EET output of activated strains, we selected 2-3 strains with different upregulation fold changes for each gene and conducted bio-electrochemical analysis in microbial fuel cells (MFCs). The output voltages of the MFCs were presented in Figure S2. The highest voltages of omcA, ribC, and bolA-activated strains were 155.91 mV, 170.36 mV, and 221.51 mV, which were remarkably higher than that of the control strain cr0 (94.47 mV) (Figure S2). Besides, the linear sweep voltammetry (LSV) was performed during the plateau of voltage to obtain the power density. The maximum power densities of the strains omcA2, ribC1, bolA2 were up to 230.46 mW/m2 and 252.96 mW/m2 and 321.67 mW/m2. The EET efficiencies showed ∼1.5, ∼1.6, and ∼2.1 times enhancement than the strain cr0 (156.67 mW/m2), respectively (Figures 5G–5I). Furthermore, corresponding phenotype changes in the activated strains were also observed. The RF production of strain ribC1 was 3.709 mg/L, 0.51 mg/L higher than that of strain cr0. The results indicated slight improvement in the RF production, leading to the enhanced ability of electron transfer (Figures 5H and 5J). We next investigated whether the biofilm of strain bolA2 was increased by utilizing confocal laser scanning microscope (CLSM) on the MFC carbon cloth. As shown in Figure 5K, the biofilm of strain bolA2 was obviously denser and thicker than that of strain cr0, causing the drastically improved EET efficiency. Collectively, endogenous genes were successfully upregulated by the PaeCascade-RpoD-mediated CRISPRa system, resulting in positive effects on the electron generation capacity of S. oneidensis MR-1 in MFCs.
Inhibition of biofilm, cell morphology-related gene expression by PaeCascade-RpoD for enhancing extracellular electron transfer efficiency
Although activators recruit the RNAp complex, transcription inhibition can still be achieved by CRISPR/dCas9-activator system when targeting the open reading frame (ORF) instead of upstream of promoter (Bhokisham et al., 2020). We then assessed whether our PaeCascade-RpoD system could also be employed as the tool of CRISPRi, and four crRNAs were designed to target the ORF of gfp (Figure 6A). As shown in Figure 6B, the expression of gfp was repressed when targeting all these four loci, from 2-fold to 2.5-fold, indicating that PaeCascade-RpoD remained the interference capability and there was also almost no position dependence as discovered for PaeCascade-mediated gene repression (Figure 3B). In consequence, it is flexible to employ PaeCascade-RpoD as both transcription activation and inhibition tool, and thus the following application utilized PaeCascade-RpoD for gene repression in S. oneidensis MR-1.
Figure 6.
PaeCascade-RpoD system also successfully enabled transcriptional inhibition in S. oneidensis MR-1
(A) The schematic diagram of gene repression by PaeCascade-RpoD.
(B) Transcriptional inhibition was enabled by PaeCascade-RpoD targeting gfp. Targeting sites of designed crRNAs of gfp in the plasmid pTarget and the inhibition efficiency of type I-F PaeCascade-RpoD system. Median GFP levels are normalized to the control strain cr0. Values and error bars indicate mean ± SEM of three replicates.
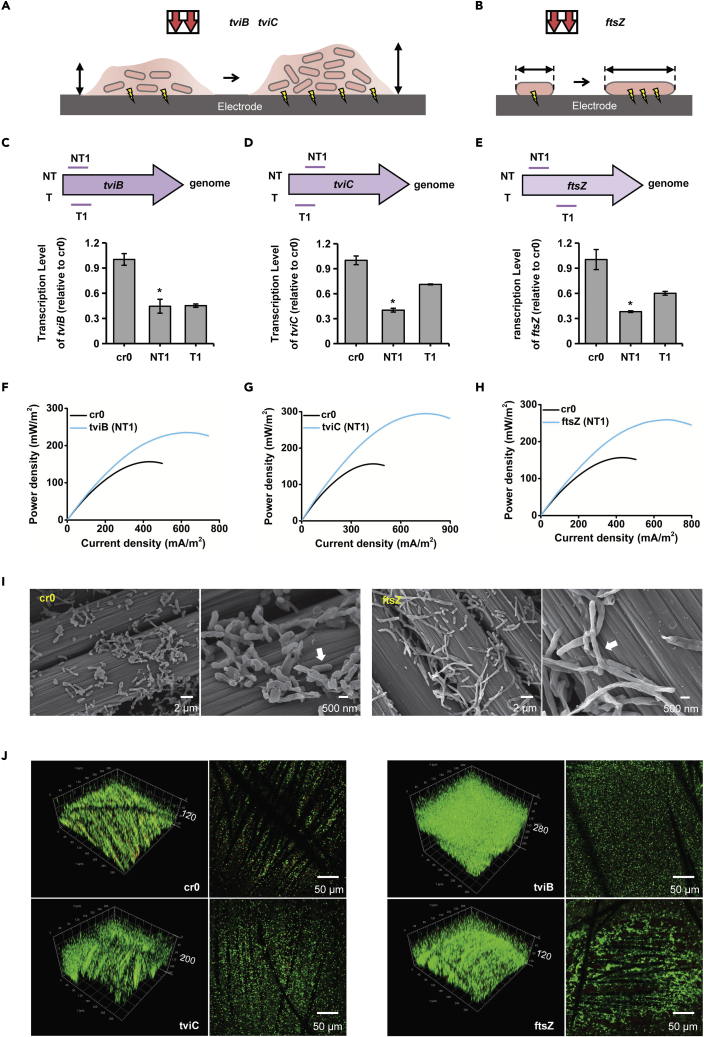
To demonstrate the application of PaeCascade-RpoD-mediated CRISPRi for enhancing EET efficiency, the expression of three genes, tviB, tviC, and ftsZ were repressed separately in S. oneidensis MR-1. Biofilm-related genes tviB and tviC were selected from the cell surface polysaccharide biosynthesis gene cluster (Kouzuma et al., 2010). It has been reported that base editing-mediated deactivation of these two genes increased the thickness and density of biofilm formed by EAMs on electrode surfaces (Chen et al., 2022), which has a positive effect on both cytochrome-mediated direct EET and shuttle-related indirect EET pathways (Edel et al., 2019). We thus conjectured that interference of tviB and tviC expression might cause thicker and/or denser biofilm, contributing to the improvement of the EET efficiency (Figure 7A). FtsZ participates in the process of cell division through self-polymerization, responsible for the formation of a Z ring in the middle of the cell (Bi et al., 1991). Accordingly, we speculated that the inhibition of ftsZ expression may lead to interfering with the formation of the Z ring, resulting in the longer cell morphology of S. oneidensis MR-1 and directly increasing the contact area between EAMs and electron acceptor (Figure 7B).
Figure 7.
Inhibition of biofilm, cell morphology-related gene expression by PaeCascade-RpoD for enhancing EET efficiency in S. oneidensis MR-1
(A and B) Schematic representation of (A) engineering the thickness and morphology of biofilm by repressing the expression of tviB or tviC, (B) engineering the cell morphology by repressing the expression of ftsZ to enhance the EET efficiency.
(C–E) Targeting sites and corresponding transcriptional inhibition efficiency of (C) tviB, (D) tviC, and (E) ftsZ by qRT-PCR. Transcription levels are normalized to the control strain cr0. Values and error bars indicate mean ± SEM of three replicates. Asterisks mean the strains selected for the bio-electrochemical analysis in microbial fuel cells.
(F–H) The power density output curves obtained by linear sweep voltammetry (LSV) with a scan rate of 0.1 mV/s of (F) tviB, (G) tviC, and (H) ftsZ-repressed strains. cr0 is the control strain. See also Figure S3.
(I) Scanning electron microscope image of the strain cr0 and repressed strain ftsZ (NT1) on anode carbon cloth. Scale bars are 2 μm for low magnification views, and 500 nm for high magnification views.
(J) Bio-image of strain cr0 and the repressed strains tviB (NT1), tviC (NT1), ftsZ (NT1) embedded on anode carbon cloth by CLSM. The number on the right of the biofilm means the bio-image thickness. All scale bars are 50 μm.
To repress the expression of tviB, tviC, and ftsZ, we designed two crRNAs (NT1 and T1) to target the ORF of each gene. As shown in Figures 7C–7E, the qRT-PCR results demonstrated that the higher transcriptional inhibition efficiency of tviB, tviC, and ftsZ-repressed strains were 1.75-, 2.49-, and 2.62-fold, between the two targets of each gene. And next these three strains were incubated in anodic chambers of MFCs for bio-electrochemical analysis. The output voltages of tviB, tviC, and ftsZ-repressed strains were 156.49 mV, 187.04 mV, and 171.54 mV, indicating a large increase over the control strain cr0 (94.47 mV) in the MFCs (Figure S3). The LSV was conducted during the plateau of voltage. The maximum power density of the strains tviB (NT1), tviC (NT1), and ftsZ (NT1) reached 234.97 mW/m2 and 294.63 mW/m2 and 259.10 mW/m2, respectively, showing ∼1.5, ∼1.9 and ∼1.7 times higher than that of the strain cr0 (156.67 mW/m2) (Figures 7F–7H). The results indicated that the PaeCascade-RpoD-mediated CRISPRi system significantly improved the EET efficiency in S. oneidensis MR-1 by repressing these three genes’ expression. Then scanning electron microscope was conducted to investigate the effect on cell morphology of repressing the expression of ftsZ. The strain ftsZ (NT1) showed obviously longer morphology than strain cr0 on carbon cloth in MFCs (Figure 7I). We also performed CLSM on the MFC carbon cloth electrodes of the cr0 and the repressed strains to detect the phenotype of the biofilm. The biofilm of strains tviB (NT1) and tviC (NT1) showed a remarkably thicker and more compact structure than cr0 strain (Figure 7J). In addition, more biomass was adhered on the carbon fiber of strains tviC (NT1) and ftsZ (NT1) than strain cr0, suggesting high-level adhesiveness of engineered strains to anodes (Figure 7J). Based on the above bio-imaging results, repression of cell division- and polysaccharide synthesis-related genes led to the expected changes in phenotypes, including cell morphology, biofilm thickness/density, and cell-anode adhesiveness, thereby enhancing the electron transfer capacity of the engineered strains in the MFCs. Therefore, the PaeCascade-RpoD-mediated CRISPRi system demonstrated the outstanding ability of transcription inhibition of native genes, and contributed to improved EET efficiency in S. oneidensis MR-1.
Simultaneous activation and inhibition by PaeCascade-RpoD in S. oneidensis MR-1
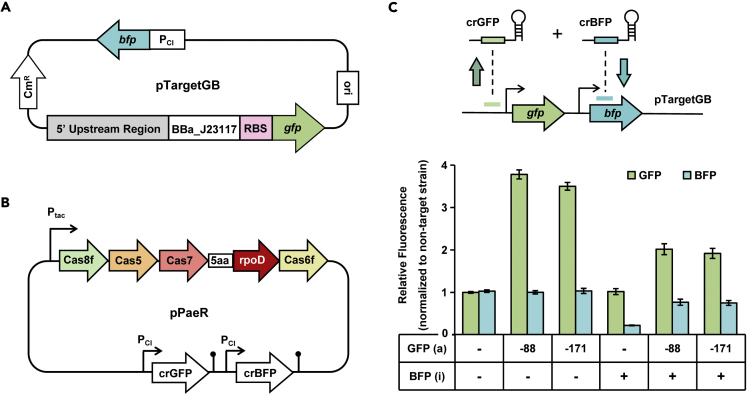
On the basis of PaeCascade-RpoD enabling sole activation or inhibition, the possibility of simultaneous modulation was further testified. We attempted to activate GFP and repress BFP at the same time, and gfp and bfp were constructed in one plasmid pTargetGB (Figure 8A). Then two crRNAs targeting gfp and bfp were assembled into another plasmid pPaeR (Figure 8B). As shown in Figure 8C, the fluorescence of gfp was increased by ∼2.0-fold or ∼1.9-fold targeting 88 bp (NT) or 171 (T) upstream of TSS, and the expression of bfp was decreased by ∼1.3-fold targeting ORF. The results indicated that PaeCascade-RpoD was able to achieve gene activation and inhibition at the same time. The fold changes in simultaneous regulation were lower than that of separate regulation (∼3.8-fold for sole activation and ∼4.5-fold for sole repression) (Figure 8C). We speculated the possible reason was that two crRNAs competed for a fixed pool of Cas proteins, leading to the insufficient supply of Cascade for activating GFP and inhibiting BFP. In sum, PaeCascade-RpoD has the potential to perform orchestra regulation easily by incorporating multiple crRNAs, providing a powerful dual-modulation tool in S. oneidensis.
Figure 8.
Simultaneous activation and inhibition by PaeCascade-RpoD in S. oneidensis MR-1
(A) Plasmid map of pTargetGB for simultaneous regulation.
(B) Plasmid map of pPaeR containing two crRNA expression cassettes for activating GFP and inhibiting BFP.
(C) Schematic diagram and simultaneous regulation efficiency of PaeCascade-RpoD. GFP (a) indicates GFP activation, and BFP (i) indicates BFP inhibition. “+” stands for performing corresponding regulation, and “-” stands for without corresponding regulation. “-88” or “-171” means the targeting position for activating GFP. Median fluorescence is normalized to the non-target strain. Values and error bars indicate mean ± SEM of three replicates.
Discussion
In this work, the transcriptional activation was achieved by CRISPRa in S. oneidensis. To our knowledge, the previous approaches to increase gene expression level in S. oneidensis MR-1 is overexpression based on plasmids and genomic knock-in (Fan et al., 2021a, 2021b; Yi and Ng, 2021). These methods are labor-intensive and time-consuming, owing to the difficulty of plasmid construction, inevitable steps of codons optimization and appropriate promoter selection to achieve specially adapted strength for the EET improvement (Cao et al., 2019). For the PaeCascade-RpoD-mediated CRISPRa system implemented in this study, only 32-bp crRNA needs to be designed and assembled into plasmid through the Golden Gate strategy to enhance the transcription level of target genes. Thus, PaeCascade-RpoD makes it easier to obtain regulated strains with high EET efficiency rapidly.
Furthermore, this novel type I-F CRISPR/PaeCascade-RpoD-mediated activation and inhibition regulation (CRISPR-PAIR) platform not only enables transcriptional activation readily, but also achieves transcriptional inhibition and even simultaneous modulation through a single system in S. oneidensis MR-1. Compared to the previous CRISPRi tools (Cao et al., 2017; Li et al., 2020) and PaeCascade system without RpoD, CRISPR-PAIR platform facilitates multi-mode and multi-gene regulation more conveniently. Besides, genetic up-regulation and down-regulation tools can be used as Amplicon and NOT-gate in genetic circuits (Santos-Moreno and Schaerli, 2020; Wall et al., 2004). In this way, combining our CRISPR-PAIR platform with biosensors could shed light on the dynamical control of the substance and energy metabolism automatically.
For the practical applications of MFCs, the use of antibiotics and inducers is detrimental. To circumvent this problem, it is a good choice to replace the inducible promoter with a constitutive promoter and insert our system into the genome of S. oneidensis MR-1. Accordingly, gene activation and inhibition are able to be implemented without the addition of antibiotics and inducible agents. Meanwhile, in the context of EET fundamental research, the CRISPR-PAIR platform is an ideal tool to identify the relationship between genes and electron transfer, offering the potential of large-scale modulation to inspire mechanistic studies of EET in S. oneidensis.
Limitations of the study
Type I-F Cascade (cas8f, cas5, cas7, and cas6f) gene sequences used in plasmids pPaeCascade and pPaeR were directly obtained from the plasmid pCsy_complex. That is to say, the sequences were not codon-optimized, which might result in the suboptimal effect of gene activation and inhibition via CRISPR-PAIR platform.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli DH5α | Lab stock | N/A |
| E. coli Trans1-T1 | Lab stock | N/A |
| E. coli WM3064 | Lab stock | N/A |
| S. oneidensis MR-1 | Lab stock | N/A |
| S. oneidensis GZ | Cheng et al., 2020 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 60% Sodium lactate solution | ACMEC Biochemical | S58050 |
| 2,6-Diaminopimelic acid | Yuanye Biology | S30747 |
| Kanamycin Sulfate | Solarbio | Cat#K8020 |
| Chloromycetin | Solarbio | Cat#C8050 |
| Potassium ferricyanide(Ⅲ) | J&K Scientific | Cat#911115 |
| Critical commercial assays | ||
| RNAprep pure Cell/Bacteria Kit | TIANGEN | Cat#DP430 |
| RevertedAid First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | Cat#K1622 |
| SYBR Green super mix | AB clonal | Cat#RM21203 |
| LIVE/DEADTM BaclightTM bacterial Viability Kit | Invitrogen (Thermo Fisher) | Cat#L7012 |
| Oligonucleotides | ||
| All oligonucleotides used as guide sequences or for qRT-PCR and plasmid construction are listed in supplemental materials. | N/A | N/A |
| Recombinant DNA | ||
| All plasmids used in this study are listed in supplemental materials. | N/A | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yingxiu Cao (caoyingxiu@tju.edu.cn).
Materials availability
All requests for strains and plasmids constructed in this study should be directed to lead contact, Yingxiu Cao (caoyingxiu@tju.edu.cn).
Experimental model and subject details
E. coli DH5α and Trans-T1 were employed for cloning and cultivated aerobically at 37°C in Luria-Bertani (LB) broth. E. coli WM3064 was used to carry out Golden Gate Assembly and transform plasmids into S. oneidensis MR-1. E. coli WM3064 was cultivated in LB broth with 0.3 mM 2,6-Diaminopimelic acid (DAP). Kanamycin (Km, 50 μg/mL) or chloramphenicol (Cm, 34 μg/mL) was added to LB broth as required. S. oneidensis MR-1 and S. oneidensis GZ strains were cultured aerobically at 30 °C in LB broth. Km (50 μg/mL), Cm (34 μg/mL), and isopropyl-β-D-thiogalactopyranoside (IPTG, 0.8 mM for Ptac) were added in the medium as required.
Method details
Plasmid construction and crRNAs design
The plasmids used in this study are listed in Table S1.
pPaeCascade was constructed by Genewiz with cas8f (csy1), cas5 (csy2), cas7 (csy3), cas6f (csy4) sequence from pCsy_complex, gift from Prof. Puping Liang (Chowdhury et al., 2017) and crRNA cassette integrated to pCYR011 (Chen et al., 2022). To facilitate rapid construction of crRNA, we introduced Golden Gate assembly strategy (Engler et al., 2008; Fang et al., 2021), so the crRNA cassette contained repeat sequences and spacer sequence with BsaI recognition sites synthesized by Genewiz (China). For Golden Gate assembly, a 17-bp base sequence is between the repeats of PaeCascade handle: GAAA-AgagaccAAAggtctcG-GTTC (lowercase letters represent the BsaI recognition site) in the crRNA expression cassette. The sticky ends of -CTTT and GTTC- were generated with the cleavage by BsaI. Besides, the designed spacer primers were annealed to form a 32-bp DNA fragment with a 4-bp sticky end, which was just complementary to -CTTT and GTTC- produced by BsaI in the crRNA expression cassette in the plasmid. Then 32-bp spacer sequence was connected to the plasmid by DNA ligation in one pot (Fang et al., 2021). pSpuCascade was constructed with cas7fv, cas5fv, cas6fv sequences and corresponding rRNA cassette synthesized by Genewiz (China) integrated to pCYR011. pGsuCascade was constructed with cas8u2, cas7, cas5 sequences and corresponding crRNA cassette synthesized by Genewiz (China) integrated to pCYR011. To generate pPaeR, the linker sequence (-ggtggtggtggttct-) and rpoD sequence were connected to C-terminus of Cas7 derived from pPaeCascade. pPaeR-cr0 were constructed by removing the spacer sequence from pPaeR utilizing 2×Seamless Cloning Mix (Biomed, China). pTarget was constructed by Genewiz with J1 upstream region (Dong et al., 2018), promoter J23117 and gfp sequence integrated to pEWTEST1 (Meitl et al., 2009). Sequences of the primers for plasmid construction, genome amplification and sequencing are listed in Table S2. Sequences of PaeR and crRNA cassette used in this study are listed in Table S3.
Fluorescence assay in S. oneidensis MR-1
GFP were employed to characterize expression intensity. The promoter Ptac was induced by 0.8 mM IPTG. When gfp was in the plasmid pTarget, chloramphenicol (Cm, 34 μg/mL) was needed to add to LB broth. For fluorescence intensity assay, every strain inoculated from a fresh colony on an LB agar plate was cultured in LB medium (2 ml) with corresponding antibiotics for 12 h as a seed culture. Then 50 μl of seed culture was added to 5 ml LB medium with the proper antibiotic and the corresponding concentration of inducer in test tubes. After 24-h incubation with constant shaking (200 rpm) at 30°C, 50 μl suspensions from each test tube were centrifuged at 5000 rpm for 8 min to remove the supernatant. Then we resuspended the culture with 500 μl of phosphate-buffered saline (PBS) and transferred 200 μl of suspension to a 96-well polystyrene plate (black plat) (Greiner bio-one μclear, Germany). Cell optical density and fluorescence intensity were detected by a Tecan Infinite200 M Plex microplate reader. Optical density was measured at 600 nm. The excitation/emission wavelength was set at 485 nm/520 nm for GFP, and 399 nm/456 nm for BFP. Assays were conducted in triplicate, and S. oneidensis MR-1 strains with pPaeR-cr0 were used as control. Relative fluorescence was calculated as the following equation.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the strains in mid-log phase through a RNAprep pure Cell/Bacteria Kit (TIANGEN, China) on the basis of the manufacturer’s instructions. Besides, cDNA was obtained via the RevertedAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, the USA). Quantitative analysis of gene expression was conducted by SYBR Green super mix (AB clonal, the USA). The gyrB gene of S. oneidensis MR-1 was utilized for normalization. Samples were tested in triplicate with the listed primers (Table S3). PCR conditions consisted of denaturing at 95°C for 1 min, and 40 cycles of denaturing at 95°C for 15 s followed by annealing and extension at 60°C for 30 s. Relative gene expression was calculated using the method 2−ΔΔCT, normalized with the reference gene gyrB.
Bio-electrochemical characterization
omcA, ribC, bolA-activated strains, and tviB, tviC, ftsZ-inhibited strains and cr0 strain were inoculated from a fresh single colony on an LB agar plate with Km (50 μg/mL) into 2 ml LB medium with Km (50 μg/mL) as a seed culture. 1 mL seed was added into 100 mL fresh LB broth with Km (50 μg/mL) and IPTG(0.8 mM) at 30°C with shaking (200 rpm). After about 10-h culture, we adjusted the concentrations of cell suspensions to the same level (OD600 = 0.5). MFCs were incubated in a 30°C incubator conducted in triplicate, and S. oneidensis MR-1 strains with pPaeR-cr0 were used as control. Dual-chamber MFCs with 140 mL as working volume separated through the Fanion 117 membrane (DuPont Inc., the USA) were employed in this study. Carbon cloth was utilized as the electrodes for cathode (2.5 cm × 3 cm) and anode (1 × 1 cm, the geometric area is 1 cm2). The anolyte was composed of M9 buffer (NaCl, 0.5 g/L; NH4Cl, 1 g/L; KH2PO4, 3 g/L; Na2HPO4, 6 g/L; CaCl2, 0.1 mM MgSO4, 1 mM), supplemented with 20 mM lactate, 5% (v/v) LB broth, 1 mg/L Kan and 1 mM IPTG. The catholyte constituted 50 mM KH2PO4, 50 mM K3[Fe (CN)6], and 50 mM K2HPO4 solution. To measure the voltage generation, we connected a 2k external resistor with the external circuits of MFCs, and the output voltages were recorded. Linear sweep voltammetry (LSV) analysis with a scan rate (0.1 mV/s) was performed on a two-electrode mode. The anode was used as the working electrode with the cathode as the reference. The electrode got the polarization curves to estimate the maximum power density. Power density (P) was calculated as P = V (output voltage) × I (current density). Both I and P were normalized to the projected area of the anode surface.
Quantification of extracellular flavin
ribC-activated strain and cr0 strain were inoculated from a fresh single colony on an LB agar plate with Km (50 μg/mL) into 2 mL LB medium with Km (50 μg/mL) as a seed culture. 0.5 mL seed was added into 50 mL fresh LB broth with Km (50 μg/mL) and IPTG (0.8 mM) at 30°C with shaking (200 rpm). After 12-h fermentation, 1 mL suspensions were centrifuged at 10000 rpm for 10 min to obtain the supernatant. Then the supernatant was filtered through membrane (0.22 mm) and riboflavin (RF) in the supernatant was determined by high-performance liquid chromatography (HPLC, Shimadzu Corporation).
Bio-imaging
For strain cr0 and ftsZ(i), after LSV synthesis was conducted, we cut the carbon cloth, soaked it in 2.5% glutaraldehyde, and put it in the refrigerator at 4°C overnight. Then after pouring out the glutaraldehyde, the carbon cloth was soaked in the PBS for three times (8 min/time). Next, the carbon cloth was soaked in 30%, 50%, 70%, 80%, and 90% ethanol solutions (8 min/time), respectively. Finally, vacuum freeze-drying was conducted for 10 h, and the bacteria on the carbon cloth were imaged by scanning electron microscope.
For strain cr0, bolA, tviB, tviC, ftsZ, anode carbon cloth was dyed by LIVE/DEADTM BaclightTM bacterial Viability Kit. Then the carbon cloth was imaged to observe biofilm thickness and morphology by Confocal Laser Scanning Microscope (CLSM).
Quantification and statistical analysis
Results are reported as values with error bars, which indicate mean ± SEM of technical triplicates in the figure legends. SEM indicates the standard error of the mean. The figures of power density output are drawn by Origin software. The CLSM bio-imaging was exported by ZEN software.
Acknowledgments
We thank Prof. Puping Liang for his gift of plasmid pCsy_complex. We thank Prof. Dongfeng Liu for his gift of chassis S. oneidensis GZ. We thank Prof. Yujie Feng and Dr. Yunfei Li for their technical support in operating the CLSM. This work was supported by the National Key Research and Development Program of China (2018YFA0901300), the National Natural Science Foundation of China (NSFC 21621004, NSFC 32071411, and NSFC 22078240), and the Young Elite Scientists Sponsorship Program by Tianjin (TJSQNTJ-2018-16).
Author contributions
Conceptualization, Y.R.C., M.J.C., and Y.X.C.; Methodology, Y.R.C. and M.J.C.; Investigation, M.J.C. and Y.R.C.; Writing - Original Draft, M.J.C., Y.R.C., H.S., and Y.X.C.; Writing - Review & Editing, M.J.C., Y.R.C., H.S., and Y.X.C.; Supervision, H.S. and Y.X.C.; Funding Acquisition, H.S. and Y.X.C.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104491.
Contributor Information
Hao Song, Email: hsong@tju.edu.cn.
Yingxiu Cao, Email: caoyingxiu@tju.edu.cn.
Supplemental information
–S4
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bhokisham N., VanArsdale E., Stephens K.T., Hauk P., Payne G.F., Bentley W.E. A redox-based electrogenetic CRISPR system to connect with and control biological information networks. Nat. Commun. 2020;11:2427. doi: 10.1038/s41467-020-16249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Dai K., Subbarao S., Beall B., Lutkenhaus J. FtsZ and cell division. Res. Microbiol. 1991;142:249–252. doi: 10.1016/0923-2508(91)90037-b. [DOI] [PubMed] [Google Scholar]
- Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel E.D., Gralnick J.A. Shuttling happens: soluble flavin mediators of extracellular electron transfer in Shewanella. Appl. Microbiol. Biotechnol. 2012;93:41–48. doi: 10.1007/s00253-011-3653-0. [DOI] [PubMed] [Google Scholar]
- Burstein D., Harrington L.B., Strutt S.C., Probst A.J., Anantharaman K., Thomas B.C., Doudna J.A., Banfield J.F. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady K.C., Bondy-Denomy J., Heussler G.E., Davidson A.R., O'Toole G.A. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 2012;194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li X., Li F., Song H. CRISPRi-sRNA: transcriptional-translational regulation of extracellular electron transfer in Shewanella oneidensis. ACS Synth. Biol. 2017;6:1679–1690. doi: 10.1021/acssynbio.6b00374. [DOI] [PubMed] [Google Scholar]
- Cao Y., Song M., Li F., Li C., Lin X., Chen Y., Chen Y., Xu J., Ding Q., Song H. A synthetic plasmid toolkit for Shewanella oneidensis MR-1. Front. Microbiol. 2019;10:410. doi: 10.3389/fmicb.2019.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Su T., Qi Q., Liang Q. Easy regulation of metabolic flux in Escherichia coli using an endogenous type I-E CRISPR-Cas system. Microb. Cell Fact. 2016;15:195. doi: 10.1186/s12934-016-0594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Fang L., Ying X., Cheng M., Wang L., Sun P., Zhang Z., Shi L., Cao Y., Song H. Development of whole genome-scale base editing toolbox to promote efficiency of extracellular electron transfer in Shewanella Oneidensis MR-1. Adv. Biol. 2022;6:e2101296. doi: 10.1002/adbi.202101296. [DOI] [PubMed] [Google Scholar]
- Chen Y., Liu J., Zhi S., Zheng Q., Ma W., Huang J., Liu Y., Liu D., Liang P., Songyang Z. Repurposing type I-F CRISPR-Cas system as a transcriptional activation tool in human cells. Nat. Commun. 2020;11:3136. doi: 10.1038/s41467-020-16880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Carter J., Rollins M.F., Golden S.M., Jackson R.N., Hoffmann C., Nosaka L., Bondy-Denomy J., Maxwell K.L., Davidson A.R., et al. Structure reveals mechanisms of viral suppressors that intercept a CRISPR RNA-guided surveillance complex. Cell. 2017;169:47–57.e11. doi: 10.1016/j.cell.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Fontana J., Patel A., Car J.M., Zalatan J.G. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 2018;9:2489. doi: 10.1038/s41467-018-04901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressaire C., Moreira R.N., Barahona S., Alves de Matos A.P., Arraiano C.M. BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio. 2015;6 doi: 10.1128/mBio.02352-14. e02352–02314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwarakanath S., Brenzinger S., Gleditzsch D., Plagens A., Klingl A., Thormann K., Randau L. Interference activity of a minimal Type I CRISPR-Cas system from Shewanella putrefaciens. Nucleic Acids Res. 2015;43:8913–8923. doi: 10.1093/nar/gkv882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edel M., Horn H., Gescher J. Biofilm systems as tools in biotechnological production. Appl. Microbiol. Biotechnol. 2019;103:5095–5103. doi: 10.1007/s00253-019-09869-x. [DOI] [PubMed] [Google Scholar]
- Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.Y., Tang Q., Li F.H., Sun H., Min D., Wu J.H., Li Y., Li W.W., Yu H.Q. Enhanced bioreduction of radionuclides by driving microbial extracellular electron pumping with an engineered CRISPR platform. Environ. Sci. Technol. 2021;55:11997–12008. doi: 10.1021/acs.est.1c03713. [DOI] [PubMed] [Google Scholar]
- Fan Y.Y., Tang Q., Li Y., Li F.H., Wu J.H., Li W.W., Yu H.Q. Rapid and highly efficient genomic engineering with a novel iEditing device for programming versatile extracellular electron transfer of electroactive bacteria. Environ. Microbiol. 2021;23:1238–1255. doi: 10.1111/1462-2920.15374. [DOI] [PubMed] [Google Scholar]
- Fang L., Fan J., Luo S., Chen Y., Wang C., Cao Y., Song H. Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids. Nat. Commun. 2021;12:4976. doi: 10.1038/s41467-021-25243-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J.K., Romine M.F., Beliaev A.S., Auchtung J.M., Driscoll M.E., Gardner T.S., Nealson K.H., Osterman A.L., Pinchuk G., Reed J.L., et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 2008;6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- Ghavami S., Pandi A. CRISPR interference and its applications. Prog. Mol. Biol. Transl. Sci. 2021;180:123–140. doi: 10.1016/bs.pmbts.2021.01.007. [DOI] [PubMed] [Google Scholar]
- Gleditzsch D., Muller-Esparza H., Pausch P., Sharma K., Dwarakanath S., Urlaub H., Bange G., Randau L. Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system. Nucleic Acids Res. 2016;44:5872–5882. doi: 10.1093/nar/gkw469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralnick J.A., Newman D.K. Extracellular respiration. Mol. Microbiol. 2007;65:1–11. doi: 10.1111/j.1365-2958.2007.05778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D.H., Ha S.C., Kim J.S. A CRISPR RNA is closely related with the size of the cascade nucleoprotein complex. Front. Microbiol. 2019;10:2458. doi: 10.3389/fmicb.2019.02458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzuma A., Meng X.Y., Kimura N., Hashimoto K., Watanabe K. Disruption of the putative cell surface polysaccharide biosynthesis gene SO3177 in Shewanella oneidensis MR-1 enhances adhesion to electrodes and current generation in microbial fuel cells. Appl. Environ. Microbiol. 2010;76:4151–4157. doi: 10.1128/AEM.00117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li Y., Sun L., Li X., Yin C., An X., Chen X., Tian Y., Song H. Engineering Shewanella oneidensis enables xylose-fed microbial fuel cell. Biotechnol. Biofuels. 2017;10:196. doi: 10.1186/s13068-017-0881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tang Q., Li Y., Fan Y.Y., Li F.H., Wu J.H., Min D., Li W.W., Lam P.K.S., Yu H.Q. Rediverting electron flux with an engineered CRISPR-ddAsCpf1 system to enhance the pollutant degradation capacity of Shewanella oneidensis. Environ. Sci. Technol. 2020;54:3599–3608. doi: 10.1021/acs.est.9b06378. [DOI] [PubMed] [Google Scholar]
- Li Y., Lin Z., Huang C., Zhang Y., Wang Z., Tang Y.J., Chen T., Zhao X. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab. Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Lian J., HamediRad M., Hu S., Zhao H. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat. Commun. 2017;8:1688. doi: 10.1038/s41467-017-01695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.F., Min D., Cheng L., Zhang F., Li D.B., Xiao X., Sheng G.P., Yu H.Q. Anaerobic reduction of 2,6-dinitrotoluene by Shewanella oneidensis MR-1: roles of Mtr respiratory pathway and NfnB. Biotechnol. Bioeng. 2017;114:761–768. doi: 10.1002/bit.26212. [DOI] [PubMed] [Google Scholar]
- Logan B.E., Rossi R., Ragab A., Saikaly P.E. Electroactive microorganisms in bioelectrochemical systems. Nat. Rev. Microbiol. 2019;17:307–319. doi: 10.1038/s41579-019-0173-x. [DOI] [PubMed] [Google Scholar]
- Luo M.L., Mullis A.S., Leenay R.T., Beisel C.L. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitl L.A., Eggleston C.M., Colberg P.J.S., Khare N., Reardon C.L., Shi L. Electrochemical interaction of Shewanella oneidensis MR-1 and its outer membrane cytochromes OmcA and MtrC with hematite electrodes. Geochem. Cosmochim. Acta. 2009;73:5292–5307. [Google Scholar]
- Mevers E., Su L., Pishchany G., Baruch M., Cornejo J., Hobert E., Dimise E., Ajo-Franklin C.M., Clardy J. An elusive electron shuttle from a facultative anaerobe. Elife. 2019;8:e48054. doi: 10.7554/eLife.48054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min D., Cheng L., Zhang F., Huang X.N., Li D.B., Liu D.F., Lau T.C., Mu Y., Yu H.Q. Enhancing extracellular electron transfer of Shewanella oneidensis MR-1 through coupling improved flavin synthesis and metal-reducing conduit for pollutant degradation. Environ. Sci. Technol. 2017;51:5082–5089. doi: 10.1021/acs.est.6b04640. [DOI] [PubMed] [Google Scholar]
- Reardon C.L., Dohnalkova A.C., Nachimuthu P., Kennedy D.W., Saffarini D.A., Arey B.W., Shi L., Wang Z., Moore D., McLean J.S., et al. Role of outer-membrane cytochromes MtrC and OmcA in the biomineralization of ferrihydrite by Shewanella oneidensis MR-1. Geobiology. 2010;8:56–68. doi: 10.1111/j.1472-4669.2009.00226.x. [DOI] [PubMed] [Google Scholar]
- Santos-Moreno J., Schaerli Y. CRISPR-based gene expression control for synthetic gene circuits. Biochem. Soc. Trans. 2020;48:1979–1993. doi: 10.1042/BST20200020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E., Kuznedelov K., Datsenko K.A., Boudry P.M., Savitskaya E.E., Medvedeva S., Beloglazova N., Logacheva M., Yakunin A.F., Severinov K. The Cas6e ribonuclease is not required for interference and adaptation by the E. coli type I-E CRISPR-Cas system. Nucleic Acids Res. 2015;43:6049–6061. doi: 10.1093/nar/gkv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Dong H., Reguera G., Beyenal H., Lu A., Liu J., Yu H.Q., Fredrickson J.K. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 2016;14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- Shi L., Squier T.C., Zachara J.M., Fredrickson J.K. Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 2007;65:12–20. doi: 10.1111/j.1365-2958.2007.05783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.V., Edel M., Gescher J., Paquete C.M. Exploring the effects of bolA in biofilm formation and current generation by Shewanella oneidensis MR-1. Front. Microbiol. 2020;11:815. doi: 10.3389/fmicb.2020.00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar K., Wang V.B., Chen X.F., Bazan G.C., Kjelleberg S., Loo S.C.J., Cao B. Membrane permeabilization underlies the enhancement of extracellular bioactivity in Shewanella oneidensis by a membrane-spanning conjugated oligoelectrolyte. Appl. Microbiol. Biotechnol. 2014;98:9021–9031. doi: 10.1007/s00253-014-5973-3. [DOI] [PubMed] [Google Scholar]
- Wall M.E., Hlavacek W.S., Savageau M.A. Design of gene circuits: lessons from bacteria. Nat. Rev. Genet. 2004;5:34–42. doi: 10.1038/nrg1244. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Manefield M., Lee M., Kouzuma A. Electron shuttles in biotechnology. Curr. Opin. Biotechnol. 2009;20:633–641. doi: 10.1016/j.copbio.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Westra E.R., van Erp P.B., Kunne T., Wong S.P., Staals R.H., Seegers C.L., Bollen S., Jore M.M., Semenova E., Severinov K., et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., van Duijn E., Bultema J.B., Waghmare S.P., Zhou K., Barendregt A., Westphal W., Heck A.J., Boekema E.J., Dickman M.J., et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. U S A. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Li M., Li Y., Cao H., Miao L., Xu Z., Higuchi Y., Yamasaki S., Nishino K., Woo P.C.Y., et al. Native CRISPR-cas-mediated genome editing enables dissecting and sensitizing clinical Multidrug-resistant P. aeruginosa. Cell Rep. 2019;29:1707–1717.e3. doi: 10.1016/j.celrep.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Yi Y.-C., Ng I.S. Redirection of metabolic flux in Shewanella oneidensis MR-1 by CRISPRi and modular design for 5-aminolevulinic acid production. Bioresour. Bioprocess. 2021;8:13–23. doi: 10.1186/s40643-021-00366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong Y.C., Cai Z., Yu Y.Y., Chen P., Jiang R., Cao B., Sun J.Z., Wang J.Y., Song H. Increase of riboflavin biosynthesis underlies enhancement of extracellular electron transfer of Shewanella in alkaline microbial fuel cells. Bioresour. Technol. 2013;130:763–768. doi: 10.1016/j.biortech.2012.11.145. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Han J., Wang B., Hu X., Li R., Shen W., Ma X., Ma L., Yi L., Yang S., et al. Characterization and repurposing of the endogenous Type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res. 2019;47:11461–11475. doi: 10.1093/nar/gkz940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Li J., Wang B., Han J., Hao Y., Wang S., Ma X., Yang S., Ma L., Yi L., et al. Endogenous type I CRISPR-cas: from foreign DNA defense to prokaryotic engineering. Front. Bioeng. Biotechnol. 2020;8:62. doi: 10.3389/fbioe.2020.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Lu Z.S., Huang Y.H., Long Z.E., Qiao Y. Nanoporous Mo2C functionalized 3D carbon architecture anode for boosting flavins mediated interfacial bioelectrocatalysis in microbial fuel cells. J. Power Sources. 2017;359:549–555. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
–S4
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.