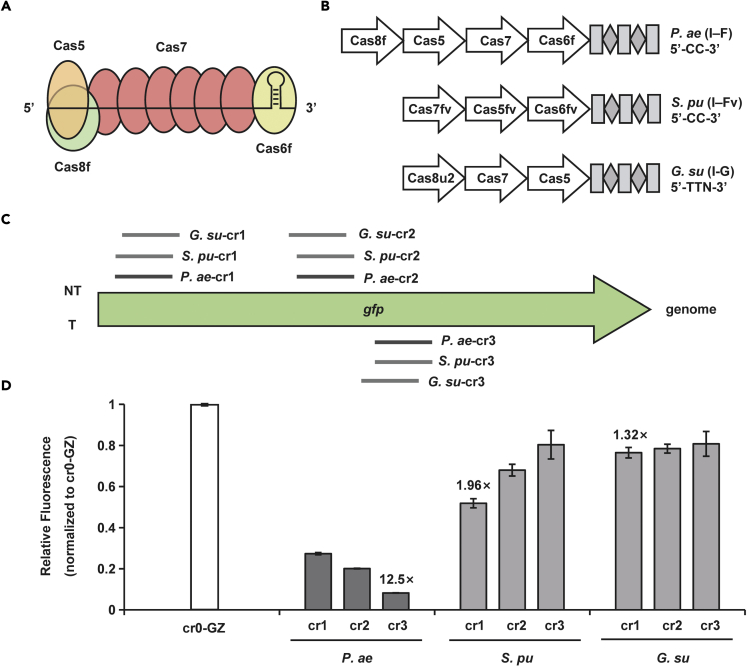
Figure 2.
To screen type I CRISPR-Cas systems in S. oneidensis
(A) The structure of PaeCascade-crRNA complex. Cas5 binds to the 5′ handle of the crRNA and Cas8f is responsible for PAM recognition (5′-CC-3′) at the 5′ end of the protospacer. Cas7 proteins function as several copies bind to the protospacer, serving as the backbone of the cascade complex. Cas6f combines with crRNA 3′ hairpin structure and mediates pre-crRNA maturation.
(B) The schematic diagram of the P. aeruginosa type I-F (P. ae I-F), S. putrefaciens type I-Fv (S. pu I-Fv), and G. sulfurreducens type I-G (G. su I-G) CRISPR-Cas systems used in this study. Cas proteins are presented with arrows. CRISPR repeats and spacers are indicated with squares and diamonds, respectively.
(C) Targeting sites of designed crRNAs binding different positions of gfp in the genome of the strain S. oneidensis GZ for three type I CRISPR-Cas systems. Three crRNAs were designed for each system.
(D) The inhibition efficiency of three type I CRISPR-Cas systems targeting gfp. The values above the bar indicate the highest repression folds of three crRNAs for each system. Median GFP levels are normalized to the control strain cr0-GZ. Values and error bars indicate mean ± standard error of mean (SEM) of three replicates.