Abstract
The yeast Williopsis mrakii produces a mycocin or yeast killer toxin designated HMK; this toxin exhibits high thermal stability, high pH stability, and a broad spectrum of activity against other yeasts. We describe construction of a synthetic gene for mycocin HMK and heterologous expression of this toxin in Aspergillus niger. Mycocin HMK was fused to a glucoamylase protein carrier, which resulted in secretion of biologically active mycocin into the culture media. A partial purification protocol was developed, and a comparison with native W. mrakii mycocin showed that the heterologously expressed mycocin had similar physiological properties and an almost identical spectrum of biological activity against a number of yeasts isolated from silage and yoghurt. Two food and feed production systems prone to yeast spoilage were used as models to assess the ability of mycocin HMK to act as a biocontrol agent. The onset of aerobic spoilage in mature maize silage was delayed by application of A. niger mycocin HMK on opening because the toxin inhibited growth of the indigenous spoilage yeasts. This helped maintain both higher lactic acid levels and a lower pH. In yoghurt spiked with dairy spoilage yeasts, A. niger mycocin HMK was active at all of the storage temperatures tested at which yeast growth occurred, and there was no resurgence of resistant yeasts. The higher the yeast growth rate, the more effective the killing action of the mycocin. Thus, mycocin HMK has potential applications in controlling both silage spoilage and yoghurt spoilage caused by yeasts.
Yeasts and molds can have both positive and negative effects on fermented products consumed by humans and animals. Yeasts are used as starter cultures in cheeses and bread, as well as wine, beer, and other alcoholic fermentation products, but they can also initiate spoilage in foods, such as yoghurt, fruit juice, salads, and mayonnaise. Indeed, spoilage yeasts have been isolated from many foods and beverages, including bread (22), wine, beer, fruit juices (56), mayonnaise (4), dairy products (15, 16, 23, 46, 60), and chocolate (28).
In ruminant feeds, yeasts are viewed as spoilage organisms (24, 38, 68) except when they are used as probiotics (12, 63, 64). Silage is a fermented feedstuff which constitutes a major proportion of winter forage for livestock, particularly in Europe and North America (61). The aim of the ensilage process is to preserve crops having high moisture contents at times of abundance by encouraging a natural acidic fermentation, but yeast growth is undesirable either during the fermentation or after exposure to air when the silo is opened for feeding. In well-preserved silages lactic acid bacteria predominate and convert the available soluble sugars to lactic acid and a smaller amount of acetic acid. This causes the pH to decrease rapidly, often to pH values below 4, and inhibits the growth of potential spoilage microorganisms, such as Clostridium spp. and enterobacteria (34). Yeasts enter silos as part of the crop epiphytic microflora, and although they tend to grow best aerobically, many yeasts are efficient fermenters of sugars and are not inhibited by low-pH conditions. Thus, they can thrive in an anaerobic environment by fermenting sugars to alcohol, acetate, and carbon dioxide. Although these products can contribute to preservation to some extent (32), the rate of the decrease in pH is slower than the rate when lactic acid bacteria dominate fermentation and lactic acid is produced. Carbon dioxide produced by yeast fermentation leads to a net loss of dry matter (34) and a corresponding reduction in nutritive value. Extensive alcohol yeast fermentation is unusual in silage and generally occurs only when the level of dry matter is greater than 40% (13). Organic acids that accumulate during ensilage, such as lactate and acetate, can be assimilated further only by aerobic metabolic pathways (34). Aerobic spoilage occurs when air permeates the silage during storage or when the silo is opened and the silage is exposed to the atmosphere. Aerobic microorganisms become active, which results in an increase in the pH as degradation of lactate, acetate, and other fermentation products occurs. This leads to progressive reduction in the nutritive value of the silage as successive populations of yeasts, acetic acid bacteria, and molds become active. Lactate-assimilating yeasts, which are capable of rapid growth in the presence of oxygen, are considered the primary initiators of aerobic spoilage (39). Thus, yeasts act as spoilage microorganisms in a number of different systems, and many chemical and biological approaches have been taken to prevent proliferation of those organisms (34, 35, 37, 59).
Killer yeasts produce antimycotic compounds to which they themselves are immune (33). Yeast killer toxins are proteinaceous compounds which are active against members of the same species or closely related species, and the activities of these toxins are analogous to the activities of bacteriocins in bacterial species. We prefer to call yeast killer toxins mycocins and killer strains mycogenic in order to emphasize the general nature of the antagonistic interactions (17). Mycocins were first found in brewing strains of Saccharomyces cerevisiae (6) and since then have been shown to occur in a large number of yeast species of agronomic, environmental, industrial, and clinical interest, including Candida, Cryptococcus, Debaryomyces, Pichia, Torulopsis, and Williopsis species (17, 42, 74, 75). Certain mycocins have also been shown to have inhibitory effects on some pathogenic gram-positive bacteria, including Staphylococcus aureus (21). Some mycocins have been investigated with a view toward exploitation for biocontrol. However, the use of these toxins as novel antimicrobial compounds can be limited by pH and temperature instability and a narrow spectrum of activity (20). A killer strain of Hansenula mrakii (reclassified as Williopsis mrakii [5]), strain LKB 169 (= NCYC 2251), produces two mycocins that have broad spectra of activity (3). The smaller mycotic peptide was purified, and its amino acid sequence was determined. This toxin is a single 88-amino-acid polypeptide (70) which is designated mycocin HMK or HM-1. It is basic with an isoelectric point of 9.1 (unlike other mycocins, which tend to be acidic) and has a molecular mass of 10.7 kDa (70). It is very thermostable, retaining biological activity after incubation at 100°C for 10 min, and is stable at pH values between 2 and 11. Mycocin HMK has 10 cysteine residues which confer exceptional stability because of multiple disulfide bonds that also appear to be essential for biological activity of the molecule. Mycocin HMK is inhibited by reducing agents, such as β-mercaptoethanol (69), and is inactivated by boiling at a high pH value. It kills intact cells by interfering with β-1,3 glucan synthesis, which makes the cell wall osmotically fragile and thus results in cell lysis and death (55). The chromosomal gene hmk encodes the HMK peptide, which is synthesized as a 125-amino-acid precursor with a 37-amino-acid N-terminal signal sequence. The leader is cleaved at a Kex2 site, which results in the 88-amino-acid mycocin. An almost identical mycocin is encoded by the hsk gene in the related species Williopsis saturnus (26, 27, 30), and this toxin has a wide spectrum of activity against other yeast species (71).
The broad spectrum of activity, the pH stability, and the temperature stability suggest that W. mrakii mycocin HMK could be a versatile antispoilage agent in a range of food and feed systems. Biocontrol studies have focused on the potential of mycocins produced by native or transformed yeasts (25). The killer characteristic has been transferred into starter yeasts in order to combat wild strains during the production of beer (73), wine (7, 18, 49), and bread (8) and also undesirable yeasts during food preservation (40). A killer strain of Kluveromyces lactis was found to prevent aerobic spoilage of silage caused by other yeasts in a model synthetic medium-based system, but the killer strain also contributed to the spoilage (29). Heterologous expression of a mycocin by bacteria could allow delivery of the toxin when inoculation with a mycogenic yeast is undesirable, and purified mycocins could have applications as biocontrol agents in both foods and feeds.
In this paper we describe construction of a synthetic gene for W. mrakii mycocin HMK. We examined expression of this gene in the heterologous host Aspergillus niger. We developed a partial purification procedure for the A. niger HMK toxin, and the effects of this toxin on yeast spoilage were investigated by studying maize silage and yoghurt.
MATERIALS AND METHODS
Strains, culture media, and plasmids.
Escherichia coli MC1022 (10) was used for transformation. The culture conditions used were the conditions described by Sambrook et al. (45). For selection we used L agar (31) supplemented with 50 μg of ampicillin per ml, 20 μg of isopropyl-β-d-thiogalactopyranoside (IPTG) per ml, and 20 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) per ml when appropriate. pUC 18 and pUC 19 vectors were used for cloning (72).
Some yeasts were obtained from the National Collection of Yeast Cultures and Express Dairies Ltd., and others were isolated from seven different silages, including mixed wheat and pea, maize, and grass silages. All yeast strains were maintained on yeast malt extract medium (Oxoid). The yeast methylene blue agar (YMBM) used for mycocin detection was based on the medium described by Philliskirk and Young (42); this medium was acidified to pH 4.1 and contained 0.1 M phosphate-citrate buffer. Initial problems were encountered due to denaturation of the agar when it was autoclaved at pH 4.5 or a lower pH value and to the formation of a green color. These problems were overcome by autoclaving the agar, glucose, and methylene blue separately from the other medium constituents. Liquid yeast cultures were grown routinely in yeast malt extract broth (Oxoid) at 25°C. Mycogenic strains were grown in YEPG broth (YMBM without agar) at 22°C for mycocin production. Yeast viable counts in the yoghurt experiments were obtained by using malt extract agar plates (Oxoid) supplemented with chloramphenicol (5 μg/ml) and streptomycin (5 μg/ml) to prevent bacterial growth. In the silage experiments total yeast counts were determined on malt extract agar containing chloramphenicol and streptomycin and acidified with 0.5% (vol/vol) lactic acid, and lactate-assimilating yeast counts were determined on lactate yeast agar plates (24). Lactate-assimilating bacteria were isolated on lactate yeast agar plates containing the antimycotic compound pimaricin (Sigma) at a concentration of 0.008% (vol/vol) and were confirmed to be bacteria by microscopic examination. They were identified as Acetobacter spp. by using methods described by Carr and Passmore (9). Viable lactic acid bacteria were plated onto MRS agar (Oxoid) supplemented with 0.008% pimaricin as an antimycotic agent. All silage extraction plates were incubated at 30°C for 3 to 5 days. Yeasts were identified by using the methods described by van der Walt and Yarrow (57).
A. niger AB4.1 was used for transformation (58). A. niger strains were maintained as spore cultures on AMMN slopes that contained or did not contain 10 mM uridine (58). Batch cultures of A. niger were grown in ACMS/N/P broth that contained or did not contain 10 mM uridine (2). The plasmids used in Aspergillus transformations were pAB4-1, which allowed growth on minimal media in the absence of uridine (58), and pIGF, a glucoamylase fusion vector used for site-specific integration of heterologous sequences into the glucoamylase gene with an adjacent Kex2 site (1).
Molecular techniques.
Standard procedures were performed as described by Sambrook et al. (45). Restriction enzymes (Life Technologies Ltd.), T4 DNA ligase (Pharmacia), and Taq polymerase (Promega) were all used according to the manufacturers' instructions. PCRs were performed with 100 ng of template DNA in a 50-μl PCR mixture containing 1× Taq buffer, each deoxynucleoside triphosphate at a concentration of 0.1 mM, 20 pmol of each primer, and 1 U of Taq polymerase. The complete double-stranded DNA sequence was determined by using a PRISM ready reaction dideoxy terminator cycle sequencing kit (ABI). Primers were synthesized with an ABI model 394 DNA synthesizer, and sequencing reactions were performed with a model 373A DNA sequencer. Sequences were determined at every stage of construction to ensure that the synthetic toxin gene and any in-frame fusions or introduced restriction sites were correct.
Design of a peptide antibody for the toxin.
A hydrophobic region of the toxin peptide designated KTM (amino acid residues 9 to 26) was selected as a likely epitope for antibody production by using the Peptidestructure computer program, which predicts secondary structure and an antigenicity index (GCG, University of Wisconsin). Polyclonal antibodies raised against this peptide were purchased from Genosys Biotechnologies Ltd. (Cambridge, United Kingdom). Final bleed antisera were divided into aliquots and stored at −20°C until they were needed.
SDS-PAGE analysis of proteins, Western blotting, and immunodetection.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by using a Mini-Protean II Dual Slab cell (Bio-Rad Laboratories) as recommended by the manufacturer. Tricine was used instead of glycine for enhanced resolution of small proteins, as described by Schagger and von Jagow (47). Proteins separated by SDS-PAGE were transferred to ProBlott membranes. Immunodetection was performed with primary antibody KTM diluted 500- to 1,000-fold in 20 ml of TTBS buffer containing 1% (wt/vol) bovine serum albumin. The secondary antibody used was A-8025 anti-rabbit immunoglobulin G (whole molecule)–alkaline phosphatase conjugate (Sigma Immuno Chemicals), which was diluted 500-fold in 20 ml of TTBS buffer containing 1% (wt/vol) bovine serum albumin. The secondary antibody was detected via a chemical reaction of the alkaline phosphatase with 5-bromo-4-chloro-3-indolylphosphate (BCIP)–nitroblue tetrazolium. The reaction was developed by incubating the preparation with a Sigma FAST tablet (Sigma Immuno Chemicals) until sufficient color developed.
Transformation.
E. coli transformation was carried out by using the calcium chloride method (45). We used a protoplast transformation system for A. niger which allows efficient introduction of nonselectable genes by cotransformation. A. niger AB4.1 requires uridine for growth. Protoplasts were cotransformed with the selection vector pAB4-1, which allowed growth on minimal media in the absence of uridine (58), and with the pIGF vector with the hmk mycocin gene inserted, which fused the heterologous toxin gene in frame with the Kex2 cleavage site of the glucoamylase coding sequence (1) and allowed secretion of the mycocin into the media.
Screening of yeasts for sensitivity to mycocin HMK.
Crude mycocin HMK-containing and mycocin HMK-free preparations were obtained by filtering batch cultures of Aspergillus strains that had been inoculated with approximately 104 spores per 100 ml and grown with agitation at 25°C for 72 h. Crude mycocin-containing W. mrakii supernatants were obtained by centrifuging and filter sterilizing 48-h cultures grown in buffered YEPG broth (initial pH, 4.1). Samples of these extracts were adjusted to pH 2.0, 2.5, 3.0, 4.5 (unadjusted), 6.0, 8.0, 9.0, and 10.0 and then boiled for 2, 5, and 10 min in order to examine thermal stability and pH stability. The extracts used to compare native Williopsis and heterologous Aspergillus mycocins were boiled for 6 min. Well diffusion plate assays (75) were performed with YMBM agar (pH 4.1) seeded with 60 μl of a fresh overnight yeast culture per 50 ml of agar. Extracts (100 μl) were placed into the wells (diameter, 8 mm) and incubated for 48 h at 22°C. Mycocin activity was scored by measuring the diameter of each inhibition zone, excluding the well.
Partial purification of heterologous mycocin HMK from Aspergillus strains.
A time course study of protein and mycocin production was carried out by using recombinant mycocin-producing and control pAB4.1-containing Aspergillus strains. Total protein contents were determined by the Bio-Rad protein assay, and mycocin activity was determined by performing well diffusion assays. Maximum mycocin production occurred at about 72 h, although the total protein levels continued to increase (data not shown); therefore, 72 h was chosen as the standard incubation time. Mycocin-producing and control Aspergillus cultures (100 ml) were grown for 72 h, and the filtered supernatants were boiled for 10 min. Total supernatant protein was precipitated with 75% ammonium sulfate and was resuspended in a reduced volume of 0.1 M citrate buffer (pH 4.1). The mycocin-containing and control solutions were ultrafiltered by using a YM 30-kDa membrane in a stirred ultrafiltration cell (model 8400; Amicon Ltd.) at a pressure of 55 lb/in2. The filtrate was concentrated by a second ultrafiltration step with a YM 3-kDa membrane (Amicon Ltd.). These two steps removed proteins having molecular masses greater than 30 kDa and less than 3 kDa (data not shown). All fractions were tested for activity by using the well diffusion assay. This partial purification procedure was developed in order to prepare the control and mycocin preparations used in the silage and yoghurt challenge experiments described below.
Quantitative assessment of HMK activity expressed in AU.
The activity of an antagonistic substance can be quantified and expressed in arbitrary units (AU) by using defined preparation and assay conditions (48). The YMBM agar well assay was used with Candida lambica NCYC 2255 as the sensitive indicator strain in order to assess and compare HMK activities in different preparations, including fresh culture filtrates and more purified concentrated extracts. Serial dilutions of the preparations were prepared by using a range of dilutions from the dilution at which the size of the inhibition zone was linear to the dilution at which no antagonistic activity was detected; 1 AU was defined as the reciprocal of the highest dilution that still resulted in a minimal level of detection.
Application of Aspergillus mycocin HMK to silage.
A 20-kg portion of mature (ensiled for 100 days) whole-crop maize silage was obtained from the center of a farm silage clamp at Trawscoed Research Farm, Institute of Grassland and Environmental Research, Aberystwyth, United Kingdom. The silage was well mixed and was divided into 1-kg portions in clean polyethylene bags. Triplicate samples of untreated silage were used to determine microbial counts and for chemical analysis of volative fatty acids and lactic acid (36). Mycocin HMK-containing preparations and control preparations from a mycocin-free A. niger culture and a distilled water control were applied as fine sprays (10 ml/kg); three replicates per treatment were used. The mycocin was added at a final concentration of 30 AU per g of silage.
Silage composition analysis.
The dry matter content, pH, and lactic acid content of silage were analyzed as described by Cussen et al. (11). Volative fatty acids were analyzed in water extracts by gas chromatography by using the method of Zhu et al. (76).
Assessment of the aerobic stability of silage.
When good-quality silage is exposed to oxygen, either through accidental ingress of air in the silo or during feeding, it can spoil following growth of microorganisms, including yeasts, bacteria, and molds (34). Considerable evidence suggests that yeasts initiate spoilage and that a temperature increase accompanies the growth of these and other spoilage microorganisms (35). Temperature increases during aerobic spoilage of silage have been correlated with other indications of growth, including carbon dioxide production, loss of dry matter, increases in pH, and changes in microbial numbers (34, 65, 66). Thus, monitoring temperature changes is the simplest and most commonly used way to assess the aerobic stability or deterioration of silage (19, 62, 64). For electronic temperature monitoring we used a Squirrel series 2000 data logger (Grant Instruments, Cambridge, United Kingdom) and specifically designed aerobic spoilage vessels (ASVs). Each 1-kg silage sample was divided into two 400-g duplicate samples, loosely packed into ASVs, and stored at 20°C. Each ASV consisted of a 30-cm section of polypropylene pipe (diameter, 8 cm) with a perforated base inserted 5 cm from the bottom of the pipe to allow release of CO2 from the spoiling silage. The vessels were wrapped in thermal insulating material and stored upright in cardboard boxes. Each ASV was fitted with a thermocouple probe that was placed in the center of the sample and connected to an electronic data logger (Squirrel series 2000). Temperatures were recorded every hour for 14 days in order to monitor the progress of microbial spoilage. After 72 h one 400-g sample from each replicate pair was destructively sampled and used for microbiological and chemical analyses.
Food challenge experiment.
Two dairy spoilage yeasts, Candida krusei D1241 and Saccharomyces cerevisiae D1247, were used for the food challenge experiment. Aliquots (10 ml) of 65% (wt/vol) plain low-fat yoghurt diluted with sterile distilled water were treated by adding 500-μl portions of a sterile 0.1 M citrate buffer or control preparation or two Aspergillus mycocin HMK preparations that resulted in 167 and 83 AU per ml. The yoghurt samples were inoculated or not inoculated with 103 D1241 or D1247 cells per ml and stored at 4, 10, and 20°C. Samples (100 μl) were removed after 1, 2, 4, 7, 14, and 21 days of storage and used to determine numbers of viable yeast cells.
RESULTS
Design and construction of a structural gene for the mycocin.
Mycocin HMK of W. mrakii NCYC 2251 has been purified previously (3), and the amino acid sequence has been determined (70); the secreted toxin is an 88-amino-acid peptide. The amino acid sequence was used to derive a DNA sequence by utilizing the lactococcal codon preferences. Six overlapping oligonucleotides were synthesized (Table 1). Primer 1 with a 5′ BamHI site was annealed with primer 6, which resulted in fragment A encoding the 5′ N-terminal portion of the peptide. Primers 2 and 5 annealed to give fragment B, which encoded the central region of the peptide. Primer 3 with the 3′ PstI site and primer 4 annealed to give fragment C, which encoded the 3′ C-terminal portion of the peptide. Fragments A, B, and C were treated with kinase and ligated. The resulting synthetic fragment was cloned into pUC18 digested with BamHI and PstI, which resulted in pFI757. Restriction digestion and sequencing confirmed that this construction resulted in the loss of the first glycine residue of the toxin, which was replaced by two alanine residues and a 5′ BamHI site and by a 3′ stop codon and a PstI site.
TABLE 1.
Primers used to construct the synthetic mycocin gene
| Primer | Sequence (5′-3′) | Description |
|---|---|---|
| 1 | GATCCGCCGCGGATGGTTATCTGATCATGTGCAAAAATTGCGATCCAAATACTGGTTCTTGCGATTGGAAACAAAATTGGAATACTTGCGTTGG | 5′ BamHI site, encodes N-terminal portion of mycocin gene |
| 2 | TATCGGTGCTAATGTTCATTGGATGGTTACTGGTGGATCCACTGATGGTAAACAAGGTTGCGCTACTATCTGGGAAGGTTCTGGTTGCGTT | Encodes central portion of mycocin gene |
| 3 | GGTCGTTCTACTACTATGTGCTGCCCAGCTAATACTTGCTGCAATATCAATACTGGTTTTTATATCCGTTCTTATCGTCGTGTTGAATAACTGCA | 3′ PstI site, encodes C-terminal portion of mycocin gene |
| 4 | GTTATTCAACACGACGATAAGAACGGATATAAAAACCAGTATTGATATTGCAGCAAGTATTAGCTGGGCAGCACATAGTAG | Complementary to primer 3 |
| 5 | TAGAACGACCAACGCAACCAGAACCTTCCCAGATAGTAGCGCAACCTTGTTTACCATCAGTGGATCCACCAGTAACCATCCAATGAACAT | Complementary to primers 2 and 3 |
| 6 | TAGCACCGATACCAACGCAAGTATTCCAATTTTGTTTCCAATCGCAAGAACCAGTATTTGGATCGCAATTTTTGCACATGATCAGATAACCATCCGCGGCG | Complementary to primers 1 and 2 |
Provision of a leader sequence for toxin secretion.
The hmk mycocin gene was fused to the glucoamylase protein for expression in A. niger. To do this, we utilized the glucoamylase fusion vector (pIGF) for site-specific integration into the glucoamylase gene with a Kex2 site adjacent to the toxin sequence (1). A PCR was used to amplify the mycocin gene from pFI757 incorporating flanking XbaI sites and a Kex2 cleavage site. The XbaI fragment was cloned in the pUC19 vector (pFI870) and was transferred into pIGF in order to obtain pFI874. A. niger AB4-1 was cotransformed with pFI874 and pAB4-1, which resulted in integration of the toxin gene in the chromosomal glucoamylase gene.
Heterologous expression of mycocin in A. niger.
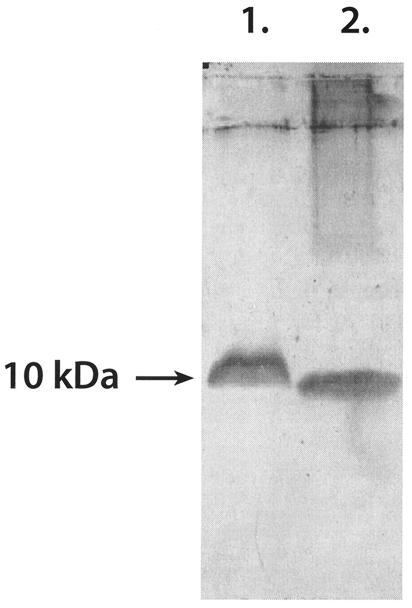
The synthetic gene encoding the W. mrakii mycocin was expressed in A. niger, and the resulting toxin was compared with native Williopsis mycocin HMK by using bioassays (Fig. 1) and Western blots (Fig. 2). Aspergillus mycocin HMK activity was stable in all of the samples at pH values between 2.0 and 10.0 before heat treatment. At pH 8.0 boiling for 10 min resulted in some loss of activity, and at pH 10.0 all activity was lost after 2 min (data not shown). Williopsis mycocin HMK and the heterologous peptide both reacted with the KTM antibodies (Fig. 2).
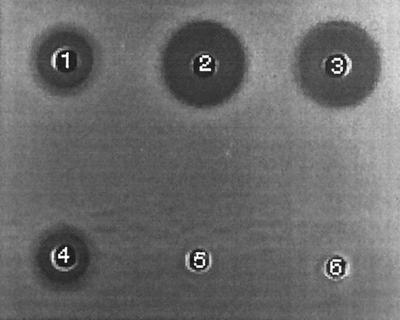
FIG. 1.
Well diffusion bioassay of crude supernatants from W. mrakii and A. niger. C. krusei NCYC 2698 was the sensitive indicator strain (originally isolated from silage) that was inoculated onto YMBM agar. Well 1, W. mrakii, untreated supernatant; well 2, A. niger toxin producer, untreated supernatant; well 3, A. niger toxin producer, supernatant boiled for 10 min; well 4, W. mrakii, supernatant boiled for 10 min; well 5, A. niger AB4.1, untreated supernatant; well 6, A. niger AB4.1, supernatant boiled for 10 min.
FIG. 2.
Western blot probed with KTM peptide antibodies. Lane 1, W. mrakii; lane 2, A. niger. Culture supernatants were concentrated 10-fold before they were loaded onto the SDS-PAGE gel.
A partial purification procedure was developed based on the growth kinetics of HMK production in A. niger. This procedure involved ultrafiltration of the mycocin-expressing Aspergillus culture supernatant, which removed proteins larger than 30 kDa and smaller than 3 kDa and concentrated the mycocin (see above).
Effect of mycocin HMK on yeasts isolated from food and silage.
When a deferred antagonism assay described by Starmer et al. (52; Lowes, unpublished data) was used, W. mrakii NCYC 2251 exhibited strong mycocin activity against 40 different yeast strains isolated from samples of seven different silages. Twelve of these silage yeasts, which were chosen based on their different patterns of sensitivity to a panel of nine different mycogenic yeasts, were used as indicator strains in a comparison of the native W. mrakii mycocin(s) and the heterologous Aspergillus mycocin in well diffusion assays (Table 2). A typical example is shown in Fig. 1, which shows the results obtained with C. krusei NCYC 2698. W. mrakii NCYC 2251 produces more than one mycocin (3), but only mycocin HMK is heat stable. A comparison of W. mrakii mycocin activities before and after heat treatment against the of silage yeast isolates revealed that there were only minimal losses of activity due to heat treatment (data not shown). A. niger mycocin HMK was thermally stable; we observed no differences in zone size when we compared the results obtained for silage yeast isolates with and without heat treatment (data not shown). The apparently greater Aspergillus mycocin activity (Table 2) than Williopsis mycocin HMK activity probably reflected higher concentrations of mycocin HMK in the Aspergillus preparations rather than increased activity.
TABLE 2.
Comparison of W. mrakii and A. niger mycocin HMK activities against silage yeast isolates
| Silage yeast isolate | Diam of inhibition zone (mm) witha:
|
|
|---|---|---|
| W. mrakii mycocin HMK | A. niger mycocin HMK | |
| 3 | 13 | 20 |
| 6 | 14 | 21 |
| 14 | 10 | 18 |
| 36 | 11 | 19 |
| 72 | 16 | 21.5 |
| 73 | 8 | 12 |
| 84 | 13 | 20 |
| 102 | 10 | 18 |
| 128 | 16.5 | 23 |
| 129 | 14 | 20 |
| 133 | 8 | 20 |
| 151 | 14 | 20 |
The diameters of the inhibition zones do not include the diameters of the wells (8 mm).
A number of yeasts responsible for yoghurt spoilage were obtained from the National Collection of Yeast Cultures. These organisms were identified as members of a variety of species, including Candida, Saccharomyces, and Torulaspora species, and were tested to determine their sensitivities to both types of mycocin HMK (Table 3). The HMK-sensitive strains C. lambica NCYC 2255 and S. cerevisiae NCYC 1006 were used as positive controls and were sensitive to both types of mycocin HMK, whereas the native producer W. mrakii NCYC 2251 was resistant. Fifty percent of the dairy yeasts were sensitive to mycocin HMK; the sensitive strains included all of the S. cerevisiae strains but only 20% of the Torulaspora delbrueckii strains. Supernatant from Aspergillus control strain AB4.1 had no effect on the growth of any of the yeast strains tested (data not shown).
TABLE 3.
Comparison of W. mrakii and A. niger mycocin HMK activities against yoghurt yeast isolates
| Yeast species | Strain | Diam of inhibition zone (mm) witha:
|
|
|---|---|---|---|
| W. mrakii mycocin HMK | A. niger mycocin HMK | ||
| Candida lambica | NCYC 2255 | 8 | 14 |
| Candida krusei | D1241 | 6 | 12 |
| Candida intermedia | D1245 | 0 | 0 |
| Saccharomyces cerevisiae | NCYC 1006 | 3.5 | 7 |
| Saccharomyces cerevisiae | D1246 | 3.5 | 8 |
| Saccharomyces cerevisiae | D1247 | 8 | 14 |
| Saccharomyces cerevisiae | D1249 | 8 | 14 |
| Torulaspora delbrueckii | D1242 | 2 | 9 |
| Torulaspora delbrueckii | D1243 | 0 | 0 |
| Torulaspora delbrueckii | D1244 | 0 | 0 |
| Torulaspora delbrueckii | D1248 | 0 | 0 |
| Torulaspora delbrueckii | D1250 | 0 | 0 |
The diameters of the inhibition zones do not include the diameters of the wells (8 mm).
Aerobic stability of maize silage.
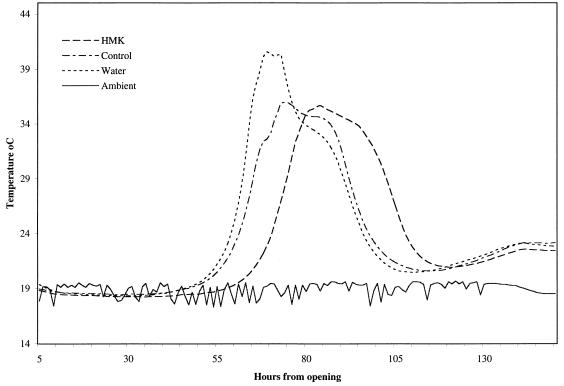
The low pH values, low numbers of yeast cells, and high lactic acid contents of the initial silage samples were typical of well-preserved, good-quality maize silage (Table 4). The silage was treated with semipurified HMK toxin from A. niger and two controls, semipurified protein filtrate from nontransformed A. niger AB4.1 and sterile distilled water. Aerobic spoilage was monitored by determining temperature changes, and the temperature profiles for the 6 days after the silage samples were exposed to air are shown in Fig. 3. Within the treatments the data were very consistent; with the HMK-treated samples there was a clear delay before the initial temperature increase occurred of approximately 15 h, while with both control groups the increase occurred within 48 h.
TABLE 4.
Microbial and chemical analysis of fresh maize silage and maize silage treated in different ways and exposed to aira
| Prepn | pH | % Dry matter | Concn (CFU/g [fresh wt]) of:
|
Concn (mM) of:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Lactic acid bacteria | Total yeasts | Lactate-assimilating yeasts | Acetic acid bacteria | Lactate | Acetate | Propionate | |||
| Fresh silage | 3.49 (0.01)b | 29.6 (0.02) | 2.76 × 107 | 1.0 × 104 | 5.4 × 103 | 5.7 × 103 | 73.4 (1.98) | 22.2 (0.25) | 0.47 (0.01) |
| Silage treated with A. niger mycocin HMK | 3.54 (0.01) | 28.5 (0.44) | NDc | 8.6 × 106 | 4.2 × 106 | 5.3 × 107 | 62.1 (3.05) | 27.3 (0.35) | 1.26 (0.07) |
| Silage treated with A. niger AB4.1 | 4.08 (0.14) | 27.4 (0.13) | ND | 1.1 × 108 | 6.3 × 107 | 2.3 × 108 | 29.9 (3.01) | 1.63d | 2.66 (0.12) |
| Water control | 4.48 (0.29) | 28.1 (0.44) | ND | 1.2 × 108 | 6.8 × 107 | 8.7 × 108 | 21.1 (1.98) | 1.60e | 2.33 (0.01) |
Triplicate samples were obtained after 72 h of exposure to air.
The values in parentheses are standard errors (n = 3).
ND, not determined.
Acetate was detected in only one of three samples.
Acetate was detected in two of three samples.
FIG. 3.
Temperature profiles for maize silage during 6 days of exposure to air. The preparations examined were an A. niger mycocin-treated preparation (30 AU of mycocin per g of silage), an A. niger AB4.1 control preparation, and a water control. The lines represent the averages based on three replicate samples per treatment. The solid line indicates the ambient temperature.
Seventy-two hours after aeration and treatment, the pH values for the two mycocin-negative control groups (A. niger AB4.1 and water controls) had risen to more than 4.0 (Table 4), and 50% of the lactate and the majority of the acetate were oxidized. In contrast, the pH values of the HMK-treated samples had hardly changed after 72 h of aerobic exposure, as reflected by greater retention of lactate and a slight increase in the acetate content compared with the silage at the start of the experiment. The numbers of yeast cells (both total and lactate assimilators) increased in all of the samples but were 10-fold lower in the mycocin-treated silage. The numbers of lactate-assimilating bacteria identified as acetic acid bacteria also increased dramatically after 72 h in all samples but were lower in the HMK-treated silage (Table 4).
Food challenge experiments.
Two yeast strains, C. krusei D1241 and S. cerevisiae D1247, were used for the food challenge experiments (Table 3). We monitored the growth of these two strains in yoghurt samples which received an initial inoculum consisting of 103 CFU per ml and were treated with two levels of mycocin HMK, as well as in mycocin-free controls. Yeast growth was monitored at three different storage temperatures, 4, 10, and 20°C (Fig. 4 and 5).
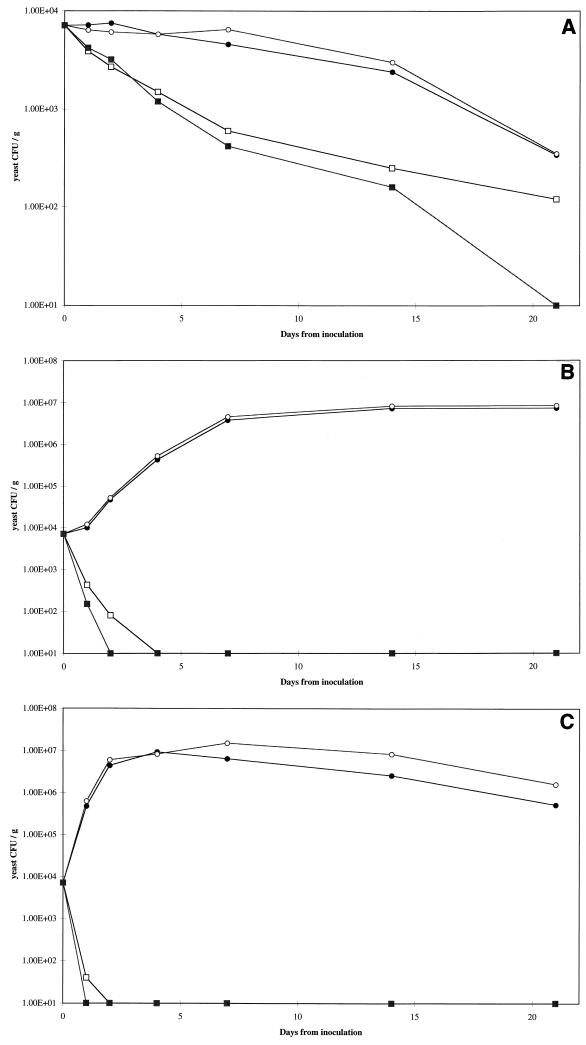
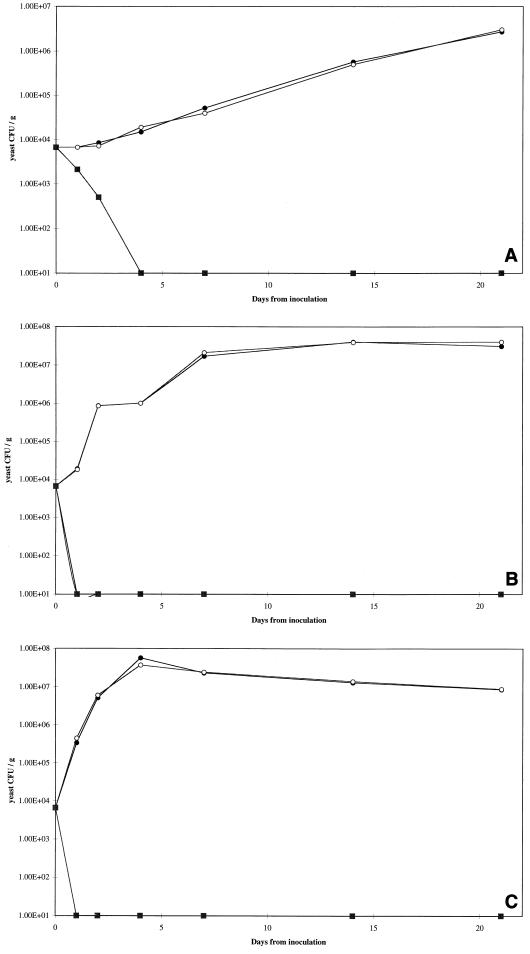
FIG. 4.
Growth curves for C. krusei D1241 in yoghurt. Preparations were stored at 4°C (A), 10°C (B), and 20°C (C). Symbols: ●, buffer control; ○, mycocin-free supernatant; □, W. mrakii HMK toxin (83 AU/ml); ■, W. mrakii HMK toxin (167 AU/ml). Yeast counts were plotted logarithmically; 1.00E+04 is equivalent to 104.
FIG. 5.
Growth curves for S. cerevisiae D1247 in yoghurt. Preparations were stored at 4°C (A), 10°C (B), and 20°C (C). Symbols: ●, buffer control; ○, mycocin-free supernatant; □, W. mrakii HMK toxin (83 AU/ml); ■, W. mrakii toxin (167 AU/ml). Yeast counts were plotted logarithmically; 1.00E+04 is equivalent to 104.
At 4°C, C. krusei D1241 did not grow, and the numbers of cells declined gradually in both control groups; however, more rapid decreases were observed in the samples treated with mycocin HMK at both concentrations (Fig. 4A). This strain grew steadily at 10°C and in the control preparations reached the stationary phase (approximately 107 CFU ml−1) within 7 days. The lower concentration of mycocin HMK reduced the number of yeast cells to below the level of detection by day 4, while the higher concentration reduced the number of yeast cells to below the level of detection by day 2 (Fig. 4B). Growth at 20°C was faster and resulted in an even more rapid rate of killing by mycocin HMK; yeasts were eliminated by day 1 and by day 2 at the two different concentrations used (Fig. 4C).
S. cerevisiae D1247 grew at 4°C throughout the 3-week incubation period, and by day 21 the cell numbers in the controls were more than 106 CFU ml−1. In the HMK-treated samples, yeast cells were undetectable within 4 days (Fig. 5A). The growth of this strain at both 10 and 20°C was rapid in the control samples, while both concentrations of HMK eliminated all yeasts within 24 h of inoculation (Fig. 5B and C). Overall, S. cerevisiae D1247 was more sensitive to the mycocin than C. krusei D1241 was, which was consistent with the results obtained in well diffusion assays (Table 3). None of the yoghurt samples to which no yeasts cells were added gave rise to yeast colonies during the experiment, irrespective of HMK treatment (data not shown). This demonstrated that the yoghurt used was not contaminated with yeasts and that the yeasts observed in the inoculated samples were the strains that were inoculated.
DISCUSSION
W. mrakii produces mycocin HMK which is very pH and heat stable and has a broad spectrum of activity (71). We found that this mycocin could be a versatile antispoilage agent in food and feed systems. In studies of the potential of mycogenic yeasts as biocontrol agents, workers have focused on the production of mycocins in situ by natural toxin producers or transformed yeasts (25). Other approaches are possible when use of a mycocin is necessary but inoculation with a mycogenic yeast is undesirable. Crude or purified mycocin could be used as an additive, which could have economic or regulatory disadvantages, and heterologous expression of a mycocin in situ could be obtained by using an organism which is either beneficial or at least not harmful and does not cause spoilage.
Heterologous expression of a synthetic mycocin HMK gene was achieved in A. niger. Native Willopsis mycocin HMK and heterologous Aspergillus mycocin HMK had similar physiological properties and molecular weights and reacted with the mycocin HMK-specific antibodies. Their spectra of biological activity against a variety of silage and yoghurt yeast strains were almost identical. The fact that multiple toxins are produced must be taken into account when comparisons with Aspergillus HMK activity are made as not all of the Willopsis killer activity is attributable to HMK. Expression in A. niger allowed us to partially purify the HMK toxin, and purification could be optimized, which resulted in an efficient and high-yielding production system for recovering this very stable peptide.
Application of heterologous mycocin HMK to mature maize silage prior to exposure to air delayed the onset of aerobic spoilage, as monitored by temperature changes. Destructive sampling 72 h after the initial aeration revealed that in the non-HMK-treated samples the numbers of lactate-assimilating and total yeast cells had increased. Virtually all of the lactate had been utilized in these samples, and the resultant increase in the pH meant that the barrier to proliferation of non-acid-tolerant spoilage organisms had been removed. By contrast, in the HMK-treated samples the numbers of yeast cells were lower, the levels of lactate were higher, and, consequently, the pH values were lower. The delay in the onset of aerobic spoilage could be attributed to the effect of the HMK toxin on the growth of yeasts that cause aerobic spoilage. The fact that aerobic deterioration was delayed rather than reduced or prevented suggests that some yeasts or other lactate-assimilating aerobes were not affected by mycocin HMK. It is possible that the mycocin was degraded by proteolytic activity in the silage, which is a more complex system than yoghurt. The concentration of mycocin HMK added to the silage was fairly low, and a higher concentration could perhaps have a more sustained effect on aerobic spoilage.
It has been shown that acetic acid bacteria are capable of initiating aerobic spoilage in maize silage (50), and six typical isolates of lactate-assimilating bacteria from the spoiled silage were identified as Acetobacter sp. isolates by metabolic characterization tests (9). Mycocin HMK had no effect on these Acetobacter strains in well diffusion tests. Thus, the apparent reduction in the number of acetic acid bacteria in HMK-treated silage could have been due to an indirect effect, perhaps impairment of a synergistic relationship between yeasts and acetic acid bacteria.
There is evidence that interactions between mycogenic yeasts and sensitive yeasts are widespread in natural habitats and are probably ecologically significant (53). Most research on mycogenic yeasts has been based on in vitro activity assays and has focused on the molecular aspects of production, the physiochemical properties of the mycocins, and the mechanisms of action. The role of killer yeasts in ecological community structure has received little attention, and it is assumed that these organisms have an advantage over sensitive competitors. When mycogenic yeasts are present in natural communities, a single killer strain usually predominates (51). Kinetic studies have shown that a killer strain can predominate in a mixed culture with a sensitive yeast strain (41, 43). It is essential to develop an understanding of the interactions of killer strains in communities if mycocins are to be used as biological control agents. Starmer and coworkers (51) found that mycogenic yeasts are widespread in natural populations of fruit and decaying vegetable matter and concluded that these organisms have an important effect on the development and composition of the yeast flora. We have isolated yeasts with mycogenic activities from silage (Lowes and Merry, unpublished data), and it is possible that they influenced the development and composition of the silage yeast population. This may explain the considerable variation in aerobic stability between different silages or even between silages prepared in the same way with identical herbage (19).
In the food challenge experiment we used two sensitive yeast strains that were inoculated into yoghurt and monitored their growth at three different storage temperatures. The growth rates were highest at the highest storage temperatures. Yeast strains vary in their ability to grow at low temperatures (16, 54), and in this study S. cerevisiae D1247 grew at 4°C, demonstrating that refrigeration alone would not prevent spoilage by this yeast. Mycocin HMK kills actively growing sensitive cells by interfering with β-1,3 glucan synthesis, which makes the cell wall osmotically fragile and results in cell lysis and death. Our experiments showed that the faster the growth rate, the more efficient the killing action of the mycocin, which prevented any further growth and destroyed the inoculated yeasts at all of the temperatures at which growth was possible; there was no resurgence of resistant yeasts.
In this study we found that mycocin HMK produced by A. niger can delay yeast spoilage in both silage and yoghurt. This toxin is effective at a low pH and a wide range of temperatures against a variety of yeast species. Silage is a product that is particularly prone to yeast contamination once it is exposed to air (38, 66, 68). Using higher levels of mycocin HMK after ensilage or, alternatively, adding purified mycocin before ensilage may delay the onset of aerobic spoilage. The influence of indigenous mycogenic yeast strains in silage is an area which requires further study. Yoghurt has a yeast spoilage problem that is associated particularly with the use of added fruits, nuts, and honey. In yoghurt, the introduced yeasts were eliminated by the mycocin at the different storage temperatures used. The use of mycocin HMK could be expanded to other fermented products and could be improved if the mycocin were expressed by compatible bacteria, such as lactic acid bacteria.
The use of mycocins to control unwanted yeasts could be expanded beyond applications in foods and feeds; they could be used as prophylactics for the treatment of fungal diseases. The medical importance of opportunistic yeast infections is increasing, particularly in immunocompromised hosts, and the problem is compounded by growing resistance to azole-based and amphotericin antifungal drugs (13). W. mrakii K500 mycocin exhibits extensive activity against a large number of clinical isolates, particularly Candida spp., and it has been proposed that this toxin could be used as a topical application to treat yeast infections (20). Mycocins have strong antigenicity and might elicit an immune response if they are used systematically, but they could be used for topical administration. pH instability makes most mycocins unsuitable for oral or intravenous use.
C. kruseii has been designated a class II pathogen because it exhibits a high level of resistance to fluconazoles and is frequently isolated from patients with suppressed immune systems that have been treated with fluconazoles for opportunistic yeast infections; obviously, it benefits from the removal of other competitor yeasts, such as Candida albicans (14, 44). Mycocin HMK from Williopsis sp. or the recombinant Aspergillus strain is very effective against C. kruseii strains from silage and yoghurt and exhibits a broad range of activity against other yeast species. It has good physiochemical properties, including pH stability and heat stability. This mycocin could provide a much-needed alternative treatment for pathogenic yeast infections.
ACKNOWLEDGMENTS
K. F. Lowes's Ph.D. studentship was jointly funded by the Institute of Food Research and the Institute of Grassland and Environmental Research.
We thank C. Bond for advice about yeast identification and E. Bakewell for help with silage microbiological and extraction techniques.
REFERENCES
- 1.Archer D B, Jeenes D J, Mackenzie D A. Strategies for improving heterologous protein production from filamentous fungi. Antonie Leeuwenhoek. 1994;65:245–250. doi: 10.1007/BF00871952. [DOI] [PubMed] [Google Scholar]
- 2.Archer D B, Jeenes D J, Mackenzie D A, Brightwell G, Lambert N, Lowe G, Radford S E, Dodson C M. Hen egg white lysozyme expressed in, and secreted from, Aspergillus niger is correctly processed and folded. Bio/Technology. 1990;8:741–745. doi: 10.1038/nbt0890-741. [DOI] [PubMed] [Google Scholar]
- 3.Ashida S, Shimazaki T, Kitano K, Hara S. New killer toxin of Hansenula mrakii. Agric Biol Chem. 1983;47:2953–2955. [Google Scholar]
- 4.Baleiras Couto M M, Hartog B J, Huis in't Veld J H J, Hofstra H, Van der Vossen J M B M. Identification of spoilage yeasts in a food-production chain by microsatellite polymerase chain reaction fingerprinting. Food Microbiol. 1996;13:59–67. [Google Scholar]
- 5.Barnett J A, Payne R W, Yarrow D. Yeasts. Characteristics and identification. 2nd ed. Cambridge, United Kingdom: Cambridge University Press; 1990. [Google Scholar]
- 6.Bevan E A, Makower M. The physiological basis of the killer character in yeast. In: Geerts S J, editor. Genetics today. Proceedings of the 11th International Congress on Genetics. Vol. 1. Oxford, United Kingdom: Pergamon Press; 1963. pp. 202–203. [Google Scholar]
- 7.Boone C, Sdicu A-M, Wagner J, Degre R, Sanchez C, Bussey H. Integration of the yeast K1 killer toxin gene into the genome of marked wine yeasts and its effect on vinification. Am J Enol Vitic. 1990;41:37–42. [Google Scholar]
- 8.Bortol A, Nudel C, Fraille E, de Torres R, Giuletti A, Spencer J F T, Spencer D. Isolation of yeast with killer activity and its breeding with an industrial baking strain by protoplast fusion. Appl Microbiol Biotechnol. 1986;24:414–416. [Google Scholar]
- 9.Carr J P G, Passmore S M. Methods for identifying acetic acid bacteria. Soc Appl Bacteriol Tech Ser. 1979;14:33–45. [Google Scholar]
- 10.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 11.Cussen R F, Merry R J, Williams A P, Tweed J K S. The effect of additives on the ensilage of forage of differing perennial ryegrass and white clover content. Grass Forage Sci. 1995;50:249–258. [Google Scholar]
- 12.Dawson K A. Yea-sacc, in the rumen: a natural fermentation modifier. In: Lyons T P, editor. Biotechnology in the feed industry. Proceedings of the Alltech 3rd Annual Symposium. Nicholasville, Ky: Alltech Technical Publications; 1990. pp. 119–125. [Google Scholar]
- 13.Driehuis F, van Wikselaar P G. The occurrence of alcoholic fermentation in high dry matter grass silages. In: Jones D I H, Jones R, Dewhurst R, Merry R J, Haigh P M, editors. Proceedings of the 11th International Silage Conference. Aberystwyth, United Kingdom: University of Wales; 1996. pp. 254–255. [Google Scholar]
- 14.Evans E G V. Causative pathogens in onychomosis and the possibility of treatment resistance: a review. J Am Acad Dermatol. 1998;38:S32–S36. doi: 10.1016/s0190-9622(98)70481-5. [DOI] [PubMed] [Google Scholar]
- 15.Fleet G H. Yeasts in dairy products. J Appl Bacteriol. 1990;68:199–211. doi: 10.1111/j.1365-2672.1990.tb02566.x. [DOI] [PubMed] [Google Scholar]
- 16.Fleet G H, Mian M A. The occurrence and growth of yeasts in dairy products. Int J Food Microbiol. 1987;4:145–155. [Google Scholar]
- 17.Golubev W I. Mycocins (killer toxins) In: Kurtzman C P, Fall J W, editors. The yeasts. A taxonomic study. Amsterdam, The Netherlands: Elsevier; 1998. pp. 55–62. [Google Scholar]
- 18.Hara S, Imura Y, Otsuka K. Breeding of useful killer wine yeasts. Am J Enol Vitic. 1980;31:28–33. [Google Scholar]
- 19.Henderson A R, Ewart J M, Robertson G M. Studies on the aerobic stability of commercial silages. J Sci Food Agric. 1979;30:223–230. [Google Scholar]
- 20.Hodgson V J, Button D, Walker G M. Anti-Candida activity of a novel killer toxin from the yeast Williopsis mrakii. Microbiology. 1995;141:2003–2021. doi: 10.1099/13500872-141-8-2003. [DOI] [PubMed] [Google Scholar]
- 21.Izgu F, Altinbay D. Killer toxins of certain yeast strains have potential growth inhibitory activity on Gram-positive pathogenic bacteria. Microbios. 1997;89:15–22. [PubMed] [Google Scholar]
- 22.Izgu F, Altinbay D, Yucelis A. Identification and killer activity of a yeast contaminating starter cultures of Saccharomyces cerevisiae strains used in the Turkish baking industry. Food Microbiol. 1997;14:125–131. [Google Scholar]
- 23.Jakobsen M, Narvhus N. Yeasts and their possible beneficial and negative effects on the quality of dairy products. Int Dairy J. 1995;6:755–768. [Google Scholar]
- 24.Jonsson A, Pahlow G. Systematic classification and characterization of yeasts growing in grass silage inoculated with Lactobacillus cultures. Anim Res Dev. 1984;20:7–22. [Google Scholar]
- 25.Kimura T, Kitamoto N, Imura Y, Kito Y. Production of HM-1 killer toxin in Saccharomyces cerevisiae transformed with the PDR4 gene and gamma-sequence-mediated multi-integration system. J Ferment Bioeng. 1995;5:423–428. [Google Scholar]
- 26.Kimura T, Kitamoto N, Matsuoka K, Nakamura K, Iimura Y, Kito Y. Isolation and nucleotide sequences of the genes encoding killer toxins for Hansenula mrakii and H. saturnus. Gene. 1993;137:265–270. doi: 10.1016/0378-1119(93)90018-x. [DOI] [PubMed] [Google Scholar]
- 27.Kimura T, Kitamoto N, Ohta Y, Kito Y, Imura Y. Structural relationships among killer toxins secreted from the killer strains of the genus Williopsis. J Ferment Bioeng. 1995;80:85–87. [Google Scholar]
- 28.Kinderlerer J L. Chrysosporium species, potential spoilage organisms of chocolate. J Appl Microbiol. 1997;83:771–778. doi: 10.1046/j.1365-2672.1997.00314.x. [DOI] [PubMed] [Google Scholar]
- 29.Kitamoto H K, Ohmomo S, Nakahara T. Selection of killer yeasts (Kluyveromyces lactis) to prevent aerobic deterioration in silage making. J Dairy Sci. 1993;76:803–811. doi: 10.3168/jds.S0022-0302(93)77404-4. [DOI] [PubMed] [Google Scholar]
- 30.Komiyama T, Ohta T, Furuichi Y, Ohta Y, Tsukada Y. Structure and activity of HY1 killer toxin from Hansenula saturnus. Biol Pharm Bull. 1995;18:1057–1059. doi: 10.1248/bpb.18.1057. [DOI] [PubMed] [Google Scholar]
- 31.Lennox E S. Transduction of linked genetic characters of the host bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 32.Lindgren S E, Dobrogosz W J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol Rev. 1990;87:149–164. doi: 10.1111/j.1574-6968.1990.tb04885.x. [DOI] [PubMed] [Google Scholar]
- 33.Magliani W, Conti S, Gerloni M, Bertolotti D, Polonelli L. Yeast killer systems. Clin Microbiol Rev. 1997;10:369–400. doi: 10.1128/cmr.10.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald P, Henderson A R, Heron S J E. The biochemistry of silage. 2nd ed. Marlow, United Kingdom: Chalcombe Publications; 1991. [Google Scholar]
- 35.Merry R J, Davies D R. Propionibacteria and their role in the biological control of aerobic spoilage in silage. Lait. 1999;79:149–164. [Google Scholar]
- 36.Merry R J, Dhanoa M S, Theodorou M K. Use of freshly cultured lactic acid bacteria as silage inoculants. Grass Forage Sci. 1995;50:112–123. [Google Scholar]
- 37.Merry R J, Lowes K F, Winters A L. Current and future approaches to biocontrol in silage. In: Jampor L, Klapil P, Chromec P, Prochazaka P, editors. Proceedings of the 8th International Conference on Forage Conservation. Pohorelice, Czech Republic: Research Institute of Animal Nutrition; 1997. pp. 17–27. [Google Scholar]
- 38.Middelhoven W J, van Baalen A H M. Development of the yeast flora of whole-crop maize during ensiling and during subsequent aerobiosis. J Sci Food Agric. 1988;42:199–207. [Google Scholar]
- 39.Pahlow G. Verbesserung der aeroben Stabilitat von Silage durch Impfpraparate. Wirtschaftseigene Futter. 1982;28:107–122. [Google Scholar]
- 40.Palpacelli V, Ciani M, Rosini G. Activity of different “killer” yeasts on strains of yeast species undesirable in the food industry. FEMS Microbiol Lett. 1991;84:75–78. doi: 10.1016/0378-1097(91)90398-t. [DOI] [PubMed] [Google Scholar]
- 41.Petering J E, Symons M R, Lanridge P, Henschke P A. Determination of killer yeast activity in fermenting grape juice by using a marked Saccharomyces wine yeast strain. Appl Environ Microbiol. 1991;57:3232–3236. doi: 10.1128/aem.57.11.3232-3236.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philliskirk G, Young T W. The occurrence of killer character in yeasts of different genera. Antonie Leeuwenhoek. 1975;41:147–151. doi: 10.1007/BF02565046. [DOI] [PubMed] [Google Scholar]
- 43.Ramon-Portugal F, Delia M L, Strehaiano P, Riba J P. Mixed culture of killer and sensitive Saccharomyces cerevisiae strains in batch and continuous fermentations. World J Microbiol Biotechnol. 1998;14:83–87. [Google Scholar]
- 44.Samaranayake Y H, Samaranayake L P. Candida krusei—biology, epidemiology, pathogenity and clinical manifestations of an emerging pathogen. J Med Microbiol. 1994;41:295–310. doi: 10.1099/00222615-41-5-295. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Sarais I, Piussi D, Aquili V, Stecchini M L. The behaviour of yeast populations in Stracchino cheese packaged under various conditions. J Food Prot. 1996;59:541–544. doi: 10.4315/0362-028X-59.5.541. [DOI] [PubMed] [Google Scholar]
- 47.Schagger H, von Jagow G. Tricine-sodium dodecyl sulphate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100KDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 48.Schillinger U, Stiles M E, Holzapfel W H. Bacteriocin production by Carnobacterium pisicola LV61. Int J Food Microbiol. 1993;20:131–147. doi: 10.1016/0168-1605(93)90106-q. [DOI] [PubMed] [Google Scholar]
- 49.Seki T, Choi E-H, Ryu D. Construction of killer wine yeast strain. Appl Environ Microbiol. 1985;49:1211–1215. doi: 10.1128/aem.49.5.1211-1215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spoelstra S F, Courtin M G, van Beers J A C. Acetic acid bacteria can initiate aerobic spoilage of whole crop maize silage. J Agric Sci. 1995;111:127–132. [Google Scholar]
- 51.Starmer W T, Ganter P F, Aberdeen V. The ecological role of killer yeasts in natural communities of yeasts. Can J Microbiol. 1987;33:783–796. doi: 10.1139/m87-134. [DOI] [PubMed] [Google Scholar]
- 52.Starmer W T, Ganter P F, Aberdeen V. Geographic distribution and genetics of killer phenotypes for the yeast Pichia kluyveri across the United States. Appl Environ Microbiol. 1992;58:990–997. doi: 10.1128/aem.58.3.990-997.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stumm C, Hermans J M H, Middelbeek E J, Croes A F, De Vries G J M L. Killer-sensitive relationships in yeasts from natural habitats. Antonie Leeuwenhoek. 1977;43:125–128. doi: 10.1007/BF00395667. [DOI] [PubMed] [Google Scholar]
- 54.Suriyarachchi V R, Fleet G H. Occurrence and growth of yeasts in yogurts. Appl Environ Microbiol. 1981;42:574–579. doi: 10.1128/aem.42.4.574-579.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takasuka T, Komiyama T, Furuichi Y, Watanabe T. Cell wall synthesis specific cytocidal effect of Hansenula mrakii toxin-1 on Saccharomyces cerevisiae. Cell Mol Biol Res. 1995;41:575–581. [PubMed] [Google Scholar]
- 56.TchangoTchango J, Tailliez R, Eb P, Njine T, Hornez J P. Heat resistance of the spoilage yeasts Candida pelliculosa and Kloeckera apis and pasteurization values for some tropical fruit juices and nectars. Food Microbiol. 1997;14:93–99. [Google Scholar]
- 57.van der Walt J P, Yarrow D. Methods for the isolation, maintenance, classification of yeasts. In: Kreger-vanRij N J W, editor. The yeasts, a taxonomic study. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. pp. 47–104. [Google Scholar]
- 58.van Hartingsveld W, Mattern I E, van Zeijl C M J, Pouwels P H. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol Gen Genet. 1987;206:71–75. doi: 10.1007/BF00326538. [DOI] [PubMed] [Google Scholar]
- 59.Weinberg Z G, Muck R E. New trends and opportunities in the development and use of inoculants for silage. FEMS Microbiol Rev. 1996;19:53–68. [Google Scholar]
- 60.Westall S, Filtenborg O. Spoilage yeasts of decorated soft cheese packed in modified atmosphere. Food Microbiol. 1998;15:243–249. [Google Scholar]
- 61.Wilkinson J M, Bolsen K K. The production of silage and hay in Europe and North America. In: Jones D I H, Jones R, Dewhurst R, Merry R J, Haigh P M, editors. Proceedings of the 11th International Silage Conference. Aberystwyth, United Kingdom: University of Wales; 1996. pp. 42–43. [Google Scholar]
- 62.Williams A G, Hoxey R P, Lowe J F. Changes in temperature and silo gas composition during ensiling, storage and feeding-out grass silage. Grass Forage Sci. 1997;52:176–189. [Google Scholar]
- 63.Williams P E V, Newbold C J. Rumen probiosis: the effects of novel microorganisms on rumen fermentation and rumen productivity. In: Haresign W, Cole D J A, editors. Recent advances in animal production. London, United Kingdom: Butterworths; 1990. pp. 211–227. [Google Scholar]
- 64.Williams P E V, Tait C A G, Innes M, Newbold C J. Effects of the inclusion of yeast culture (Saccharomyces cerevisiae plus growth medium) in the diet of dairy cows on milk yield and forage degradation and fermentation patterns in the rumen of steers. J Anim Sci. 1991;69:3016–3026. doi: 10.2527/1991.6973016x. [DOI] [PubMed] [Google Scholar]
- 65.Woolford M K. The silage fermentation. New York, N.Y: Marcel Dekker; 1984. [Google Scholar]
- 66.Woolford M K. A review: the detrimental effects of air on silage. J Appl Bacteriol. 1990;68:101–116. doi: 10.1111/j.1365-2672.1990.tb02554.x. [DOI] [PubMed] [Google Scholar]
- 67.Woolford M K, Bolsen K K, Peart L A. Studies on the aerobic deterioration of whole-crop cereal silages. J Agric Sci. 1982;98:529–535. [Google Scholar]
- 68.Woolford M K, Wilkie A C. Investigations into the role of specific micro-organisms in the aerobic deterioration of maize silage. J Agric Sci. 1984;102:97–104. [Google Scholar]
- 69.Yamamoto T, Hiratani T, Hirata H, Imai M, Yamaguchi H. Killer toxin from Hansenula mrakii selectively inhibits cell wall synthesis in a sensitive yeast. FEBS Lett. 1986;197:50–54. doi: 10.1016/0014-5793(86)80296-4. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto T, Imai M, Tachibana K, Mayumi M. Application of monoclonal antibodies to the isolation and characterization of a killer toxin secreted by Hansenula mrakii. FEBS Lett. 1986;195:253–257. doi: 10.1016/0014-5793(86)80170-3. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto T, Uchida K, Yamaguchi H. In vitro activity of the killer toxin from yeast Hansenula mrakii against yeasts and moulds. J Antibiot. 1988;41:398–403. doi: 10.7164/antibiotics.41.398. [DOI] [PubMed] [Google Scholar]
- 72.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains—nucleotide sequences of M13mp18 and pUC 19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 73.Young T W. The genetic manipulation of killer character into brewing yeast. J Inst Brew. 1981;87:292–295. [Google Scholar]
- 74.Young T W. The yeasts. London, United Kingdom: Academic Press; 1987. Killer yeasts; pp. 131–164. [Google Scholar]
- 75.Young T W, Yagiu M. A comparison of the killer character in different yeasts and its classification. Antonie Leeuwenhoek. 1978;44:59–77. doi: 10.1007/BF00400077. [DOI] [PubMed] [Google Scholar]
- 76.Zhu W-Y, Theodorou M K, Longland A C, Nilsen B B, Dijkstra J, Trinci A P J. Growth and survival of anaerobic fungi in batch and continuous flow cultures. Anaerobe. 1966;2:29–37. [Google Scholar]