Abstract
Human tumor viruses cause various human cancers that account for at least 15% of the global cancer burden. Among the currently identified human tumor viruses, two are small DNA tumor viruses: human papillomaviruses (HPVs) and Merkel cell polyomavirus (MCPyV). The study of small DNA tumor viruses (adenoviruses, polyomaviruses, and papillomaviruses) has facilitated several significant biological discoveries and established some of the first animal models of virus-associated cancers. The development and use of preclinical in vivo models to study HPVs and MCPyV and their role in human cancer is the focus of this review. Important considerations in the design of animal models of small DNA tumor virus infection and disease, including host range, cell tropism, choice of virus isolates, and the ability to recapitulate human disease, are presented. The types of infection-based and transgenic model strategies that are used to study HPVs and MCPyV, including their strengths and limitations, are also discussed. An overview of the current models that exist to study HPV and MCPyV infection and neoplastic disease are highlighted. These comparative models provide valuable platforms to study various aspects of virus-associated human disease and will continue to expand knowledge of human tumor viruses and their relationship with their hosts.
Keywords: DNA tumor Viruses, Human papillomaviruses, Merkel cell polyomavirus, Animal models, Infection models
1. Introduction
Viruses are the etiological agents of a wide variety of diseases, yet the pathological consequences of viral infection are not always immediately evident. Across virus species, there is a broad range of time that exists between initial virus infection and the onset of disease. For instance, the world is currently in the grips of global pandemic caused by a virus that triggers acute disease within days after infection. There are other types of viruses that become latent in their hosts, yet sporadically reactivate to cause recurrent disease. Other viruses can establish lifelong, asymptomatic infections in the host that can ultimately lead to cancer. The latter example refers to a subset of viruses known as tumor viruses, a classification that reflects their oncogenic potential.
The field of tumor virology officially began over a century ago, when, in 1911, Peyton Rous discovered a transmissible agent, a retrovirus now known as Rous sarcoma virus, that caused sarcomas in chickens [1,2]. Over the next 50 years, a number of additional viruses were discovered that caused benign tumors and/or cancers in mammals, further strengthening the causal link between viruses and cancer [reviewed in 3]. In 1964, a herpesvirus, Epstein-Barr virus, was discovered in Burkitt's lymphoma cells and became the first known human tumor virus [4]. There are now seven human tumor viruses identified with strong causal links to human cancers (Table 1) [5]: Epstein-Barr virus (EBV), hepatitis B virus (HBV), human papillomavirus (HPV), human T cell lymphotropic virus 1 (HTLV-1), hepatitis C virus (HCV), Kaposi's sarcoma-associated herpesvirus (KSHV), and Merkel cell polyomavirus (MCPyV). This group consists of two RNA viruses (HTLV-1 and HCV) and five DNA viruses (EBV, HBV, HPV, KSHV, MCPyV) that together cause approximately 15% of the global cancer burden [[6], [7], [8]].
Table 1.
Human tumor viruses.
| Virus | Family | Genome | Year Discovered | Cell type targeted | Associated Cancer(s) | References |
|---|---|---|---|---|---|---|
| RNA Viruses | ||||||
| Human T-lymphotropic virus-1 (HTLV-1) | Retroviridae | +ssRNA | 1980 | T cells | Adult T cell leukemia | [304] |
| Hepatitis C virus (HCV) | Flaviviridae | +ssRNA | 1989 | Hepatocytes | Hepatocellular carcinoma, lymphoma | [305] |
| DNA Viruses | ||||||
| Epstein-Barr virus (EBV) | Herpesviridae | dsDNA | 1964 | B cells, epithelial cells, T and NK cells | Burkitt's lymphoma, nasopharyngeal carcinomas, lymphoproliferative diseases | [4] |
| Hepatitis B virus (HBV) | Hepadnaviridae | ssDNA, dsDNA | 1965 | Hepatocytes | Hepatocellular carcinoma | [306] |
| Human papillomavirus (HPV16, HPV18) | Papillomaviridae | dsDNA | 1983–84 | Keratinocytes | Cervical cancer and other anogenital cancers, oropharyngeal cancer | [55,56] |
| Kaposi's sarcoma herpesvirus (KSHV) | Herpesviridae | dsDNA | 1994 | B cells, endothelial cells | Kaposi's sarcoma, primary effusion lymphoma | [307] |
| Merkel cell polyomavirus (MCPyV) | Polyomaviridae | dsDNA | 2008 | Keratinocytes? | Merkel cell carcinoma | [82] |
| Fibroblasts? | ||||||
| Merkel cells? | ||||||
Since its inception, the tumor virology field has heavily relied on animal models to study the virus-host interface and oncogenic properties of these tumor viruses. Many of the first tumor viruses discovered infected rodents and other animals, thus providing in vivo research models. In addition to Rous sarcoma virus being studied in avian species [1], other early tumor virus models included cottontail rabbit papillomavirus [9], mouse mammary tumor virus [10], murine leukemia virus [11], and murine polyomavirus [12], among others. With the discovery of human tumor viruses, animal models continued to provide invaluable comparative platforms to study all facets of tumor virology including causality and the role of viral and host factors, co-carcinogens, and environmental co-factors in viral oncogenesis. Certain models provide opportunities to study fundamental aspects of virus interactions with their hosts that underlie their oncogenic potential, such as host immune responses, viral persistence, and cellular transformation. Some preclinical models recapitulate the full spectrum of neoplastic disease progression of virus-induced cancers, allowing researchers to study potential treatments and mechanisms that underlie metastasis, therapy resistance, and recurrence.
A group of tumor viruses known as small DNA tumor viruses has had a large impact on our collective understanding of virology and the molecular mechanisms underlying oncogenesis that are relevant not just to virally induced cancers but also to other cancers. Animal models for these viruses have played important roles in this discovery process. This review will focus on preclinical animal models used to study infection and viral oncology of two small DNA tumor viruses currently understood to have the strongest links to human cancers: specifically, human papillomaviruses (HPVs) and Merkel cell polyomavirus (MCPyV).
1.1. Overview of small DNA tumor viruses
There are three main groups of viruses that are classified as small DNA tumor viruses: adenoviruses, papillomaviruses, and polyomaviruses. Members of the Adenoviridae, Papillomaviridae, and Polyomaviridae families are considered ‘small’ due to their small genome sizes compared to other DNA tumor viruses with much larger genomes, such as members of the Herpesviridae family (e.g., EBV, KSHV). For over 50 years, the study of small DNA tumor viruses has not only fostered our understanding of tumor virology but has facilitated some of the most seminal discoveries in modern cell and molecular biology [[13], [14], [15], [16]]. Prototypic small DNA tumor viruses such as simian vacuolating virus 40 (SV40), murine polyomavirus (MPyV), and adenovirus type 12 (Ad12) became workhorses in the laboratory leading to scientific breakthroughs such as the discovery of the tumor suppressor p53 [17,18], the structure and function of the tumor suppressor pRB [reviewed in 19], RNA splicing [20,21], non-homologous end joining [22], nuclear localization signals [23], polyadenylation sequences [24], tyrosine phosphorylation [25], and PI3K signaling [26]. Studies of SV40 and adenovirus also helped identify DNA replication factors [27] and molecular mechanisms of DNA replication [28,29], and also helped facilitate recombinant DNA technology and genetic cloning [[30], [31], [32]]. Fundamental principles of tumor virology, such as oncogene addiction [33] and the targeting of cellular tumor suppressors by tumor viruses [34,35], were borne out of studies on small DNA tumor viruses. Moreover, many of the first tumor virus animal models were generated with small DNA tumor viruses, including cottontail rabbit papillomavirus [9], murine polyomavirus [12,36], and adenoviruses [[37], [38], [39]]. The contributions that the study of small DNA tumor viruses have made to our collective scientific understanding is vast and quite simply remarkable.
1.2. Small DNA tumor viruses and human cancer
Not all small DNA tumor viruses are oncogenic in humans. To date, the only small DNA tumor viruses considered human tumor viruses are certain genotypes of human papillomaviruses (HPVs) and one human polyomavirus, Merkel cell polyomavirus (MCPyV). This section will discuss the current understanding of the association between small DNA tumor viruses and human cancers.
1.2.1. Adenoviruses and human cancer
Adenoviruses were discovered in the early 1950's in the tonsil and adenoid tissues of both healthy individuals and individuals with acute respiratory illness [40,41]. These viruses infect epithelial cells within the respiratory and gastrointestinal tracts, as well as corneal epithelia. Adenoviruses can claim the distinction of being the first human viruses to cause tumors and cancers in animals [38], including hamsters [37,38], mice [39], rats [42,43], and baboons [44]. Human adenoviruses can also transform a variety of cell types in vitro, yielding some of the most widely used human cell lines in molecular biology like the human embryonic kidney cell line HEK293 [45]. However, no definitive etiological link between human adenoviruses and human cancers has been established. Several other articles discuss the prolific transforming and tumorigenic potential of adenoviruses in various model systems [[37], [38], [39],[46], [47], [48], [49], [50], [51], [52], [53]] and clearly justify the classification of human adenoviruses as tumor viruses. However, given the lack of evidence supporting a causal role of adenoviruses in human cancers, animal models of human adenoviruses will not be discussed at length in this review.
1.2.2. Papillomaviruses and human cancer
In 1976, Harald zur Hausen published a brief note highlighting epidemiological similarities between papillomavirus-induced genital warts and human cervical cancers [54]. His laboratory subsequently reported the discovery of HPV types 16 and 18 (HPV16, HPV18), DNA of which were present in a significant proportion of human cervical and other anogenital cancers [55,56]. These paradigm-altering studies established a strong etiological link between these small DNA tumor viruses and human cancers. Papillomaviridae is a large and diverse family of viruses with over 220 formally accepted genotypes that infect humans [[57], [58], [59]]. Papillomaviruses exhibit tropism for stratified squamous epithelia, which is present in the skin, oral cavity, upper respiratory tract, and the anogenital tract that includes the cervix, vagina, vulva, anus, and penis. The large number of anatomical target sites and broad diversity of HPV genotypes underlies their high prevalence. HPVs remain the most common sexually transmitted infection in the United States despite the availability of prophylactic vaccines that protect against infection by several common, sexually transmitted HPV genotypes including ones that cause cancer [60]. HPVs are informally classified as either low-risk or high-risk depending on their oncogenic potential and as either cutaneous or mucosal based on their epithelial tropism [61]. Low-risk HPVs cause benign warts (papillomas) of the skin, oral/respiratory tract, and anogenital tract, whereas high-risk HPVs are associated with progressive neoplastic disease that can lead to cancer. While most of the formally recognized high-risk HPVs are mucosotropic members of the alpha genus that cause cancers of the anogenital tract and oral cavity, there are also cutaneotropic HPVs of the beta genus with compelling associations with some skin cancers [62]. Since their discovery as human cancer-causing viruses, it is now recognized that high-risk HPVs are etiologically linked to at least 5% of human cancers worldwide [7].
1.2.3. Polyomaviruses and human cancer
There are 13 known polyomaviruses that infect humans [63,64]. Most human polyomaviruses (HPyVs) are ubiquitous in the population and cause mild or asymptomatic disease in their hosts but can cause significant disease in immunosuppressed (e.g. transplant patients) and immunocompromised individuals. The identification of HPyVs as human carcinogens has taken a circuitous route. SV40 was discovered in 1960 in monkey kidney cells used in production of the human poliovirus vaccine [65]. After several reports were published demonstrating its oncogenic potential in preclinical settings [[66], [67], [68], [69]], concern mounted that SV40, as a contaminant of certain poliovirus vaccines, could be oncogenic in humans. While the intense scientific scrutiny of SV40 that ensued yielded some of the most important discoveries in modern molecular biology, there has yet to be a direct role established for SV40 in human cancers [reviewed in 70]. Two additional HPyVs, human JC polyomavirus (JCPyV) [71,72] and BK polyomavirus (BKPyV) [73] were discovered in a brain demyelinating disease and renal nephropathy, respectively, and have clear etiological roles in causing acute disease in immunosuppressed/immunocompromised individuals. Although both JCPyV and BKPyV have been implicated in human cancers of the central nervous system and renourinary tract, respectively [reviewed in 74,75,76], they have not yet been officially classified as human tumor viruses. In the last several years, new HPyVs such as human polyomaviruses 6 and 7 (HPyV6, HPyV7) [77], trichodysplasia spinulosa-associated polyomavirus (TSPyV) [78], Lyon IARC polyomavirus [79] and others [80] have been discovered. Thus far, however, data for only one HPyV, Merkel cell polyomavirus, has been sufficient to classify it as a causative agent of human cancer.
Merkel cell polyomavirus (MCPyV) was discovered in 2008, when Feng and colleagues sought to determine whether Merkel cell carcinoma (MCC), a rare cutaneous neuroendocrine cancer, has a viral etiology. Risk factors for MCC include immunosuppression, advanced age, and ultraviolet exposure [81], all of which can increase susceptibility to viral infection. Using a panel of MCC tissue samples, digital transcriptome subtraction analysis identified several polyomavirus-related transcripts that led to their identifying MCPyV (or MCV) [82]. Subsequent studies have confirmed that at least 80% of MCC tumors are MCPyV-positive and contain integrated sequences of the MCPyV viral genome, leading to continued expression of virally encoded, truncated large tumor (LT) and full-length small tumor (ST) antigens in MCC tumors. Clonal integration of the viral genome and continued expression of these viral proteins, which were found to be required for the growth of MCC-derived cell lines [83], provided strong evidence that MCPyV is an etiological agent of MCC.
The cell tropism and transmission route for many mammalian polyomaviruses remains unclear [63]. Many have tropism for epithelial cells at various anatomical sites, and others like murine polyomavirus (MPyV) infect a variety of different cell types in vivo [84]. MCPyV can be detected in skin swabs from healthy individuals [77] suggesting it likely infects a resident cell type in the skin, although the exact cell type remains unclear. Normal Merkel cells arise from epidermal progenitors [85,86], prompting speculation that MCPyV infects epidermal keratinocytes. However, Liu et al. reported that productive MCPyV infection can be achieved in human and animal dermal fibroblasts [87,88], which is intriguing since MCC commonly arises in the dermis. Ongoing research efforts seek to illuminate the relationship of MCPyV with the skin, its life cycle and replication, and the mechanisms underlying its oncogenic potential [89].
1.3. Common features of small DNA tumor viruses
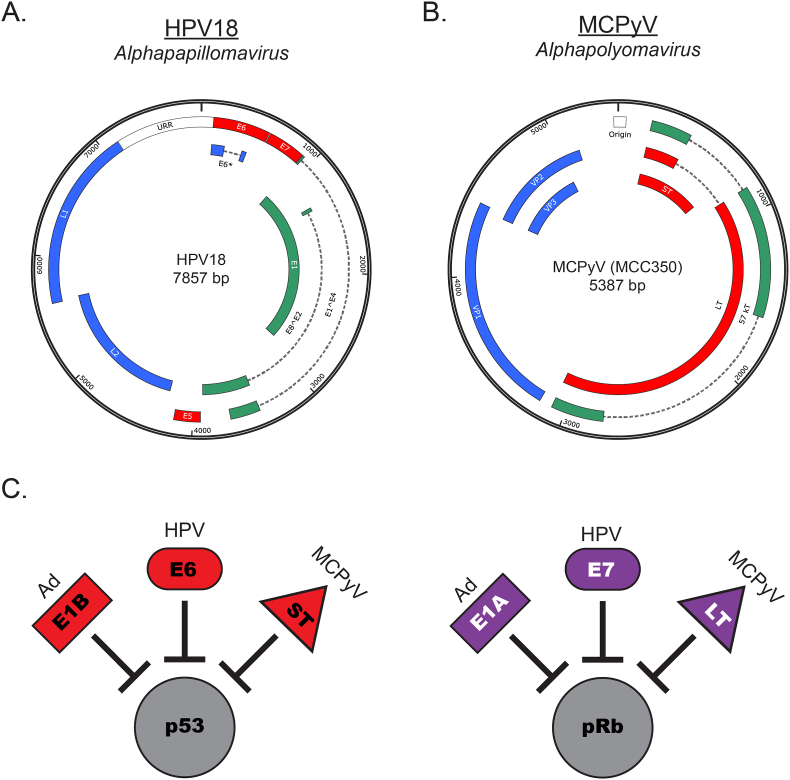
There are several similarities between HPVs and MCPyV, the two small DNA tumor viruses linked to human cancers [59,63]. Both viruses are non-enveloped, icosahedral viruses between 40 and 45 nm (polyomaviruses) and 55 nm (papillomaviruses) in size that contain a circular, double-stranded DNA viral genome 5–8 kilobase pairs in length (Fig. 1A and 1B). During viral replication of both viruses, a temporally regulated gene expression cascade yields ‘early’ and ‘late’ proteins [90,91]. Aside from being directly involved in viral replication, the early genes also help create a cellular environment conducive for viral replication by driving cell cycle entry [[92], [93], [94], [95], [96], [97]], which is necessary for the infected host cell to support viral DNA replication and progeny virus production. When removed from the context of viral replication, many of these same functions moonlight as oncogenic properties. Thus, the HPV and MCPyV encoded oncoproteins are all ‘early’ viral proteins: the E5, E6, and E7 proteins of HPVs [98] and the ST and LT antigens of MCPyV [97].
Fig. 1.
Similarities among small DNA tumor viruses. Viral genomes of small DNA tumor viruses classified as human tumor viruses are shown for A. high-risk human papillomavirus type 18 (HPV18) and B. Merkel cell polyomavirus isolate (MCPyV) MCC350. Early viral genes are green, late viral genes are blue, and viral oncoproteins are shown in red. C. The small DNA tumor viruses express viral early proteins that share common targets, including the tumor suppressors p53 and pRb. Ad: adenovirus; HPV: human papillomavirus; MCPyV: Merkel cell polyomavirus. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Persistent infections are a common feature of small DNA tumor viruses with oncogenic potential. HPV infections are extremely common, yet most are asymptomatic and usually cleared [99]. MCPyV is also a ubiquitous virus that is likely acquired during early childhood [[100], [101], [102], [103]]. It is well-established that HPVs can establish infections that persist throughout the lifetime of the host [[104], [105], [106]]. MCPyV also seems to become a persistent part of the natural skin flora given its detection in skin swabs of healthy individuals [77]. The persistent presence of HPV and MCPyV viral DNA in hosts, together with other factors and conditions that are not yet fully elucidated, coalesce to promote the relatively rare event of cancer development. Viral genome integration in these cancers is a common (though not absolutely necessary in the case of HPV) hallmark of both HPV and MCPyV-mediated malignant progression [82,107,108]. Integration renders the viruses replication-incompetent yet preserves early region DNA sequences that encode viral oncoproteins. In the case of HPV, viral genome integration provides a selective growth advantage to cells [109] by increasing and stabilizing viral transcripts encoding the potent HPV oncogenes E6 and E7 [108,110]. Integration of the MCPyV genome preserves viral DNA sequences of the early region and thus expression of ST and LT in human MCCs [82]. During viral genome integration and/or malignant progression, truncating mutations occur within LT antigen [111] that give rise to truncated LT proteins that lack the C-terminal domain which is involved in viral replication and confers growth-inhibitory functions [112]. Viral integration and continued expression of viral oncoproteins are primary drivers of HPV- and MCPyV-associated carcinogenesis as highlighted by the fact these virus-associated cancers are ‘oncogene addicted’ to viral oncoprotein expression [83,113].
Among the most striking commonalities among the small DNA tumor viruses are the parallel functions of their viral oncoproteins. Adenoviruses, papillomaviruses, and polyomaviruses all express highly multifunctional early viral proteins, yet those with transformation capacity have converged to share at least two common targets for inactivation: the cellular tumor suppressor proteins, p53 and pRb [15,51,98,114,115] (Fig. 1C). Using direct or indirect mechanisms, the high-risk HPV E6 protein and MCPyV ST antigen inactivate the p53 tumor suppressor pathway [[116], [117], [118]], whereas the high-risk HPV E7 and MCPyV truncated LT antigen inactivate the pRb tumor suppressor pathway [19,111,114,115,[119], [120], [121], [122]]. While they are not associated with human cancers, adenovirus E1 proteins also target p53 and pRb for degradation and/or inactivation [51,52,123,124], further substantiating the analogous mechanisms of small DNA tumor viruses. While there are certainly other functions of the HPV E6/E7 and MCPyV T antigen proteins that contribute to human carcinogenesis [114,[125], [126], [127]], the existence of these common mechanisms and cellular targets among small DNA tumor viruses draws attention to their likely importance in driving cancer development. These molecular clues helped provide important context and guidance in the development of preclinical models used to study small DNA tumor virus-associated human carcinogenesis.
2. Considerations in the design and use of small DNA tumor virus preclinical models
Comparative animal models are one of the most valuable tools available to researchers seeking to learn more about the multiple facets of human disease in intact, living systems. As methodology evolves and new models are developed, or when a new human tumor virus is discovered as was recently the case with MCPyV [82], there are several important aspects to consider in the design and use of animal models of small DNA tumor virus action.
2.1. Host range
The host range of a virus is defined by the host species it can infect and in which a fully productive virus infection can occur. Most viruses have a very strict host range and fail to establish and/or complete productive infections in any host other than those with which they co-speciated. This is largely the case for small DNA tumor viruses, where the restricted host range has relegated researchers to studying virus infection and disease in laboratory animals that are their natural hosts. Host range is mainly a consideration for natural infection models, although issues such as species-specific restriction factors and/or processes can also affect the extent to which a transgenic animal model accurately mimics human disease.
2.2. Cell tropism
Tropism is defined by the cell or tissue type that a virus naturally and preferentially targets for infection. In natural infection models wherein the administered virus can infect its target cell, considerations may be necessary if technical and ethical limitations arise in achieving access to or infection of specific population of cells. For instance, mouse polyomavirus (MPyV)-based murine models of neurotropic JCPyV-induced disease required techniques for intracranial infection that resulted in serious ethical considerations upon development of severe neurological phenotypes [128]. On the other hand, in natural infection models where there is broad cellular tropism, issues such as lateral transmission and reinfection can create confounding variables that threaten interpretation of otherwise well-controlled experiments. In the development of transgenic animal models in particular, cell and tissue tropism are especially important considerations since the genetic design will determine the spatial and temporal expression of the viral oncoproteins.
2.3. Virus types/isolates and association with disease
The virus types/isolates used when creating and using models of virally induced disease are an important consideration. In the case of models of HPV-associated cancers, for example, one would want to utilize high-risk HPV types that cause the majority of HPV-associated cancers across anatomical sites [[129], [130], [131]]. When developing models of MCPyV-associated carcinogenesis, one might consider the MCC-specific T antigen mutations that arise in the early region of the integrated viral genome [111] and seek to use MCC-derived MCPyV isolates. As further sequencing is performed on MCC tumors, it is possible that MCPyV isolates that are more prevalent in human cancers will emerge, as was the case with high-risk HPV16 and HPV18 [129,130,132].
2.4. Reproduction of key aspects of virus-induced disease
According to the National Cancer Institute, an animal model is “… an animal with a disease either the same as or like a disease in humans … used to study the development and progression of diseases and to test new treatments before they are given to humans” [133]. The ability to recapitulate aspects of human disease accurately and reproducibly is at the crux of all preclinical models. As will be discussed in the next section, modern genetics have made it possible for investigators to control the spatial and temporal expression of both viral and host factors in animals with high levels of precision. While limitations certainly exist, the extent to which a laboratory animal model mirrors the anatomical aspects of the virus-induced human cancer is important. Ideally, models of virus-induced cancer closely recapitulate the neoplastic process and ultimately give rise to cancers that have similar histopathology and biomarker expression as in humans. Meeting these standards can often require significant optimization, but achieving a model that recapitulates viral carcinogenesis as accurately as possible is key to relevant translational studies.
3. Types of animal models to study small DNA tumor virus-associated human cancers
This review will focus on two main types of animal models that are used to study HPVs and MCPyV and their association with human disease: infection models and transgenic models. There are other valuable in vivo models to study virus-induced cancers including xenografts, patient-derived xenografts, and humanized mice, but they will not be discussed in this review. The following section provides a brief and generic overview of the design, strengths, and limitations of infection and transgenic models.
3.1. Infection models
Both papillomaviruses and polyomaviruses have rigid species specificity [59,63]. Consequently, animals commonly used as laboratory model systems do not support productive HPV or HPyV infections. This species specificity has largely limited productive infection models to those natural hosts with a cognate virus, giving rise to papillomavirus and polyomavirus infection models in various rodent and livestock species, among others [[134], [135], [136], [137], [138]]. These models utilize either pseudoviruses or natural viruses for infection (Fig. 2A). Pseudoviruses, which contain reporter plasmids encapsidated in viral capsid proteins and are therefore non-replicating, are a useful surrogate model to study early steps in virus attachment and cell entry in vivo [139,140]. Natural infection models involve the preparation of viral stocks from naturally occurring lesions on host animals or the in vitro generation of ‘quasivirus’ species using the viral genome and a packaging cell line [141,142]. These natural or engineered virus stocks can then be applied to their natural host at susceptible sites corresponding to those infected in humans. Infection models can be used to study transforming infections, as is the case with the cottontail rabbit papillomavirus model [reviewed in 143] and animal models used to study adenovirus [reviewed in [48]]. The ideal infection model, however, will support the full spectrum of events including productive infection, viral persistence, and neoplastic disease progression.
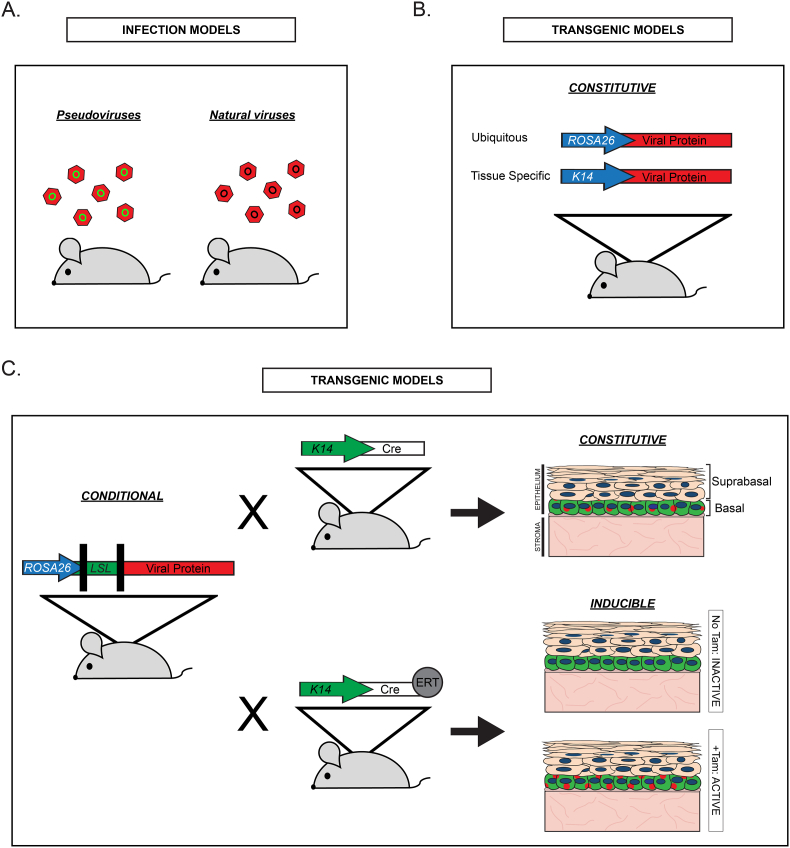
Fig. 2.
Overview of common types of animal models used to study the role of small DNA tumor viruses in human cancers. A. Infection models used to study the papillomavirus life cycle and pathogenesis. Pseudoviruses contain plasmid DNA, often expressing reporter proteins like GFP (green circles), encapsidated by the HPV capsid proteins in vitro. Natural viruses, such as those that infect laboratory hosts, are often used to establish animal models of papillomavirus infection and disease. B. Constitutive transgenic models express transgenes (Viral Protein; red) driven by either ubiquitous (e.g., ROSA26) or cell/tissue-specific (e.g., keratin 14; K14) promoters (shown as arrows). C. Conditional transgenic models often employ recombinase systems, such as the Cre/loxP system, to direct either constitutive or inducible transgene expression. Using Cre/loxP as an example, conditional transgene cassettes include a promoter (blue arrow) and a transgene (Viral Protein; red) separated by a transcriptional stop sequence flanked by two loxP sequences (loxP-stop-loxP; LSL; green). Presence of the LSL will prevent viral protein expression in animals expressing this cassette. To achieve tissue-specific constitutive expression (top right), mice carrying the conditional allele can be crossed with mice that express Cre recombinase driven by a tissue-specific promoter. The example shown here is K14Cre, which will direct expression of the viral protein (red dots) in K14-positive cells (shown in green) in basal cells of the stratified epithelium. To achieve tissue-specific inducible expression (bottom right), mice carrying the conditional allele can be crossed with mice that express a Cre recombinase fusion protein (e.g., Cre-ERT) driven by a cell-specific promoter. The Cre-ERT fusion protein is inactive until bound by its ligand (tamoxifen; Tam). In the absence of Tam, there is no viral protein expression in K14-positive cells. Upon administration of Tam, the viral protein (red) is now expressed only in K14-positive cells (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1.1. Strengths and limitations
The first and most obvious strength of infection models is that they support the full productive life cycle in its natural host. This not only provides a platform to longitudinally study virus-host interactions throughout the course of infection and pathogenesis, but also makes studies of the host immune response possible. Infection models involve the inherent mechanisms of spatial and temporal regulation of gene expression that occur as they do during a natural virus infection. Because a common characteristic of both papillomaviruses [105] and polyomaviruses [144] is that they establish persistent infections, those models that support persistence provide the important opportunity to study factors involved in viral persistence and potential pharmacological inhibitors of persistent viral infections. The same is true of viral genome integration, which occurs during both HPV and MCPyV infection-mediated carcinogenesis.
Infection models do have some limitations. As with most models of infection, there are inherent safety and containment considerations when working with infectious agents. Broad anatomical susceptibility, especially within immunodeficient animal strains, can introduce multiple confounding variables that complicate interpretation and limit control of the experimental design. Animal models of infection can also be limited by anatomical, histological, and molecular dissimilarities to humans and lead to inaccurate extrapolations of human equivalence.
3.2. Transgenic models
Transgenic animals are genetically engineered to express genetic material from another source, such as viral genes. A small DNA tumor virus was used to develop what is considered the first transgenic mouse model in 1974 when researchers injected the SV40 T antigen genes into mouse embryos resulting in the detection of viral DNA in multiple tissues [145]. Today, transgenic models of virus-associated carcinogenesis help establish causality, provide insight into viral and host protein functions involved in tumorigenesis and malignant progression, and help determine which viral proteins are sufficient and/or necessary for cancer.
The most common species used to generate transgenic models is the laboratory mouse, Mus musculus. Mice are the most well-characterized and well-supported laboratory animal with respect to the availability of technical reagents and protocols [146] and at least 80% of their genes have a human ortholog [147]. The most common approach for generating transgenic mice is through direct injection of DNA sequences into the pronucleus of fertilized oocytes [[148], [149], [150]] resulting in the random integration of DNA into the mouse genome and transgene expression in all cells in which the transgene promoter is active. More recent advances allow a DNA construct to be specifically introduced (‘knock-in’) at a permissive locus through the use homologous recombination. A commonly targeted locus for this type of knock-in approach is ROSA26, a genetic locus with no identified essential genes that drives stable, predictable, and ubiquitous expression in mice [151]. As discussed below, the use of tissue-specific promoters and/or recombinase technologies enable conditional expression that can help circumvent otherwise detrimental types of expression. More on the history of transgenic models and the genetic strategies used in their development can be found in other reviews [for example 152,153].
3.2.1. Constitutive transgenic models
Most of the transgenic models used to study small DNA tumor viruses involve either random insertion or targeted knock-in designs that achieve viral oncoprotein expression in relevant tissues of transgenic animals (Fig. 2). Constitutive transgenic models drive either constitutive expression of the transgene in a ubiquitous manner (e.g., the ROSA26 promoter) or in a cell- or tissue-specific manner (e.g., the keratin 14 promoter) (Fig. 2B). A benefit of transgenic models with constitutive expression is that they can provide relatively rapid assessment of phenotype development induced by a transgene. However, the strength of the promoter of choice can lead to expression that is either too strong or too weak, resulting in nonphysiologically relevant effects.
3.2.2. Conditional transgenic models
Conditional transgenic models expand the ways in which transgene expression can be regulated, allowing both temporal and spatial regulation (Fig. 2C). The most common features of conditional models are elements of the Cre/loxP recombination system. This system incorporates expression of a Cre recombinase enzyme, which recognizes DNA sequences known as loxP sequences [154]. One example of the Cre/loxP system involves introducing transcription/translation termination or ‘stop’ sequences flanked by loxP sequences (often referred to as loxP-stop-loxP, or LSL, sequences; Fig. 2C) between the promoter and gene of interest. These mice will only express the gene of interest when crossed with transgenic mice expressing Cre recombinase under the direction of specific promoters. For example, if a conditional model of viral protein expression is crossed with a Cre driven by a keratin 14 (K14) promoter (K14Cre), this will result in constitutive expression of the viral protein in K14-positive cells. The FLP/FRT system is another recombination system similar to Cre/loxP that can be used to manipulate transgene expression. Conditional transgene models provide broad flexibility in directing viral oncoprotein expression to a variety of different cell/tissue types by simply breeding with mice that drive Cre from any given cell/tissue-specific promoter. For this reason, conditional models are especially useful when studying viruses with unknown tissue/cell tropism, as expression can be readily targeted to new cell types as new knowledge surfaces.
3.2.3. Inducible transgenic models
Inducible transgenic models are those that can induce expression of a transgene at some point in time, thus enabling temporal regulation of viral transgene expression (Fig. 2C). One of the most common approaches for creating inducible models uses Cre recombinase proteins fused to a mutated ligand-binding domain (LBD) of steroid receptors, most frequently the estrogen receptor (ER), which cannot be regulated by its natural ligands but is activated by 4-hydroxytamoxifen (CreERT) [reviewed in [154]]. In the absence of ligand, Cre fusion protein is excluded from the nucleus. However, when mice are treated systemically or topically with tamoxifen (Tam), the active Cre fusion protein is transported to the nucleus where it can cleave at loxP sites. A more recent version, CreERT2, is more efficiently induced by tamoxifen and has reduced ‘leaky’ activity in the absence of its ligand [155]. The expression of inducible CreERT is usually driven by a cell-specific promoter such as K14. Using K14CreERT as an example, Cre recombinase will only be expressed in K14-positive cells upon tamoxifen treatment. Another frequently used inducible system is the tetracycline-based system which uses the tetracycline operator (tetO) and transactivator protein (tTA). Cre recombinase can also be delivered to mice using engineered adenovirus or lentivirus vectors expressing Cre [156] to achieve temporal and tissue-specific expression.
3.2.4. Strengths and limitations
A main strength of transgenic models is that they allow precise tissue-specific and/or temporal regulation of viral protein expression. Transgenic models can help test the individual contributions of viral oncoproteins and their specific functions to the oncogenic process, and these models can be easily combined with transgenic or genetically engineered mouse models that manipulate host factors. Transgenic models of human tumor viruses also provide well-defined and controlled platforms to test mechanistic hypotheses and potential therapeutic treatments. Limitations of transgenic models include the lack of viral infection/replication and the expression of viral proteins from a heterologous promoter that may alter natural in vivo expression patterns. The viral proteins may also be expressed in a way that prevents their being seen as foreign antigens, thus limiting the ability to study some aspects of the host immune response during neoplastic progression.
4. Preclinical models of HPV infection and disease
Researchers have established various preclinical models, including both infection and transgenic models, to study HPV infection and disease. The following section will provide an overview of the current models frequently used in HPV research.
4.1. Infection models
4.1.1. Pseudoviruses
The development of HPV pseudoviruses [157,158] harnessed the knowledge that the HPV capsid proteins L1 and L2 self-assemble into virus-like particles [[159], [160], [161]] to package and assemble infectious particles in vitro that can subsequently be used to infect animals. HPV pseudoviruses have been used to study early stages of viral infection like cell attachment and cell entry. Despite the strong species specificity of papillomaviruses, pseudoviruses packaged with HPV capsid proteins can infect the murine female reproductive tract [162], suggesting that species restrictions exist downstream of cell attachment and cell entry. This cervicovaginal pseudovirus infection model has been used to test vaccines [163], antiviral compounds that prevent viral entry [164,165], and molecular studies of viral attachment and entry mechanisms [[166], [167], [168], [169], [170]]. Capsid proteins from different HPV or animal papillomavirus types can be used to generate pseudoviruses to better understand the similarities and differences in attachment and entry mechanisms between viruses [171]. The availability of pseudovirus-based infection models continues to help scientists better understand the early interactions between host cells and HPV virus particles, an event necessary to the development of HPV-associated cancers.
4.1.2. Rodent models
Rodent models of papillomavirus infection have greatly enhanced our understanding of HPV pathogenesis, tissue tropism, and carcinogenic properties. The first natural infection model used to study papillomaviruses was established in the 1930s with the discovery of cottontail rabbit papillomavirus (CRPV) [9], which causes horn-like keratinous protrusions on rabbits. Soon thereafter, Rous and colleagues found that CRPV causes skin malignancies in rabbits [172], making CRPV the first mammalian model of a virus-induced cancer. The CRPV model became instrumental in understanding cutaneotropic papillomaviruses and their oncogenic potential in the skin [173]. Rabbit oral papillomavirus (ROPV) and canine oral papillomavirus (COPV) infect the oral mucosa and anogenital tissues similar to the alpha genus high-risk HPVs that are oncogenic in humans, thus making these important models in understanding mucosotropic papillomavirus pathogenesis [[174], [175], [176]]. Animal models of CRPV, ROPV, and COPV infection were used to test early vaccine candidates, thus paving the way for the current availability of highly effective prophylactic HPV vaccines [136,177,178]. Mastomys natalensis papillomavirus 1 (MnPV1) and Mastomys coucha papillomavirus 2 (McPV2), which persistently infect the multimammate rat species Mastomys natalensis and Mastomys coucha, respectively, exhibit dual tropism for cutaneous and mucosal sites [179,180]. The Mastomys models have been used to study viral persistence [181], cutaneous tumorigenesis [182], the role of environmental factors like UV in papillomavirus-associated cutaneous disease and ‘hit-and-run’ oncogenesis [183], and other key aspects of papillomavirus pathogenesis [184]. There are several reviews that provide further information on preclinical rodent papillomavirus models and their contributions to our understanding of virus-host interactions and pathogenesis of papillomaviruses [136,137,185].
4.1.3. MmuPV1
Until recently, the HPV field lacked a murine papillomavirus that could harness the potential of an infection-based mouse model of papillomavirus pathogenesis. In 2011, a murine papillomavirus (MmuPV1) isolated from a cutaneous papilloma present on the skin of immunodeficient NMR1-FoxN1nu/nu mice was reported [186]. Additional murine papillomaviruses were discovered soon thereafter in the skin of a house mouse (Mus musculus) and wood mouse (Apodemus sylvaticus) [187], although most studies thus far have utilized MmuPV1. The MmuPV1 genome is circular dsDNA that is organized similarly to other papillomaviruses. MmuPV1 does not express an E5 protein and this, along with other characteristics of its transcriptional map, make it more similar to beta genus cutaneotropic HPVs than mucosotropic alpha HPVs [188]. Notably, MmuPV1 exhibits expanded tropism in mice and causes disease in both cutaneous [[189], [190], [191], [192], [193], [194]] and mucosal [138,189,[195], [196], [197], [198], [199], [200], [201]] epithelia of immunodeficient and immunocompetent mice. This feature has facilitated the establishment of MmuPV1 infection-based models in all anatomical sites infected by HPVs in humans and these models are being used to study several aspects of pathogenesis and disease [reviewed in 138, 185, 202].
Although MmuPV1 shares less than 50% sequence similarity with high-risk HPVs like HPV16 [203], the MmuPV1 E6 and E7 proteins retain many activities of high-risk HPV oncoproteins that are associated with transformation [reviewed in [138]]. For example, MmuPV1 E7 binds pRb [204] and PTPN14 [205], both targets of high-risk HPV E7 [121,206]. MmuPV1 E6 inhibits Notch and TGF-beta (TGF-β) pathways to delay differentiation and promote proliferation [207,208] and contains a putative PDZ-binding motif [187]. It has recently been demonstrated that the MmuPV1 genome integrates in benign skin warts [209], and although integration has not yet been reported in mucosal tissues, its capacity to integrate recapitulates a key aspect of HPV-induced malignant disease.
Many MmuPV1-infection models are being used to study multiple steps of infection-mediated carcinogenesis. MmuPV1 pseudoviruses have been used to study and compare virus entry mechanisms and species/tissue tropism in mice [171,[210], [211], [212]]. We have also established an MmuPV1-based model of natural sexual transmission that results in MmuPV1 infections in both male and female genital tissues in immunocompetent mice [200]. Many researchers are using MmuPV1 to study the host immune response to papillomaviruses. Early studies found that various strains and genetic backgrounds of mice exhibited different susceptibilities to MmuPV1 infection [186,190,193,194,213,214]. By comparing these differential sensitivities using a variety of approaches including immune cell depletion, adoptive transfers, and the induction of chronic immunosuppression, it was determined that complete T-cell deficiency makes mice more susceptible to MmuPV1 infection and disease [190,192,194,214,215]. We identified a host factor, stress keratin 17 (K17), that is upregulated during MmuPV1 infection and prevents T-cell recruitment to protect against MmuPV1-induced cutaneous disease [216]. We have observed similar results in a MmuPV1-infection based model of cervicovaginal carcinogenesis (unpublished). Through the use of genetically engineered keratin 17 germline knockout mice (K17KO) [217] we helped establish the role of K17 and characterized the immune profile of MmuPV1-infected cutaneous lesions in the presence and absence of this host factor. MmuPV1 infection-based models will undoubtedly continue to provide insight into the host immune response and mechanisms underlying papillomavirus-driven immune evasion.
Persistent infection with mucosotropic high-risk HPVs is a major risk factor for cancer progression in humans [[104], [105], [106]] and MmuPV1 infection models allow in vivo longitudinal studies of papillomavirus persistence and carcinogenesis. Persistent infections with MmuPV1 are readily established in the mucosal epithelia of the cervicovaginal epithelium [200,213,218] and to a lesser but still significant degree in the oral cavity [201,218] and anal canal [195,198,218]. Consistent with the link between persistent infections and HPV-associated cancers in humans, MmuPV1-infected mice that establish persistent infections in the mucosal epithelia also develop SCC. In heterozygous FoxN1nu/+ mice infected in the female reproductive tract with MmuPV1, Cladel and colleagues observed carcinoma in situ development several months after infection [218] and we have observed SCC development in wild-type FVB/N mice after 6 months of MmuPV1 infection [213]. Similar to our findings in HPV16 transgenic mice [[219], [220], [221], [222], [223]], we found that estrogen treatment significantly increases the incidence of high-grade dysplasia and SCC in MmuPV1-infected mice [213]. Using wild-type FVB/N mice, we have also established MmuPV1-induced carcinogenesis models for anal cancer [195] and oral cancer [201,224]. These MmuPV1 infection-based models of papillomavirus pathogenesis, persistence, and cancer development will be valuable in furthering our understanding of infection-mediated de novo neoplastic progression and may help identify novel therapeutic interventions across all stages of HPV-mediated disease.
4.2. Transgenic models
The establishment of preclinical models of HPV-associated disease became more imperative following the discovery of HPVs in genital cancers in the early 1980s [55,56]. These findings prompted researchers to develop experimental systems in which to perform studies needed to firmly establish and interrogate the mechanisms of the causal link between HPVs and human cancers. However, in the absence of a murine papillomavirus, which was not discovered until 2011 [186], researchers proceeded with the development of transgenic models of the high-risk, mucoscotropic HPVs starting in the early 1990s. As will be discussed in this section, these transgenic models involve the targeted expression of high-risk HPV oncogenes to the stratified epithelia that, when combined with cofactors and cocarcinogens, largely recapitulated many aspects of HPV-mediated carcinogenesis in humans (Table 2).
Table 2.
Preclinical transgenic models used to study human cancers caused by high-risk alpha genus HPVs.
| Virus | Protein expression | Cell type/tissue targeted | Other genes/cofactors incorporated in model | Disease development and anatomical site | Model Development References |
|---|---|---|---|---|---|
| HPV16 | E6/E7 | α-crystallin (epithelial tissues) | None | Tumors: Ocular lens, SCC: Skin | [227,232] |
| HPV16 | E7 | interstitial retinol-binding protein (IRBP) (retinal cells) | p53 deficiency | Apoptosis-induced retinal degeneration (p53-WT animals); photoreceptor tumors (p53-null animals) | [231] |
| HPV16 Early Region | Early region | K14+ cells | Estrogen, DMBA | SCC: Cervical [219], Penile [252] | [235] |
| HPV16 | E6 | K14+ cells | Estrogen | SCC: Cervical [258] | [237] |
| HPV16 | E7 | K14+ cells | Estrogen, 4NQO, DMBA | SCC: Cervical [223], Oral [308], Anal [254] | [238] |
| HPV16 | E7 | Conditional (tetracycline system) | K5-tTA, doxycyline | Cervical SCCs require continuous E7 expression | [267,268] |
| HPV16 | E6/E7 | K14+ cells | Estrogen, 4NQO, DMBA | SCC: Cervical [223], Oral [250], Anal [251] | [237,238] |
| HPV16 | E5 | K14+ cells | Estrogen, DMBA/TPA | SCC: Cervical [253], Skin [239] | [236] |
| HPV16 | E6/E7 | Conditional (Adeno-Cre) | Mutant Kras | SCC: Cervical | [270] |
| HPV18 | E6/E7 | K1+ cells | None | Verrucous lesions, papillomas: skin | [264] |
| HPV18 | E6/E7 | α-crystallin (epithelial tissues) | WT, mutant p53; WT pRb | Microphakia: ocular lens | [266] |
| HPV18 | B-globin | HPV18 URR | Estrogen, progesterone | Hormonal regulation of URR activity | [265] |
| HPV18 | E7 | K14+ cells | None | Cataracts of ocular lens | [263] |
4.2.1. Alpha papillomaviruses
Following the successful demonstration that mice could be genetically engineered to express regions of the bovine papillomavirus type 1 (BPV1) viral genome in the late 1980s [225,226], the field recognized the opportunity to apply this technology to study high-risk alpha mucosotropic HPVs and their role in human cancers. The first transgenic mouse model to direct expression of HPV viral oncoproteins to epithelial cells used the α-crystallin promoter to express HPV16 E6 and E7 in the ocular lens, an epithelial tissue [227]. These mice developed hyperplasia and tumors of the lens, phenotypes that were subsequently found to result from E7-induced inactivation of pRb [228] and E6-dependent inhibition of apoptosis and other p53-dependent and independent activities of E6 [229,230]. Around the same time, HPV16 E7 expression was targeted to retinal cells in the eye to inactivate pRb, but these mice only developed tumors in p53-null mice [231]. The α-crystallin promoter also resulted in ectopic expression of HPV16 E6 and E7 in the murine epidermis and these transgenic mice developed preneoplastic lesions that progressed to carcinomas, thus providing the first in vivo evidence that high-risk HPV oncoproteins have oncogenic potential in an experimental model system [232]. This first generation of HPV transgenic mice demonstrated the feasibility of generating high-risk HPV transgenic mice and provided key evidence supporting the oncogenicity of both E6 and E7-related functions in vivo.
Around the same time that early HPV transgenic models were being developed, Fuchs and colleagues reported their use of the human keratin 14 (K14) promoter for tissue-specific transgene expression in basal cells of the stratified squamous epithelia [233,234]. This molecular advancement paved the way for the next generation of HPV transgenic models driving constitutive expression of the entire HPV16 early region [235] or the individual HPV16 oncoproteins E5 [236], E6 [237], or E7 [238]. Early studies with K14-driven HPV16 transgenic mice evaluated skin phenotypes and revealed that each of the HPV16 oncoproteins could induce skin cancers by functioning as tumor promoters [239,240]. Subsequent studies in HPV transgenic mice have been particularly instrumental in understanding HPV-mediated oncogenesis in mucosal epithelia. Using K14-HPV16 transgenic mice, Arbeit and colleagues discovered that treatment with the female hormone estrogen was required for carcinogenesis and potentiated progressive neoplastic disease molecularly and histopathologically similar to the HPV-mediated neoplastic disease process in women [219,241]. In studies of K14E6 and K14E7 single transgenic mice and K14E6/E7 double transgenic mice, we discovered that E7 alone can drive cervical carcinogenesis when combined with estrogen treatment and that E6 acts to drive the development of larger cancers [223]. Additional studies found that estrogen contributes to the onset and maintenance of HPV-driven cancer in transgenic mice [242], a process that requires expression of the estrogen receptor alpha (ERα) [221]. Chung and colleagues revealed that estrogen signaling in the stromal compartment is both required and sufficient for cervical carcinogenesis in HPV16 transgenic mice [220,243]. We also found that the HPV16 E6/E7 oncoproteins, both alone and synergistically with estrogen, globally alter gene expression in the cervical stroma of HPV16 mice and induce inflammation-associated gene expression including several CXCR2 chemokine receptor ligands [244]. A survey of gene expression patterns in human cervical cancers across the spectrum of disease also supports a role for stromal estrogen signaling [245]. These studies reveal just one facet of HPV-mediated cervical carcinogenesis that has been illuminated through the use of HPV transgenic mice. This research has also identified the estrogen signaling pathway as a potential target for therapeutic treatment using FDA-approved anti-estrogen drugs, which prevent, treat, and reduce recurrence of estrogen-induced cervical cancers in HPV16 transgenic mice [246,247]. Ongoing studies using HPV transgenic mice continue to explore the dynamic and bidirectional interaction between HPV, estrogen, and the cervical microenvironment during viral oncogenesis [244,248]. In addition to preclinical models of HPV-associated cervical cancer, HPV16 transgenic mice have also been used to establish preclinical models of HPV-driven neoplastic disease and cancer development in the oral cavity [249,250], anal canal [251], and penis [252]. HPV16 transgenic mice have been used to establish the potency of individual HPV oncoproteins [223,239,240,242,249,253,254], explore the role of host factors and signaling pathways [[219], [220], [221],243,250,[255], [256], [257], [258], [259], [260], [261]], and evaluate potential therapeutic treatments [246,247,259,262]. Several transgenic models have also been developed using HPV type 18 (HPV18), another high-risk mucosotropic HPV (Table 2). Many of the phenotypes that develop in HPV18-based transgenic models largely mirror those seen in HPV16 models [[263], [264], [265], [266]].
Other transgenic models have been designed to allow temporal regulation of high-risk HPV oncoproteins. Our laboratory developed HPV16 E7 transgenic mice in which E7 and luciferase gene expression is under the control of a bidirectional promoter controlled by the tetracycline response element (Bi-L E7) [267]. Crossing these mice to a strain of mice that expresses a tetracycline transactivator driven by basal cell-specific keratin 5 (K5) generated Bi-LE7/K5-tTA bitransgenic mice that express luciferase and HPV16 E7 in the stratified squamous epithelia. Upon administration of doxycycline, E7 expression is repressed. This transgenic model was used to demonstrate that continuous expression of E7 is required for cervical cancer maintenance [[267], [268], [269]], highlighting the potential efficacy of therapeutic strategies targeting E7 protein expression in HPV-associated cancers. A more recent model developed by the Schreiber group expresses a conditional transgene containing a loxP-stop-loxP sequence between the K14 promoter and the E6/E7 genes (E6/E7LSL) in which tissue specific expression of the HPV16 E6 and E7 proteins can be induced through the delivery of Cre-expressing adenovirus vectors [270]. This exciting new model is therefore able to induce focal expression of the HPV oncoproteins, more closely mimicking focal neoplastic lesions in HPV-infected human tissues and facilitating future studies on virus-host interactions and immune regulation in the absence of chronic overexpression of HPV oncoproteins.
4.2.2. Beta papillomaviruses
Beta genus HPVs are currently classified as Group 3 carcinogens by the International Agency for Research on Cancer (IARC), meaning there is inadequate evidence of carcinogenicity in humans [6]. However, these viruses have attracted great interest due to a growing body of evidence that suggests they contribute to the development of cutaneous SCC [62,[271], [272], [273], [274]]. Two beta HPVs, HPV5 and HPV8, were isolated from cutaneous SCC that developed in patients with a rare hereditary disease called epidermodysplasia verucciformis (EV) [275]. This finding caused speculation that beta HPVs, which are rather common in the population [276,277], may be etiological factors in cutaneous skin cancers. To date, the EV-associated HPVs have some of the strongest evidence for causality and for this reason they are classified as “possibly carcinogenic” by IARC [6]. A prevailing theory is that beta HPVs are involved in cancer initiation but their functions then become unnecessary, possibly due to a synergy with ultraviolet (UV) exposure, leading to an inability to detect viral DNA in the resulting tumors, or rather a ‘hit-and-run’ hypothesis [278]. Several transgenic animal models have been developed to study the following beta HPVs: HPV5 [279], HPV8 [[280], [281], [282], [283]], HPV20/27 [284], HPV38 [285,286], and HPV49 [287]. Most of these models use epithelial-specific promoters (K14, K10) to drive expression in the skin and many have used UV exposure as a co-factor. To mimic the ‘hit-and-run’ hypothesis, one model combines chronic UV exposure with temporal expression of HPV38 oncoproteins regulated by the Cre/loxP system to show that tumor growth was not affected upon HPV protein loss after they had been established [288]. These transgenic models of beta HPV oncoprotein expression will prove valuable as the field continues to interrogate their link to human cancer.
5. Preclinical models of MCPyV infection and disease
5.1. Infection models
The current ambiguity regarding MCPyV cell tropism and host range has complicated efforts to generate natural infection animal models [89,94]. Several groups have demonstrated that MCPyV pseudoviruses can achieve entry into a variety of human and animal cell types in vitro, including epithelial cells and fibroblasts [87,102,289]. Multiple groups have also established in vitro replication assays using MCPyV viral genomes, although infectious progeny production and serial transmission is either weak or undetected [90,96,[290], [291], [292]]. Liu and colleagues have achieved MCPyV replication in dermal fibroblasts both in vitro and ex vivo [87,88]. Among a panel of primary dermal fibroblasts isolated from several rodent and primate species, only those from chimpanzees and humans supported the fully MCPyV life cycle and production of infectious progeny virus and rat fibroblasts supported early gene expression [87]. Therefore, while there is currently no natural MCPyV infection animal model, this study provided insight into potential suitable in vivo hosts. Undoubtedly, the ability to produce infectious MCPyV virions in vitro using a viral genome isolated from healthy skin [289,291] and the use of chimeric mammalian polyomaviruses [87] are important technical advancements. These tools will facilitate continuing studies of the natural life cycle and host range of MCPyV in the skin that can inform the development of future natural infection animal models.
5.2. Transgenic models
The molecular pathogenesis of MCPyV-induced MCC is coming into better focus [114], although there is still a great deal to learn. Despite the lack of a MCPyV natural infection model, several transgenic mouse models have been developed to study the MCPyV viral LT and ST antigens (Table 3). The MCPyV T antigens are the logical choice to study in this context given their continued expression in MCC tumors and high degree of functional similarity to other tumor virus oncoproteins. The apparent tropism of MCPyV for the skin and its link to a cutaneous cancer have so far steered investigators to design models that target the viral T antigens to epithelial keratinocytes or Merkel cells using DNA sequences from MCC tumor-derived MCPyV isolates in order to replicate expression of the ST and truncated LT antigens found in MCC.
Table 3.
Preclinical transgenic models used to study human cancers caused by MCPyV.
| Virus | Protein expression | Cell type/tissue targeted | Other genes/cofactors used in model | Disease development and anatomical site | References |
|---|---|---|---|---|---|
| MCPyV MCC350 isolate | ST | K5+ basal cells in pre-term embryos | None | Skin: Proliferation, hyperplasia, dysplastic epithelium | [293] |
| MCPyV MCC350 isolate | ST | Conditional (K5+ cells; induced in postnatal animals) | None | Skin: Carcinoma in situ | [293] |
| MCPyV codon-optimized | ST | Conditional (inducible ubiquitous expression and Atoh1+ cells) | Conditional p53 deletion | Liver, spleen tumors with ubiquitous expression; Merkel cell proliferation with Atoh1-specific expression | [296] |
| MCPyV MCCw168 isolate | tLT | K5+ basal cells | None | None | [295] |
| MCPyV MCCw168 isolate | Early region; tLT + ST | Conditional (constitutive expression in K14+ cells) | None | Skin: hyperplasia, proliferation, E2F-dependent gene expression, spontaneous tumor development | [298] |
| MCPyV MCC350 isolate | ST | K5+ basal cells | Atoh1 | Skin: intraepidermal MCC-like tumors | [295] |
| MCPyV MCCw168 isolate | tLT | K5+ basal cells | Atoh1 | Skin: ectopic Merkel cells (effect of Atoh1 expression) | [295] |
| MCPyV MCCw168 and MCC350 isolates | tLT + ST | K5+ basal cells | Atoh1 + conditional p53 deletion | Skin: MCC development | [303] |
5.2.1. Single MCPyV T antigen transgenic models
The first MCPyV transgenic model was reported in 2015 by Verhaegen and colleagues [293]. In this model, MCPyV ST (MCC350 isolate) expression in the skin was achieved with a keratin 5 (K5) promoter that directs expression to the basal cells of the stratified squamous epithelium. They evaluated both constitutive ST expression (K5-sTAg) in pre-term embryos to avoid potential issues with lethality and inducible ST expression using Cre-loxP technology (K5-loxP-eGFP-loxP-sTAg mice crossed to a tamoxifen-inducible K5-CreERT2 mice) to evaluate expression in post-natal adults. Both prenatal and postnatal ST transgenic mice developed several prominent phenotypes in the skin consistent with epithelial transformation, including hyperplasia, dysplasia, increased proliferation and apoptosis, and activation of the DNA damage response. Postnatal ST induction also induced lesions histologically consistent with squamous cell carcinoma in situ. Two additional K5-driven constitutive models were created with a PP2A-binding ST mutant (K5-STL142A) and a large T stabilizing domain (LSD) [294] mutant (K5-ST91-95A), revealing that the prenatal ST-induced phenotypes were independent of PP2A binding but required the LSD domain. The investigators have used a similar approach to generate transgenic mice expressing truncated LT only (K5-tLTAg), but these mice developed no apparent phenotypes [295]. Although the ST model helped establish that MCPyV ST is oncogenic in vivo, notably K5-sTAg mice did not develop MCC.
Shuda and colleagues developed MCPyV transgenic models to evaluate the effects of both ubiquitous and Merkel cell-specific expression of ST [296]. In this model, a loxP-stop-loxP (LSL) cassette was cloned upstream of a codon-optimized ST cDNA and introduced into the murine ROSA26 locus, generating ROSAST mice. To achieve ubiquitous ST expression, ROSAST mice were crossed with a tamoxifen-inducible ubiquitous Cre line (Ubc-CreERT2). Ubiquitous ST expression in Ubc-CreERT2-ROSAST mice developed epidermal and dermal hyperplasia and hyperkeratosis on the ears. Ubiquitous ST expression in conditional p53-null mice caused poorly differentiated tumors in the liver and spleen, demonstrating the oncogenic potential of ST. To drive expression of ST in Merkel cells, ROSAST mice were crossed with mice expressing Cre recombinase driven by Atoh1 (Atoh1-CreERT2), a transcription factor involved in Merkel cell specification and development [297]. When ST expression in Atoh1-positive cells was induced during embryogenesis via tamoxifen administration to pregnant dams, the number of Merkel cells increased in the touch domes of prenatal mice but no tumors developed over the course of several months. This effect was not seen when tamoxifen was administered to adult mice, indicating that ST expression in adult Merkel cells does not induce their proliferation. Combined expression of ST with p53-loss in Atoh1-positive Merkel cells did not increase their proliferation or induce tumorigenesis. These studies provided the first evidence that MCPyV ST expression in Merkel cells, even when combined with p53 loss, is insufficient to induce tumorigenesis or MCC development in vivo, at least under the conditions used. Neither Ubc-CreERT2-ROSAST nor Atoh1-CreERT2-ROSAST mice developed MCC.
5.2.2. Combined MCPyV ST and truncated LT transgenic models
We developed a conditional, tissue-specific transgenic MCPyV mouse model that expresses both ST and truncated LT [298]. These targeted knock-in mice express a transgene that contains the ROSA26 promoter upstream of the entire early region of the MCPyV MCC168 isolate, with a loxP-stop-loxP (LSL) sequence between the promoter and MCPyV early region (ROSA26-LSL-MCPyV168). Therefore, expression of the MCPyV T antigens will only be achieved by expression of Cre recombinase and removal of the LSL sequence. Like other MCPyV model designs, this allows both spatial and temporal control of T antigen expression. K14Cre mice were used to drive constitutive expression of ST and truncated LT to the basal cells of the skin and other stratified epithelia. K14Cre-MCPyV168 transgenic mice developed several overt phenotypes within 8–10 days of birth, including ruffled fur and small body size, that progressed to include severe skin thickening, cataracts, and alopecia. Similar to other MCPyV transgenic models, as well as to the K14E6/E7 HPV16 transgenic model [237,238] that we incorporated in our studies as a benchmark for well-validated viral oncoprotein action, we observed several histological and molecular phenotypes in the skin consistent with neoplastic progression. These phenotypes included epithelial hyperplasia, increased proliferation, activation of the DNA damage response, and increased E2F-dependent gene expression in keratinocytes and Merkel cells. By 2–3 months of age, approximately half of the K14Cre-MCPyV168 transgenic mice spontaneously developed skin tumors, a phenotype that was only observed in mice on a pure FVB/N genetic background. In a subsequent study, we expressed the MCPyV T antigens in mice homozygous for an RbΔLXCXE knock-in allele that attenuates LT-pRb interactions through LT's LXCXE motif and found that the majority of the MCPyV T antigen-induced phenotypes were in large part attributable to the LXCXE-dependent interaction between truncated LT and the tumor suppressor pRb [299]. Using a classical, multi-stage model for squamous cell carcinoma development, we found that the MCPyV T antigens synergized with the tumor initiator DMBA, but not with the tumor promoter TPA, to cause tumors in K14Cre-MCPyV168 transgenic mice [300]. This study established that the MCPyV tumor antigens function primarily as tumor promoters, similar to that seen with the HPV16 oncoproteins E6 and E7 in this model. In all of our studies thus far, we have not observed MCC development in K14Cre-MCPyV168 transgenic mice.
5.2.3. MCPyV T antigen + Atoh1 transgenic models
A major advantage of using genetically engineered mouse models is that they can be readily combined with other models, such as those that modulate host factor expression. Verhaegen et al. generated transgenic mice with combined expression of the neuroendocrine cell fate determinant Atoh1 (K5-Atoh1-IRES-GFP), which induces Merkel cell development [301], and MCPyV T antigens in the epidermis [295]. In pre-term embryos, they observed that combinations of ST + Atoh1 and ST + LT + Atoh1, but not ST + LT or LT + Atoh1, expression induced the development of intraepidermal clusters of cells that histologically resembled MCC. These highly proliferative cells expressed several Merkel cell markers, including SOX2, K8, and K20. The authors concluded that epithelial expression of ST combined with overexpression of the Merkel cell specification factor Atoh1 is sufficient to induce MCC-like tumor development. These in vivo findings are consistent with a recent in vitro study that showed co-expression of ST and a Merkel cell differentiation factor Gli1 in keratinocytes can drive Merkel cell gene expression [302].
More recently, the first transgenic mouse model of MCC development was reported [303]. For this model, Verhaegen and colleagues employed various inducible and conditional transgene targeting strategies to achieve tightly controlled tissue-specific expression of the MCPyV T antigens, Atoh1, and p53 deficiency in the skin. Transgenic mice that express MCPyV ST, truncated LT, and Atoh1 under the control of doxycycline-inducible (tetO) elements were generated. In order to direct both temporal and tissue-specific expression of these factors, these mice were crossed with transgenic mice carrying a hormone-inducible Cre allele (K5-CreERT2) and a Cre-inducible allele regulating expression of the tetracycline transactivator protein rtTA (R26-LSL-rtTA). Using this elegant and innovative approach, the administration of tamoxifen and doxycline to these multi-allelic ‘SLA’ adult mice (ST + LT + Atoh1) induced expression of the MCPyV T antigens and Atoh1 in K5-positive basal cells of the skin. Following transgene induction, cellular aggregates expressing multiple Merkel cell and proliferation markers were evident in areas proximal to the hair follicle bulge. However, these aggregates failed to progress to MCC tumors due to what investigators postulated was p53-dependent cell death based on molecular evidence of apoptosis in the SLA-initiated cellular aggregates. Therefore, they generated ‘SLAP’ mice (ST + LT + Atoh1+p53) carrying a floxed allele for p53, resulting in hemizygous p53 expression upon recombination (Trp53WT/fl). Remarkably, SLAP mice developed cutaneous tumors within 11–22 weeks post-induction with gross similarities to human MCC tumors. Histological analysis revealed that tumors in SLAP mice arose within the dermal compartment of the skin, consistent with human MCC tumors, and genetic analysis indicated loss of heterozygosity for the p53 gene within all tumors analyzed. Even more compelling, transcriptomic analysis of global gene expression patterns revealed that the tumors arising in SLAP mice shared a high degree of similarity with human MCC tumors and expressed several neuroendocrine and other Merkel cell-specific genes. Moreover, the SLAP tumors and human MCCs clustered together in hierarchical clustering analyses away from normal skin and other types of skin cancers such as basal cell carcinoma. Taken together, this work reveals that the combined effects of MCPyV T antigen expression, Atoh1 overexpression, and p53 deficiency appear to be sufficient to drive de novo MCC development in the skin of mice and is, to date, the most accurate representation of human MCPyV-positive MCC achieved in a transgenic mouse model.
6. Conclusions and future directions
The study of small DNA tumor viruses has a long and storied history of seminal contributions to our fundamental understanding of cellular and molecular biology, virus-host interactions, and mechanisms of viral oncogenesis. Two small DNA tumor viruses, high-risk alpha HPVs and MCPyV, are considered human tumor viruses because of their role in causing human cancers. Preclinical animal models of HPVs and MCPyV, including infection models and transgenic murine models, have provided essential in vivo systems to study all facets of their causal relationship with human cancers. Moving forward, it will be interesting to observe future experimental and genetic manipulations to both infection and transgenic models that result in comparative models that recapitulate human disease with ever increasing precision. This future repertoire of preclinical models will provide a readily adaptable set of tools that can be adapted and applied if and when future small DNA tumor viruses are discovered.
Author statement
Megan E. Spurgeon: Conceptualization, Investigation, Writing-Original Draft, Writing-Review & Editing, Funding acquisition.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
I would like to acknowledge Paul F. Lambert for providing critical evaluation of the manuscript. I offer my regret to those colleagues whose work was not referenced due to space limitations. Megan E. Spurgeon is supported by grants from the National Institutes of Health (CA211246, CA210807, CA022443).
References
- 1.Rous P.A. Transmissible avian neoplasm. (Sarcoma of the common fowl.) J. Exp. Med. 1910;12:696–705. doi: 10.1084/jem.12.5.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rous P.A. Sarcoma of the fowl transmissible by an agent separable from the tumor cells. J. Exp. Med. 1911;13:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore P.S., Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein M.A., Achong B.G., Barr Y.M. Virus particles in cultured lymphoblasts from burkitt's lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- 5.White M.K., Pagano J.S., Khalili K. Viruses and human cancers: a long road of discovery of molecular paradigms. Clin. Microbiol. Rev. 2014;27:463–481. doi: 10.1128/CMR.00124-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L., et al. A review of human carcinogens--Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/s1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 7.de Martel C., Ferlay J., Franceschi S., Vignat J., Bray F., Forman D., Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 8.Parkin D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 9.Shope R.E., Hurst E.W. Infectious papillomatosis of rabbits : with a note on the histopathology. J. Exp. Med. 1933;58:607–624. doi: 10.1084/jem.58.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bittner J.J. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 11.Gross L. Spontaneous" leukemia developing in C3H mice following inoculation in infancy, with AK-leukemic extracts, or AK-embrvos. Proc. Soc. Exp. Biol. Med. 1951;76:27–32. [PubMed] [Google Scholar]
- 12.Gross L. A filterable agent, recovered from Ak leukemic extracts, causing salivary gland carcinomas in C3H mice. Proc. Soc. Exp. Biol. Med. 1953;83:414–421. doi: 10.3181/00379727-83-20376. [DOI] [PubMed] [Google Scholar]
- 13.DiMaio D. Small size, big impact: how studies of small DNA tumour viruses revolutionized biology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374 doi: 10.1098/rstb.2018.0300. 20180300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howley P.M., Livingston D.M. Small DNA tumor viruses: large contributors to biomedical sciences. Virology. 2009;384:256–259. doi: 10.1016/j.virol.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert P.F. The interwoven story of the small DNA tumor viruses. Virology. 2009;384:255. doi: 10.1016/j.virol.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Pipas J.M. DNA tumor viruses and their contributions to molecular biology. J. Virol. 2019;93 doi: 10.1128/JVI.01524-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane D.P., Crawford L.V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 18.Linzer D.I., Levine A.J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 19.DeCaprio J.A. How the Rb tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology. 2009;384:274–284. doi: 10.1016/j.virol.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Berget S.M., Moore C., Sharp P.A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chow L.T., Gelinas R.E., Broker T.R., Roberts R.J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 22.Wilson J.H., Berget P.B., Pipas J.M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol. Cell Biol. 1982;2:1258–1269. doi: 10.1128/mcb.2.10.1258-1269.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanford R.E., Butel J.S. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 1984;37:801–813. doi: 10.1016/0092-8674(84)90415-x. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981;24:251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- 25.Eckhart W., Hutchinson M.A., Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979;18:925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- 26.Whitman M., Kaplan D.R., Schaffhausen B., Cantley L., Roberts T.M. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–242. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 27.Challberg M.D., Kelly T.J., Jr. Adenovirus DNA replication in vitro: origin and direction of daughter strand synthesis. J. Mol. Biol. 1979;135:999–1012. doi: 10.1016/0022-2836(79)90524-2. [DOI] [PubMed] [Google Scholar]
- 28.Li J.J., Kelly T.J. Simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. U. S. A. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waga S., Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 30.Cohen S.N., Chang A.C., Boyer H.W., Helling R.B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. U. S. A. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danna K., Nathans D. Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae. Proc. Natl. Acad. Sci. U. S. A. 1971;68:2913–2917. doi: 10.1073/pnas.68.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mertz J.E., Davis R.W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc. Natl. Acad. Sci. U. S. A. 1972;69:3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fried M. Cell-transforming ability of a temperature-sensitive mutant of polyoma virus. Proc. Natl. Acad. Sci. U. S. A. 1965;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeCaprio J.A., Ludlow J.W., Figge J., Shew J.Y., Huang C.M., Lee W.H., Marsilio E., Paucha E., Livingston D.M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 35.Whyte P., Buchkovich K.J., Horowitz J.M., Friend S.H., Raybuck M., Weinberg R.A., Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 36.Gross L. The fortuitous isolation and identification of the polyoma virus. Cancer Res. 1976;36:4195–4196. [PubMed] [Google Scholar]
- 37.Huebner R.J., Rowe W.P., Lane W.T. Oncogenic effects in hamsters of human adenovirus types 12 and 18. Proc. Natl. Acad. Sci. U. S. A. 1962;48:2051–2058. doi: 10.1073/pnas.48.12.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trentin J.J., Yabe Y., Taylor G. The quest for human cancer viruses. Science. 1962;137:835–841. doi: 10.1126/science.137.3533.835. [DOI] [PubMed] [Google Scholar]
- 39.Yabe Y., Samper L., Bryan E., Taylor G., Trentin J.J. Oncogenic effect of human adenovirus type 12, in mice. Science. 1964;143:46–47. doi: 10.1126/science.143.3601.46. [DOI] [PubMed] [Google Scholar]
- 40.Hilleman M.R., Werner J.H. Recovery of new agent from patients with acute respiratory illness. Proc. Soc. Exp. Biol. Med. 1954;85:183–188. doi: 10.3181/00379727-85-20825. [DOI] [PubMed] [Google Scholar]
- 41.Rowe W.P., Huebner R.J., Gilmore L.K., Parrott R.H., Ward T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953;84:570–573. doi: 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 42.Javier R., Raska K., Jr., Macdonald G.J., Shenk T. Human adenovirus type 9-induced rat mammary tumors. J. Virol. 1991;65:3192–3202. doi: 10.1128/JVI.65.6.3192-3202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi S., Mukai N. Retinoblastoma-like tumors induced by human adenovirus type 12 in rats. Cancer Res. 1974;34:1646–1651. [PubMed] [Google Scholar]
- 44.Mukai N., Kalter S.S., Cummins L.B., Matthews V.A., Nishida T., Nakajima T. Retinal tumor induced in the baboon by human adenovirus 12. Science. 1980;210:1023–1025. doi: 10.1126/science.7434012. [DOI] [PubMed] [Google Scholar]
- 45.Graham F.L., Smiley J., Russell W.C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 46.Berk A.J. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 47.Bernards R., Van der Eb A.J. Adenovirus: transformation and oncogenicity. Biochim. Biophys. Acta. 1984;783:187–204. doi: 10.1016/0167-4781(84)90029-0. [DOI] [PubMed] [Google Scholar]
- 48.Bertzbach L.D., Ip W.H., Dobner T. Animal models in human adenovirus research. Biology. 2021;10 doi: 10.3390/biology10121253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Endter C., Dobner T. Cell transformation by human adenoviruses. Curr. Top. Microbiol. Immunol. 2004;273:163–214. doi: 10.1007/978-3-662-05599-1_6. [DOI] [PubMed] [Google Scholar]
- 50.Graham F.L., Rowe D.T., McKinnon R., Bacchetti S., Ruben M., Branton P.E. Transformation by human adenoviruses. J. Cell. Physiol. Suppl. 1984;3:151–163. doi: 10.1002/jcp.1041210418. [DOI] [PubMed] [Google Scholar]
- 51.Ip W.H., Dobner T. Cell transformation by the adenovirus oncogenes E1 and E4. FEBS Lett. 2020;594:1848–1860. doi: 10.1002/1873-3468.13717. [DOI] [PubMed] [Google Scholar]
- 52.Tessier T.M., Dodge M.J., MacNeil K.M., Evans A.M., Prusinkiewicz M.A., Mymryk J.S. Almost famous: human adenoviruses (and what they have taught us about cancer) Tumour Virus Res. 2021;12 doi: 10.1016/j.tvr.2021.200225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turnell A.S., Mymryk J.S. Roles for the coactivators CBP and p300 and the APC/C E3 ubiquitin ligase in E1A-dependent cell transformation. Br. J. Cancer. 2006;95:555–560. doi: 10.1038/sj.bjc.6603304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.zur Hausen H. Condylomata acuminata and human genital cancer. Cancer Res. 1976;36:794. [PubMed] [Google Scholar]
- 55.Boshart M., Gissmann L., Ikenberg H., Kleinheinz A., Scheurlen W., zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984;3:1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durst M., Gissmann L., Ikenberg H., zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl. Acad. Sci. U. S. A. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernard H.U., Burk R.D., Chen Z., van Doorslaer K., zur Hausen H., de Villiers E.M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Villiers E.M., Fauquet C., Broker T.R., Bernard H.U., zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 59.Van Doorslaer K., Chen Z., Bernard H.U., Chan P.K.S., DeSalle R., Dillner J., Forslund O., Haga T., McBride A.A., Villa L.L., et al. ICTV virus taxonomy profile: Papillomaviridae. J. Gen. Virol. 2018;99:989–990. doi: 10.1099/jgv.0.001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Satterwhite C.L., Torrone E., Meites E., Dunne E.F., Mahajan R., Ocfemia M.C., Su J., Xu F., Weinstock H. Sexually transmitted infections among US women and men: prevalence and incidence estimates. Sex. Transm. Dis. 2008;40:187–193. doi: 10.1097/OLQ.0b013e318286bb53. 2013. [DOI] [PubMed] [Google Scholar]
- 61.Cogliano V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Cancer W.H.O. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6:204. doi: 10.1016/s1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 62.Rollison D.E., Viarisio D., Amorrortu R.P., Gheit T., Tommasino M. An emerging issue in oncogenic virology: the role of beta human papillomavirus types in the development of cutaneous squamous cell carcinoma. J. Virol. 2019;93 doi: 10.1128/JVI.01003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moens U., Calvignac-Spencer S., Lauber C., Ramqvist T., Feltkamp M.C.W., Daugherty M.D., Verschoor E.J., Ehlers B. Ictv report, C. ICTV virus taxonomy profile: Polyomaviridae. J. Gen. Virol. 2017;98:1159–1160. doi: 10.1099/jgv.0.000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polyomaviridae Study Group of the International Committee on Taxonomy of, V. Calvignac-Spencer S., Feltkamp M.C., Daugherty M.D., Moens U., Ramqvist T., Johne R., Ehlers B. A taxonomy update for the family Polyomaviridae. Arch. Virol. 2016;161:1739–1750. doi: 10.1007/s00705-016-2794-y. [DOI] [PubMed] [Google Scholar]
- 65.Sweet B.H., Hilleman M.R. The vacuolating virus, S.V. 40. Proc. Soc. Exp. Biol. Med. 1960;105:420–427. doi: 10.3181/00379727-105-26128. [DOI] [PubMed] [Google Scholar]
- 66.Eddy B.E., Borman G.S., Berkeley W.H., Young R.D. Tumors induced in hamsters by injection of rhesus monkey kidney cell extracts. Proc. Soc. Exp. Biol. Med. 1961;107:191–197. doi: 10.3181/00379727-107-26576. [DOI] [PubMed] [Google Scholar]
- 67.Eddy B.E., Borman G.S., Grubbs G.E., Young R.D. Identification of the oncogenic substance in rhesus monkey kidney cell culture as simian virus 40. Virology. 1962;17:65–75. doi: 10.1016/0042-6822(62)90082-x. [DOI] [PubMed] [Google Scholar]
- 68.Rabson A.S., Kirschstein R.L. Induction of malignancy in vitro in newborn hamster kidney tissue infected with simian vacuolating virus (SV40) Proc. Soc. Exp. Biol. Med. 1962;111:323–328. doi: 10.3181/00379727-111-27780. [DOI] [PubMed] [Google Scholar]
- 69.Shein H.M., Enders J.F. Transformation induced by simian virus 40 in human renal cell cultures. I. Morphology and growth characteristics. Proc. Natl. Acad. Sci. U. S. A. 1962;48:1164–1172. doi: 10.1073/pnas.48.7.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poulin D.L., DeCaprio J.A. Is there a role for SV40 in human cancer? J. Clin. Oncol. 2006;24:4356–4365. doi: 10.1200/JCO.2005.03.7101. [DOI] [PubMed] [Google Scholar]
- 71.Padgett B.L., Walker D.L., ZuRhein G.M., Eckroade R.J., Dessel B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 72.Zurhein G., Chou S.M. Particles resembling papova viruses in human cerebral demyelinating disease. Science. 1965;148:1477–1479. doi: 10.1126/science.148.3676.1477. [DOI] [PubMed] [Google Scholar]
- 73.Gardner S.D., Field A.M., Coleman D.V., Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 74.Maginnis M.S., Atwood W.J. JC virus: an oncogenic virus in animals and humans? Semin. Cancer Biol. 2009;19:261–269. doi: 10.1016/j.semcancer.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Papadimitriou J.C., Randhawa P., Rinaldo C.H., Drachenberg C.B., Alexiev B., Hirsch H.H. BK polyomavirus infection and renourinary tumorigenesis. Am. J. Transplant. 2016;16:398–406. doi: 10.1111/ajt.13550. [DOI] [PubMed] [Google Scholar]
- 76.Starrett G.J., Buck C.B. The case for BK polyomavirus as a cause of bladder cancer. Curr. Opin. Virol. 2019;39:8–15. doi: 10.1016/j.coviro.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schowalter R.M., Pastrana D.V., Pumphrey K.A., Moyer A.L., Buck C.B. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Meijden E., Janssens R.W., Lauber C., Bouwes Bavinck J.N., Gorbalenya A.E., Feltkamp M.C. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gheit T., Dutta S., Oliver J., Robitaille A., Hampras S., Combes J.D., McKay-Chopin S., Le Calvez-Kelm F., Fenske N., Cherpelis B., et al. Isolation and characterization of a novel putative human polyomavirus. Virology. 2017;506:45–54. doi: 10.1016/j.virol.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeCaprio J.A., Garcea R.L. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 2013;11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harms P.W., Harms K.L., Moore P.S., DeCaprio J.A., Nghiem P., Wong M.K.K., Brownell I. International Workshop on Merkel Cell Carcinoma Research Working, G. The biology and treatment of Merkel cell carcinoma: current understanding and research priorities. Nat. Rev. Clin. Oncol. 2018;15:763–776. doi: 10.1038/s41571-018-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Houben R., Shuda M., Weinkam R., Schrama D., Feng H., Chang Y., Moore P.S., Becker J.C. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J. Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dawe C.J., Freund R., Mandel G., Ballmer-Hofer K., Talmage D.A., Benjamin T.L. Variations in polyoma virus genotype in relation to tumor induction in mice. Characterization of wild type strains with widely differing tumor profiles. Am. J. Pathol. 1987;127:243–261. [PMC free article] [PubMed] [Google Scholar]
- 85.Morrison K.M., Miesegaes G.R., Lumpkin E.A., Maricich S.M. Mammalian Merkel cells are descended from the epidermal lineage. Dev. Biol. 2009;336:76–83. doi: 10.1016/j.ydbio.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Keymeulen A., Mascre G., Youseff K.K., Harel I., Michaux C., De Geest N., Szpalski C., Achouri Y., Bloch W., Hassan B.A., et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J. Cell Biol. 2009;187:91–100. doi: 10.1083/jcb.200907080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu W., Krump N.A., MacDonald M., You J. Merkel cell polyomavirus infection of animal dermal fibroblasts. J. Virol. 2018;92 doi: 10.1128/JVI.01610-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu W., Yang R., Payne A.S., Schowalter R.M., Spurgeon M.E., Lambert P.F., Xu X., Buck C.B., You J. Identifying the target cells and mechanisms of merkel cell polyomavirus infection. Cell Host Microbe. 2016;19:775–787. doi: 10.1016/j.chom.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spurgeon M.E., Lambert P.F. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. Virology. 2013;435:118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng H., Kwun H.J., Liu X., Gjoerup O., Stolz D.B., Chang Y., Moore P.S. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng Z.M., Baker C.C. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front. Biosci. 2006;11:2286–2302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Doorbar J. The papillomavirus life cycle. J. Clin. Virol. 2005;32(Suppl 1):S7–S15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 93.Graham S.V. Human papillomavirus: gene expression, regulation and prospects for novel diagnostic methods and antiviral therapies. Future Microbiol. 2010;5:1493–1506. doi: 10.2217/fmb.10.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krump N.A., You J. From merkel cell polyomavirus infection to merkel cell carcinoma oncogenesis. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.739695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwun H.J., Guastafierro A., Shuda M., Meinke G., Bohm A., Moore P.S., Chang Y. The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J. Virol. 2009;83:12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neumann F., Borchert S., Schmidt C., Reimer R., Hohenberg H., Fischer N., Grundhoff A. Replication, gene expression and particle production by a consensus Merkel Cell Polyomavirus (MCPyV) genome. PLoS One. 2011;6 doi: 10.1371/journal.pone.0029112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wendzicki J.A., Moore P.S., Chang Y. Large T and small T antigens of Merkel cell polyomavirus. Curr. Opin. Virol. 2015;11:38–43. doi: 10.1016/j.coviro.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moody C.A., Laimins L.A. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 99.Rodriguez A.C., Schiffman M., Herrero R., Hildesheim A., Bratti C., Sherman M.E., Solomon D., Guillen D., Alfaro M., Morales J., et al. Longitudinal study of human papillomavirus persistence and cervical intraepithelial neoplasia grade 2/3: critical role of duration of infection. J. Natl. Cancer Inst. 2010;102:315–324. doi: 10.1093/jnci/djq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carter J.J., Paulson K.G., Wipf G.C., Miranda D., Madeleine M.M., Johnson L.G., Lemos B.D., Lee S., Warcola A.H., Iyer J.G., et al. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J. Natl. Cancer Inst. 2009;101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kean J.M., Rao S., Wang M., Garcea R.L. Seroepidemiology of human polyomaviruses. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pastrana D.V., Tolstov Y.L., Becker J.C., Moore P.S., Chang Y., Buck C.B. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tolstov Y.L., Pastrana D.V., Feng H., Becker J.C., Jenkins F.J., Moschos S., Chang Y., Buck C.B., Moore P.S. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int. J. Cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bodily J., Laimins L.A. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 2011;19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Della Fera A.N., Warburton A., Coursey T.L., Khurana S., McBride A.A. Persistent human papillomavirus infection. Viruses. 2021;13 doi: 10.3390/v13020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moscicki A.B., Schiffman M., Burchell A., Albero G., Giuliano A.R., Goodman M.T., Kjaer S.K., Palefsky J. Updating the natural history of human papillomavirus and anogenital cancers. Vaccine. 2012;30(Suppl 5):F24–F33. doi: 10.1016/j.vaccine.2012.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McBride A.A., Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Warburton A., Redmond C.J., Dooley K.E., Fu H., Gillison M.L., Akagi K., Symer D.E., Aladjem M.I., McBride A.A. HPV integration hijacks and multimerizes a cellular enhancer to generate a viral-cellular super-enhancer that drives high viral oncogene expression. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jeon S., Allen-Hoffmann B.L., Lambert P.F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jeon S., Lambert P.F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shuda M., Feng H., Kwun H.J., Rosen S.T., Gjoerup O., Moore P.S., Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cheng J., Rozenblatt-Rosen O., Paulson K.G., Nghiem P., DeCaprio J.A. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J. Virol. 2013;87:6118–6126. doi: 10.1128/JVI.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.DeFilippis R.A., Goodwin E.C., Wu L., DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 2003;77:1551–1563. doi: 10.1128/jvi.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.DeCaprio J.A. Molecular pathogenesis of merkel cell carcinoma. Annu. Rev. Pathol. 2021;16:69–91. doi: 10.1146/annurev-pathmechdis-012419-032817. [DOI] [PubMed] [Google Scholar]
- 115.Munger K., Baldwin A., Edwards K.M., Hayakawa H., Nguyen C.L., Owens M., Grace M., Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Levine A.J. The common mechanisms of transformation by the small DNA tumor viruses: the inactivation of tumor suppressor gene products: p53. Virology. 2009;384:285–293. doi: 10.1016/j.virol.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 117.Park D.E., Cheng J., Berrios C., Montero J., Cortes-Cros M., Ferretti S., Arora R., Tillgren M.L., Gokhale P.C., DeCaprio J.A. Dual inhibition of MDM2 and MDM4 in virus-positive Merkel cell carcinoma enhances the p53 response. Proc. Natl. Acad. Sci. U. S. A. 2019;116:1027–1032. doi: 10.1073/pnas.1818798116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scheffner M., Werness B.A., Huibregtse J.M., Levine A.J., Howley P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 119.Helt A.M., Galloway D.A. Mechanisms by which DNA tumor virus oncoproteins target the Rb family of pocket proteins. Carcinogenesis. 2003;24:159–169. doi: 10.1093/carcin/24.2.159. [DOI] [PubMed] [Google Scholar]
- 120.Hesbacher S., Pfitzer L., Wiedorfer K., Angermeyer S., Borst A., Haferkamp S., Scholz C.J., Wobser M., Schrama D., Houben R. RB1 is the crucial target of the Merkel cell polyomavirus Large T antigen in Merkel cell carcinoma cells. Oncotarget. 2016;7:32956–32968. doi: 10.18632/oncotarget.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Munger K., Werness B.A., Dyson N., Phelps W.C., Harlow E., Howley P.M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Roman A., Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Miller D.L., Myers C.L., Rickards B., Coller H.A., Flint S.J. Adenovirus type 5 exerts genome-wide control over cellular programs governing proliferation, quiescence, and survival. Genome Biol. 2007;8:R58. doi: 10.1186/gb-2007-8-4-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.White E. Regulation of the cell cycle and apoptosis by the oncogenes of adenovirus. Oncogene. 2001;20:7836–7846. doi: 10.1038/sj.onc.1204861. [DOI] [PubMed] [Google Scholar]
- 125.Doorbar J., Egawa N., Griffin H., Kranjec C., Murakami I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015;25(Suppl 1):2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Krump N.A., You J. Molecular mechanisms of viral oncogenesis in humans. Nat. Rev. Microbiol. 2018;16:684–698. doi: 10.1038/s41579-018-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 128.Frost E.L., Lukacher A.E. The importance of mouse models to define immunovirologic determinants of progressive multifocal leukoencephalopathy. Front. Immunol. 2014;5:646. doi: 10.3389/fimmu.2014.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.D'Souza G., Kreimer A.R., Viscidi R., Pawlita M., Fakhry C., Koch W.M., Westra W.H., Gillison M.L. Case-control study of human papillomavirus and oropharyngeal cancer. N. Engl. J. Med. 2007;356:1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 130.Hoots B.E., Palefsky J.M., Pimenta J.M., Smith J.S. Human papillomavirus type distribution in anal cancer and anal intraepithelial lesions. Int. J. Cancer. 2009;124:2375–2383. doi: 10.1002/ijc.24215. [DOI] [PubMed] [Google Scholar]
- 131.Munoz N., Bosch F.X., Castellsague X., Diaz M., de Sanjose S., Hammouda D., Shah K.V., Meijer C.J. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int. J. Cancer. 2004;111:278–285. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 132.Munoz N., Hernandez-Suarez G., Mendez F., Molano M., Posso H., Moreno V., Murillo R., Ronderos M., Meijer C., Munoz A., et al. Persistence of HPV infection and risk of high-grade cervical intraepithelial neoplasia in a cohort of Colombian women. Br. J. Cancer. 2009;100:1184–1190. doi: 10.1038/sj.bjc.6604972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Institute, N.C. NCI Dictionary of Cancer Terms. Availabe online: https://www.cancer.gov/publications/dictionaries/cancer-terms (accessed on December 17).
- 134.Barth H., Solis M., Kack-Kack W., Soulier E., Velay A., Fafi-Kremer S. In vitro and in vivo models for the study of human polyomavirus infection. Viruses. 2016;8 doi: 10.3390/v8100292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Campo M.S. Animal models of papillomavirus pathogenesis. Virus Res. 2002;89:249–261. doi: 10.1016/s0168-1702(02)00193-4. [DOI] [PubMed] [Google Scholar]
- 136.Christensen N.D. Cottontail rabbit papillomavirus (CRPV) model system to test antiviral and immunotherapeutic strategies. Antivir. Chem. Chemother. 2005;16:355–362. doi: 10.1177/095632020501600602. [DOI] [PubMed] [Google Scholar]
- 137.Christensen N.D., Budgeon L.R., Cladel N.M., Hu J. Recent advances in preclinical model systems for papillomaviruses. Virus Res. 2017;231:108–118. doi: 10.1016/j.virusres.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Spurgeon M.E., Lambert P.F. Mus musculus papillomavirus 1: a new frontier in animal models of papillomavirus pathogenesis. J. Virol. 2020:94. doi: 10.1128/JVI.00002-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Buck C.B., Thompson C.D. Production of papillomavirus-based gene transfer vectors. Curr. Protoc. Cell Biol. 2007 doi: 10.1002/0471143030.cb2601s37. (Chapter 26), Unit 26 21. [DOI] [PubMed] [Google Scholar]
- 140.Gee G.V., O'Hara B.A., Derdowski A., Atwood W.J. Pseudovirus mimics cell entry and trafficking of the human polyomavirus JCPyV. Virus Res. 2013;178:281–286. doi: 10.1016/j.virusres.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Porter S.S., McBride A.A. Human papillomavirus quasivirus production and infection of primary human keratinocytes. Curr. Protoc. Microbiol. 2020;57:e101. doi: 10.1002/cpmc.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pyeon D., Lambert P.F., Ahlquist P. Production of infectious human papillomavirus independently of viral replication and epithelial cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9311–9316. doi: 10.1073/pnas.0504020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cladel N.M., Peng X., Christensen N., Hu J. The rabbit papillomavirus model: a valuable tool to study viral-host interactions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374 doi: 10.1098/rstb.2018.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Imperiale M.J., Jiang M. Polyomavirus persistence. Annu. Rev. Virol. 2016;3:517–532. doi: 10.1146/annurev-virology-110615-042226. [DOI] [PubMed] [Google Scholar]
- 145.Jaenisch R., Mintz B. Simian virus 40 DNA sequences in DNA of healthy adult mice derived from preimplantation blastocysts injected with viral DNA. Proc. Natl. Acad. Sci. U. S. A. 1974;71:1250–1254. doi: 10.1073/pnas.71.4.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Justice M.J., Dhillon P. Using the mouse to model human disease: increasing validity and reproducibility. Dis. Model Mech. 2016;9:101–103. doi: 10.1242/dmm.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mouse Genome Sequencing C., Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 148.Brinster R.L., Chen H.Y., Trumbauer M., Senear A.W., Warren R., Palmiter R.D. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Costantini F., Lacy E. Introduction of a rabbit beta-globin gene into the mouse germ line. Nature. 1981;294:92–94. doi: 10.1038/294092a0. [DOI] [PubMed] [Google Scholar]
- 150.Gordon J.W., Ruddle F.H. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214:1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- 151.Zambrowicz B.P., Imamoto A., Fiering S., Herzenberg L.A., Kerr W.G., Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lampreht Tratar U., Horvat S., Cemazar M. Transgenic mouse models in cancer research. Front. Oncol. 2018;8:268. doi: 10.3389/fonc.2018.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Smith H.W., Muller W.J. Transgenic mouse models--a seminal breakthrough in oncogene research. Cold Spring Harb. Protoc. 2013;2013:1099–1108. doi: 10.1101/pdb.top069765. [DOI] [PubMed] [Google Scholar]
- 154.Kuhn R., Torres R.M. Cre/loxP recombination system and gene targeting. Methods Mol. Biol. 2002;180:175–204. doi: 10.1385/1-59259-178-7:175. [DOI] [PubMed] [Google Scholar]
- 155.Indra A.K., Warot X., Brocard J., Bornert J.M., Xiao J.H., Chambon P., Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.DuPage M., Dooley A.L., Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Buck C.B., Pastrana D.V., Lowy D.R., Schiller J.T. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 2004;78:751–757. doi: 10.1128/jvi.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Unckell F., Streeck R.E., Sapp M. Generation and neutralization of pseudovirions of human papillomavirus type 33. J. Virol. 1997;71:2934–2939. doi: 10.1128/JVI.71.4.2934-2939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kirnbauer R., Booy F., Cheng N., Lowy D.R., Schiller J.T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. U. S. A. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rose R.C., Bonnez W., Reichman R.C., Garcea R.L. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J. Virol. 1993;67:1936–1944. doi: 10.1128/JVI.67.4.1936-1944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Volpers C., Schirmacher P., Streeck R.E., Sapp M. Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology. 1994;200:504–512. doi: 10.1006/viro.1994.1213. [DOI] [PubMed] [Google Scholar]
- 162.Roberts J.N., Buck C.B., Thompson C.D., Kines R., Bernardo M., Choyke P.L., Lowy D.R., Schiller J.T. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat. Med. 2007;13:857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- 163.Mejia A.F., Culp T.D., Cladel N.M., Balogh K.K., Budgeon L.R., Buck C.B., Christensen N.D. Preclinical model to test human papillomavirus virus (HPV) capsid vaccines in vivo using infectious HPV/cottontail rabbit papillomavirus chimeric papillomavirus particles. J. Virol. 2006;80:12393–12397. doi: 10.1128/JVI.01583-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Buck C.B., Thompson C.D., Roberts J.N., Muller M., Lowy D.R., Schiller J.T. Carrageenan is a potent inhibitor of papillomavirus infection. PLoS Pathog. 2006;2:e69. doi: 10.1371/journal.ppat.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang H.S., Buck C.B., Lambert P.F. Inhibition of gamma secretase blocks HPV infection. Virology. 2010;407:391–396. doi: 10.1016/j.virol.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Day P.M., Lowy D.R., Schiller J.T. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J. Virol. 2008;82:12565–12568. doi: 10.1128/JVI.01631-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Day P.M., Thompson C.D., Schowalter R.M., Lowy D.R., Schiller J.T. Identification of a role for the trans-Golgi network in human papillomavirus 16 pseudovirus infection. J. Virol. 2013;87:3862–3870. doi: 10.1128/JVI.03222-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Day P.M., Weisberg A.S., Thompson C.D., Hughes M.M., Pang Y.Y., Lowy D.R., Schiller J.T. Human papillomavirus 16 capsids mediate nuclear entry during infection. J. Virol. 2019;93 doi: 10.1128/JVI.00454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huang H.S., Lambert P.F. Use of an in vivo animal model for assessing the role of integrin alpha(6)beta(4) and syndecan-1 in early steps in papillomavirus infection. Virology. 2012;433:395–400. doi: 10.1016/j.virol.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Selinka H.C., Giroglou T., Sapp M. Analysis of the infectious entry pathway of human papillomavirus type 33 pseudovirions. Virology. 2002;299:279–287. doi: 10.1006/viro.2001.1493. [DOI] [PubMed] [Google Scholar]
- 171.Johnson K.M., Kines R.C., Roberts J.N., Lowy D.R., Schiller J.T., Day P.M. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J. Virol. 2009;83:2067–2074. doi: 10.1128/JVI.02190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rous P., Beard J.W. The progression to carcinoma of virus-induced rabbit papillomas (shope) J. Exp. Med. 1935;62:523–548. doi: 10.1084/jem.62.4.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Brandsma J.L. The cottontail rabbit papillomavirus model of high-risk HPV-induced disease. Methods Mol. Med. 2005;119:217–235. doi: 10.1385/1-59259-982-6:217. [DOI] [PubMed] [Google Scholar]
- 174.Harvey S.B., Cladel N.M., Budgeon L.R., Welsh P.A., Griffith J.W., Lang C.M., Christensen N.D. Rabbit genital tissue is susceptible to infection by rabbit oral papillomavirus: an animal model for a genital tissue-targeting papillomavirus. J. Virol. 1998;72:5239–5244. doi: 10.1128/JVI.72.6.5239-5244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sundberg J.P., Junge R.E., el Shazly M.O. Oral papillomatosis in New Zealand white rabbits. Am. J. Vet. Res. 1985;46:664–668. [PubMed] [Google Scholar]
- 176.Watrach A.M., Small E., Case M.T. Canine papilloma: progression of oral papilloma to carcinoma. J. Natl. Cancer Inst. 1970;45:915–920. [PubMed] [Google Scholar]
- 177.Doorbar J. Model systems of human papillomavirus-associated disease. J. Pathol. 2016;238:166–179. doi: 10.1002/path.4656. [DOI] [PubMed] [Google Scholar]
- 178.Kirnbauer R. Papillomavirus-like particles for serology and vaccine development. Intervirology. 1996;39:54–61. doi: 10.1159/000150475. [DOI] [PubMed] [Google Scholar]
- 179.Amtmann E., Volm M., Wayss K. Tumour induction in the rodent Mastomys natalensis by activation of endogenous papilloma virus genomes. Nature. 1984;308:291–292. doi: 10.1038/308291a0. [DOI] [PubMed] [Google Scholar]
- 180.Nafz J., Schafer K., Chen S.F., Bravo I.G., Ibberson M., Nindl I., Stockfleth E., Rosl F. A novel rodent papillomavirus isolated from anogenital lesions in its natural host. Virology. 2008;374:186–197. doi: 10.1016/j.virol.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 181.Nafz J., Kohler A., Ohnesorge M., Nindl I., Stockfleth E., Rosl F. Persistence of Mastomys natalensis papillomavirus in multiple organs identifies novel targets for infection. J. Gen. Virol. 2007;88:2670–2678. doi: 10.1099/vir.0.82955-0. [DOI] [PubMed] [Google Scholar]
- 182.Tan C.H., Tachezy R., Van Ranst M., Chan S.Y., Bernard H.U., Burk R.D. The Mastomys natalensis papillomavirus: nucleotide sequence, genome organization, and phylogenetic relationship of a rodent papillomavirus involved in tumorigenesis of cutaneous epithelia. Virology. 1994;198:534–541. doi: 10.1006/viro.1994.1064. [DOI] [PubMed] [Google Scholar]
- 183.Hasche D., Stephan S., Braspenning-Wesch I., Mikulec J., Niebler M., Grone H.J., Flechtenmacher C., Akgul B., Rosl F., Vinzon S.E. The interplay of UV and cutaneous papillomavirus infection in skin cancer development. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hasche D., Rosl F. Mastomys species as model systems for infectious diseases. Viruses. 2019;11 doi: 10.3390/v11020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Uberoi A. Lambert, P.F. Rodent papillomaviruses. Viruses. 2017:9. doi: 10.3390/v9120362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ingle A., Ghim S., Joh J., Chepkoech I., Bennett Jenson A., Sundberg J.P. Novel laboratory mouse papillomavirus (MusPV) infection. Vet. Pathol. 2011;48:500–505. doi: 10.1177/0300985810377186. [DOI] [PubMed] [Google Scholar]
- 187.Schulz E., Gottschling M., Ulrich R.G., Richter D., Stockfleth E., Nindl I. Isolation of three novel rat and mouse papillomaviruses and their genomic characterization. PLoS One. 2012;7 doi: 10.1371/journal.pone.0047164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xue X.Y., Majerciak V., Uberoi A., Kim B.H., Gotte D., Chen X., Cam M., Lambert P.F., Zheng Z.M. The full transcription map of mouse papillomavirus type 1 (MmuPV1) in mouse wart tissues. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cladel N.M., Budgeon L.R., Cooper T.K., Balogh K.K., Hu J., Christensen N.D. Secondary infections, expanded tissue tropism, and evidence for malignant potential in immunocompromised mice infected with Mus musculus papillomavirus 1 DNA and virus. J. Virol. 2013;87:9391–9395. doi: 10.1128/JVI.00777-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Handisurya A., Day P.M., Thompson C.D., Bonelli M., Lowy D.R., Schiller J.T. Strain-specific properties and T cells regulate the susceptibility to papilloma induction by Mus musculus papillomavirus 1. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Handisurya A., Day P.M., Thompson C.D., Buck C.B., Pang Y.Y., Lowy D.R., Schiller J.T. Characterization of Mus musculus papillomavirus 1 infection in situ reveals an unusual pattern of late gene expression and capsid protein localization. J. Virol. 2013;87:13214–13225. doi: 10.1128/JVI.02162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jiang R.T., Wang J.W., Peng S., Huang T.C., Wang C., Cannella F., Chang Y.N., Viscidi R.P., Best S.R.A., Hung C.F., et al. Spontaneous and vaccine-induced clearance of Mus Musculus papillomavirus 1 infection. J. Virol. 2017;91 doi: 10.1128/JVI.00699-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sundberg J.P., Stearns T.M., Joh J., Proctor M., Ingle A., Silva K.A., Dadras S.S., Jenson A.B., Ghim S.J. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Uberoi A., Yoshida S., Frazer I.H., Pitot H.C., Lambert P.F. Role of ultraviolet radiation in papillomavirus-induced disease. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Blaine-Sauer S., Shin M.K., Matkowskyj K.A., Ward-Shaw E., Lambert P.F. A novel model for papillomavirus-mediated anal disease and cancer using the mouse papillomavirus. mBio. 2021;12 doi: 10.1128/mBio.01611-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cladel N.M., Budgeon L.R., Balogh K.K., Cooper T.K., Hu J., Christensen N.D. A novel pre-clinical murine model to study the life cycle and progression of cervical and anal papillomavirus infections. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cladel N.M., Budgeon L.R., Balogh K.K., Cooper T.K., Hu J., Christensen N.D. Mouse papillomavirus MmuPV1 infects oral mucosa and preferentially targets the base of the tongue. Virology. 2016;488:73–80. doi: 10.1016/j.virol.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hu J., Budgeon L.R., Cladel N.M., Balogh K., Myers R., Cooper T.K., Christensen N.D. Tracking vaginal, anal and oral infection in a mouse papillomavirus infection model. J. Gen. Virol. 2015;96:3554–3565. doi: 10.1099/jgv.0.000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Joh J., Ghim S.J., Chilton P.M., Sundberg J.P., Park J., Wilcher S.A., Proctor M.L., Bennett Jenson A. MmuPV1 infection and tumor development of T cell-deficient mice is prevented by passively transferred hyperimmune sera from normal congenic mice immunized with MmuPV1 virus-like particles (VLPs) Exp. Mol. Pathol. 2016;100:212–219. doi: 10.1016/j.yexmp.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 200.Spurgeon M.E., Lambert P.F. Sexual transmission of murine papillomavirus (MmuPV1) in Mus musculus. Elife. 2019;8 doi: 10.7554/eLife.50056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wei T., Buehler D., Ward-Shaw E., Lambert P.F. An infection-based murine model for papillomavirus-associated head and neck cancer. mBio. 2020;11 doi: 10.1128/mBio.00908-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hu J., Cladel N.M., Budgeon L.R., Balogh K.K., Christensen N.D. The mouse papillomavirus infection model. Viruses. 2017;9 doi: 10.3390/v9090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Joh J., Jenson A.B., King W., Proctor M., Ingle A., Sundberg J.P., Ghim S.J. Genomic analysis of the first laboratory-mouse papillomavirus. J. Gen. Virol. 2011;92:692–698. doi: 10.1099/vir.0.026138-0. [DOI] [PubMed] [Google Scholar]
- 204.Wei T., Grace M., Uberoi A., Romero-Masters J.C., Lee D., Lambert P.F., Munger K. The Mus musculus papillomavirus type 1 E7 protein binds to the retinoblastoma tumor suppressor: implications for viral pathogenesis. mBio. 2021;12 doi: 10.1128/mBio.02277-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.White E.A., Munger K., Howley P.M. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. mBio. 2016;7 doi: 10.1128/mBio.01530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hatterschide J., Bohidar A.E., Grace M., Nulton T.J., Kim H.W., Windle B., Morgan I.M., Munger K., White E.A. PTPN14 degradation by high-risk human papillomavirus E7 limits keratinocyte differentiation and contributes to HPV-mediated oncogenesis. Proc. Natl. Acad. Sci. U. S. A. 2019;116:7033–7042. doi: 10.1073/pnas.1819534116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Meyers J.M., Grace M., Uberoi A., Lambert P.F., Munger K. Inhibition of TGF-beta and NOTCH signaling by cutaneous papillomaviruses. Front. Microbiol. 2018;9:389. doi: 10.3389/fmicb.2018.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Meyers J.M., Uberoi A., Grace M., Lambert P.F., Munger K. Cutaneous HPV8 and MmuPV1 E6 proteins target the NOTCH and TGF-beta tumor suppressors to inhibit differentiation and sustain keratinocyte proliferation. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Yu L., Majerciak V., Xue X.Y., Uberoi A., Lobanov A., Chen X., Cam M., Hughes S.H., Lambert P.F., Zheng Z.M. Mouse papillomavirus type 1 (MmuPV1) DNA is frequently integrated in benign tumors by microhomology-mediated end-joining. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Day P.M., Thompson C.D., Lowy D.R., Schiller J.T. The HPV16 and MusPV1 papillomaviruses initially interact with distinct host components on the basement membrane. Virology. 2015;481:79–94. doi: 10.1016/j.virol.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Handisurya A., Day P.M., Thompson C.D., Buck C.B., Kwak K., Roden R.B., Lowy D.R., Schiller J.T. Murine skin and vaginal mucosa are similarly susceptible to infection by pseudovirions of different papillomavirus classifications and species. Virology. 2012;433:385–394. doi: 10.1016/j.virol.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Peh W.L., Middleton K., Christensen N., Nicholls P., Egawa K., Sotlar K., Brandsma J., Percival A., Lewis J., Liu W.J., et al. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 2002;76:10401–10416. doi: 10.1128/jvi.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Spurgeon M.E., Uberoi A., McGregor S.M., Wei T., Ward-Shaw E., Lambert P.F. A novel in vivo infection model to study papillomavirus-mediated disease of the female reproductive tract. mBio. 2019;10 doi: 10.1128/mBio.00180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang J.W., Jiang R., Peng S., Chang Y.N., Hung C.F., Roden R.B. Immunologic control of Mus musculus papillomavirus type 1. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Joh J., Chilton P.M., Wilcher S.A., Zahin M., Park J., Proctor M.L., Ghim S.J., Jenson A.B. T cell-mediated antitumor immune response eliminates skin tumors induced by mouse papillomavirus, MmuPV1. Exp. Mol. Pathol. 2017;103:181–190. doi: 10.1016/j.yexmp.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 216.Wang W., Uberoi A., Spurgeon M., Gronski E., Majerciak V., Lobanov A., Hayes M., Loke A., Zheng Z.M., Lambert P.F. Stress keratin 17 enhances papillomavirus infection-induced disease by downregulating T cell recruitment. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hobbs R.P., Batazzi A.S., Han M.C., Coulombe P.A. Loss of Keratin 17 induces tissue-specific cytokine polarization and cellular differentiation in HPV16-driven cervical tumorigenesis in vivo. Oncogene. 2016;35:5653–5662. doi: 10.1038/onc.2016.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cladel N.M., Budgeon L.R., Balogh K.K., Cooper T.K., Brendle S.A., Christensen N.D., Schell T.D., Hu J. Mouse papillomavirus infection persists in mucosal tissues of an immunocompetent mouse strain and progresses to cancer. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-17089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Arbeit J.M., Howley P.M., Hanahan D. Chronic estrogen-induced cervical and vaginal squamous carcinogenesis in human papillomavirus type 16 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2930–2935. doi: 10.1073/pnas.93.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chung S.H., Shin M.K., Korach K.S., Lambert P.F. Requirement for stromal estrogen receptor alpha in cervical neoplasia. Horm. Cancer. 2013;4:50–59. doi: 10.1007/s12672-012-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chung S.H., Wiedmeyer K., Shai A., Korach K.S., Lambert P.F. Requirement for estrogen receptor alpha in a mouse model for human papillomavirus-associated cervical cancer. Cancer Res. 2008;68:9928–9934. doi: 10.1158/0008-5472.CAN-08-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Elson D.A., Riley R.R., Lacey A., Thordarson G., Talamantes F.J., Arbeit J.M. Sensitivity of the cervical transformation zone to estrogen-induced squamous carcinogenesis. Cancer Res. 2000;60:1267–1275. [PubMed] [Google Scholar]
- 223.Riley R.R., Duensing S., Brake T., Munger K., Lambert P.F., Arbeit J.M. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 2003;63:4862–4871. [PubMed] [Google Scholar]
- 224.Wei T., Choi S., Buehler D., Lee D., Ward-Shaw E., Anderson R.A., Lambert P.F. Role of IQGAP1 in papillomavirus-associated head and neck tumorigenesis. Cancers. 2021;13 doi: 10.3390/cancers13092276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lindgren V., Sippola-Thiele M., Skowronski J., Wetzel E., Howley P.M., Hanahan D. Specific chromosomal abnormalities characterize fibrosarcomas of bovine papillomavirus type 1 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5025–5029. doi: 10.1073/pnas.86.13.5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sippola-Thiele M., Hanahan D., Howley P.M. Cell-heritable stages of tumor progression in transgenic mice harboring the bovine papillomavirus type 1 genome. Mol. Cell Biol. 1989;9:925–934. doi: 10.1128/mcb.9.3.925-934.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Griep A.E., Herber R., Jeon S., Lohse J.K., Dubielzig R.R., Lambert P.F. Tumorigenicity by human papillomavirus type 16 E6 and E7 in transgenic mice correlates with alterations in epithelial cell growth and differentiation. J. Virol. 1993;67:1373–1384. doi: 10.1128/JVI.67.3.1373-1384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.McCaffrey J., Yamasaki L., Dyson N.J., Harlow E., Griep A.E. Disruption of retinoblastoma protein family function by human papillomavirus type 16 E7 oncoprotein inhibits lens development in part through E2F-1. Mol. Cell Biol. 1999;19:6458–6468. doi: 10.1128/MCB.19.9.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Pan H., Griep A.E. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- 230.Pan H., Griep A.E. Temporally distinct patterns of p53-dependent and p53-independent apoptosis during mouse lens development. Genes Dev. 1995;9:2157–2169. doi: 10.1101/gad.9.17.2157. [DOI] [PubMed] [Google Scholar]
- 231.Howes K.A., Ransom N., Papermaster D.S., Lasudry J.G., Albert D.M., Windle J.J. Apoptosis or retinoblastoma: alternative fates of photoreceptors expressing the HPV-16 E7 gene in the presence or absence of p53. Genes Dev. 1994;8:1300–1310. doi: 10.1101/gad.8.11.1300. [DOI] [PubMed] [Google Scholar]
- 232.Lambert P.F., Pan H., Pitot H.C., Liem A., Jackson M., Griep A.E. Epidermal cancer associated with expression of human papillomavirus type 16 E6 and E7 oncogenes in the skin of transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1993;90:5583–5587. doi: 10.1073/pnas.90.12.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Vassar R., Fuchs E. Transgenic mice provide new insights into the role of TGF-alpha during epidermal development and differentiation. Genes Dev. 1991;5:714–727. doi: 10.1101/gad.5.5.714. [DOI] [PubMed] [Google Scholar]
- 234.Vassar R., Rosenberg M., Ross S., Tyner A., Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1989;86:1563–1567. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Arbeit J.M., Munger K., Howley P.M., Hanahan D. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J. Virol. 1994;68:4358–4368. doi: 10.1128/JVI.68.7.4358-4368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Genther Williams S.M., Disbrow G.L., Schlegel R., Lee D., Threadgill D.W., Lambert P.F. Requirement of epidermal growth factor receptor for hyperplasia induced by E5, a high-risk human papillomavirus oncogene. Cancer Res. 2005;65:6534–6542. doi: 10.1158/0008-5472.CAN-05-0083. [DOI] [PubMed] [Google Scholar]
- 237.Song S., Pitot H.C., Lambert P.F. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J. Virol. 1999;73:5887–5893. doi: 10.1128/JVI.73.7.5887-5893.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Herber R., Liem A., Pitot H., Lambert P.F. Squamous epithelial hyperplasia and carcinoma in mice transgenic for the human papillomavirus type 16 E7 oncogene. J. Virol. 1996;70:1873–1881. doi: 10.1128/JVI.70.3.1873-1881.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Maufort J.P., Williams S.M., Pitot H.C., Lambert P.F. Human papillomavirus 16 E5 oncogene contributes to two stages of skin carcinogenesis. Cancer Res. 2007;67:6106–6112. doi: 10.1158/0008-5472.CAN-07-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Song S., Liem A., Miller J.A., Lambert P.F. Human papillomavirus types 16 E6 and E7 contribute differently to carcinogenesis. Virology. 2000;267:141–150. doi: 10.1006/viro.1999.0106. [DOI] [PubMed] [Google Scholar]
- 241.Brake T., Connor J.P., Petereit D.G., Lambert P.F. Comparative analysis of cervical cancer in women and in a human papillomavirus-transgenic mouse model: identification of minichromosome maintenance protein 7 as an informative biomarker for human cervical cancer. Cancer Res. 2003;63:8173–8180. [PubMed] [Google Scholar]
- 242.Brake T., Lambert P.F. Estrogen contributes to the onset, persistence, and malignant progression of cervical cancer in a human papillomavirus-transgenic mouse model. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2490–2495. doi: 10.1073/pnas.0409883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Son J., Park Y., Chung S.H. Epithelial oestrogen receptor alpha is dispensable for the development of oestrogen-induced cervical neoplastic diseases. J. Pathol. 2018;245:147–152. doi: 10.1002/path.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Spurgeon M.E., Lambert P.F. Human papillomavirus and the stroma: bidirectional crosstalk during the virus life cycle and carcinogenesis. Viruses. 2017;9 doi: 10.3390/v9080219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.den Boon J.A., Pyeon D., Wang S.S., Horswill M., Schiffman M., Sherman M., Zuna R.E., Wang Z., Hewitt S.M., Pearson R., et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E3255–E3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chung S.H., Lambert P.F. Prevention and treatment of cervical cancer in mice using estrogen receptor antagonists. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19467–19472. doi: 10.1073/pnas.0911436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Spurgeon M.E., Chung S.H., Lambert P.F. Recurrence of cervical cancer in mice after selective estrogen receptor modulator therapy. Am. J. Pathol. 2014;184:530–540. doi: 10.1016/j.ajpath.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chung S.H., Franceschi S., Lambert P.F. Estrogen and ERalpha: culprits in cervical cancer? Trends Endocrinol. Metabol. 2010;21:504–511. doi: 10.1016/j.tem.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jabbar S., Strati K., Shin M.K., Pitot H.C., Lambert P.F. Human papillomavirus type 16 E6 and E7 oncoproteins act synergistically to cause head and neck cancer in mice. Virology. 2010;407:60–67. doi: 10.1016/j.virol.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Strati K., Pitot H.C., Lambert P.F. Identification of biomarkers that distinguish human papillomavirus (HPV)-positive versus HPV-negative head and neck cancers in a mouse model. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14152–14157. doi: 10.1073/pnas.0606698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Stelzer M.K., Pitot H.C., Liem A., Schweizer J., Mahoney C., Lambert P.F. A mouse model for human anal cancer. Cancer Prev. Res. 2010;3:1534–1541. doi: 10.1158/1940-6207.CAPR-10-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Medeiros-Fonseca B., Mestre V.F., Estevao D., Sanchez D.F., Canete-Portillo S., Fernandez-Nestosa M.J., Casaca F., Silva S., Brito H., Felix A., et al. HPV16 induces penile intraepithelial neoplasia and squamous cell carcinoma in transgenic mice: first mouse model for HPV-related penile cancer. J. Pathol. 2020;251:411–419. doi: 10.1002/path.5475. [DOI] [PubMed] [Google Scholar]
- 253.Maufort J.P., Shai A., Pitot H.C., Lambert P.F. A role for HPV16 E5 in cervical carcinogenesis. Cancer Res. 2010;70:2924–2931. doi: 10.1158/0008-5472.CAN-09-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Thomas M.K., Pitot H.C., Liem A., Lambert P.F. Dominant role of HPV16 E7 in anal carcinogenesis. Virology. 2011;421:114–118. doi: 10.1016/j.virol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Carchman E.H., Matkowskyj K.A., Meske L., Lambert P.F. Dysregulation of autophagy contributes to anal carcinogenesis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0164273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nyman P.E., Buehler D., Lambert P.F. Loss of function of canonical Notch signaling drives head and neck carcinogenesis. Clin. Cancer Res. 2018;24:6308–6318. doi: 10.1158/1078-0432.CCR-17-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Park J.W., Nickel K.P., Torres A.D., Lee D., Lambert P.F., Kimple R.J. Human papillomavirus type 16 E7 oncoprotein causes a delay in repair of DNA damage. Radiother. Oncol. 2014;113:337–344. doi: 10.1016/j.radonc.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Shai A., Brake T., Somoza C., Lambert P.F. The human papillomavirus E6 oncogene dysregulates the cell cycle and contributes to cervical carcinogenesis through two independent activities. Cancer Res. 2007;67:1626–1635. doi: 10.1158/0008-5472.CAN-06-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Shin M.K., Payne S., Bilger A., Matkowskyj K.A., Carchman E., Meyer D.S., Bentires-Alj M., Deming D.A., Lambert P.F. Activating mutations in Pik3ca contribute to anal carcinogenesis in the presence or absence of HPV-16 oncogenes. Clin. Cancer Res. 2019;25:1889–1900. doi: 10.1158/1078-0432.CCR-18-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Shin M.K., Sage J., Lambert P.F. Inactivating all three rb family pocket proteins is insufficient to initiate cervical cancer. Cancer Res. 2012;72:5418–5427. doi: 10.1158/0008-5472.CAN-12-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Spurgeon M.E., den Boon J.A., Horswill M., Barthakur S., Forouzan O., Rader J.S., Beebe D.J., Roopra A., Ahlquist P., Lambert P.F. Human papillomavirus oncogenes reprogram the cervical cancer microenvironment independently of and synergistically with estrogen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E9076–E9085. doi: 10.1073/pnas.1712018114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Stelzer M.K., Pitot H.C., Liem A., Lee D., Kennedy G.D., Lambert P.F. Rapamycin inhibits anal carcinogenesis in two preclinical animal models. Cancer Prev. Res. 2010;3:1542–1551. doi: 10.1158/1940-6207.CAPR-10-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ghim S., Jenson A.B., Bubier J.A., Silva K.A., Smith R.S., Sundberg J.P. Cataracts in transgenic mice caused by a human papillomavirus type 18 E7 oncogene driven by KRT1-14. Exp. Mol. Pathol. 2008;85:77–82. doi: 10.1016/j.yexmp.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Greenhalgh D.A., Wang X.J., Rothnagel J.A., Eckhardt J.N., Quintanilla M.I., Barber J.L., Bundman D.S., Longley M.A., Schlegel R., Roop D.R. Transgenic mice expressing targeted HPV-18 E6 and E7 oncogenes in the epidermis develop verrucous lesions and spontaneous, rasHa-activated papillomas. Cell Growth Differ. 1994;5:667–675. [PubMed] [Google Scholar]
- 265.Michelin D., Gissmann L., Street D., Potkul R.K., Fisher S., Kaufmann A.M., Qiao L., Schreckenberger C. Regulation of human papillomavirus type 18 in vivo: effects of estrogen and progesterone in transgenic mice. Gynecol. Oncol. 1997;66:202–208. doi: 10.1006/gyno.1997.4745. [DOI] [PubMed] [Google Scholar]
- 266.Nakamura T., Williams-Simons L., Westphal H. A human papillomavirus type 18 E6/E7 transgene sensitizes mouse lens cells to human wild-type p53-mediated apoptosis. Oncogene. 1997;14:2991–2998. doi: 10.1038/sj.onc.1201155. [DOI] [PubMed] [Google Scholar]
- 267.Jabbar S.F., Abrams L., Glick A., Lambert P.F. Persistence of high-grade cervical dysplasia and cervical cancer requires the continuous expression of the human papillomavirus type 16 E7 oncogene. Cancer Res. 2009;69:4407–4414. doi: 10.1158/0008-5472.CAN-09-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Jabbar S.F., Park S., Schweizer J., Berard-Bergery M., Pitot H.C., Lee D., Lambert P.F. Cervical cancers require the continuous expression of the human papillomavirus type 16 E7 oncoprotein even in the presence of the viral E6 oncoprotein. Cancer Res. 2012;72:4008–4016. doi: 10.1158/0008-5472.CAN-11-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Park S., Park J.W., Pitot H.C., Lambert P.F. Loss of dependence on continued expression of the human papillomavirus 16 E7 oncogene in cervical cancers and precancerous lesions arising in fanconi anemia pathway-deficient mice. mBio. 2016;7 doi: 10.1128/mBio.00628-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Bottinger P., Schreiber K., Hyjek E., Krausz T., Spiotto M.T., Steiner M., Idel C., Booras H., Beck-Engeser G., Riederer J., et al. Cooperation of genes in HPV16 E6/E7-dependent cervicovaginal carcinogenesis trackable by endoscopy and independent of exogenous estrogens or carcinogens. Carcinogenesis. 2020;41:1605–1615. doi: 10.1093/carcin/bgaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hampras S.S., Reed R.A., Bezalel S., Cameron M., Cherpelis B., Fenske N., Sondak V.K., Messina J., Tommasino M., Gheit T., et al. Cutaneous human papillomavirus infection and development of subsequent squamous cell carcinoma of the skin. J. Skin Cancer. 2016 doi: 10.1155/2016/1368103. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Howley P.M., Pfister H.J. Beta genus papillomaviruses and skin cancer. Virology. 2015;479–480:290–296. doi: 10.1016/j.virol.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lambert P.F., Munger K., Rosl F., Hasche D., Tommasino M. Beta human papillomaviruses and skin cancer. Nature. 2020;588:E20–E21. doi: 10.1038/s41586-020-3023-0. [DOI] [PubMed] [Google Scholar]
- 274.Orth G. Epidermodysplasia verruciformis: a model for understanding the oncogenicity of human papillomaviruses. Ciba Found. Symp. 1986;120:157–174. doi: 10.1002/9780470513309.ch11. [DOI] [PubMed] [Google Scholar]
- 275.Orth G., Jablonska S., Favre M., Croissant O., Jarzabek-Chorzelska M., Rzesa G. Characterization of two types of human papillomaviruses in lesions of epidermodysplasia verruciformis. Proc. Natl. Acad. Sci. U. S. A. 1978;75:1537–1541. doi: 10.1073/pnas.75.3.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hampras S.S., Giuliano A.R., Lin H.Y., Fisher K.J., Abrahamsen M.E., Sirak B.A., Iannacone M.R., Gheit T., Tommasino M., Rollison D.E. Natural history of cutaneous human papillomavirus (HPV) infection in men: the HIM study. PLoS One. 2014;9 doi: 10.1371/journal.pone.0104843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Hampras S.S., Rollison D.E., Giuliano A.R., McKay-Chopin S., Minoni L., Sereday K., Gheit T., Tommasino M. Prevalence and concordance of cutaneous beta human papillomavirus infection at mucosal and cutaneous sites. J. Infect. Dis. 2017;216:92–96. doi: 10.1093/infdis/jix245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hasche D., Vinzon S.E., Rosl F. Cutaneous papillomaviruses and non-melanoma skin cancer: causal agents or innocent bystanders? Front. Microbiol. 2018;9:874. doi: 10.3389/fmicb.2018.00874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Buitrago-Perez A., Hachimi M., Duenas M., Lloveras B., Santos A., Holguin A., Duarte B., Santiago J.L., Akgul B., Rodriguez-Peralto J.L., et al. A humanized mouse model of HPV-associated pathology driven by E7 expression. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Heuser S., Hufbauer M., Steiger J., Marshall J., Sterner-Kock A., Mauch C., Zigrino P., Akgul B. The fibronectin/alpha3beta1 integrin axis serves as molecular basis for keratinocyte invasion induced by betaHPV. Oncogene. 2016;35:4529–4539. doi: 10.1038/onc.2015.512. [DOI] [PubMed] [Google Scholar]
- 281.Marcuzzi G.P., Hufbauer M., Kasper H.U., Weissenborn S.J., Smola S., Pfister H. Spontaneous tumour development in human papillomavirus type 8 E6 transgenic mice and rapid induction by UV-light exposure and wounding. J. Gen. Virol. 2009;90:2855–2864. doi: 10.1099/vir.0.012872-0. [DOI] [PubMed] [Google Scholar]
- 282.Pfefferle R., Marcuzzi G.P., Akgul B., Kasper H.U., Schulze F., Haase I., Wickenhauser C., Pfister H. The human papillomavirus type 8 E2 protein induces skin tumors in transgenic mice. J. Invest. Dermatol. 2008;128:2310–2315. doi: 10.1038/jid.2008.73. [DOI] [PubMed] [Google Scholar]
- 283.Schaper I.D., Marcuzzi G.P., Weissenborn S.J., Kasper H.U., Dries V., Smyth N., Fuchs P., Pfister H. Development of skin tumors in mice transgenic for early genes of human papillomavirus type 8. Cancer Res. 2005;65:1394–1400. doi: 10.1158/0008-5472.CAN-04-3263. [DOI] [PubMed] [Google Scholar]
- 284.Michel A., Kopp-Schneider A., Zentgraf H., Gruber A.D., de Villiers E.M. E6/E7 expression of human papillomavirus type 20 (HPV-20) and HPV-27 influences proliferation and differentiation of the skin in UV-irradiated SKH-hr1 transgenic mice. J. Virol. 2006;80:11153–11164. doi: 10.1128/JVI.00954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Dong W., Kloz U., Accardi R., Caldeira S., Tong W.M., Wang Z.Q., Jansen L., Durst M., Sylla B.S., Gissmann L., et al. Skin hyperproliferation and susceptibility to chemical carcinogenesis in transgenic mice expressing E6 and E7 of human papillomavirus type 38. J. Virol. 2005;79:14899–14908. doi: 10.1128/JVI.79.23.14899-14908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Viarisio D., Mueller-Decker K., Kloz U., Aengeneyndt B., Kopp-Schneider A., Grone H.J., Gheit T., Flechtenmacher C., Gissmann L., Tommasino M. E6 and E7 from beta HPV38 cooperate with ultraviolet light in the development of actinic keratosis-like lesions and squamous cell carcinoma in mice. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Viarisio D., Muller-Decker K., Zanna P., Kloz U., Aengeneyndt B., Accardi R., Flechtenmacher C., Gissmann L., Tommasino M. Novel ss-HPV49 transgenic mouse model of upper digestive tract cancer. Cancer Res. 2016;76:4216–4225. doi: 10.1158/0008-5472.CAN-16-0370. [DOI] [PubMed] [Google Scholar]
- 288.Viarisio D., Muller-Decker K., Accardi R., Robitaille A., Durst M., Beer K., Jansen L., Flechtenmacher C., Bozza M., Harbottle R., et al. Beta HPV38 oncoproteins act with a hit-and-run mechanism in ultraviolet radiation-induced skin carcinogenesis in mice. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Schowalter R.M., Reinhold W.C. Buck, C.B. Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Liu W., Krump N.A., Buck C.B., You J. Merkel cell polyomavirus infection and detection. JoVE. 2019 doi: 10.3791/58950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Schowalter R.M., Pastrana D.V., Buck C.B. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Siebels S., Czech-Sioli M., Spohn M., Schmidt C., Theiss J., Indenbirken D., Gunther T., Grundhoff A., Fischer N. Merkel cell polyomavirus DNA replication induces senescence in human dermal fibroblasts in a kap1/trim28-dependent manner. mBio. 2020;11 doi: 10.1128/mBio.00142-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Verhaegen M.E., Mangelberger D., Harms P.W., Vozheiko T.D., Weick J.W., Wilbert D.M., Saunders T.L., Ermilov A.N., Bichakjian C.K., Johnson T.M., et al. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J. Invest. Dermatol. 2015;135:1415–1424. doi: 10.1038/jid.2014.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Kwun H.J., Shuda M., Feng H., Camacho C.J., Moore P.S., Chang Y. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe. 2013;14:125–135. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Verhaegen M.E., Mangelberger D., Harms P.W., Eberl M., Wilbert D.M., Meireles J., Bichakjian C.K., Saunders T.L., Wong S.Y., Dlugosz A.A. Merkel cell polyomavirus small T antigen initiates merkel cell carcinoma-like tumor development in mice. Cancer Res. 2017;77:3151–3157. doi: 10.1158/0008-5472.CAN-17-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Shuda M., Guastafierro A., Geng X., Shuda Y., Ostrowski S.M., Lukianov S., Jenkins F.J., Honda K., Maricich S.M., Moore P.S., et al. Merkel cell polyomavirus small T antigen induces cancer and embryonic merkel cell proliferation in a transgenic mouse model. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Maricich S.M., Wellnitz S.A., Nelson A.M., Lesniak D.R., Gerling G.J., Lumpkin E.A., Zoghbi H.Y. Merkel cells are essential for light-touch responses. Science. 2009;324:1580–1582. doi: 10.1126/science.1172890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Spurgeon M.E., Cheng J., Bronson R.T., Lambert P.F., DeCaprio J.A. Tumorigenic activity of merkel cell polyomavirus T antigens expressed in the stratified epithelium of mice. Cancer Res. 2015;75:1068–1079. doi: 10.1158/0008-5472.CAN-14-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Spurgeon M.E., Cheng J., Ward-Shaw E., Dick F.A., DeCaprio J.A., Lambert P.F. Merkel cell polyomavirus large T antigen binding to pRb promotes skin hyperplasia and tumor development. PLoS Pathog. 2022;18(5) doi: 10.1371/journal.ppat.1010551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Spurgeon M.E., Liem A., Buehler D., Cheng J., DeCaprio J.A., Lambert P.F. The merkel cell polyomavirus T antigens function as tumor promoters in murine skin. Cancers. 2021;13 doi: 10.3390/cancers13020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ostrowski S.M., Wright M.C., Bolock A.M., Geng X., Maricich S.M. Ectopic Atoh1 expression drives Merkel cell production in embryonic, postnatal and adult mouse epidermis. Development. 2015;142:2533–2544. doi: 10.1242/dev.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kervarrec T., Samimi M., Hesbacher S., Berthon P., Wobser M., Sallot A., Sarma B., Schweinitzer S., Gandon T., Destrieux C., et al. Merkel cell polyomavirus T antigens induce merkel cell-like differentiation in GLI1-expressing epithelial cells. Cancers. 2020;12 doi: 10.3390/cancers12071989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Verhaegen M.E., Harms P.W., Van Goor J.J., Arche J., Patrick M.T., Wilbert D., Zabawa H., Grachtchouk M., Liu C.J., Hu K., et al. Direct cellular reprogramming enables development of viral T antigen-driven Merkel cell carcinoma in mice. J. Clin. Invest. 2022:132. doi: 10.1172/JCI152069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Poiesz B.J., Ruscetti F.W., Gazdar A.F., Bunn P.A., Minna J.D., Gallo R.C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Choo Q.L., Kuo G., Weiner A.J., Overby L.R., Bradley D.W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 306.Blumberg B.S., Alter H.J., Visnich S. A "new" antigen in leukemia sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 307.Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 308.Strati K., Lambert P.F. Role of Rb-dependent and Rb-independent functions of papillomavirus E7 oncogene in head and neck cancer. Cancer Res. 2007;67:11585–11593. doi: 10.1158/0008-5472.CAN-07-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]