Highlights
-
•
89% of investigated Omicron cases were symptomatic with mild symptoms.
-
•
64% of investigated Omicron cases were vaccinated with two doses.
-
•
9% and 8.3% of cases reported loss of taste and smell, respectively.
-
•
2% of investigated Omicron cases were hospitalized, none admitted to intensive care or deceased.
Keywords: SARS-CoV-2, COVID-19, Surveys and questionnaires, Epidemiology, Signs and symptoms
Abstract
Objectives
We aimed to investigate the first Omicron cases detected in France in order to assess case characteristics and provide supporting information on the possible impact of this variant on the healthcare system.
Methods
A standardized questionnaire was used to collect information from confirmed and probable Omicron cases.
Results
Median age of 468 investigated cases was 35 years, 376 were symptomatic (89%); 64% were vaccinated with two doses and 7% had received three doses. Loss of smell and taste were reported by 8.3% and 9% of cases, respectively. Seven cases were hospitalized, three of those were unvaccinated (including two with reported precondition). No admissions to intensive care and no deaths were reported.
Conclusions
Our results confirm a mild clinical presentation among the first Omicron cases detected in France and highlight the importance for the national COVID-19 surveillance system to quickly detect and adapt to the emergence of a new variant.
1. Introduction
The World Health Organization classified Omicron as a variant of concern (VOC) on 26 November 2021, following its detection in South Africa and its important genetic divergence compared to previously described variants [1], [2]. Rapid detection and characterization of this emerging variant was a major challenge worldwide [3]. In France, the first case was confirmed on the island of La Réunion (Indian Ocean) on 29 November 2021. The aim of this investigation was to assess the characteristics of the first cases infected with Omicron, including demographics, travel history, clinical presentation, vaccine status and outcome, in order to provide supporting information on the possible impact on the healthcare system.
2. Methods
Between 23 November 2021 and 11 January 2022, epidemiologists at the regional units of Santé publique France in collaboration with the Regional Health Agencies investigated 468 confirmed and probable Omicron cases. Cases were included consecutively, as soon as laboratory results were available for the regional units. A standardized questionnaire was used to obtain information about the cases, including demographics (age, sex), travel history (dates of travel and countries traveled), clinical symptoms (date of symptom onset or absence of symptoms; fever, feverish feeling, cough, asthenia/fatigue, myalgia, headache, shortness of breath, diarrhea, ageusia, anosmia, runny nose, sore throat, nausea/vomiting, acute respiratory distress syndrome, abnormal lung auscultation or other) and outcome (hospitalization and intensive care admissions, dates of admission and discharge, and death), preconditions (hypertension, obesity, diabetes, chronic respiratory disease, renal insufficiency, cancer, immunosuppression, liver disease, heart disease, neuromuscular pathology, pregnancy or other), previous SARS-CoV-2 infection and vaccination status (number of doses and date of administration). Questionnaires were administered by staff of the regional health offices, who interviewed Omicron cases by phone or interrogated the attending physician in case of hospitalization.
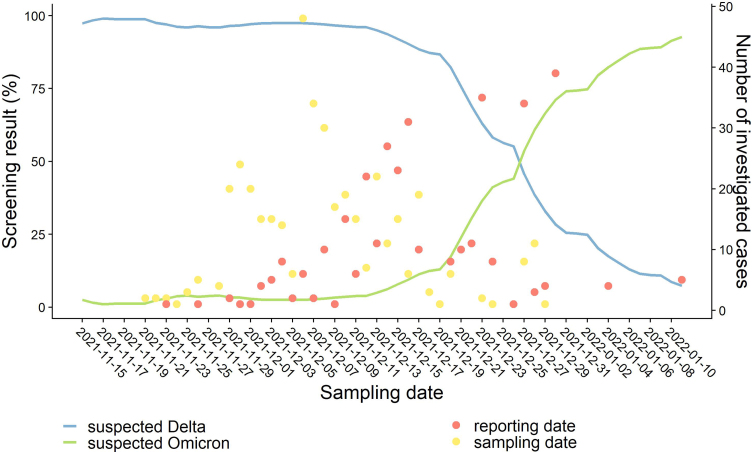
At the time of the survey, the French RT-qPCR (reverse transcription followed by quantitative PCR) screening strategy targeted the E484K and L452R mutations in the Spike protein and the absence of both mutations in the sample was considered to be a suspected Omicron infection (Fig. 1). Additional RT-qPCR screening of Spike protein substitutions K417N, S371L-S373P and Q493R, or deletion at position 69/70 was performed by some laboratories, and the detection of one of these was considered as a probable Omicron infection. We defined a confirmed Omicron case as a person with a SARS-CoV-2 infection and whole genome sequencing (WGS) of the virus by laboratories from the EMERGEN Consortium confirming Omicron [4].
Fig. 1.
Number of investigated Omicron cases and screening results for suspected Delta and Omicron. Lines indicate the proportion of RT-qPCR screening test results corresponding to suspected Delta (blue line, detection of the mutation L452R) and Omicron (green line, absence of the mutations E484K and L452R) variants, over time (7-day moving average). Dots indicate the number of investigated cases by sampling (yellow dots) and reporting date (red dots).
All Omicron cases consecutively included in this study were confirmed cases, except for 45 probable cases detected in two French overseas territories (Guadeloupe and Martinique) due to delays in confirmation by sequencing.
3. Results
As of 17 January 2022, 468 Omicron cases were investigated in 17 out of 18 different regions in France, including four overseas territories.
The sampling dates ranged from 21 November to 29 December 2021 in a period of Delta predominance (Fig. 1). Onset dates ranged from 20 November to 30 December 2021, and the median reporting delay after sampling date was 8 days (Fig. 1). Symptomatic cases were reported after a median of 9 days after symptom onset (Table 1). Ninety-five cases were investigated after being reported as part of a cluster, though the total number of clusters in this period is not known.
Table 1.
Characteristics of Omicron cases (n = 468). Proportions, medians and interquartile range were calculated per available information, respective denominators are provided in brackets beside the variable name.
| Characteristics | n | % |
|---|---|---|
| Sex (n = 357) | ||
| Female | 196 | 55 |
| Male | 161 | 45 |
| Age (n = 445) | 11 | 2 |
| 0–9 | ||
| 10–14 | 17 | 4 |
| 15–19 | 25 | 6 |
| 20–29 | 113 | 25 |
| 30–39 | 108 | 24 |
| 40–49 | 67 | 15 |
| 50–59 | 63 | 14 |
| 60–69 | 29 | 7 |
| 70–79 | 9 | 2 |
| 80+ | 3 | 1 |
| Symptomatic infection (n = 421) | ||
| Yes | 376 | 89 |
| No | 45 | 11 |
| Risk factors (n = 283) | ||
| Yes | 45 | 16 |
| No | 238 | 84 |
| Hospitalization (n = 294) | ||
| Yes | 7 | 2 |
| No | 287 | 98 |
| Intensive care (n = 292) | ||
| Yes | 0 | 0 |
| No | 292 | 100 |
| Travel history (n = 407) | ||
| Yes | 148 | 36 |
| No | 259 | 64 |
| Previous SARS-CoV-2 infection (n = 278) | ||
| Yes | 39 | 14 |
| No | 239 | 86 |
| Vaccination status (n = 394) | ||
| Vaccinated one dose | 20 | 5 |
| Vaccinated two doses | 254 | 64 |
| Vaccinated three doses | 28 | 7 |
| Unvaccinated | 92 | 23 |
| Temporal variables | Median (days) | IQR (days) |
|---|---|---|
| Delay between symptom onset and reporting (n = 271) | 9 | 7–13 |
| Duration of symptoms (n = 141) | 4 | 2–7 |
| Delay between symptom onset and hospitalization (n = 4) | 8 | 4–12 |
| Delay between travel return and symptom onset (n = 64) | 1 | 0–2 |
| Delay between last vaccination and symptom onset (n = 209) | 151 | 102–167 |
| Vaccinated one dose (n = 14) | 128 | 6–159 |
| Vaccinated two doses (n = 175) | 153 | 3–168 |
| Vaccinated three doses (n = 20) | 10 | 2–84 |
n/a: not applicable; AZ: AstraZeneca; IQR: interquartile range.
The age of the cases ranged from 2 months to 91 years (median: 35 years) and 55% were female (Table 1). Travel history or contact with a person returning from travel was reported by 148 (36%) cases (including 45 from South Africa and 27 from European countries). Most cases developed symptoms few days after returning from travel (median of 1 day).
In addition, 254 cases (64%) had received 2 doses of vaccination and 28 cases (7%) had received a third dose, including three immunocompromised individuals. The median delay between the last vaccination and symptom onset was 151 days, with a median of 153 days after 2 doses and 10 days after 3 doses (Table 1).
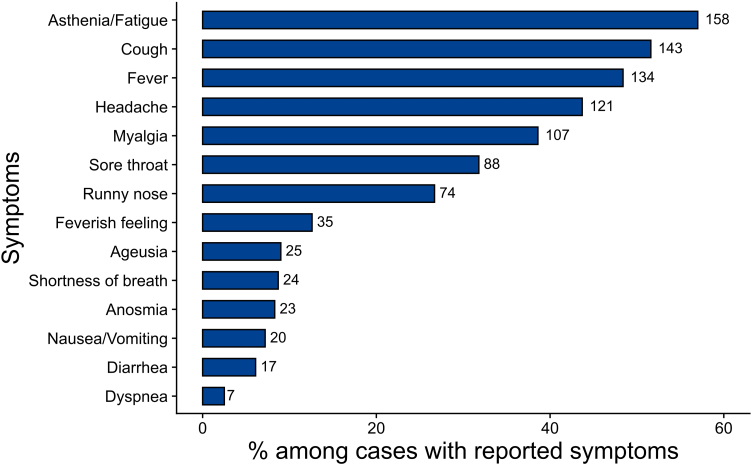
The majority of cases was symptomatic (89%), and reported mostly mild symptoms (Fig. 2). Asthenia/fatigue, cough, fever, headache, myalgia, sore throat or runny nose were the most frequently reported symptoms. Loss of smell (anosmia) and taste (ageusia) were reported by 8.3% (n = 23) and 9% (n = 25) of cases, respectively.
Fig. 2.
Proportion of symptoms reported by symptomatic cases (n = 277). Multiple symptoms could be reported by one case. Number of cases exhibiting a symptom are given beside the bars.
The duration of signs varied from 1 to 30 days (median 4 days, Table 1). Forty-five cases (16%) had at least one risk factor and 39 of these were symptomatic. Thirty-nine cases (14%) had a history of SARS-CoV-2 infection and 36 of these were symptomatic.
Seven cases were hospitalized (2%), none in intensive care. Of these seven cases, three were unvaccinated (including two reporting a previous infection), one had received 3 doses of vaccine and no information was available for the other three cases. These cases ranged in age from 26 to 71 years, and three reported a precondition (two of which were unvaccinated). One hospitalized case had a 15-day history of symptoms (including fever, asthenia, myalgia/hammering, headache, cough, nausea/vomiting and low blood pressure) and had neuromuscular pathology. Two cases were hospitalized for one day, two cases were still hospitalized at the time of the investigation and information was missing for two further cases. Cases were hospitalized after a median of 8 days following symptom onset (Table 1).
4. Discussion
Recent weeks have shown a rapid and global shift in the circulation of variants, with Omicron becoming dominant over Delta in several countries [5], [6], [7]. At the time of our investigation, Omicron represented around 10% of the cases in France that were sequenced in the framework of random genomic surveillance mid-December 2021, increasing to 41% one week later and reaching 70% by the end of December 2021 [8].
Similar to what was initially observed in South Africa and in the UK [9], [10], our investigation confirms a mild clinical presentation of Omicron cases. Data from the UK showed that loss of taste and smell were reported as rarely for Omicron as in our study, and were found to be less commonly reported at the end of December 2021 as compared to the start of December 2021 (loss of taste/smell: 13%/11% compared to 44%/44%) [11]. Fewer patients were hospitalized during the Omicron wave in South Africa, and fewer patients presented with severe disease, compared to previous waves [9]. The proportion of hospitalizations in our study population was as low (2%) as also shown in a Danish study with 1.1% of hospitalized cases, including one admission to intensive care [6]. Preliminary analyses and predictions from other countries suggest a reduced risk of hospitalization for Omicron compared to other variants after three doses of vaccine (up to 81%) [12], [13], [14]. However, data from this descriptive study and international preliminary investigation should be interpreted with caution as the majority of Omicron infections are still detected in a young population, with generally lower rates of comorbidities and therefore less at risk of developing severe forms. Cases were added to our investigation after obtaining a confirmatory sequencing result, including symptomatic and asymptomatic individuals, but it is likely that the proportion of symptomatic individuals undergoing testing may be higher. In addition, the investigation did not allow for longitudinal follow-up of patient outcomes, hence the results on hospitalization, intensive care admission, and vital status for all cases may be underestimated.
Even if this reduced risk of hospitalization among Omicron-infected cases is confirmed in the general population, it may have an increased impact on public health due to the increased transmission of this variant. Modelling data by the Pasteur Institute suggested that an important circulation of Omicron might still lead to the saturation of the French hospital system [15]. While hospitalizations have been increasing since the end of November 2021, as of 18 January, 26 593 COVID-19 cases were hospitalized in France with 3 894 patients in intensive care [8]. Hospitalizations trends are closely monitored.
According to international studies, Omicron's competitive advantage over Delta may be due to its higher transmissibility, but more importantly, its escape from the immune response. Vaccine efficacy against symptomatic forms and hospitalizations, however, seems to be maintained against Omicron after a booster dose [2], [10], [12], [16], [17], [18]. The post-infection or post-vaccination T-cell response, which remains effective against Omicron, may also confer protection against severe forms [19], [20].
As of January 2022, most cases in France were vaccinated with an mRNA vaccine. In our study, the low number of Omicron infected individuals who have received a third dose might be explained by the effectiveness of the third dose to prevent infection. However, the majority of these cases were also younger on average, hence fully vaccinated at a later time point, and part of this population was not yet eligible for the booster dose at the time of investigation. At the time of median onset date of the investigated cases (6 December 2021), the proportion of adults, who were vaccinated with a booster dose, was 21%, and the proportion of individuals older than 64-years-old was 50% [21].
5. Conclusion
The increased transmission and greater immune escape of Omicron are both worrying, although results of our investigation and other preliminary data suggest a reduced severity compared to previously circulating variants. A risk of overburdening the hospital system with patients infected with Omicron remains, although intensive care units are not yet saturated. Caution is required and continued vaccination efforts as well as reinforcing barrier measures are necessary to reduce the impact of Omicron on the health care system.
At the time of the emergence of a new variant, such as Omicron, a rapid detection and characterization of Omicron cases is essential to inform decision making in terms of health care capacities, testing strategy and control measures. The French public health system, including laboratories for testing and genomic surveillance through the EMERGEN Consortium, and local, regional and national authorities, showed their ability to quickly react and adapt to the emergence of a new variant.
Human and animal rights
The authors declare that the work described has not involved experimentation on humans or animals.
Informed consent
The authors declare that this report does not contain any personal information that could lead to the identification of the patient(s) and/or volunteers.
Disclosure of interest
The authors declare the following financial or personal relationships that could be viewed as influencing the work reported in this paper:
Laurent Andreoletti, Reims, has received grants from PHRC (Programme Hospitalier de Recherche Clinique) APHP (Assistance Publique–Hôpitaux de Paris) and FFC (Fédération Française de Cardiologie); he is on the board of RICAI (Réunion interdisciplinaire de Chimiothérapie anti-infectieuse) and ECCMID (European Congress of Clinical Microbiology and Infectious Diseases).
Slim Fourati, Créteil, has received payments or honoraria for lectures or presentations from ABBOTT, MSD and CEPHEID; he holds leadership roles in the CSS5 INSERM and ANRS (l’Agence nationale de recherches sur le sida) boards or committees.
All other authors declare that they have no competing interest.
Funding
Santé publique France, the French national public health agency.
Caisse nationale d’assurance maladie (Cnam), the national health insurance funds.
“Enhancing Whole Genome Sequencing (WGS) and/or Reverse Transcription Polymerase Chain Reaction (RT-PCR) national infrastructures and capacities to respond to the COVID-19 pandemic in the European Union and European Economic Area” Grant Agreement ECDC/HERA/2021/007 ECD. 12221.
Author contributions
All authors attest that they meet the current International Committee of Medical Journal Editors (ICMJE) criteria for Authorship.
Anna Maisa: conceptualization, methodology, writing – original draft, writing – review & editing, vizualisation.
Guillaume Spaccaferri: conceptualization, methodology, writing – review & editing.
Lucie Fournier: conceptualization, methodology, software, formal analysis, data curation, writing – review & editing, vizualisation.
Justine Schaeffer: conceptualization, methodology, software, formal analysis, data curation, writing – review & editing, vizualisation.
Joël Deniau: methodology, sofware, writing – review & editing.
Patrick Rolland: writing – review & editing, supervision.
Bruno Coignard: writing – review & editing, supervision, project management.
Acknowledgement
We would like to acknowledge the tireless work of the regional health authorities, the clinicians and laboratories, who have contributed to the sample and data collection, as well as the individuals interviewed for this investigation.
Contributor Information
A. Maisa, Email: Anna.Maisa@santepubliquefrance.fr.
G. Spaccaferri, Email: guillaume.spaccaferri@santepubliquefrance.fr.
L. Fournier, Email: Lucie.FOURNIER@santepubliquefrance.fr.
J. Schaeffer, Email: Justine.SCHAEFFER@santepubliquefrance.fr.
J. Deniau, Email: joel.deniau@santepubliquefrance.fr.
P. Rolland, Email: Patrick.ROLLAND@santepubliquefrance.fr.
B. Coignard, Email: Bruno.COIGNARD@santepubliquefrance.fr.
The regional COVID-19 investigation team:
A. Andrieu, O. Broustal, S. Chene, S. Chent, E. Fougère, G. Gbaguidi, M. Hamidouche, A. Lamy, Q. Mano, B. Mastrovito, A. Mercier, G. Modenesi, G. Picard, J. Prudhomme, F. Rapilly, A. Riondel, M. Rivière, B. Villegas Ramirez, A. Zhu-Soubise, and M. Zurbaran
The EMERGEN consortium:
A. Amzert, L. Andreoletti, A. Bal, R. Beaurepere, S. Behillil, L. Belec, C. Bernard, L. Bocket, L. Bouri, T. Bourlet, C. Bressollette-Bodin, S. Brichler, C. Brugerolles, S. Cado, V. Calvez, N. Capron, S. Castelain, J. Castro-Alvarez, M.-L. Chaix, C. Charpentier, D. Che, C. Chillou, P. Colson, P. Coudene, A. Crinquette, A. De Rougemont, H. Delagrèverie, C. Delamare, T. Denecker-Berardino, D. Descamps, M. Desroches, G. Destras, G. Dos Santos, A. Ducancelle, S. Ducreux, T. Duret, V. Enouf, S. Fafi-Kremer, C. Felici, S. Fourati, P.-E. Fournier, C. Gaudy, H. Germain, V. Giordanengo, O. Gorge, S. Haim-Boukobza, C. Henquell, A. Holstein, L. Houhamdi, J. Izopet, V. Jacomo, A. Jacques, M.-C. Jaffar-Bandjee, M. Jimenez, L. Josset, S. Kemeny, M.-E. Lafon, A. Le Bars, G. Le Corguille, Q. Lepiller, A. Levasseur, N. Leveque, B. Lina, C. Madelaine, C. Malabat, S. Marque-Juillet, T. Martin-Dunavit, P. Mavingui, A. Merens, I. Messak, L. Morand-Joubert, X. Naudot, P. Neybecker, J.-M. Pawlotsky, L. Pilorge, J.-C. Plantier, C. Poggi, M. Pretet, C. Ragot, H. Raoul, S. Rogez, A.-M. Roque-Afonso, B. Roquebert, D. Rousset, F. Rozenberg, C. Sagot, S. Sahnoune, D. Salgado, O. Sand, C. Saudemont, E. Schvoerer, E. Simon-Loriere, R. Stephan, J. Sudour, V. Thibault, E. Tuaillon, A. Vabret, E. Vallee, S. Van Der Werf, J. Van Helden, L. Verdurme, A. Vignola, D. Wilkinson, and Y. Yazdanpanah
References
- 1.WHO . 2021. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Last accessed 22/01/2022. Available from: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern. [Google Scholar]
- 2.Willett B.J., Grove J., MacLean O., Wilkie C., Logan N., De Lorenzo G., et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. medRxiv. 2022 [2022.01.03.21268111] [Google Scholar]
- 3.WHO . 2021. Weekly epidemiological update on COVID-19-30 November 2021. Last accessed 22/01/2022. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---30-november-2021. [Google Scholar]
- 4.Santé publique France . 2022. Consortium EMERGEN. [Available from: https://www.santepubliquefrance.fr/emergen. [Google Scholar]
- 5.Grabowski F., Kochańczyk M., Lipniacki T. Omicron strain spreads with the doubling time of 3.2–3.6 days in South Africa province of Gauteng that achieved herd immunity to Delta variant. medRxiv. 2021 [2021.12.08.21267494] [Google Scholar]
- 6.Espenhain L., Funk T., Overvad M., Edslev S.M., Fonager J., Ingham A.C., et al. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark. Eurosurveillance. 2021;26(50) doi: 10.2807/1560-7917.ES.2021.26.50.2101146. [2101146] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnard R.C., Davies N.G., Pearson C.A.B., Jit M., Edmunds W.J. Projected epidemiological consequences of the Omicron SARS-CoV-2 variant in England, December 2021 to April 2022. medRxiv. 2021 [2021.12.15.21267858] [Google Scholar]
- 8.Santé publique France . 2022. COVID-19: point épidémiologique du 20 janvier 2022. Last accessed 22/01/2022. Available from: https://www.santepubliquefrance.fr/maladies-et-traumatismes/maladies-et-infections-respiratoires/infection-a-coronavirus/documents/bulletin-national/covid-19-point-epidemiologique-du-20-janvier-2022. [Google Scholar]
- 9.Maslo C., Friedland R., Toubkin M., Laubscher A., Akaloo T., Kama B. Characteristics and outcomes of hospitalized patients in south africa during the covid-19 omicron wave compared with previous waves. JAMA. 2021;30:e2124868. doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UKHSA . 2022. SARS-CoV-2 variants of concern and variants under investigation in England Technical briefing 34. Last accessed 22/01/2022. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1046853/technical-briefing-34-14-january-2022.pdf. [Google Scholar]
- 11.Vihta K.-D., Pouwels K.B., Peto T.E., Pritchard E., House T., Studley R., et al. Omicron-associated changes in SARS-CoV-2 symptoms in the United Kingdom. medRxiv. 2022 doi: 10.1093/cid/ciac613. [2022.01.18.22269082] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.UKHSA . 2021. SARS-CoV-2 variants of concern and variants under investigation in England Technical briefing: Update on hospitalisation and vaccine effectiveness for Omicron VOC-21NOV-01 (B.1.1.529) Last accessed 22/01/2022. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1044481/Technical-Briefing-31-Dec-2021-Omicron_severity_update.pdf. [Google Scholar]
- 13.Ulloa A.C., Buchan S.A., Daneman N., Brown K.A. Early estimates of SARS-CoV-2 Omicron variant severity based on a matched cohort study, Ontario, Canada. medRxiv. 2021 [2021.12.24.21268382] [Google Scholar]
- 14.Israel A., Schäffer A.A., Merzon E., Green I., Magen E., Golan-Cohen A., et al. COVID-19 severity prediction based on patient risk factors and number of vaccines received. medRxiv. 2022 doi: 10.3390/microorganisms10061238. [2021.12.31.21268575] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andronico A., Tran Kiem C., Bosetti P., Paireau J., Emergen C., Fontanet A., et al. 2021. Impact du variant Omicron sur l’épidémie COVID-19 et son contrôle en France métropolitaine durant l’hiver 2021–2022. Last accessed 22/01/2022. Available from: https://modelisation-covid19.pasteur.fr/variant/Institut_Pasteur_Impact_dOmicron_sur_lepidemie_francaise_20211227.pdf. [Google Scholar]
- 16.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., et al. Effectiveness of COVID-19 vaccines against the Omicron (B.1.1.529) variant of concern. medRxiv. 2021 [2021.12.14.21267615] [Google Scholar]
- 17.Khoury D.S., Steain M., Triccas J., Sigal A., Davenport M.P., Cromer D. Analysis:a meta-analysis of early results to predict vaccine efficacy against omicron. medRxiv. 2021 [2021.12.13.21267748] [Google Scholar]
- 18.Collie S., Champion J., Moultrie H., Bekker L.-G., Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in South Africa. N Engl J Med. 2022;386:494–496. doi: 10.1056/NEJMc2119270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed S.F., Quadeer A.A., McKay M.R. SARS-CoV-2 T cell responses are expected to remain robust against Omicron. bioRxiv. 2021 doi: 10.3390/v14010079. [2021.12.12.472315] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., et al. SARS-CoV-2 spike T cell responses induced upon vaccination or infection remain robust against Omicron. medRxiv. 2021 [2021.12.26.21268380. [Google Scholar]
- 21.Santé publique France . 2022. GEODES - Géo données en Santé publique. Last accessed 22/01/2022. Available from: https://geodes.santepubliquefrance.fr/#c=indicator&view=map2. [Google Scholar]