Abstract
One of the most important reasons underlying the resistance of tumors is the immune suppression induced by cancer cells. Myeloid-derived suppressor cells (MDSCs), which exerts pivotal functions in immunosuppression, is a key participator in tumor microenvironment and a novel target for cancer therapy. Here curcumin (Cur) was employed as a specific MDSCs repressor to inhibit the number and function of MDSCs. Moreover, a novel self-assembled nano-filament system was generated through the conjugation of Cur and a self-assembled peptide. In vivo study demonstrated the powerful antitumor effect of curcumin-loaded nano-filaments (Nano-Cur) with delayed tumor growth and longer survival. The immune status of tumor microenvironment (TME) was well improved by Nano-Cur treatment with increased T cell proliferation and activation as well as enhanced production of inflammatory mediators such as GM-CSF and IL-6, which revealed that Nano-Cur contributed to relieve the tumor burden by regulating and improving the TME. Furthermore, flow cytometry analysis implied the lower MDSCs levels under Nano-Cur treatment, which indicated that the anticancer effect of Nano-Cur may be associated with the inhibition of recruitment and accumulation of MDSCs in the TME. Therefore, Nano-Cur may be a novel therapeutic approach for lung cancer, and extensive studies of mechanisms are required to better understand how TME affects tumor progression and provide new insights into anticancer therapeutics.
Keywords: Curcumin, MDSC, Self-assemble, Nano-filaments
Graphical abstract
Schematic illustration of the anticancer effect of Nano-Cur by suppressing MDSCs in TME.
1. Introduction
Lung cancer ranked leadingly among the most dangerous cancers worldwide, which can be mainly divided into two groups: small-cell lung carcinomas (SCLC) and non-small cell lung carcinomas (NSCLC). The morbidity and mortality of lung cancer continue to head the list of cancers [1,2]. According to the type and staging of cancer, a relatively appropriate treatment option can be chosen from surgery, chemotherapy, radiotherapy, molecular targeted therapy, and immunotherapy [3]. However, unless the lung tumors are localized without invading and spreading, their responses to these therapies are still poor. Therefore, in the field of cancer therapy, more efficient and effective approaches for cancer treatment are being studied, among which nanomedicine deserves our attention. Cancer treatment efficacy can be significantly improved for the capability of nanotechnology to decrease the toxicity of anticancer drugs and optimize the therapeutic index of free drugs [4].
Immunotherapy has revolutionized cancer therapy, aiming at harnessing the immune system to mount effective antitumor responses [5,6]. The goal of immunotherapy is not simply limited to activating the immune system against the tumor, but also to relieve the immunosuppressive tumor microenvironment (TME) [7]. TME is complex and continuously evolving. In addition to stromal cells, fibroblasts, and endothelial cells, the TME comprises innate (macrophages, neutrophils, dendritic cells, innate lymphoid cells, myeloid-derived suppressor cells (MDSCs), and NK cells) and adaptive immune cells (T cells and B cells) [8]. MDSCs served as a major constitute of the tumor microenvironment. MDSCs are generated from the bone marrow and tumor-bearing hosts, and later would be recruited to the peripheral lymphoid organs and tumor to participate in the formation of the tumor microenvironment [9]. MDSCs can be divided into two subgroups of cells with either a monocytic (M-MDSCs) or a neutrophilic (G-MDSCs) morphology [10,11]. The immunosuppressive capabilities of MDSCs are performed by the inhibition of T-cell proliferation and activation, suppressive cytokines, and high levels of arginase, nitric oxide, reactive oxygen species (ROS), and prostaglandin E2 [12,13]. A range of transcription factors, such as Irf8 and Stat3, are also reported to implicated in MDSCs expansion [14,15]. In addition, MDSCs promote immune tolerance in cancer by inducing NK cell anergy [16]. MDSCs can also recruit Tregs by expressing CD40 to shape the immunosuppressive microenvironment [17].
The previous study has demonstrated that in the peripheral blood of patients with advanced stage of NSCLC, MDSC level is up-regulated significantly along with fewer CD8+ T cells [18]. Moreover, in the lung cancer microenvironment, MDSCs are associated with a worse prognosis and a negative effect on chemotherapy and immunotherapy [19]. In breast cancers, MDSCs favor tumor progression through suppression of T cells and induction of breast cancer-related osteolysis as osteoclast progenitors [12]. Therefore, as an important regulatory immune cell population in tumor microenvironment., MDSCs exert crucial functions for promoting the growth and metastasis of cancer and weakening the effect of MDSCs irrelevant anticancer immunotherapy, which suggests the application of MDSCs as an indicator of tumor progression and a target of immunotherapy for cancer treatment [20].
A previous study has investigated the potential role of curcumin, which is a polyphenol extracted and purified from the Curcuma longa rootstock, for treating colorectal cancer [21]. It is noted that curcumin is effective in enhancing the effect of chemotherapy and radiotherapy. The possible mechanism may include the inhibition of NF-kB pathway [22]. Nevertheless, the bad solubility of curcumin largely limited its potential application in previous studies [23]. Curcumin was dissolved in corn oil and administered orally with low bioavailability. To improve the solubility of curcumin and increase the tumor retention effect, nano-drug delivery system was employed to deliver curcumin [24,25]. As reported in our earlier studies, curcumin-loaded nanoparticles were prepared by using methoxy polyethylene glycol–poly (caprolactone) (mPEG–PCL) as the amphiphilic polymer matrix. Curcumin was incorporated into mPEG–PCL-based nanoparticles with high encapsulation efficiency due to its lipophilicity [26]. In vitro preliminary study showed that there was some increase of tumor cell retention time but it was still not satisfying.
In this article, we primarily aimed to use nanoplatforms to restrain the development of lung cancer for its capacity to effectively restrict anticancer drugs at the tumor sites. Recent advance in self-assembled nano-filaments has shed light on how to improve the loading efficiency and tumor retention effect of nano-drug delivery systems, especially based on peptide [27,28]. Peptides were utilized as the construction carriers for drug assembly, which confers several advantages, including good biocompatibility and degradability [29]. Hao Wang's group developed a polymer-peptide conjugates (PPCs) synthesis strategy, which could induce nanoparticle self-assembly and optimized cancer therapeutic efficacy both in vitro and in vivo [30]. Bing Xu et al. reported enzyme-guided assembly of peptides for specifically eliminating ALP-overexpressed tumor [31]. To our best knowledge, the dipeptide diphenylalanine had a benign for self-assembly. In the current study, we conjugated curcumin with peptide FFE-ss-EE. The structure of “Curcumin-FFE-ss-EE” (called Nano-curcumin, Nano-Cur) has several advantages. Firstly, it can self-assemble into nano-filaments in PBS. It is reported that nano-filaments have enhanced tumor retention ability than nanoparticles. Secondly, the loading efficiency of curcumin was significantly improved [32]. Thirdly, curcumin itself was incorporated into the peptide, which made it ema part of hydrogelator and release the drug molecules persistently from the hydrogel.
In the study, our objective is to employ this Nano-Cur to achieve well anti-tumor effects on lung cancer. According to our results, the use of Nano-Cur effectively inhibited the progression of lung cancer in in vivo models with a significantly decreased tumor burden and lower histological scores. Additionally, Nano-Cur treatment dramatically suppressed MDSCs recruitment and immunosuppressive function both in in vitro and in vivo models. Altogether, this study indicates that Nano-Cur provides an effective approach to treat lung cancer through an inhibition of MDSCs, which may help widen the chemotherapeutic and immunotherapeutic modality for lung cancer treatment.
2. Materials and methods
2.1. Materials
Cell line of Lewis lung carcinoma was obtained from the Cell Bank at the China Academy of Science. C57BL/6 (6 weeks of age) mice were purchased from the Model Animal Research Center of Nanjing University and kept under specific pathogen-free conditions. All procedures were approved by the Committee the Ethics of Animal Experiments of Nanjing University Medical School. Curcumin was purchased from Sigma-Aldrich. Curcumin-peptide (cur-peptide) conjugates was synthesized according to our previous report [22]. Dulbecco's Modified Eagle's Medium, FBS and Penicillin-Streptomycin Solution were purchased from Gibco. Anti-Ki-67 antibody was purchased from Roche (Basel, Switzerland). Tumor dissociation kit, Myeloid-Derived Suppressor Cell Isolation Kit, recombinant protein of granulocyte-macrophage colony stimulating factor (GM-CSF) and murine interleukin 6 (IL-6) were purchased from Miltenyi Biotec (Bergisch Gladbach, Germany). Anti-mouse-CD11b antibody, anti-mouse-CD11c antibody, anti-mouse-CD4 antibody, anti-mouse-CD8 antibody, anti-mouse-CD11b antibody, anti-mouse-Ly6C antibody, anti-mouse-Ly6G antibody, anti-mouse-CD45 antibody, anti-mouse-CD4 antibody, anti-mouse-CD25 antibody, anti-mouse-Foxp3 antibody, anti-mouse-IL-17 antibody, and the ELISA kits for G-CSF, IL-1α, IL-6, IFN-γ,GM-CSF,TNF-α, CXCL1 were purchased from Biolegend (San Diego, CA, US). TRIzol Reagent was purchased from Invitrogen (USA).
2.2. Construction of nano-filaments
We first synthesized nano-filaments and its control compound [22]. In briefly, 1 mg of conjugates were added in 0.1 mL of PBS buffer solution containing Na2CO3 in a ratio of 1:3, which were used to neutralize the compounds with a final pH value of 7.4. And then 0.1 mL of PBS buffer solution containing 4 equiv. of glutathione (GSH) and Na2CO3 (which were needed to neutralize GSH) was added. The hydrogel composed of entangled nano-filaments would form after being kept at 37 °C for 1 h.
2.3. In vitro drug release, rheology, morphology, and uptake study
In vitro release and rheology were reported in our previous study [22]. A JEM 1010 transmission electron microscope (JEOL, USA, Inc., Peabody, MA) was used to observe the morphology of the filament at an accelerating voltage of 80 kV. Digital images were generated and obtained by the AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA). In the uptake study, nano-cur was incubated with LLC cells for 2 h and then observed by TEM.
2.4. Lung cancer models and tumor histopathology
Cells were cultured in Dulbecco's Modified Eagle's Medium containing 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin, 37 °C, 5% CO2 incubator in the closed-culture. For tumor model establishment, 5 × 105 LLC cells supplemented in 100 μl of PBS were injected subcutaneously into mice. For LLC-Blank group (blank nano-filament), LLC-Cur group, and LLC-Nano-Cur group, blank nano-filaments, free Cur, and Nano-Cur were injected intratumorally, respectively. The diameters of tumor mass and bodyweight of mice were measured and recorded every three days. Tumor volumes were calculated as length × (width)2 × 1/2. After mice were euthanized, tumor and spleen were extracted from every mouse and weighed. Paraffin-embedded tumor samples were sectioned and stained with hematoxylin and eosin (H&E). A professional pathologist was invited to assess the tumor grade, to whom mouse genotype and treatment were kept unknown, according to previous clinical and pathological scoring criteria [33]. For immunohistochemical staining, tumor sections were stained with the anti-Ki-67. Images of stained sections were reviewed under a light microscope. To evaluate the proliferation index, we referred to the standard that positive cells/tumor cells with brown staining were counted in 10 consecutive areas.
2.5. Flow cytometric analysis
The spleen, peripheral blood, and tumor tissues were taken out from each mouse. Tumor tissues were cut and ground into small pieces and digested by enzymes following the protocol. The gentlMASCTM dissociator was used to dissociate the samples and pass them through a 200-mesh sieve with a diameter of 74 μm. Tumor dissociation kit and a gentlMASCTM dissociator were utilized to extract suspension of single-cell from tumor tissue. The splenocytes were washed out from spleen by 1 × PBS and then transmitted through the 200-mesh sieve to generate the single-cell suspension. Red blood cells were removed by ACK Lysing Buffer. All single-cell suspension was processed by FACS after washing. We used the following antibodies for flow cytometric detection: Anti-CD11b (#101211), anti-CD11c (#117305), anti-CD4 (#100405), anti-CD8 (#140403), anti-CD11b (#101205), anti-Ly6C (#128015), anti-Ly6G (#127607), anti-CD45 (#103129), anti-CD4 (#100516), CD25 (#102012), Foxp3 (#320107), IL-17 (#146303). FlowJo software (Treestar, Inc., San Carloos, CA) was utilized to analyze the data.
2.6. Quantitative RT-PCR analysis
TRIzol Reagent was used to extract RNA from tumor tissue or primary MDSCs following the Manufacturer's instructions. We used oligo (dT) primer to reverse the mRNAs into cDNAs. Q-PCR assays were performed on the platform of ABI vii 7 detection system (Applied Biosystems) or StepOne Plus using SYBR Green PCR master mix solution. The reaction conditions were under pre-denaturation in a temperature of 95 °C for 10 min, and then experienced 40 cycles of 95 °C for 15 s, 60 °C for 1 min. Formula of 2−ΔΔCT was utilized to measure the relative expressive level of the genes with GAPDH used as an internal control.
2.7. ELISA
The ELISA kits for IL-1α, IL-6, G-CSF, IFN-γ, GM-CSF, CXCL1 and TNF-α were used to detect GM-CSF, G-CSF, IL-6, IL-1α, TNF-α, IFNγand CXCL1. Experiments were performed following the instructions.
2.8. Extraction of MDSCs and MDSCs functional assay
A Specific kit for MDSC isolation (#130-094-538) was utilized to isolated the MDSC from tumor tissue in mice. The T cells extracted from spleen (105 cells/well) were co-cultured with previously-isolated MDSCs at different ratios in the 96-well plate, with the addition of 1 μg/mL plate-bound anti-CD3 monoclonal antibody (mAb) and 0.5 μg/mL soluble anti-CD28 mAb for 72 h. Cells were treated by 0.5 μCi/well [3H] thymidine for the last 16 h of incubation. For evaluating proliferative level, an LS 6500 Multi-Purpose Scintillation Counter (Beckman Coulter, California, US) was used measuring [3H] thymidine incorporation in triplicate.
2.9. Isolation of bone marrow-derived MDSCs
Tibiae and flushing femurs were used for isolating BM cells from mice. Afterwards, the extracted BM cells were then centrifuged. Culture medium with the addition of Nano-Cur or free Cur was used to resuspend the debris of BM cells. Parallelly, we used GM-CSF and IL-6 with the concentration of 40 ng/mL as the positive control. The culture is sustained for 5 days.
2.10. Statistical analysis
Prism 5 (Graphpad Software, Inc, San Diego, CA) was used to perform the statistical analysis for the experimental data. Approaches of Student's t-test (two-tailed) or two-way ANOVA were used for analyzing the significance of differences between groups, with all data expressing as means ± SD. P value < 0.05 were considered as statistically significant. At least three repeated experiments were performed for all the in vivo and in vitro investigations.
3. Results
3.1. In vitro inhibition of the function and differentiation of MDSCs by nano-cur
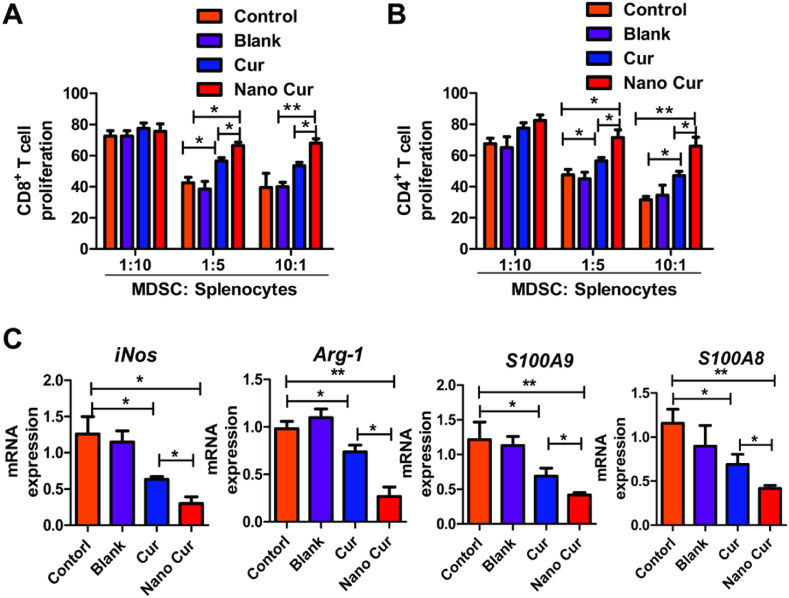
As reported in our previous study, Nano-Cur was characterized by TEM with a diameter of about 30 nm and length of micrometers. The drug loading content of curcumin in Nano-Cur was nearly 40%, demonstrating a high loading efficiency of self-assemble supramolecular nano-filaments [22]. In vitro studies with bone marrow cells were first performed to evaluate the effect of Nano-Cur in mediating the accumulation and function of MDSCs. Bone marrow cells were acquired and differentiated into MDSCs with the stimulation of GM-CSF and IL-6. External supplement of Nano-Cur markedly reduced the percentage of MDSCs, mainly G-MDSCs (Fig. 1A–C). Co-culture of MDSCs and T cells with Nano-Cur contributed to the significantly higher proliferation of both CD8+ and CD4+ T cells as compared to free Cur (Fig. 2A and B). Meanwhile, the expression of iNOS, Arg-1, S100 Calcium Binding Protein A9 (S100A9) and S100 Calcium Binding Protein A8 (S100A8) was substantially reduced upon Nano-Cur treatment (Fig. 2C).
Fig. 1.
The in vitro inhibition of MDSCs differentiation by Nano-Cur. Bone marrow cells from C57BL/6 mice were stimulated with GM-CSF and IL6 for 6 days. Cur and Nano-Cur were added during differentiation. (A) Typical flow cytometry images of MDSCs (CD11b+Gr1+ cells) in various groups. (B) The proportions of MDSCs in different treatment groups were determined by flow cytometry. (C) Quantitative analysis of MDSC differentiation in tumors from mice in various groups. All experiments were performed with three replicates. Error bars, SD. ∗ Represents P < 0.05, and ∗∗ represents P < 0.01.
Fig. 2.
The in vitro inhibitory effect of Nano-Cur on the function of MDSCs. Cur and Nano-Cur were added during differentiation. Differentiated MDSCs were co-cultured with CD8+ T cells (A) or CD4+ T cells (B). [3H] thymidine in-corporation was used to assess the immunosuppressive capabilities of MDSCs. (C) The transcriptional expression of Arg-1, iNOS, S100A9 and S100A8 in differentiated MDSCs were determined by qPCR. All experiments were performed with three replicates. Error bars, SD. ∗ Represents P < 0.05, and ∗∗ represents P < 0.01.
3.2. Inhibition of lung cancer growth by nano-cur
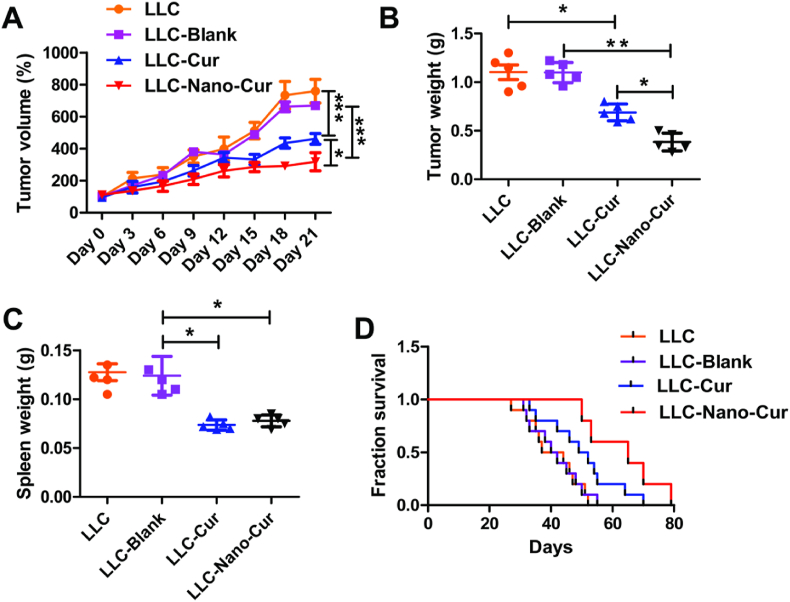
To further assess the anti-cancer effect of Nano-Cur in lung cancer, Lewis lung cancer models were established by injecting LLC cells subcutaneously into C57BL/6 (6 weeks) mice. Mice were then randomly divided into four groups following different treatments: LLC, LLC-Blank, LLC-Cur, LLC-Nano-Cur. As shown in Fig. 3A and B, either the treatment of Cur or Nano-Cur resulted in obvious inhibition of tumor growth, compared with control group, respectively. Moreover, Nano-Cur treatment inhibited the growth of tumor with a higher efficiency than free Cur. The weight of spleen was also measured in each mouse. Upon LLC injection, the spleen is enlarged, while treatment of Cur and Nano-Cur reversed the splenomegaly in tumor-bearing mice (Fig. 3C). In addition, The Kaplan-Meier (KM) analysis demonstrated the most effective benefit of Nano-Cur on increasing overall survival among all the treatment groups (Fig. 3D).
Fig. 3.
The in vivo inhibitory effect of Nano-Cur on the lung cancer growth in LLC mouse model. 5 × 105 LLC cells supplemented in 100 μl of PBS were subcutaneously injected into the right flank of the mice (n = 8 per group). Mice models were sacrificed on day 21. The growth curves of xenograft tumors (A) and tumor weights (B) were measured in mice of different treatment groups as previously mentioned. Spleen were isolated from each mouse and spleen weight were measured (C). Kaplan-Meier curves for survival analysis (D). All experiments were repeated at three times and the data shown represents one of them. Error bars, SD. ∗ Represents P < 0.05, ∗∗ represents P < 0.01, and ∗∗∗ Represents P < 0.001.
Tumor malignancy is closely associated with the proliferation of tumor cells. Fast progression requires a high proliferative rate, which could be characterized by a histological study including H&E and immunohistochemical (IHC) staining. Histological scores and Ki-67 staining are two of the main indicators of tumor proliferation in clinic. As shown in Fig. 4A and B, tumors in Nano-Cur group had lower histological scores than the other three groups. The results revealed that lower scores were significantly related with histological types and lower tumor grade. Tumor nodules from each group were excised and examined for the expression of Ki-67 by IHC. It is noted that Nano-Cur or Cur alone suppressed the expression of Ki-67 more effectively than saline. Moreover, as compared to the group of free Cur, positive staining of Ki-67 was almost inhibited to a much lower degree in the group of Nano-Cur treatment (Fig. 4C and D), implying a significant association of the low ki-67 expression with good DFS and OS. These data suggest that Nano-Cur treatment can significantly inhibit the growth of lung cancer than free Cur.
Fig. 4.
The in vivo inhibitory effect of Nano-Cur on the tumor proliferation in LLC mouse model. Mice were treated as described in Fig. 2. (A–B) The typical histological images and histologic scores of lung cancers in different groups. H&E staining were performed on the acquired tumor tissues. (C–D) The images of the immunohistochemistry analysis and the quantitative results of Ki67 expressive degree of the tumors from different groups. The standard of Ki67 positivity was determined as the mean value of Ki67 positive cells in 10 randomly selected areas. Data represent one of three independent experiments. Error bars, SD. ∗ Represents P < 0.05, ∗∗ represents P < 0.01, and ∗∗∗ Represents P < 0.001.
3.3. Nano-cur regulation on the immune cells within the tumor environment
The tumor microenvironment remains to be the main battlefield between the tumor and host immunity, whereas the cross-talk between them seems to be important [[34], [35], [36], [37]]. Immune cells infiltrated in tumor microenvironment not only receives the signals from tumor cells but also influence the malignant biological behavior of tumor cells. As parts of immune cells in tumor microenvironment, CD4+ and CD8+ T cells, the most powerful lymphocytes, form the basis of cell-mediated immunity and are of great significance in inducing immune response against tumor [38,39].
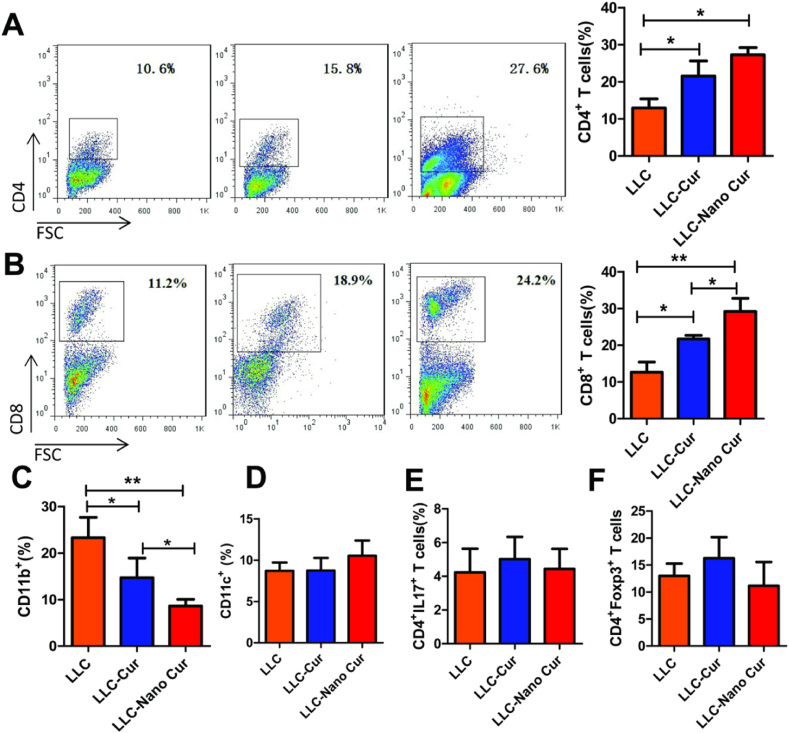
Therefore, the proportions of immune cells in the tumor microenvironment of each group were further detected. As shown in Fig. 5A and B, Cur-treatment can increase the expression of both CD4+ T cells and CD8+ T cells in tumor environment. Of note, Nano-Cur treatment induced more CD8+ T cells than Cur-treated mice. Moreover, the innate and adaptive immune cells were also detected in the mice model. As shown in Fig. 5C, myeloid cells (CD11b+ cells) were significantly decreased upon free-Cur and Nano-Cur treatment. However, the proportion of DC cells (CD11c+), Treg cells and Th17 cells showed no difference among those three groups (Fig. 5D–F).
Fig. 5.
The in vivo up-regulation of the infiltration level of T cells and myeloid cells in tumor microenvironment by Nano-Cur. Mice were treated as described in Fig. 2 (n = 8 per group). Tumor tissues were isolated. The proportion of CD4+ cells (A), CD8+ cells (B), CD11b+ cells (C), CD11c+ cells (D), CD4+IL17+ cells (E) and CD4+CD25+Foxp3+ cells (F) in tumor tissues were determined by flow cytometry. Data represent one of three repeated experiments. Error bars, SD. ∗ Represents P < 0.05, ∗∗ represents P < 0.01, and ∗∗∗ Represents P < 0.001.
As mentioned above, strong infiltration of MDSCs along with fewer CD8+ T cells remains to be one of the main factors for tumor resistance. Nano-Cur effectively initiated the increase of CD8+ T cells, which demonstrated the potential to affect the proportion of immune cells in tumor microenvironment and would definitely boost the immune system through improving active immune response.
3.4. Influence on the expression of a variety of cytokines by nano-cur
The interaction between immune cells and tumor cells relies on the messengers including cytokines, lymphokines, etc. Cytokines excreted from immune cells could activate other immune cells or stromal cells in the tumor microenvironment and facilitate the recognition of tumors by cytotoxic immune cells, which leads to the consequent tumor inhibition [40]. To further understand the possible mechanism of immune response recovery by Nano-cur, primary tumor cells were extracted from mice and then cultured for 24 h. Multiple cytokine detection assays showed that the expression of parts of the detected cytokines, such as IL-6, GM-CSF, TNF-alpha, and CXCL-1, were significantly lower in either Cur or Nano-Cur treatment group than in LLC group (Fig. 6A and B). Moreover, Nano-Cur exhibited stronger inhibitory effect on the expression of these cytokines than free Cur (Fig. 6B). As confirmed by qPCR and ELISA, Nano-Cur significantly inhibited the release of GM-CSF, IL-6 and TNF-α, when compared with free Cur (Fig. 6B and C).
Fig. 6.
The enhanced production of IL-6 and GM-CSF in the tumor tissue by Nano-Cur treatment. Mice were treated as described in Fig. 2 (n = 8 per group). (A) Primary tumor cells from LLC group and LCC-Nano-Cur group were isolated and cultured from 24 h. Cytokine production were detected using multiple cytokine detection assay. (B) The production of GM-CSF, IL-6, TNF-α, IFN-g, IL-1a, G-CSF, and CXCL1 were detected using ELISA. (C) Relative mRNA expressions of GM-CSF, IL-6 and TNF-α were detected by qPCR. Data represent one of three independent experiments. Error bars, SD. ∗ Represents P < 0.05, ∗∗ represents P < 0.01, and ∗∗∗ Represents P < 0.001.
As a kind of myeloid cells, MDSCs are a main obstacle for hampering anti-cancer immunity [12,13]. A variety of pro-inflammatory mediators such as GM-CSF and IL-6 can be released by tumor and host cells to invigorate MDSCs and enhance their accumulation and immunosuppressive effects [38]. As GM-CSF and IL-6 are critical modulators to maintain the differentiation, proliferation and function of MDSCs, reverse of their high level in tumor microenvironment by Nano-Cur enables the re-establishment of active immune response.
3.5. Inhibition of the recruitment and function of MDSCs in tumor tissues by nano-cur
Since Nano-Cur effectively reduced the expression of GM-CSF and IL-6, the infiltration of MDSCs in the spleen, peripheral blood and tumor tissues of each group was examined to analyze the potential influence of these cytokines. Both free Cur and Nano-Cur treatment induced subsequent down-regulation of MDSCs in tumor and peripheral blood. Notably, significant differences were observed in the frequency of MDSCs between the group of Nano-Cur and free Cur, which demonstrated superior effect of Nano-Cur in restricting the accumulation of MDSCs (Fig. 7A and B). Based on the expression of surface antigens, two main subpopulations of MDSCs can be distinguished in mice, G-MDSCs and M-MDSCs. Emerging findings suggest that G-MDSCs and M-MDSCs may inhibit T cell immune function via different mechanisms. The G-MDSCs repress the immune function of T cells by over-expressing reactive oxygen species (ROS), whereas M-MDSCs was found to produce large amounts of nitric oxide through high expression of iNOS [39]. As shown in Fig. 7C and D, the accumulation of G-MDSCs in tumor tissues, instead of M-MDSCs, were significantly inhibited by either free Cur or Nano-Cur treatment.
Fig. 7.
The in vivo inhibitory effect of Nano-Cur on the accumulation of MDSCs in lung cancer. Mice were treated as illustrated in Fig. 2 (n = 8 per group). Tumor tissues were isolated from each mouse. (A) The typical flow cytometry images of MDSCs (CD11b+Gr1+ cells) in tumor tissues from mice in various groups. (B) The proportions of MDSCs in spleen, blood and tumor tissues in mice. (C) The typical flow cytometry images of G-MDSCs (CD11b+Ly6G+ cells) and M-MDSCs (CD11b+Ly6C+ cells) in tumor tissues from mice in various groups. (D) The quantitative analysis of G-MDSCs and M-MDSCs proportions in tumors from mice in different treatment groups. The shown data is derived from one of the three repeated experiments. Error bars, SD. ∗ Represents P < 0.05, ∗∗ represents P < 0.01, and ∗∗∗ Represents P < 0.001.
MDSCs can inhibit the immune system by multiple mechanisms, mostly through production of arginase 1 (Agr-1), nitric oxidase synthase2 (Nos2) and several immunosuppressive cytokines, thereby inhibiting the expansion of CD8+ and CD4+ T cells [40]. Thus, the function of MDSCs could be evaluated by the expression of these cytokines and the proliferation of T cells. The primary MDSCs were isolated from tumor tissues and co-cultured with different types of T cells. As presented in Fig. 8A and B, the primary MDSCs derived from LLC tumor-bearing mice inhibited the proliferation of both CD8+ T cells and CD4+ T cells, while this immunosuppressive function was significantly reversed by the treatment of either Nano-Cur or free Cur, presenting as more proliferation of both CD8+ T cells and CD4+ T cells, compared with control group. Importantly, Nano-Cur showed significantly stronger stimulatory effect in the proliferation of T cells than free Cur. In the other hand, the transcriptional expression of iNOS, Arg-1, S100A9, and S100A8 in primary MDSCs from mice with Cur or Nano-Cur treatment was significantly lower than that from control treatment group (Fig. 8C). Moreover, Nano-Cur led to significantly lower expression level of Arg-1, S100A9 and S100A8 than free Cur, which maybe parts of the explanation for its restoration of superior antitumor immunity.
Fig. 8.
The in vivo inhibitory effect of Nano-Cur on the function of MDSCs in lung cancer. Mice were treated as described in Fig. 2 (n = 8 per group). Primary MDSCs were derived from tumor tissues in mice and were then co-cultured with CD8+ T (A) or CD4+ T cells (B). The immunosuppressive function of MDSCs was assessed using [3H] thymidine in-corporation. (C) The transcriptional expression of iNOS, Arg-1, S100A9 and S100A8 in tumor tissues were detected by qPCR. All experiments were repeated for at least three times. Error bars, SD. ∗ Represents P < 0.05, ∗∗ represents P < 0.01, and ∗∗∗ Represents P < 0.001.
4. Discussion
In recent studies, accumulating evidence indicates that tumor progression highly relied on the formation of Tumor Microenvironment (TME) [41,42]. The immunosuppressive status, which favors cancer development, is conducted by crosstalk between tumor cells and host cells, where MDSCs play a critical central role in the immune suppression of TME through the inhibition of T cell proliferation and activation, NK cell activation and the induction of Tregs expansion [9]. In our study, the total MDSCs and MDSCs subsets in lung cancer tissue and primary bone marrow cells using the same markers. MDSCs are comprised of two groups of cells with either a monocytic (M-MDSCs) or a neutrophilic (G-MDSCs) morphology. The CD11b+Gr1+ cells in lung cancer tissue and primary bone marrow cells belong to total MDSCs. The CD11b+Ly6G+ cells and CD11b+Ly6C+ cells in lung cancer tissue and primary bone marrow cells belong to G-MDSC and M-MDSC, respectively.
The G-MDSC increased activity of STAT3 and NADPH, which results in high levels of ROS and low levels of NO production. ROS induces post-translational modification of T-cell receptors and may cause antigen-specific T-cell unresponsiveness. However, the M-MDSC has upregulated expression of STAT1 and iNOS and increased levels NO but little ROS production. NO, which is produced by the metabolism of l-arginine by iNOS, suppresses T-cell function through a variety of different mechanisms that involve the inhibition of JAK3 and STAT5, the inhibition of MHC class II expression and the induction of T-cell apoptosis. Additionally, both subsets have elevated level of ARG1 activity that causes T-cell suppression through depletion of arginine [43,44]. A clinical trial on patients with extensive small-cell lung cancer found that depletion of MDSCs increased the CD8+ T cell levels and improved the immune response to vaccination [45]. Besides, it is reported that the increase of circulating MDSCs was associated with a heavier metastatic tumor burden and a higher clinical cancer stage [46]. Altogether, MDSCs play a pivotal role in the cancerous progression, making targeting on MDSCs a promising approach for the treatment of lung cancer.
Earlier studies have demonstrated the potential inhibitory capacities of Curcumin on MDSC [47]. However, small molecules of Curcumin are easy to flow out from the tumor, resulting in a poor bioavailability of Curcumin and largely limiting its therapeutic efficiency. To overcome this problem, Curcumin is generally encapsulated into nanoparticles to increase the tumor retention time, but its effect is still not satisfying [[48], [49], [50]]. In this report, curcumin was conjugated with a self-assembled peptide and formed a nano-filament structure. It is reported in a previous study that nano-formulation with fiber structure has longer tumor retention time than spherical nanoparticles [51]. Moreover, the nan-filaments would be mainly accumulated in tumor site due to the shape and EPR effect. and then metabolized and excreted by liver and kidney [52].
In this study, the application of Nano-Cur led to significant inhibitory effect on the lung tumor growth with lower Ki-67 index and histological scores. Additionally, the immune status of TME was well improved by Nano-Cur treatment with increased proliferation and activation of T cells and reduced production of inflammatory mediators such as GM-CSF and IL-6, which revealed that Nano-Cur relieved the tumor burden by regulating and improving the TME. Furthermore, Flow cytometry analysis implied the lower MDSCs level of Nano-Cur treatment group than that of control treatment group, which indicated that the anticancer effect of Nano-Cur may be associated with the inhibition of infiltration and differentiation of MDSCs in the TME.
5. Conclusion
In conclusion, we report a novel way to suppress the development of lung cancer by affecting MDSCs through curcumin loaded nano-filaments. In TME, MDSCs facilitate tumor progression by the up-regulation of Arg1 and iNOS, overproduction of ROS, and release of IL-10, which results in the inhibition of T-cell activation. The Nano-Cur exerts anticancer effect by suppressing MDSCs in TME. Together with all experimental findings, we propose the Nano-Cur as a new approach for lung cancer therapy, and extensive studies of mechanisms are required to better understand how TME affect tumor progression and provide new insights into anticancer therapeutics.
CRediT author Statement
Tingting Wang: Conceptualization, Methodology, Investigation, Writing-Original draft, Funding acquisition. Jia Wang: Investigation, Methodology, Validation.Hui Jiang: Investigation, Methodology. Mengnan Ni: Software, Validation. Yifan Zou: Form analysis, Software. Yanlong Chen: Methodology, Data curation. Ting Wu: Form analysis, Data curation. Dan Ding: Resources, Visualization. Huae Xu: Writing-Reviewing and editing, Supervision, Project administration, Funding acquisition. Xiaolin Li: Conceptualization, Writing-Reviewing and editing, Supervision, Project administration, Funding acquisition.
Data availability statement
The data are available from the corresponding authors on reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work is supported by the National Natural Science Foundation of China (82073308, 81871942, 81773211, and 31900983) and the High-level startup fund of Nanjing Medical University (KY109RC2019010).
Contributor Information
Huae Xu, Email: xuhuae@njmu.edu.cn.
Xiaolin Li, Email: lxl@njmu.edu.cn.
References
- 1.Bironzo P., Di Maio M. A review of guidelines for lung cancer. J. Thorac. Dis. 2018;10(Suppl 13):S1556–S1563. doi: 10.21037/jtd.2018.03.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y., Yang Q., Wang S. MicroRNAs: a new key in lung cancer. Cancer. Chemother. Pharmacol. 2014;74(6):1105–1111. doi: 10.1007/s00280-014-2559-9. [DOI] [PubMed] [Google Scholar]
- 3.Lemjabbar-Alaoui H., Hassan O.U., Yang Y.W., Buchanan P. Lung cancer: biology and treatment options. Biochim. Biophys. Acta. 2015;1856(2):189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W., Chen T., Xu H., Ren B., Cheng X., Qi R., Liu H., Wang Y., Yan L., Chen S., Yang Q., Chen C. Curcumin-loaded solid lipid nanoparticles enhanced anticancer efficiency in breast cancer. Molecules. 2018;23(7):1578. doi: 10.3390/molecules23071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilmi M., Vienot A., Rousseau B., Neuzillet C. Immune therapy for liver cancers. Cancers. 2019;12(1):77. doi: 10.3390/cancers12010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finck A., Gill S.I., June C.H. Cancer immunotherapy comes of age and looks for maturity. Nat. Commun. 2020;11(1):3325. doi: 10.1038/s41467-020-17140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barar J. Targeting tumor microenvironment: the key role of immune system. Bioimpacts. 2012;2(1):1–3. doi: 10.5681/bi.2012.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinshaw D.C., Shevde L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marvel D., Gabrilovich D.I. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J. Clin. Invest. 2015;125(9):3356–3364. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kusmartsev S., Nagaraj S., Gabrilovich D.I. Tumor associated CD8+ T-cell tolerance induced by bone marrow derived immature myeloid cells. J. Immunol. 2005;175(7):4583–4592. doi: 10.4049/jimmunol.175.7.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Youn J.I., Nagaraj S., Collazo M., Gabrilovich D.I. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol. 2008;181(8):5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shou D., Wen L., Song Z., Yin J., Sun Q., Gong W. Suppressive role of myeloid-derived suppressor cells (MDSCs) in the microenvironment of breast cancer and targeted immunotherapies. Oncotarget. 2016;7(39):64505–64511. doi: 10.18632/oncotarget.11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., Mandruzzato S., Murray P.J., Ochoa A., Ostrand-Rosenberg S., Rodriguez P.C., Sica A., Umansky V., Vonderheide R.H., Gabrilovich D.I. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condamine T., Gabrilovich D.I. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 2011;32(1):19–25. doi: 10.1016/j.it.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonda N., Simonato F., Peranzoni E., Calì B., Bortoluzzi S., Bisognin A., Wang E., Marincola F.M., Naldini L., Gentner B., Trautwein C., Sackett S.D., Zanovello P., Molon B., Bronte V. miR-142-3p prevents macrophage differentiation during cancer-induced myelopoiesis. Immunity. 2013;38(6):1236–1249. doi: 10.1016/j.immuni.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Law A.M.K., Valdes-Mora F., Gallego-Ortega D. Myeloid-derived suppressor cells as a therapeutic target for cancer. Cells. 2020;9(3):561. doi: 10.3390/cells9030561. Published 2020 Feb 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan P.Y., Ma G., Weber K.J., et al. Immune stimulatory receptor CD40 is required for T-cell suppression and T regulatory cell activation mediated by myeloid-derived suppressor cells in cancer. Cancer Res. 2010;70(1):99–108. doi: 10.1158/0008-5472.CAN-09-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adah D., Hussain M., Qin L., Qin L., Zhang J., Chen X. Implications of MDSCs-targeting in lung cancer chemo-immunotherapeutics. Pharmacol. Res. 2016;110:25–34. doi: 10.1016/j.phrs.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Chen J., Ye Y., Liu P., Yu W., Wei F., Li H., Yu J. Suppression of T cells by myeloid-derived suppressor cells in cancer, Hum. Immunol. 2017;78(2):113–119. doi: 10.1016/j.humimm.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Chesney J.A., Mitchell R.A., Yaddanapudi K. Myeloid-derived suppressor cells-a new therapeutic target to overcome resistance to cancer immunotherapy. J. Leukoc. Biol. 2017;102(3):727–740. doi: 10.1189/jlb.5VMR1116-458RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yallapu M.M., Jaggi M., Chauhan S.C. Curcumin nanomedicine: a road to cancer therapeutics. Curr. Pharmaceut. Des. 2013;19(11):1994–2010. doi: 10.2174/138161213805289219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H., Wang T., Yang C., Li X., Liu G., Yang Z., Singh P.K., Krishnan S., Ding D. Supramolecular nanofibers of curcumin for highly amplified radiosensitization of colorectal cancers to ionizing radiation. Adv. Funct. Mater. 2018;28(14):1707140. doi: 10.1002/adfm.201707140. [DOI] [Google Scholar]
- 23.Zhao S., Pi C., Ye Y., Zhao L., Wei Y. Recent advances of analogues of curcumin for treatment of cancer. Eur. J. Med. Chem. 2019;180:524–535. doi: 10.1016/j.ejmech.2019.07.034. [DOI] [PubMed] [Google Scholar]
- 24.Wong K.E., Ngai S.C., Chan K.G., Lee L.H., Goh B.H., Chuah L.H. Curcumin nanoformulations for colorectal cancer: a review. Front. Pharmacol. 2019;5(10):152. doi: 10.3389/fphar.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naksuriya O., Okonogi S., Schiffelers R.M., Hennink W.E. Curcumin nanoformulations: a review of pharmaceutical properties and preclinical studies and clinical data related to cancer treatment. Biomaterials. 2014;35(10):3365–3383. doi: 10.1016/j.biomaterials.2013.12.090. [DOI] [PubMed] [Google Scholar]
- 26.Yin H., Zhang H., Liu B. Superior anticancer efficacy of curcumin-loaded nanoparticles against lung cancer. Acta Biochim. Biophys. Sin. 2013;45(8):634–640. doi: 10.1093/abbs/gmt063. [DOI] [PubMed] [Google Scholar]
- 27.Liao H.S., Lin J., Liu Y., Huang P., Jin A., Chen X. Self-assembly mechanisms of nanofibers from peptide amphiphiles in solution and on substrate surfaces. Nanoscale. 2016;8(31):14814–14820. doi: 10.1039/c6nr04672j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L.L., Qiao Z.Y., Wang L., Wang H. Programmable construction of peptide-based materials in living subjects: from modular design and morphological control to theranostics. Adv. Mater. 2019;31(45) doi: 10.1002/adma.201804971. [DOI] [PubMed] [Google Scholar]
- 29.Wang H., Feng Z., Xu B. Assemblies of peptides in a complex environment and their applications. Angew Chem. Int. Ed. Engl. 2019;58(31):10423–10432. doi: 10.1002/anie.201814552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Lin Y.X., Qiao Z.Y., An H.W., Qiao S.L., Wang L., Rajapaksha R.P., Wang H. Self-assembled autophagy-inducing polymeric nanoparticles for breast cancer interference in-vivo. Adv. Mater. 2015;27(16):2627–2634. doi: 10.1002/adma.201405926. [DOI] [PubMed] [Google Scholar]
- 31.Feng Z., Han X., Wang H., Tang T., Xu B. Enzyme-instructed peptide assemblies selectively inhibit bone tumors. Inside Chem. 2019;5(9):2442–2449. doi: 10.1016/j.chempr.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng S., Ou H., Gao Z., Zhang J., Li C., Liu Q., Ding D. HCPT-peptide prodrug with tumor microenvironment-responsive morphology transformable characteristic for boosted bladder tumor chemotherapy. J. Contr. Release. 2021;330:715–725. doi: 10.1016/j.jconrel.2020.12.042. [DOI] [PubMed] [Google Scholar]
- 33.Wang T., Chen L., Yang T., Wang L., Zhao L., Zhang S., Ye Z., Chen L., Zheng Z., Qin J. Cancer risk among children conceived by fertility treatment. Int. J. Cancer. 2019;144(12):3001–3013. doi: 10.1002/ijc.32062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batista S., Gregório A.C., Hanada Otake A., Couto N., Costa-Silva B. The gastrointestinal tumor microenvironment: an updated biological and clinical perspective. JAMA Oncol. 2019;22:6240505. doi: 10.1155/2019/6240505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagamu H., Kitano S., Yamaguchi O., Yoshimura K., Horimoto K., Kitazawa M., Fukui K., Shiono A., Mouri A., Nishihara F., Miura Y., Hashimoto K., Murayama Y., Kaira K., Kobayashi K. CD4+ T-cell immunity in the peripheral blood correlates with response to anti-PD-1 therapy. Cancer Immunol. Res. 2020;8(3):334–344. doi: 10.1158/2326-6066.CIR-19-0574. [DOI] [PubMed] [Google Scholar]
- 36.Li H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C.J., van den Braber M., Rozeman E.A., Haanen J.B.A.G., Blank C.U., Horlings H.M., David E., Baran Y., Bercovich A., Lifshitz A., Schumacher T.N., Tanay A., Amit I. Dysfunctional CD8+ T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176(4):775–789. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sim G.C., Radvanyi L. The IL-2 cytokine family in cancer immunotherapy. Cytokine. Growth. Factor. Rev. 2014;25(4):377–390. doi: 10.1016/j.cytogfr.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Kumar V., Patel S., Tcyganov E., Gabrilovich D.I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37(3):208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar V., Patel S., Tcyganov E., Gabrilovich D.I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37(3):208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker K.H., Beury D.W., Ostrand-Rosenberg S. Myeloid-derived suppressor cells: critical cells driving immune suppression in the tumor microenvironment. Adv. Cancer Res. 2015;128:95–139. doi: 10.1016/bs.acr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh H., Watanabe M. Myeloid-Derived suppressor cells and therapeutic strategies in cancer. Mediat. Inflamm. 2015:159269. doi: 10.1155/2015/159269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youn J.I., Nagaraj S., Collazo M., Gabrilovich D.I. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J. Immunol. 2008;181(8):5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis R.J., Van Waes C., Allen C.T. Overcoming barriers to effective immunotherapy: MDSCs, TAMs, and Tregs as mediators of the immunosuppressive microenvironment in head and neck cancer. Oral Oncol. 2016;58:59–70. doi: 10.1016/j.oraloncology.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diaz-Montero C.M., Salem M.L., Nishimura M.I., Garrett-Mayer E., Cole D.J., Montero A.J. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol. Immunother. 2009;58(1):49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanmugam M.K., Rane G., Kanchi M.M., Arfuso F., Chinnathambi A., Zayed M.E., Alharbi S.A., Tan B.K., Kumar A.P., Sethi G. The multifaceted role of curcumin in cancer prevention and treatment. Molecules. 2015;20(2):2728–2769. doi: 10.3390/molecules20022728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murali M.Y., Prashanth K.B.N., Meena J., Subhash C.C. Therapeutic applications of curcumin nanoformulations. AAPS J. 2015;17(6):1341–1356. doi: 10.1208/s12248-015-9811-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klippstein R., Wang J.T., El-Gogary R.I., Bai J., Mustafa F., Rubio N., Bansal S., Al-Jamal W.T., Al-Jamal K.T. Passively targeted curcumin-loaded PEGylated PLGA nanocapsules for colon cancer therapy in vivo. Small. 2015;11(36):4704–4722. doi: 10.1002/smll.201403799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohanty C., Sahoo S.K. The in vitro stability and in vivo pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials. 2010;31(25):6597–6611. doi: 10.1016/j.biomaterials.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 51.Geng Y., Dalhaimer P., Cai S., Tsai R., Tewari M., Minko T., Discher D.E. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2007;2(4):249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Liu J., Xu H., et al. Novel tumor-targeting, self-assembling peptide nanofiber as a carrier for effective curcumin delivery. Int. J. Nanomed. 2014;9:197–207. doi: 10.2147/IJN.S55875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are available from the corresponding authors on reasonable request.